Abstract
Efforts to understand autoimmunity have been pursued relentlessly for several decades. It has become apparent that the immune system evolved multiple mechanisms for controlling self-reactivity, and defects in one or more of these mechanisms can lead to breakdown of tolerance. Among the multitude of lesions associated with disease, the most common appear to affect peripheral rather than central tolerance. The initial trigger for both systemic and organ-specific autoimmune disorders likely involves recognition of self or foreign molecules, especially nucleic acids, by innate sensors. This recognition, in turn, triggers inflammatory responses and engagement of previously quiescent autoreactive T and B cells. Here, we summarize the most prominent autoimmune pathways and identify key issues that require resolution to fully understand pathogenic autoimmunity.
The distinction between foreign and self by the immune system is not absolute, and under certain circumstances this system can be misdirected against the very entity it is intended to protect. Accordingly, aberrant responses against self are implicated in >80 inflammatory disorders, collectively defined as autoimmune diseases.
Autoreactivity ranges from a low “physiologic” level of self-reactivity essential for lymphocyte selection and immune system homeostasis, to an intermediate level of autoimmunity that manifests as circulating autoantibodies and minor tissue infiltrates without clinical consequences, to pathogenic autoimmunity associated with immune-mediated organ injury. Autoimmune diseases have high prevalence (~7–9%) in the population, preferentially afflict women, strike at the prime of life, and cause significant morbidity and mortality. Based on the extent of tissues involved, these diseases are divided into organ-specific (e.g. type I diabetes (T1D), multiple sclerosis (MS), inflammatory bowel diseases (IBDs), myasthenia gravis) and systemic (e.g. systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), Sjögren’s syndrome) and can be mediated by autoantibodies or cytotoxic T cells, but in all instances helper T cells are required.
Most autoimmune diseases exhibit clinical heterogeneity, a polygenic nature, and multifactorial contributions often requiring both genetic and environmental factors1. While autoimmune diseases involve both innate and adaptive immune responses, the so-called autoinflammatory diseases are associated with monogenic mutations resulting in over-activation of the innate immune system without participation of the adaptive system2. Generally, genetic susceptibility results from the additive effects of several common risk variants, each with small effect sizes that alone are insufficient3,4. These common variants probably persisted because of a survival advantage related to improved responses to infections and, not unexpectedly, they exhibit significant variation among ethnic groups.
Several hundred loci associated with autoimmunity have been identified, including >100 in RA, MS, and IBDs3. Overlapping loci across diseases frequently encompassing immune-related genes suggested common mechanistic pathways, although the specific risk allele within a locus can differ depending on the disease. Among known genetic predisposing factors, certain MHC haplotypes exert the strongest associations across most autoimmune diseases, but several other genes, including PTPN22, CTLA4, IL23R and TYK2, have been frequently implicated. Rare monogenic autoimmune diseases have also been identified with mutations in AIRE, FOXP3, IFIH1, DNASE1, TREX1, C1Q, or C4A, many of which have provided clues to our understanding of autoimmune pathogenesis. For most loci, however, the actual risk alleles remain unknown because of linkage disequilibrium, extensive heterogeneity, and incomplete sequence information. Moreover, most risk variants occur in poorly-defined noncoding regions, which has challenged efforts to understand their effects on gene function.
Central tolerance is inefficient
A key question is how an immensely diverse antigen recognition system, primarily created to detect and eliminate offending pathogens, avoids eliciting destructive anti-self responses. The main mechanisms of tolerance are exercised centrally, in the thymus for T cells and the fetal liver and bone marrow for B cells. However, is central tolerance infallible, and do escaping self-reactive cells contribute to autoimmune disease pathogenesis?
The prevailing view has been that negative selection eliminates autoreactive T cells with high fidelity. Yet early5 and more definitive recent studies have shown significant leakage in this process (Fig. 1). For example, analyses with peptide-MHC tetramers showed that the frequency and avidity of peripheral blood CD8+ T cells specific for diverse virus-derived peptides in healthy individuals not previously infected with these viruses were in the same range as T cells recognizing self-peptides, while the frequency of CD8+ T cells specific for SMCY, a Y chromosome-encoded antigen, was reduced by only 2/3 in males vs. females6. Incomplete deletion of SMCY-specific CD8+ T cells was also observed in male non-transgenic mice. Moreover, only ~60% deletion of Cre-specific CD4+ T cells was detected in the thymus and periphery of mice transgenic for ubiquitous Cre expression, and impressively no deletion was detected when Cre expression was restricted to pancreas, lung or intestine7. Therefore, it was surmised that negative selection “prunes” the repertoire with an efficiency proportional to the level of self-antigen expression in the thymus, but does not completely eliminate self-reactive T cells6–8.
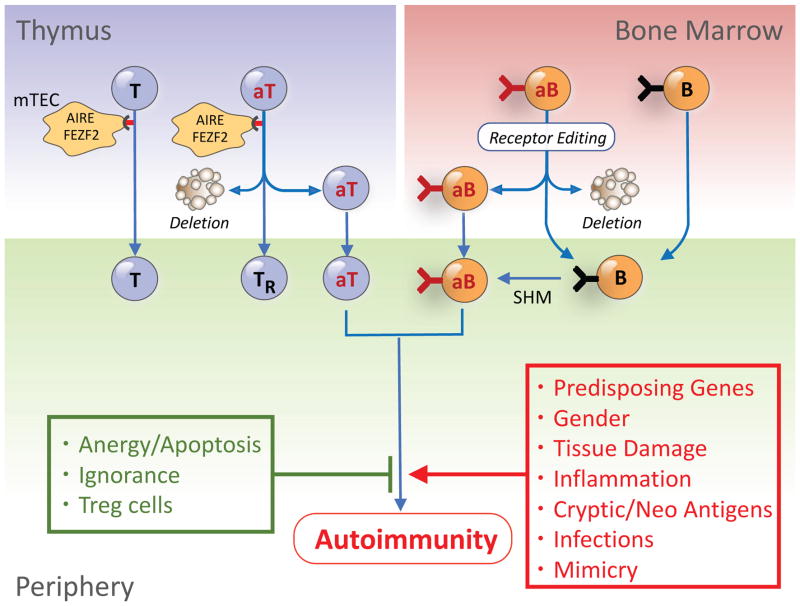
Figure 1. Escape of autoreactive T and B cells from central tolerance and engagement in the periphery.
During differentiation, T and B cell precursors with self-reactivity are positively selected in the thymic cortex and bone marrow, respectively, and those with low avidity for self are exported to the periphery. In contrast, autoreactive T cells (aT) with high avidity for self-antigens expressed by medullary thymic epithelial cells (mTEC) under the control of AIRE or FEZF2 are deleted or differentiate to Treg cells (TR), while autoreactive B cells (aB) are deleted or receptor-edited. Central tolerance, however, is incomplete, and some autoreactive T and B cells are exported to the periphery. The exported cells are normally controlled by peripheral tolerance mechanisms, including inhibitory molecules, anergy, ignorance and suppression by Treg cells. However, in genetically-predisposed individuals, tissue damage, inflammation, and presentation of sequestered, cryptic, neo self-antigens or microbial mimics might provoke break of tolerance and autoimmunity.
