Abstract
Background
Circulating cell-free DNA (cfDNA) and its integrity index may represent a rapid and noninvasive “liquid biopsy” biomarker, which gives important complementary information for diagnosis, prognosis, and treatment stratification in cancer patients. The aim of our study was to evaluate the possible role of cfDNA and its integrity index as a complementary tool for endometrial cancer (EC) management.
Methods
Alu-quantitative real-time PCR (qPCR) analysis wasprformed on 60 serum samples from preoperative EC patients randomly recruited. Both cfDNA content and DNA integrity index were measured by qPCR-Alu115 (representing total cfDNA) and qPCR-Alu247 (corresponding to high molecular weight DNA) and correlated with clinicopathologic characteristics. Lymphovascular space invasion (LVSI) was detected by hematoxylin and eosin staining. In case of doubt, LVSI status was further evaluate by immunohistochemistry using anti-CD31 and anti-CD34 antibodies.
Results
Total cfDNA content significantly increases in high grade EC. A significant decrease of DNA integrity index was detected in the subset of hypertensive and obese high grade EC. Serum DNA integrity was higher in samples with LVSI. The ordinal regression analysis predicted a significant correlation between decreased integrity index values and hypertension specifically in tumors presenting LVSI.
Conclusions
Our study supports the utility of serum DNA integrity index as a noninvasive molecular biomarker in EC. We show that a correlation analysis between cfDNA quantitative and qualitative content and clinicopathologic features, such as blood pressure level, body mass index (BMI) and LVSI status, could represent a potential predictive signature to help stratification approaches in EC.
Electronic supplementary material
The online version of this article (doi:10.1186/s13046-018-0688-4) contains supplementary material, which is available to authorized users.
Keywords: Endometrial cancer, Circulating cell-free DNA, DNA integrity index, Liquid biopsy, Lymphovascular space invasion, Hypertension
Background
Worldwide, endometrial cancer (EC) is the second most common gynecological cancer and the sixth most common tumor among reproductive and postmenopausal women [1]. The main risk factors of EC are family history of EC and of certain gynecological diseases, alcohol consumption, lack of physical activity, and disorders characteristic of metabolic syndrome. EC is classified in two broad histologic types: type I (endometrioid) and type II (non-endometrioid) as established by the International Federation of Gynecology and Obstetrics (FIGO). Treatment of the diversity of this cancer presenting in the clinic is still not sufficiently personalized. Several studies showed that combination of both clinicopathologic and molecular parameters appear to represent an improvement over either system alone in the diagnosis, prognosis, and management of cancer.
Recent progress in the analysis of blood samples for circulating cell-free DNA (cfDNA) provides a rapid, cost-effective, and noninvasive “liquid biopsy” tool, which gives important complementary information on diagnosis, therapeutic targets and drug resistance mechanisms in cancer patients [2].
CfDNA is a highly fragmented double-stranded molecule including nuclear and mitochondrial DNA that is released in the blood stream through processes of apoptosis, necrosis, and secretion, autophagy and necroptosis [3–6].
The presence of cfDNA within the plasma was first reported by Mandel and Metais in 1948 in the blood of healthy individuals [7]. In 1965, Bendich and colleagues hypothesized that cancer derived cfDNA could be involved in metastasis [8]. Because of technological limitations, only several years later the first experimental evidence supporting that cfDNA in cancer patients does indeed contain tumor DNA was provided [9].
Cancer specific somatic genetic alterations can be detected in cfDNA [5, 10]. These biological characteristics discriminate cfDNA from normal cell-free DNA and assure cfDNA as a specific biomarker that provides personalized information to detect residual disease or monitor tumor progression during therapy. However, currently there are several technical difficulties challenging the practical application in cancer screening and clinical management, mainly because of the technical complexity and high cost associated with this kind of analysis.
The variability of cfDNA levels in cancer patients is likely associated with tumor burden, stage, vascularity, cellular turnover, and response to therapy with highest levels reported in cancer patients with advanced and metastatic disease [11–15]. However, cfDNA content is also elevated in various other disorders, such as infectious and autoimmune diseases, stroke, infarction and trauma [16], thus more specific approaches and accurate methodologies are needed to determine the source of cfDNA.