A prime example of autoimmunity due to inadequate central deletion of autoreactive T cells is the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED or APS-1) syndrome, a rare autosomal recessive disease caused by mutations of the autoimmune regulator (AIRE) gene9,10. AIRE, a transcription regulator that binds to and activates superenhancers, mediates the promiscuous expression of peripheral tissue-restricted self-molecules in a stochastic manner in individual medullary thymic epithelial cells11,12. AIRE also regulates genes involved in antigen presentation and production of chemokines that modulate the density and function of thymic DCs as well as regulatory T (Treg) cell development13. Interestingly, B cells migrating into the thymus also express AIRE and contribute to T cell repertoire selection14. APECED is characterized by T cell-mediated destruction of multiple endocrine organs with considerable heterogeneity in phenotype, suggesting contributions by additional predisposing genes and environmental factors. Recent studies identified patients with dominant-negative monoallelic AIRE mutations associated with later onset, milder disease and reduced penetrance, but with a higher frequency in mixed populations15,16. A syndrome similar to APECED develops in AIRE-deficient mice, and reduced AIRE expression in heterozygous mice exacerbated T1D and collagen-arthritis. FEZF2 is another transcription factor that controls thymic expression of tissue-restricted antigens mostly non-overlapping with those affected by AIRE, and targeted Fezf2-deficiency in thymic epithelial cells also results in autoantibodies and inflammatory infiltrates in various organs17.
Like T cells, some autoreactive B cells escape central tolerance. Thus, large fractions of early immature B cells (~55 to 75%) in humans display autoreactivity, and this frequency progressively declines to ~40% in bone marrow immature B cells and peripheral transitional B cells, and finally to ~20% in mature naïve B cells18,19. These decreases occur at several checkpoints, starting with receptor editing and apoptosis early in ontogeny, followed by anergy induction prior to or immediately after emigration to the periphery20. Despite these checkpoints, polyspecific autoreactive B cells are present in the peripheral repertoire, and polyspecific natural autoantibodies are detectable in normal individuals21. Natural autoantibodies are typically germline-encoded, of the IgM isotype, non-pathogenic, and may act as transporters for disposal of cell debris or as a defense mechanism by preventing microbe dissemination to vital organs. It has been suggested, however, that polyspecific B cells may undergo somatic hypermutation and class switching to produce high-affinity IgG pathogenic autoantibodies. This is supported by the high frequency of polyspecific B cell clones in SLE, RA, T1D, Sjogren’s syndrome, and MS19,20,22, but it is unclear how such cells contribute to these distinct disease phenotypes.
Activation of escaped autoreactive cells
Because of the significant escape of autoreactive cells from central tolerance, several critical questions arise: Are there more escaping T and B cells in individuals with autoimmune diseases? Under what circumstances are these cells activated and mediate pathogenicity? How are their potentially damaging effects normally averted? Four mechanisms contribute to the control of escaping autoreactive T and B cells: inhibitory molecules, anergy, ignorance, and active suppression (Fig. 1). Several inhibitory molecules (e.g. CTLA-4, PD-1, LAG-3, TIM3, VISTA, TIGIT, FcγRIIb, certain Siglecs) are expressed on the surface of T and B cells to curtail excessive immune responses, both normal and anti-self. Deficiency of some of these molecules leads to autoimmunity, providing strong evidence that autoreactive lymphocytes are present in the peripheral repertoire but are normally under control23–28. Importantly, blockade of these inhibitory molecules by specific antibodies has recently emerged as an effective anti-tumor approach, referred to as “checkpoint immunotherapy”29. However, as expected, a wide range of immune-related adverse events due to unchecked autoreactivity frequently occurs30.
T cell anergy, an acquired state of functional unresponsiveness, is a consequence of TCR engagement in the absence of costimulatory signals31. Recent thymic emigrants to the periphery exhibit increased susceptibility to anergy in the absence of inflammation32. The anergic state is controlled by molecules that negatively regulate proximal TCR signaling, in conjunction with active transcriptional silencing, particularly at the IL2 locus, and induction of regulatory factors. Anergic CD4+ T cells with distinct phenotypic and gene expression programs may convert to Treg cells that, in turn, can promote anergy of pathogenic CD4+ T cells and inhibit autoimmunity33. However, T cell anergy is not a long-lived state and can be reversed under inflammatory conditions.
Approximately 5–7% of peripheral B cells appear to be in an anergic state, and transitional T3 B cells in the spleen may be anergic rather than arrested at an intermediate developmental stage34. Because of the short half-life of anergic B cells (~5 days vs. 40 days for follicular B cells), the frequency of newly emerging B cells that undergo anergy is estimated to be much higher, perhaps up to 50%. Upon stimulation, anergic B cells show impaired activation, proliferation and antibody secretion due to inefficient signal transduction and intracellular Ca2+ upregulation35. The anergic state is controlled by continuous low-level interaction with antigen and by a negative feedback circuitry partly mediated by the tyrosine kinase Lyn, the tyrosine phosphatase SHP-1, and the inositol phosphatase SHIP-1, and conditional B cell deficiency of any of these molecules promotes systemic autoimmunity in mice36,37. Anergic B cells, however, are not deleted and could potentially serve as a self-reactive reservoir. Indeed, reversion of IgMlo anergic B cells under inflammatory conditions has been suggested to contribute to autoimmune syndromes in humans with RA, SLE and T1D.
An issue not fully addressed is how potential acquisition of self-reactivity by somatically hypermutated B cells is controlled. One potential mechanism is that autoreactive B cells may compete poorly for cognate T cell help essential for B cell survival in germinal centers. In addition, B cells expressing BCR with specificity for high-density membrane antigens may be deleted by a Fas-dependent mechanism.
Autoreactive T and B cells exported to the periphery may also remain quiescent due to ignorance of tissue-specific antigens sequestered behind anatomic barriers. This concept is especially applicable to tissues defined as immunologically-privileged sites, such as eye, brain and testis. However, sequestration of peripheral tissue antigens can be broken by infectious agents or other causes of tissue damage, leading to engagement of ignorant autoreactive cells and disease development. Such an event is contingent on several factors, including the nature and dose of the antigen, number of exposures, frequency of activated T cells, and upregulation of MHC and costimulatory molecules in the afflicted tissues.
Exported self-reactive lymphocytes can be activated by several other mechanisms. Thus, recognition of cryptic determinants not adequately presented in the thymus or bone marrow may be enhanced in the periphery under inflammatory conditions38. Another trigger might be recognition of neo-self antigens generated by mutations, post-translational and chemical modifications, or covalent cross-linking of different self-peptides and formation of hybrid epitopes39,40. Molecular mimicry by foreign antigens with sufficient sequence or conformational similarity to self-antigens can also result in activation of non-tolerant lymphocytes41,42. Another mechanism by which microbes can promote autoimmunity is co-capture of self-antigens together with viral antigens by B cells, leading to self-antigen presentation, T cell engagement and disease43. Examples of the above mechanisms have been reported in experimental models, but information on their involvement in the pathogenesis of human autoimmune diseases is limited.
The conundrum of strongly self-reactive Treg cells
Several cell types exert suppressive activities on innate and adaptive immune responses, of which the CD4+CD25+FOXP3+ Treg cell subset is considered the most relevant44,45. Treg cells primarily develop in the thymus (natural Treg) but can also be generated in the periphery (induced Treg). Generation of thymic Treg cells is constrained by a niche defined by antigen presentation and interleukin 2 (IL-2) production by thymic DCs, as well as by a feedback competition for IL-2 by mature Treg cells that recirculate to the thymus46,47. Treg cells target all major immunocyte subsets, and cell-to-cell contact is necessary for the suppressive effect, documented by several approaches including in vivo imaging showing co-clusters of Treg and activated autoreactive T cells in secondary lymphoid tissues48. Suppression is mediated by inhibitory molecules (CTLA-4, IL-10, TGF-β, IL-35), cytolysis, interference with metabolic processes, or modulation of DC maturation and function. Metabolic signatures differ between human Treg and conventional T cells during activation and expansion49,50. Interestingly, Treg cells generated in the perinatal period persist and effectively inhibit autoimmunity throughout life51. The overall frequency of the polyclonal Treg cell population is approximately 5–15% of CD4+ T cells, but the ratio between antigen-specific Treg and effector T cells decreases during an ongoing immune response, presumably to improve anti-pathogen immunity52. Treg cells may also promote tissue repair in response to inflammatory factors released from damaged cells53 and exert a regenerative effect in the CNS54 and skeletal muscle55. These findings suggest an additional important function of Treg cells beyond suppression.
FOXP3 is essential for Treg cell development and function, and mutations in this transcription factor cause the Scurfy phenotype in mice and the immunodysregulation, polyendocrinopathy, enteropathy (IPEX) syndrome in humans44,45. Interestingly, Treg cell-specific superenhancers were shown to be required for Treg development56. Treg cells also exhibit high activity of PP2A, a serine-threonine phosphatase involved in controlling the mTORC1 pathway, and specific ablation of PP2A in Treg cells caused a severe multi-organ autoimmune disorder57.
Notably, thymus-derived Treg cells express TCRs with higher avidity for self-peptide–MHCII than conventional T cells. Accordingly, in a model of neuroinflammation, conventional T cells engineered to express myelin oligodendrocyte glycoprotein (MOG)-specific TCRs derived from Treg cells exhibited higher functional avidity and were more pathogenic than natural conventional T cells, whereas Treg cells expressing MOG-specific TCRs from conventional T cells suppressed disease less efficiently than natural Treg cells58. High self-reactivity of Treg cells is further supported by the finding that TCRs displayed by conventional T cells infiltrating target lesions in Aire−/− mice were frequently expressed by FOXP3+ Treg cells in Aire+/+ mice59. Thus, AIRE appears to promote both deletion of high affinity autoreactive T cells and differentiation of intermediate affinity clones to FOXP3+ Treg cells for peripheral self-antigens.
The extent to which numerical or functional abnormalities in Treg cells contribute to human autoimmune diseases has been difficult to ascertain due to considerable variation across studies in patient selection and undefined antigen specificities of Treg cells. Nevertheless, encouraging results have been reported in various experimental models of autoimmunity using Treg cell expansion in vivo or adoptive transfer of in vitro-propagated Treg cells60,61. Application of these findings to the treatment of human autoimmune diseases, however, has been limited62, and some concerns have been raised because of the potential conversion of Treg cells to pathology-inducing effectors under inflammatory conditions63,64. Moreover, certain issues pertaining to the biology of Treg cells need further clarification, including the mechanisms by which these self-reactive cells escape central deletion, the molecular programs that confer the ability to inhibit autoimmune responses while allowing conventional responses, and the specific abnormalities contributing to the pathogenesis of human autoimmune diseases.
Nucleic acid sensing as initial trigger of autoimmunity
The study of autoimmune diseases has long centered on the adaptive immune system. However, the discovery that innate cells express a broad spectrum of sensors for foreign and self-ligands has shifted the focus in recent years to the innate immune system, the engagement of which precedes and ignites adaptive responses65–67. Thus, endosomal and cytosolic sensors that recognize foreign and self-nucleic acids have been directly implicated in the pathogenesis of autoimmune diseases68. These endosomal sensors include TLR3 for dsRNA, TLR7 and TLR8 for ssRNA, and TLR9 for DNA, whereas the cytosolic sensors include the helicases RIG-I for uncapped 5′-triphosphate RNA and MDA5 for long dsRNA, as well as multiple DNA sensors, of which the cGAS-cGAMP-STING pathway appears the most relevant69,70 (Fig. 2). Responses by these sensors induce the production of type I interferon (IFN-I) and pro-inflammatory cytokines (e.g. IL-1, IL-6, IL12, TNF).
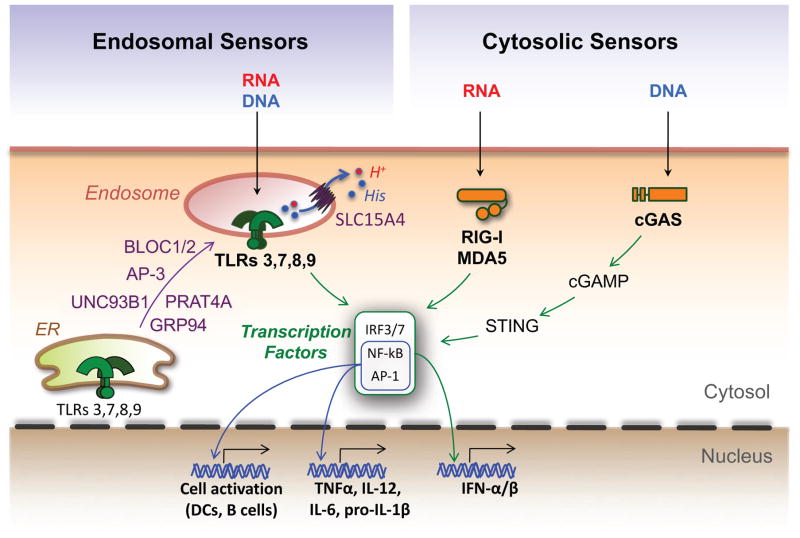
Figure 2. Engagement of endosomal or cytosolic nucleic acid sensors as central events in inflammatory responses.
Nucleic acid sensors are critical innate immune receptors that reside either in endolysosomes or the cytosol. Upon recognition of specific ligands, they initiate a signaling cascade resulting in the activation of several transcription factors that promote cell activation and production of type I interferons (IFN-I) and inflammatory cytokines. Two ER molecules, GRP94 and PRAT4A, act in concert to ensure proper folding of TLRs 3, 7, 8 and 9 and exit from the ER, while UNC93B1 mediates TLR transport to endolysosomes, where ligand recognition takes place. Other proteins participating in TLR trafficking and/or function are the adaptor protein 3 (AP-3), the biogenesis of lysosome organelle complex 1 and 2 (BLOC1/2), and the solute carrier 15A4 (SLC15A4), a molecule known to transport protons (H+), histidine (His) and selected peptides from endosomes to the cytosol. In the cytosol, RNA is sensed by the helicases RIG-I and MDA5, while DNA is primarily sensed by the cyclic GMP-AMP synthase (cGAS). Engagement of cGAS leads to synthesis of the second messenger cGAMP that interacts with the stimulator of interferon genes (STING) to promote inflammatory responses.
Retrospectively, the early findings of high concentrations of IFN-I in serum and dominance of IFN-I-inducible transcripts in PBMCs of lupus patients were the initial hints for a role of innate sensors in autoimmunity71. In addition, in vitro studies showed that complexes of lupus serum IgG with necrotic or apoptotic materials induced IFN-I production by plasmacytoid dendritic cells (pDCs) and promoted TLR9-dependent B cell proliferation72,73. Documentation of the primary role of IFN-I, specifically IFN-α, was provided by disease reduction in lupus-predisposed mice lacking IFNAR or treated with an IFNAR-blocking antibody, while IFN-β deficiency was ineffective74,75. Concurrent studies showed that Tlr9 deletion reduced anti-DNA autoantibody titers, but not overall disease, whereas Tlr7 deletion reduced both anti-RNP autoantibodies and kidney disease76, suggesting TLR7 is more pathogenic than TLR9, likely due to stronger signaling or higher availability of cell death-derived RNA-containing microparticles. Interestingly, a duplication of the Tlr7 gene due to a translocation from the X to Y chromosome enhanced disease in male BXSB lupus mice77,78. Autoimmunity in Tlr7 transgenic mice was reported to be dependent on B cell autophagy79, and defects in non-canonical autophagy or in the engulfment and clearance of dying cells have been associated with lupus-like autoimmunity in mice80. Disease reduction was more evident in Tlr7/9 double-deleted mice76 and especially in Unc93b1 mutants, in which defective TLR trafficking from ER to endolysosomes compromises responses to nucleic acids81. TLR responses to nucleic acids are also impaired by mutations in AP-3, BLOC-1 or BLOC-2, molecules critical for lysosome-related organelle trafficking and biogenesis in diverse cell types82, but the role of these molecules in autoimmunity has not yet been assessed. Notably, Unc93b1 inactivation in lupus mice reduced not only anti-nuclear antibodies (ANA) but also the broad spectrum of autoantibodies against several self-antigens (cardiolipin, myeloperoxidase, β2-glycoprotein, erythrocytes), implying that nucleic acids are potent endogenous adjuvants for autoimmune responses against diverse nucleic acid-associated self-molecules83. Disease development required engagement of endosomal TLRs in both B cells and pDCs83–86. Further studies in lupus mice with a mutation of SLC15A4, an endosomal proton-histidine transporter required for TLR responses, but not development, of pDCs82, showed that these cells contribute to disease mainly through production of IFN-I and proinflammatory cytokines84. Expression of SLC15A4 in B cells was also crucial for TLR7-triggered IFN-I and autoantibody production in a pristane-induced mouse lupus model87. Surprisingly, in the MRL-lpr model, DCs that are normally critical for adaptive immune responses were not required for the initial activation of T and B cells but rather for their expansion and the ensuing tissue damage, and kidney disease was dependent on signaling by the TLR adaptor MyD88 in B cells but not in DCs88,89.