It has been shown that in necrosis DNA fragments are generated more randomly, have a size larger than 10,000 base pairs and could be distinguished from shorter fragments, with a 180–200 base pairs or multiples of this unit in length, produced by physiological apoptosis [17]. The degree of cfDNA integrity, called integrity index, is based on the ratio between long and short cfDNA fragments and has been recently proposed as a promising specific oncological biomarker because of its high sensitivity [6, 18].
In recent years, Alu-quantitative real-time PCR (qPCR) is the most common method used to detect DNA integrity [19–21]. The most commonly used primers for Alu-PCR are Alu115 and Alu247 [20, 21]. DNA integrity is generally calculated as a ratio of longer to shorter DNA fragments, or longer to total cfDNA content [20, 21].
In this study, we measured the concentration of cfDNA by direct qPCR of Alu repeats in serum samples from EC patients and healthy women and analysed the relationship between cfDNA results and clinicopathologic characteristics including hypertension, obesity, and LVSI. Then, we assessed the degree of cfDNA fragmentation by DNA integrity assay calculated as qPCR-Alu247 value /qPCR-Alu115 value of each sample to evaluate its potential as tool in EC stratification and management.
Methods
Patient cohort
All healthy volunteers and EC patients were recruited at the Regina Elena National Cancer Institute. We collected serum samples from 60 EC patients and as controls from 22 age-matched healthy volunteers with no gynecological conditions. Patients included in this study were not previously selected, but randomly chosen and recruited between 2014 and 2017. According to the histologic grade, we analyzed samples from 12 G1, 30 G2 and 18 G3 ECs. Table 1 depicts clinicopathologic characteristics of patients enrolled in this study.
Table 1.
Clinicopathologic features of our cohort of 60 endometrial cancer patients
| Endometrial cancer (grade) | G1 | G2 | G3 |
|---|---|---|---|
| No of cases | 12 | 30 | 18 |
| Age: years Median (range) |
57 (48–71) |
57 (30–73) |
61 (52–78) |
| BMI > 30% | 41.7 | 45.4 | 28.7 |
| Type I (%) Type II (%) |
100.0 0.0 |
96.7 3.3 |
66.7 33.3 |
| FIGO stage (2009) (%) | |||
| IA | 100.0 | 73.1 | 13.3 |
| IB | 0.0 | 19.2 | 26.7 |
| II | 0.0 | 7.7 | 20.0 |
| IIIA | 0.0 | 0.0 | 6.7 |
| IIIB | 0.0 | 0.0 | 13.3 |
| IIIC1 | 0.0 | 0.0 | 20.0 |
| IIIC2 | 0.0 | 0.0 | 0.0 |
| IVA | 0.0 | 0.0 | 0.0 |
| IVB | 0.0 | 0.0 | 0.0 |
| MI > 50% (%) | 0 | 31.1 | 88.3 |
| LVSI (%) | 0.0 | 6.6 | 77.7 |
| Lymph node metastases (%) | 0.0 | 0 | 17.5 |
| Hypertension (%) | 50.0 | 33.3 | 58.8 |
| BMI ≥ 30 (%) | 16.6 | 40 | 27.7 |
Ethical approval
Experimental protocol was approved by the Ethics Committee of the Regina Elena National Cancer Institute (Rome, Italy) (RS: 2021/2017), and performed in accordance with the relevant guidelines and regulations. Written informed consent was obtained from all patients and healthy volunteers. Information about patients was obtained by reviewing their medical charts.
Sample processing
Venous blood of cancer patients was obtained before surgery and before the beginning of any treatment. Blood samples were collected in Vacutainer tubes without anticoagulant and processed within 1–4 h. After collection the blood was allowed to clot at room temperature. The blood serum was separated by centrifugation at 1000–2000 x g for 10 min in a refrigerated centrifuge and stored at − 80 °C.
Serum preparation
Serum preparation for qPCR was performed as described by Umetani et al. (20). Briefly, serum proteins which might hinder the qPCR results by binding to template DNA or DNA polymerase were deactivated by mixing 20 μL of each serum sample with 20 μL of a preparation buffer that contained 2.5% of tween 20, 50 mmol/L Tris, and 1 mmol/L EDTA. This mixture was digested with proteinase K (20 μg) solution for 50 min (Promega) at 56 °C, followed by 5 min of heat deactivation at 95 °C. After subsequent centrifugation at 10,000×g for 5 min, Supernatants were aliquoted and stored at − 80°.