Further evidence for a central role of self-nucleic acid recognition in systemic autoimmunity was the finding that gain-of-function mutations of MDA5 (encoded by Ifih1) promoted lupus-like disease in mice90. Moreover, spontaneous oligomerization of MAVS, the main signaling adaptor downstream of MDA5 and RIG-I, was observed in peripheral blood lymphocytes of some SLE patients and correlated with increased IFN-I production and mitochondrial oxidative stress91. Notably, this phenomenon was reduced in sub-Saharan African patients expressing the MAVS-C79F variant with a milder disease. Moreover, accumulation of extracellular or intracellular nucleic acids due to defects in DNase I (or its homologue DNASE1L3), DNase II, or DNase III (TREX1) is associated in mice and humans with various forms of autoimmunity68,92. Lupus-like autoimmunity in Trex1 mutant mice has been attributed to defective digestion of DNA derived from endogenous retroelements93, and a recent study suggested that Ro60, a major autoantigen in SLE and Sjogren’s syndrome, is associated with RNA that is derived from endogenous Alu retroelements and promotes TLR-dependent IFN-I production94. Finally, neutrophil extracellular traps have been shown to induce endolysosomal TLR and IFN-I responses95,96, and recent studies implicated oxidized mitochondrial DNA extruded from anti-RNP autoantibody-activated neutrophils97,98. Several accessory molecules (LL37, HMGB1, RAGE) and uptake of immune complexes by FcγR have been identified as major mechanisms for the access of self-nucleic acids to endolysosomal sensors68. Notably, in addition to IgG autoantibodies, DNA-reactive IgE can also enhance IFN-I responses by pDCs and contribute to SLE immunopathology99.
Overall, a unified concept has emerged in which the autoimmune pathologic processes are initiated by the engagement of innate sensors by nucleic acids (Fig. 3). Most of the cited examples relate to SLE, but this mechanism appears to be applicable to a broad spectrum of systemic and organ-specific autoimmune diseases (e.g. RA, Sjogren’s syndrome, polyomyositis/dermatomyositis, psoriasis, T1D, autoimmune thyroiditis, and neuromyelitis optica)68. Self-nucleic acids acting under sterile conditions are frequently the initial trigger, but microbial nucleic acids alone or together with self-nucleic acids from damaged tissues may also contribute. Thus, nucleic acid sensing, an essential evolutionarily-acquired mechanism to protect against pathogens, can under certain circumstances be a major mediator of pathogenic autoimmunity.
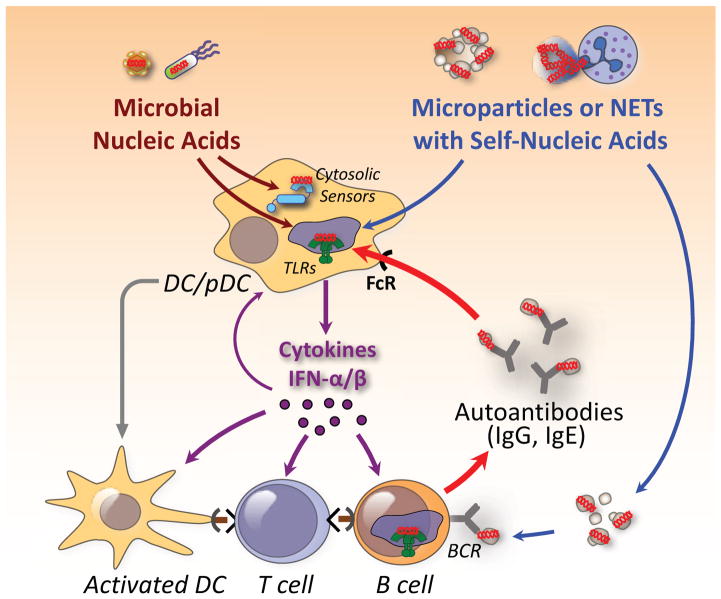
Figure 3. Pathways by which self and foreign nucleic acid sensors promote autoimmunity.
It is postulated that self-nucleic acids in microparticles released from dying cells or in neutrophil extracellular traps (NETs) gain access to acidified endolysosomal compartments of pDCs, DCs, and antigen-specific B cells. TLR engagement and production of inflammatory cytokines causes upregulation of MHC and costimulatory molecules in these cells, antigen presentation, and engagement of autoreactive T cells. Complexes of autoantibodies (IgG, IgE) with nucleic acid-associated molecules are taken up through the FcR and amplify and sustain the inflammatory response. In certain instances, microbial nucleic acids alone or in conjunction with self-nucleic acids released from damaged tissues may constitute the initial trigger.
The hidden microbial “self” and autoimmunity
The microbiota, an ecosystem of microorganisms residing in mucosal surfaces and skin in a mutually-beneficial coexistence with the host, influences numerous physiologic processes, including organismal evolution, longevity, metabolism and immune system development and function. However, a large body of recent publications has now revealed that disturbances in this ecosystem, referred to as dysbiosis, can lead to a plethora of pathologic processes, including autoimmune diseases affecting not only the gut, the largest niche for the microbiota, but also several distant organs100,101 (Fig. 4).
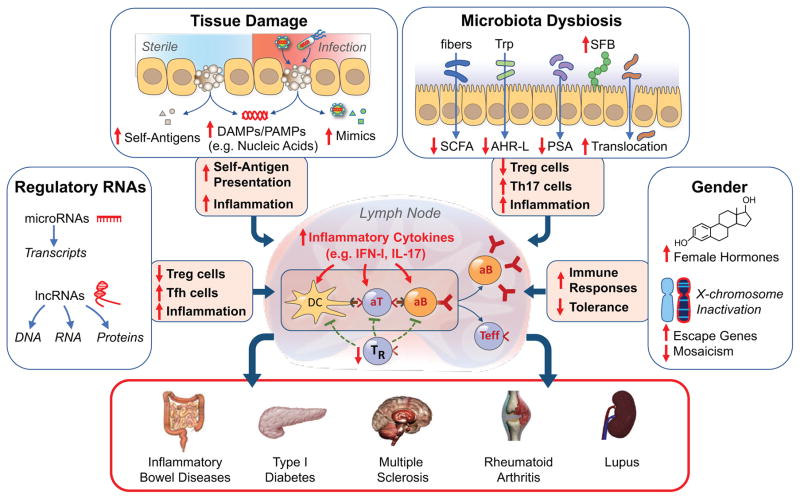
Figure 4. The multiple pathways to autoimmunity.
We posit that autoimmunity may result from disturbances in multiple processes acting singly or in combination. Tissue damage under sterile conditions or due to infections may lead to availability of nucleic acids and other damage- or pathogen-associated molecular patterns (DAMPs, PAMPs), presentation of self-antigens to non-tolerant lymphocytes, and induction of inflammatory responses. Microbiota dysbiosis may result in displacement of beneficial commensals, reductions of several anti-inflammatory factors (short chain fatty acids, SCFA; Aryl hydrocarbon receptor ligands, AHR-L; polysaccharide A, PSA), expansion of adherent bacteria (e.g. segmented filamentous bacteria, SFB in mice), damage of the mucosal/epithelial barrier, and translocation of bacteria and inflammatory products to mesenteric lymph nodes. These effects lead to engagement of TLRs and other innate sensors, production of inflammatory cytokines, reduction in Treg cells (TR), expansion of TH17 and other effector cells, and production of autoantibodies, resulting in organ-specific or systemic autoimmune diseases. Additional autoimmunity-contributing factors may include abnormalities in non-coding regulatory RNAs, gender-associated hormonal effects, and incomplete X chromosome inactivation. These processes require a predisposing genetic background for the pathogenic phenotype to be expressed.
IBDs, encompassing Crohn’s disease and ulcerative colitis, were the first to be associated with dysbiosis. Initial studies showed significant taxonomic shifts in gut microbiota of individuals with these syndromes, including decreases in beneficial subtypes of Clostridia and Bacteroides fragilis102. Fermentation of dietary fibers by Clostridia clusters IV, XIVa and XVIII produces short-chain fatty acids (SCFA), particularly butyrate, which exert significant anti-inflammatory functions, promote peripheral Treg cell generation, and are key nutrients for colonocytes103–106. Moreover, subtypes of B. fragilis and certain other intestinal bacteria produce capsular polysaccharide A (PSA), which provides immunoprotection through induction of IL-10-producing Treg cells, and administration of B. fragilis or even PSA alone corrected immune defects in germ-free mice and had protective effects in models of colitis107,108. Further, the intestinal tissues of IBD patients often show downregulation of the aryl hydrocarbon receptor (AHR), which, in response to bacteria-derived indole metabolites, induces production of IL-22 by group 3 innate lymphoid cells (ILC3)109. This effect promotes expression of antimicrobial proteins by epithelial cells, limits expansion of commensal segmented filamentous bacteria (SFB), and reduces activation of inflammatory TH17 cells in mice110,111. IBD patients also show increases in Escherichia coli and other bacteria strains with epithelium adhesive properties that promote inflammatory TH17 responses, and NOD2 mutations linked to Crohn’s disease are sometimes associated with compositional shifts to inflammation-promoting gut microbiota100.
Complex microbiota disturbances have been implicated in the pathogenesis of T1D in both the NOD mouse and humans112. Interestingly, longitudinal studies in children showed reductions in microbiota diversity and butyrate-producing bacteria in the time period between seroconversion to autoantibody positivity and diagnosis, although autoantibody-positive individuals without overt disease showed no significant differences in bacteria diversity compared to controls112. Another study showed that the dominant gut microbial taxa of infants from countries where early-onset T1D is common produced an immunoinhibitory lipopolysaccharide (LPS), whereas the dominant bacteria of infants from regions where early-onset T1D is less prevalent produced an immunostimulatory LPS113. Notably, injection of immunostimulatory, but not immunoinhibitory, LPS delayed onset and reduced incidence of diabetes in NOD mice. These results suggest that early innate immune stimulation may reduce autoimmune disease predisposition and provide a potential mechanistic explanation for the “hygiene hypothesis” of autoimmunity.
Dysbiosis has also been implicated in diseases at sites distant to the gut, including a spectrum of neurologic disorders such as anxiety, stress, migraines, depression, neurodegenerative and neuroinflammatory diseases. With regard to neuroinflammation, the main supporting evidence is that germ-free mice are refractory to experimental autoimmune encephalomyelitis (EAE), but susceptibility is restored by colonization with TH17-promoting SFB. In contrast, PSA-expressing B. fragilis or other bacteria that metabolize dietary fiber to SCFA or tryptophan to AHR agonists reduced neuroinflammation in the EAE model114,115. Regarding multiple sclerosis, studies are limited and, although changes in certain gut bacteria species have been associated with disease, there is lack of consensus for the specific bacteria implicated116.
Microbiota disturbances also reportedly affect arthritis in mice and humans. For example, gut microbiota is required for the development of arthritis in the K/BxN, SKG, IL1rn−/−, and HLA-B27 models, while dysbiosis has been reported in humans with RA, reactive arthritis, psoriatic arthritis and spondyloarthritis117. In RA, contributions by oral and gut microbiota, particularly Porphyromonas gingivalis and Prevotella copri, have been implicated117. Interestingly, P. gingivalis expresses peptidylarginine deiminase (PAD) and the cysteine-like proteases gingipains that may promote citrullination of PAD and self-antigens and induction of anti-citrullinated peptide antibodies (ACPA) that are almost pathognomonic for RA118. However, additional studies did not confirm enrichment of P. copri and detected P. gingivalis and antibodies to this microbe in non-RA individuals with periodontal disease119. Moreover, another periodontitis-associated microbe (Aggregatibacter actinomycetemcomitans) may contribute to ACPA production in RA by triggering dysregulated activation of citrullinating enzymes in neutrophils and release of citrullinated self-antigens120.
The findings as a whole provide compelling evidence for the astonishing ways dysbiosis can affect a wide spectrum of autoimmune diseases. The strongest evidence is derived from studies in experimental models, while findings in humans are circumstantial, largely based on often discordant surveys of microbiota ecosystems, and cause and effect relationships have not been clearly established. Moreover, there are several limitations and pitfalls in interpreting changes in the microbiota of humans that should be considered before integrating patient-specific data with a given disease. This is particularly relevant since gut microbiota is highly dynamic, exhibits daily cyclical fluctuations related to circadian rhythms and variable bacteria growth rates121,122, and is affected by diet and medications. In addition, the mechanisms by which dysbiosis contributes to autoimmune diseases distant to the gut need more detailed characterization. Nonetheless, despite these caveats, dietetic and other interventions to correct dysbiosis have been advocated as a new therapeutic approach for a broad spectrum of disorders.
Non-coding RNAs as modulators of autoimmunity
Estimates indicate that only ~2% of the mammalian genome encodes proteins, while the vast majority (75–90%) is transcribed as non-coding RNA, including microRNAs (miRNA, 18–23 nucleotides) and long non-coding RNAs (lncRNA, ≥200 nucleotides) with significant effects on both the innate and adaptive immune systems. Functionally, miRNA bind to mRNA targets and mediate gene silencing mostly by translational repression and transcript degradation123, while lncRNA are transcribed in a cell-specific manner and act biochemically by interacting with proteins, DNA or RNA124,125 (Fig. 4).
Recent evidence implicated diverse miRNA in inflammatory and autoimmune processes. For example, transgenic overexpression of the polycistronic miR-17~92 cluster in mice caused lymphoproliferation and lupus-like autoimmunity characterized by accumulation of TFH cells and reduced expression of the tumor suppressor phosphatase PTEN and the anti-apoptotic Bim126. Further studies with specific members of the miR-17~92 cluster showed that miR-19 suppressed expression of PTEN and played a key role in regulating central B cell tolerance, while miR-17 controlled early B cell development127. Defects in B cell tolerance were also noted in mice reconstituted with hematopoietic stem cells transduced with miR-148a, and overexpression of this miRNA accelerated autoimmunity in a lupus mouse model128. Deletions of miR181a, miR185 or Dicer (a molecule required for miRNA biogenesis) were also reported to promote autoimmunity in mouse models, while expression of miR146a in Treg cells was required for inhibition of pathogenic TH1 responses and maintenance of immune tolerance123. Translation of these findings to human autoimmune diseases is at an early stage, but increases or decreases of certain miRNAs and lncRNAs have been detected, sometimes correlating with disease severity129–131.