Quantitative PCR of Alu repeats
0.2 μL of supernatant was used as a template for each quantitative real-time polymerase chain reaction (qRT-PCR) using SYBR Green Master Mix (Applied Biosystems, CA, USA) followed by evaluation of the average of CT values from triplicate reactions from Real Time PCR software.
The sequences of the Alu115 primers were forward: 5-CCTGAGGTCAGGAGTTCGAG-3 and reverse: 5-CCCGAGTAGCTGGGATTACA-3; Alu247 primers were forward: 5-GTGGCTCACGCCTGTAATC-3 and reverse: 5-CAGGCTGGAGTGCAGTGG-3. A negative control (without template) was performed in each plate. All qPCR assays were performed in a blind fashion without knowledge of specimen identity. Mean values were calculated from triplicate reactions.
Measurements of cfDNA values
The absolute equivalent amount of cfDNA in each sample was determined by use of a calibration curve with serial dilutions (15 ng-0.015 pg) of genomic DNA obtained from peripheral blood leukocytes of a healthy donor volunteer. A negative control (without template) was run in each reaction plate.
Measurement of cfDNA integrity index
DNA integrity was calculated as the ratio of qPCR results using the 2 primer sets: qPCR-Alu247/qPCR-Alu115, where qPCR-Alu115 and qPCR-Alu 247 are the Alu-qPCR results obtained with the Alu115 and Alu247 primers, respectively. Because the annealing sites of Alu115 are within the Alu247 annealing sites, DNA integrity value would be 1.0 when template DNA is not truncated and 0.0 when all template DNA is truncated into fragments smaller than 247 bp. Because the Alu115 primers can amplify most fractions of circulating DNA, qPCR-Alu results obtained with Alu115 primers represent the absolute amount of DNA. The fit of the standard curve (R2) was higher than 0,99 for both Alu sequences.
Statistical analysis
The examined variables were not normally distributed, as verified by the Shapiro–Wilk test, thus the non parametric U–Mann–Whitney was applied to perform a two-by-two comparison between the groups. The predictive capability (i.e., diagnostic performance) of cfDNA content was investigated by means of the area under the ROC (Receiver-Operating Characteristics) curve (AUC). Cut-offs were extrapolated from the curve. Likelihood ratio Chi-square and P-values were determined using logistic ordinal regression for the prediction of LVSI status and hypertension, given the levels of cfDNA (assessed as qPCR-Alu115 and qPCR-Alu247) and DNA integrity index values (assessed as qPCR-Alu247/ qPCR-Alu115). P ≤ .05 was considered statistically significant.
Immunohistochemistry
Tumors from hysterectomy specimens taken immediately before surgery were formalin fixed and paraffin embedded. Three μm thick sections were cut from paraffin blocks using a cryostat and mounted onto histological glass slides and stained iwith hematoxylin and eosin. LVSI was diagnosed when viable tumor nests were observed within endothelial-lined spaces with or without intraluminal red cells or lymphocytes. In case of doubt, LVSI status was further assessed by immunohistochemical staining using the following primary antibodies: anti-CD31 mouse monoclonal antibody (M0823) (DAKO) and anti-CD34 mouse monoclonal antibody (NCL-L-END) (Leica). A pH 6 buffer was used for the two antibodies as antigen retrieval according to the manufacturer’s protocol. Staining was performed using an automated immunostainer (Bond-III, Leica). Stained slides were reviewed by experienced gynaecological histopathologists to confirm FIGO (2009) stage, histological subtype, grade, depth of myometrial invasion, and the presence or absence of LVSI.
Results
CfDNA content and integrity modulation in EC
CfDNA content evaluated by qPCR-Alu115 in G1 EC (median = 12.45 ng/ml; range 3.33–68.46) was very similar to that obtained from healthy specimens (median = 18.90 ng/ml; range 0.68–64.23), whereas a significant increase of total cfDNA content in G2 EC and G3 EC was detected (Table 2). Analysis of Alu247 values revealed an increased level of longer DNA fragments in sera of higher compared to lower EC grade (Table 2). We also calculated the serum DNA integrity index as qPCR-Alu247 value/qPCR-Alu115 value. Our results showed that cfDNA integrity was significantly higher only in G3 EC as shown by the higher ratio of long to total cfDNA (Table 2). Higher cfDNA and DNA integrity index values were detected in more advanced stages (Additional file 1: Table S1). However, this increase was not significant (P > 0.5).