Gender bias in autoimmunity
It has long been recognized that most autoimmune diseases exhibit considerable gender dimorphism with higher incidence in females132. Two major factors are thought to contribute to this dimorphism: gonadal hormones and direct X chromosome effects (Fig. 4). The contribution of sex hormones has been suggested by the observations that gender bias is more evident after puberty, and that estrogens enhance while androgens suppress immune responses and autoimmunity in lupus-predisposed mice. Female hormones exert broad effects on the expression of multiple immunologically-relevant genes, including inflammatory cytokines and TLR signaling molecules133,134. Estrogens also interfere with B cell tolerance135, and T cell tolerance may also be affected since AIRE expression in thymic epithelium was reported to be downregulated by estrogens and upregulated by androgens136,137. Sex hormones and the microbiota also influence each other, and gender differences in microbiota composition may also contribute to gender bias in autoimmunity138,139.
The gonad-independent role of the X chromosome was first demonstrated in a model using XX and XY mice bearing either ovaries or testes, in which susceptibility to EAE and pristane-induced lupus was dependent on XX regardless of whether the mice developed as females or males140. Three interconnected mechanisms have been proposed to explain direct X chromosome contributions to autoimmunity: escape from X-inactivation, loss of mosaicism, and aneuploidy. X chromosome inactivation is a major epigenetic event that ensures gene dosage compensation in females compared to males. Because this process is random, either the maternal or the paternal X chromosome is inactivated in each cell, resulting in mosaicism in females. X chromosome inactivation is variable and incomplete, with up to ~15% of genes (~200 genes) expressed by both X chromosomes in humans, resulting in increased expression in females vs. males141,142. Interestingly, the degree of escape from X inactivation varies among tissues, individuals and ethnic groups143,144. Depending on the nature of the genes that escape inactivation, the potential effect on autoimmunity may be detrimental if the escaped genes are involved in immune activation (e.g. DDX3X, BTK and TLR7), or beneficial if involved in immunoregulation (e.g. FOXP3). The effect of incomplete X inactivation may be amplified in females with loss of mosaicism in certain tissues or cell populations as well as in individuals with X aneuploidy, e.g. Kleinefelter patients, wherein all but one of the X chromosomes are inactivated145. Because the female bias is not uniform in all autoimmune diseases, it will be of interest to determine the underlying pathogenic processes that influence this bias, and to elucidate why males appear to require a higher cumulative genetic risk for disease development146.
Concluding remarks
This review summarizes the major triggers and pathways involved in the pathogenesis of autoimmune diseases. Each is based on a distinct mechanistic principle, but likely more than one pathway may contribute to a given disease. These pathways have been well established in experimental models, but involvement in human disorders is frequently tentative. Nonetheless, the available data highlight the outstanding progress made thus far in our quest to define the pathogenesis of these highly complex and heterogeneous syndromes, and clearly novel diagnostic and therapeutic approaches will be forthcoming. Application of new technologies may even allow specific elimination of autoreactive cells without broad suppression of the entire immune system, as is currently the practice. An indication of the feasibility of antigen-specific therapies in autoimmunity was provided by recent studies showing depletion of autoantigen-specific B cells using cytotoxic T cells expressing chimeric antigen receptors147 and expansion of autoantigen-specific Treg cells for passive transfer using nanoparticles displaying disease-relevant self-peptides/MHC complexes148. Furthermore, as genetic predisposition is a prerequisite for most autoimmune diseases, it is expected that advances in defining the role of genetic variations in these syndromes will be transformative in our approach to diagnosis and treatment.
Acknowledgments
The work of the authors is supported by Awards AR065919 and AR068910 (National Institute of Arthritis and Musculoskeletal and Skin Diseases) to A.N.T., HL114408 (National Heart, Lung and Blood Institute) to D.H.K., and AI121525 and AI117563 (National Institute of Allergy and Infectious Diseases) to R.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. The authors thank B. Beutler, D. Burton, L. Teyton and D. Nemazee for manuscript review and advice. Due to space limitations, some relevant references could not be cited and the authors apologize for these omissions.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Kono DH, Theofilopoulos AN. Kelley and Firestein’s Textbook of Rheumatology. 10. Elsevier; Philadelphia: 2017. Autoimmunity; pp. 301–317. [Google Scholar]
- 2.Park H, Bourla AB, Kastner DL, Colbert RA, Siegel RM. Lighting the fires within: the cell biology of autoinflammatory diseases. Nat Rev Immunol. 2012;12:570–580. doi: 10.1038/nri3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez-Arcelus M, Rich SS, Raychaudhuri S. Autoimmune diseases - connecting risk alleles with molecular traits of the immune system. Nat Rev Genet. 2016;17:160–174. doi: 10.1038/nrg.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt KA, et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature. 2013;498:232–235. doi: 10.1038/nature12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouneaud C, Kourilsky P, Bousso P. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 2000;13:829–840. doi: 10.1016/s1074-7613(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 6.Yu W, et al. Clonal Deletion Prunes but Does Not Eliminate Self-Specific alphabeta CD8(+) T Lymphocytes. Immunity. 2015;42:929–941. doi: 10.1016/j.immuni.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Legoux FP, et al. CD4+ T Cell Tolerance to Tissue-Restricted Self Antigens Is Mediated by Antigen-Specific Regulatory T Cells Rather Than Deletion. Immunity. 2015;43:896–908. doi: 10.1016/j.immuni.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra D, et al. Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nat Immunol. 2016;17:187–195. doi: 10.1038/ni.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 10.Anderson MS, Su MA. AIRE expands: new roles in immune tolerance and beyond. Nat Rev Immunol. 2016;16:247–258. doi: 10.1038/nri.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal K, Yoshida H, Benoist C, Mathis D. The transcriptional regulator Aire binds to and activates super-enhancers. Nat Immunol. 2017;18:263–273. doi: 10.1038/ni.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meredith M, Zemmour D, Mathis D, Benoist C. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol. 2015;16:942–949. doi: 10.1038/ni.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lei Y, et al. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med. 2011;208:383–394. doi: 10.1084/jem.20102327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamano T, et al. Thymic B Cells Are Licensed to Present Self Antigens for Central T Cell Tolerance Induction. Immunity. 2015;42:1048–1061. doi: 10.1016/j.immuni.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Oftedal BE, et al. Dominant Mutations in the Autoimmune Regulator AIRE Are Associated with Common Organ-Specific Autoimmune Diseases. Immunity. 2015;42:1185–1196. doi: 10.1016/j.immuni.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Anderson MS, Casanova JL. More than Meets the Eye: Monogenic Autoimmunity Strikes Again. Immunity. 2015;42:986–988. doi: 10.1016/j.immuni.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Takaba H, et al. Fezf2 Orchestrates a Thymic Program of Self-Antigen Expression for Immune Tolerance. Cell. 2015;163:975–987. doi: 10.1016/j.cell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 19.Meffre E. The establishment of early B cell tolerance in humans: lessons from primary immunodeficiency diseases. Ann N Y Acad Sci. 2011;1246:1–10. doi: 10.1111/j.1749-6632.2011.06347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemazee D. Mechanisms of central tolerance for B cells. Nat Rev Immunol. 2017;17:281–294. doi: 10.1038/nri.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avrameas S. Natural autoantibodies: from ‘horror autotoxicus’ to ‘gnothi seauton’. Immunol Today. 1991;12:154–159. doi: 10.1016/S0167-5699(05)80045-3. [DOI] [PubMed] [Google Scholar]
- 22.Chamberlain N, et al. Rituximab does not reset defective early B cell tolerance checkpoints. J Clin Invest. 2016;126:282–287. doi: 10.1172/JCI83840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson AM, Sharpe AH. Taming tissue-specific T cells: CTLA-4 reins in self-reactive T cells. Nat Immunol. 2010;11:109–111. doi: 10.1038/ni0210-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat Immunol. 2013;14:1212–1218. doi: 10.1038/ni.2762. [DOI] [PubMed] [Google Scholar]