Table 2.
CfDNA levels in 12 G1, 30 G2 and 18 G3 endometrial cancer serum samples
| qPCR-Alu115 (ng/ml) Md. (range) |
P –Value Mann-Whithey |
qPCR-Alu247 (ng/ml) Md. (range) |
P –Value Mann-Whithey |
qPCR-Alu247/ qPCR-Alu115 Md. (range) |
P –Value Mann-Whithey |
|
|---|---|---|---|---|---|---|
| G1 | 12.45 (3.33–68.46) |
0.03 | 2.19 (0.43–11.07) |
0.10 | 0.14 (0.09–0.47) |
0.63 |
| G2 | 28.45 (1.62–160.89) |
5.87 (0.08–22.35) |
0.14 (0.05–0.27) |
|||
| G3 | 29.03 (0.21–175.54 |
0.02 | 4.43 (0.51–36.15) |
0.06 | 0.21 (0.10–0.96) |
0.04 |
Median (Md), maximum and minimum (range), and Mann–Whitney U test for cfDNA values obtained by qPCR-Alu115, qPCR-Alu247, and qPCR-Alu247/qPCR-Alu115 (DNA integrity index) in G1, G2 and G2 EC
Correlation between cfDNA content and inflammatory diseases in EC
In order to obtain an optimal cut-off that best discriminated between high (G2 and G3) and low grade (G1) EC, we performed ROC analysis by comparing values from qPCR-Alu115, qPCR-Alu247 and DNA integrity index. All three markers showed a low predictive accuracy, indicating that this method is not sufficient by itself to differentiate high grade from low grade EC patients (Additional file 2: Table S2). To assess if chronic inflammatory diseases, such as hypertension and obesity, correlated with the amount of cfDNA released in EC, we clustered samples from hypertensive and non hypertensive patients, and from patients with BMI < 30 and BMI ≥30. The percentage of hypertensive and obese EC patients is shown in Table 1. We observed a trend, even if not significant, towards higher total cfDNA levels in hypertensive and obese patients (Table 3). DNA integrity index was significantly lower in hypertensive and obese patients (Fig. 1a and b, and Table 3). Cluster analysis based on EC grading revealed that a significant down-modulation of DNA integrity index occurred specifically in samples from hypertensive and obese compared, respectively, with normotensive and normal weight high grade EC patients. (Fig. 1c and d). The logistic regression test was applied to analyse the relationship between DNA integrity indexes and hypertension and overweight. The model predicted no direct correlation (P > .05).
Table 3.
Association between cfDNA measurements and blood pressure levels (non-hypertensive and hypertensive), body mass index or lymphovascular space invasion in 60 endometrial cancer patients
| LSVI and blood pressure status in ECs | qPCR-Alu115 (ng/ml) Median Mean |
P –Value Mann-Whithey |
qPCR-Alu247 (ng/ml) Median Mean |
P –Value Mann-Whithey |
DNA integrity index Median Mean |
P –Value Mann-Whithey |
|---|---|---|---|---|---|---|
| LSVI negative (LVSI-) | 23.98 48.9 |
0.54 | 3.58 7.2 |
0.25 | 0.14 0.16 |
0.03 |
| LSVI positive (LVSI+) | 30.61 49.4 |
6.56 11.1 |
0.22 0.27 |
|||
| Non-hypertensive | 26.34 32.99 |
0.38 | 4.89 7.4 |
0.78 | 0.19 0.23 |
0.04 |
| Hypertensive | 42.97 53.35 |
4.84 12.5 |
0.13 0.15 |
|||
| LSVI- non-hypertensive | 22.05 43.7 |
0.58 | 4.43 7.06 |