- 25.Pincetic A, et al. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat Immunol. 2014;15:707–716. doi: 10.1038/ni.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceeraz S, et al. VISTA deficiency accelerates the development of fatal murine lupus nephritis. Arthritis Rheumatol. 2016 doi: 10.1002/art.40020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt H, et al. Siglec-H protects from virus-triggered severe systemic autoimmunity. J Exp Med. 2016;213:1627–1644. doi: 10.1084/jem.20160189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michot JM, et al. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur J Cancer. 2016;54:139–148. doi: 10.1016/j.ejca.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Fathman CG, Lineberry NB. Molecular mechanisms of CD4+ T-cell anergy. Nat Rev Immunol. 2007;7:599–609. doi: 10.1038/nri2131. [DOI] [PubMed] [Google Scholar]
- 32.Friesen TJ, Ji Q, Fink PJ. Recent thymic emigrants are tolerized in the absence of inflammation. J Exp Med. 2016;213:913–920. doi: 10.1084/jem.20151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalekar LA, et al. CD4(+) T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat Immunol. 2016;17:304–314. doi: 10.1038/ni.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarkoni Y, Getahun A, Cambier JC. Molecular underpinning of B-cell anergy. Immunol Rev. 2010;237:249–263. doi: 10.1111/j.1600-065X.2010.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zikherman J, Parameswaran R, Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489:160–164. doi: 10.1038/nature11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Getahun A, Beavers NA, Larson SR, Shlomchik MJ, Cambier JC. Continuous inhibitory signaling by both SHP-1 and SHIP-1 pathways is required to maintain unresponsiveness of anergic B cells. J Exp Med. 2016;213:751–769. doi: 10.1084/jem.20150537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamagna C, Hu Y, DeFranco AL, Lowell CA. B cell-specific loss of Lyn kinase leads to autoimmunity. J Immunol. 2014;192:919–928. doi: 10.4049/jimmunol.1301979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad S, Starck SR, Shastri N. Presentation of Cryptic Peptides by MHC Class I Is Enhanced by Inflammatory Stimuli. J Immunol. 2016 doi: 10.4049/jimmunol.1502045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doyle HA, Mamula MJ. Autoantigenesis: the evolution of protein modifications in autoimmune disease. Curr Opin Immunol. 2012;24:112–118. doi: 10.1016/j.coi.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delong T, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351:711–714. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldstone MB. Molecular mimicry and autoimmune disease. Cell. 1987;50:819–820. doi: 10.1016/0092-8674(87)90507-1. [DOI] [PubMed] [Google Scholar]
- 42.Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanderson NS, et al. Cocapture of cognate and bystander antigens can activate autoreactive B cells. Proc Natl Acad Sci U S A. 2017;114:734–739. doi: 10.1073/pnas.1614472114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morikawa H, Sakaguchi S. Genetic and epigenetic basis of Treg cell development and function: from a FoxP3-centered view to an epigenome-defined view of natural Treg cells. Immunol Rev. 2014;259:192–205. doi: 10.1111/imr.12174. [DOI] [PubMed] [Google Scholar]
- 45.Li MO, Rudensky AY. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol. 2016;16:220–233. doi: 10.1038/nri.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiault N, et al. Peripheral regulatory T lymphocytes recirculating to the thymus suppress the development of their precursors. Nat Immunol. 2015;16:628–634. doi: 10.1038/ni.3150. [DOI] [PubMed] [Google Scholar]
- 47.Weist BM, Kurd N, Boussier J, Chan SW, Robey EA. Thymic regulatory T cell niche size is dictated by limiting IL-2 from antigen-bearing dendritic cells and feedback competition. Nat Immunol. 2015;16:635–641. doi: 10.1038/ni.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Z, et al. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature. 2015;528:225–230. doi: 10.1038/nature16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Procaccini C, et al. The Proteomic Landscape of Human Ex Vivo Regulatory and Conventional T Cells Reveals Specific Metabolic Requirements. Immunity. 2016;44:406–421. doi: 10.1016/j.immuni.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newton R, Priyadharshini B, Turka LA. Immunometabolism of regulatory T cells. Nat Immunol. 2016;17:618–625. doi: 10.1038/ni.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015;348:589–594. doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su LF, Del Alcazar D, Stelekati E, Wherry EJ, Davis MM. Antigen exposure shapes the ratio between antigen-specific Tregs and conventional T cells in human peripheral blood. Proc Natl Acad Sci U S A. 2016;113:E6192–E6198. doi: 10.1073/pnas.1611723113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arpaia N, et al. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dombrowski Y, et al. Regulatory T cells promote myelin regeneration in the central nervous system. Nature neuroscience. 2017 doi: 10.1038/nn.4528. [DOI] [PMC free article] [PubMed]
- 55.Kuswanto W, et al. Poor Repair of Skeletal Muscle in Aging Mice Reflects a Defect in Local, Interleukin-33-Dependent Accumulation of Regulatory T Cells. Immunity. 2016;44:355–367. doi: 10.1016/j.immuni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitagawa Y, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol. 2017;18:173–183. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Apostolidis SA, et al. Phosphatase PP2A is requisite for the function of regulatory T cells. Nat Immunol. 2016;17:556–564. doi: 10.1038/ni.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kieback E, et al. Thymus-Derived Regulatory T Cells Are Positively Selected on Natural Self-Antigen through Cognate Interactions of High Functional Avidity. Immunity. 2016;44:1114–1126. doi: 10.1016/j.immuni.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 59.Malchow S, et al. Aire Enforces Immune Tolerance by Directing Autoreactive T Cells into the Regulatory T Cell Lineage. Immunity. 2016;44:1102–1113. doi: 10.1016/j.immuni.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyara M, Ito Y, Sakaguchi S. TREG-cell therapies for autoimmune rheumatic diseases. Nat Rev Rheumatol. 2014;10:543–551. doi: 10.1038/nrrheum.2014.105. [DOI] [PubMed] [Google Scholar]
- 61.Spence A, Klementowicz JE, Bluestone JA, Tang Q. Targeting Treg signaling for the treatment of autoimmune diseases. Curr Opin Immunol. 2015;37:11–20. doi: 10.1016/j.coi.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visperas A, Vignali DA. Are Regulatory T Cells Defective in Type 1 Diabetes and Can We Fix Them? J Immunol. 2016;197:3762–3770. doi: 10.4049/jimmunol.1601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo J, Zhou X. Regulatory T cells turn pathogenic. Cellular & molecular immunology. 2015;12:525–532. doi: 10.1038/cmi.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 66.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 67.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Theofilopoulos AN, Kono DH, Beutler B, Baccala R. Intracellular nucleic acid sensors and autoimmunity. J Interferon Cytokine Res. 2011;31:867–886. doi: 10.1089/jir.2011.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 70.Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 71.Crow MK. Type I interferon in the pathogenesis of lupus. J Immunol. 2014;192:5459–5468. doi: 10.4049/jimmunol.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ronnblom L. The type I interferon system in the etiopathogenesis of autoimmune diseases. Upsala journal of medical sciences. 2011;116:227–237. doi: 10.3109/03009734.2011.624649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santiago-Raber ML, et al. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197:777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baccala R, et al. Anti-IFN-alpha/beta Receptor Antibody Treatment Ameliorates Disease in Lupus-Predisposed Mice. J Immunol. 2012;189:5976–5984. doi: 10.4049/jimmunol.1201477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Christensen SR, et al. Toll-like Receptor 7 and TLR9 Dictate Autoantibody Specificity and Have Opposing Inflammatory and Regulatory Roles in a Murine Model of Lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 78.Subramanian S, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U S A. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weindel CG, et al. B cell autophagy mediates TLR7-dependent autoimmunity and inflammation. Autophagy. 2015;11:1010–1024. doi: 10.1080/15548627.2015.1052206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martinez J, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533:115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Kono DH, et al. Endosomal TLR signaling is required for anti-nucleic acid and rheumatoid factor autoantibodies in lupus. Proc Natl Acad Sci U S A. 2009;106:12061–12066. doi: 10.1073/pnas.0905441106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blasius AL, et al. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koh YT, et al. Role of Nucleic Acid-Sensing TLRs in Diverse Autoantibody Specificities and Anti-Nuclear Antibody-Producing B Cells. J Immunol. 2013;190:4982–4990. doi: 10.4049/jimmunol.1202986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baccala R, et al. Essential requirement for IRF8 and SLC15A4 implicates plasmacytoid dendritic cells in the pathogenesis of lupus. Proc Natl Acad Sci U S A. 2013;110:2940–2945. doi: 10.1073/pnas.1222798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rowland SL, et al. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J Exp Med. 2014;211:1977–1991. doi: 10.1084/jem.20132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sisirak V, et al. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J Exp Med. 2014;211:1969–1976. doi: 10.1084/jem.20132522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kobayashi T, et al. The histidine transporter SLC15A4 coordinates mTOR-dependent inflammatory responses and pathogenic antibody production. Immunity. 2014;41:375–388. doi: 10.1016/j.immuni.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 88.Teichmann LL, et al. Dendritic cells in lupus are not required for activation of T and B cells but promote their expansion, resulting in tissue damage. Immunity. 2010;33:967–978. doi: 10.1016/j.immuni.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teichmann LL, Schenten D, Medzhitov R, Kashgarian M, Shlomchik MJ. Signals via the adaptor MyD88 in B cells and DCs make distinct and synergistic contributions to immune activation and tissue damage in lupus. Immunity. 2013;38:528–540. doi: 10.1016/j.immuni.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Funabiki M, et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 91.Buskiewicz IA, et al. Reactive oxygen species induce virus-independent MAVS oligomerization in systemic lupus erythematosus. Sci Signal. 2016;9:ra115. doi: 10.1126/scisignal.aaf1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sisirak V, et al. Digestion of Chromatin in Apoptotic Cell Microparticles Prevents Autoimmunity. Cell. 2016;166:88–101. doi: 10.1016/j.cell.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hung T, et al. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science. 2015;350:455–459. doi: 10.1126/science.aac7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garcia-Romo GS, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lande R, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Caielli S, et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med. 2016;213:697–713. doi: 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lood C, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016;22:146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Henault J, et al. Self-reactive IgE exacerbates interferon responses associated with autoimmunity. Nat Immunol. 2016;17:196–203. doi: 10.1038/ni.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 101.Ruff WE, Kriegel MA. Autoimmune host-microbiota interactions at barrier sites and beyond. Trends Mol Med. 2015;21:233–244. doi: 10.1016/j.molmed.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 106.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 107.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 108.Neff CP, et al. Diverse Intestinal Bacteria Contain Putative Zwitterionic Capsular Polysaccharides with Anti-inflammatory Properties. Cell Host Microbe. 2016;20:535–547. doi: 10.1016/j.chom.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Monteleone I, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248. 248e231. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 110.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 111.Qiu J, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12:154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 113.Vatanen T, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:1551. doi: 10.1016/j.cell.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 114.Haghikia A, et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity. 2015;43:817–829. doi: 10.1016/j.immuni.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 115.Rothhammer V, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Glenn JD, Mowry EM. Emerging Concepts on the Gut Microbiome and Multiple Sclerosis. J Interferon Cytokine Res. 2016;36:347–357. doi: 10.1089/jir.2015.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scher JU, Littman DR, Abramson SB. Microbiome in Inflammatory Arthritis and Human Rheumatic Diseases. Arthritis Rheumatol. 2016;68:35–45. doi: 10.1002/art.39259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wegner N, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and alpha-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010;62:2662–2672. doi: 10.1002/art.27552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang X, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 120.Konig MF, et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci Transl Med. 2016;8:369ra176. doi: 10.1126/scitranslmed.aaj1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 122.Korem T, et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015;349:1101–1106. doi: 10.1126/science.aac4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16:279–294. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- 124.Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med. 2014;20:623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 126.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lai M, et al. Regulation of B-cell development and tolerance by different members of the miR-17 approximately 92 family microRNAs. Nat Commun. 2016;7:12207. doi: 10.1038/ncomms12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gonzalez-Martin A, et al. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat Immunol. 2016;17:433–440. doi: 10.1038/ni.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garo LP, Murugaiyan G. Contribution of MicroRNAs to autoimmune diseases. Cell Mol Life Sci. 2016;73:2041–2051. doi: 10.1007/s00018-016-2167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu GC, et al. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14:798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 131.Castellanos-Rubio A, et al. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91–95. doi: 10.1126/science.aad0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rubtsova K, Marrack P, Rubtsov AV. Sexual dimorphism in autoimmunity. J Clin Invest. 2015;125:2187–2193. doi: 10.1172/JCI78082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Markle JG, Fish EN. SeXX matters in immunity. Trends Immunol. 2014;35:97–104. doi: 10.1016/j.it.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 134.Ter Horst R, et al. Host and Environmental Factors Influencing Individual Human Cytokine Responses. Cell. 2016;167:1111–1124 e1113. doi: 10.1016/j.cell.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grimaldi CM, Jeganathan V, Diamond B. Hormonal regulation of B cell development: 17 beta-estradiol impairs negative selection of high-affinity DNA-reactive B cells at more than one developmental checkpoint. J Immunol. 2006;176:2703–2710. doi: 10.4049/jimmunol.176.5.2703. [DOI] [PubMed] [Google Scholar]
- 136.Dragin N, et al. Estrogen-mediated downregulation of AIRE influences sexual dimorphism in autoimmune diseases. J Clin Invest. 2016;126:1525–1537. doi: 10.1172/JCI81894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhu ML, et al. Sex bias in CNS autoimmune disease mediated by androgen control of autoimmune regulator. Nat Commun. 2016;7:11350. doi: 10.1038/ncomms11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Markle JG, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 139.Yurkovetskiy L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smith-Bouvier DL, et al. A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med. 2008;205:1099–1108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 142.Brooks WH. X chromosome inactivation and autoimmunity. Clin Rev Allergy Immunol. 2010;39:20–29. doi: 10.1007/s12016-009-8167-5. [DOI] [PubMed] [Google Scholar]
- 143.Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang Y, et al. Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol Biol Evol. 2013;30:2588–2601. doi: 10.1093/molbev/mst148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Harris VM, et al. Klinefelter’s syndrome (47,XXY) is in excess among men with Sjogren’s syndrome. Clin Immunol. 2016;168:25–29. doi: 10.1016/j.clim.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hughes T, et al. Analysis of autosomal genes reveals gene-sex interactions and higher total genetic risk in men with systemic lupus erythematosus. Ann Rheum Dis. 2012;71:694–699. doi: 10.1136/annrheumdis-2011-200385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ellebrecht CT, et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science. 2016;353:179–184. doi: 10.1126/science.aaf6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clemente-Casares X, et al. Expanding antigen-specific regulatory networks to treat autoimmunity. Nature. 2016;530:434–440. doi: 10.1038/nature16962. [DOI] [PubMed] [Google Scholar]