0.70 | 0.14 0.17 |
0.77 |
| LSVI- hypertensive | 28.45 60.5 |
3.63 8.29 |
0.13 0.14 |
|||
| LSVI+ non-hypertensive | 29.02 39.1 |
0.44 | 6.11 8.90 |
0.55 | 0.23 0.32 |
0.05 |
| LSVI+ hypertensive | 71.53 66.4 |
14.01 14.6 |
0.14 0.18 |
|||
| BMI < 30 | 20.35 37.59 |
0.25 | 5.10 8.51 |
0.76 | 0.17 0.22 |
0.01 |
| BMI ≥ 30 | 26.96 46.47 |
3.53 7.13 |
0.11 0.12 |
|||
| LSVI- BMI < 30 | 16.81 29.60 |
0.16 | 3.18 6.86 |
0.50 | 0.14 0.17 |
0.17 |
| LSVI- BMI ≥ 30 | 28.45 62.66 |
3.63 7.78 |
0.11 0.12 |
|||
| LSVI+ BMI < 30 | 32.75 51.43 |
0.82 | 6.82 12.13 |
0.63 | 0.22 0.30 |
0.29 |
| LSVI+ BMI ≥ 30 | 26.78 43.12 |
4.25 7.88 |
0.21 0.18 |
Median, mean and Mann–Whitney U test for cfDNA values obtained from qPCR-Alu115, qPCR-Alu247 and DNA integrity index (qPCR-Alu247/qPCR-Alu115) in samples from hypertensive and non-hypertensive patients, from patients with BMI < 30 and ≥30, and from LVSI negative (LVSI-) and positive (LVSI+) tumors
LVSI Limphovascular Space Invasion, BMI Body Mass Index
Fig. 1.
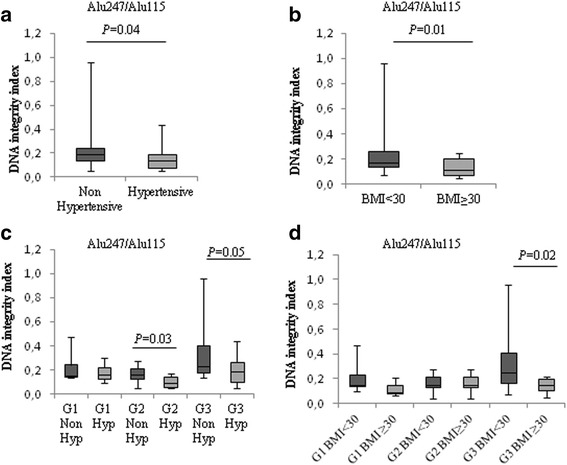
CfDNA levels related to hypertension and obesity in EC. Box-plots of DNA integrity index values in non-hypertensive and hypertensive (a), and normal weight (BMI < 30) and obese (BMI ≥ 30) EC patients (b). Cluster analysis of DNA integrity index values from non hypertensive (Non Hyp) and hypertensive (Hyp) (c), or normal weight (BMI < 30) and obese (BMI ≥ 30) (d) in G1, G2 and G3 EC samples. The upper border of the box indicates the upper quartile (75th percentile) while the lower border indicates the lower quartile (25th percentile), and the horizontal line in the box the median. P values, Mann–Whitney U test
Correlation between cfDNA integrity index with LVSI in high grade EC
We also investigated the possible involvement of LVSI in cfDNA release and integrity. LVSI was assessed by morphology and, in case of doubt, also by immunohistochemistry using anti-CD31 and anti-CD34 antibodies. We grouped serum samples from EC patients with or without LVSI. A trend toward increased cfDNA levels, evaluated by qPCR with both Alu115 and Alu247 couple of primers, was observed in samples with LVSI (Table 3). A significant increase was observed for DNA integrity index values in LSVI+ serum samples (Fig. 2a and Table 3). Cluster analysis of samples from hypertensive and non-hypertensive EC patients, or from patients with BMI < 30 and ≥30 showed that the higher DNA integrity index was encountered in LSVI+ samples derived from non-hypertensive patients. The other subgroups showed comparable values (Fig. 2b and Table 3). Based on these results, to better determine the relationship between DNA integrity index values and hypertension in LVSI + EC samples, we performed a logistic regression test for the prediction of LVSI status and hypertension, given the values of DNA integrity index. Decreased DNA integrity values significantly correlated with the hypertensive status in high grade ECs with the presence of LVSI (likelihood ratio chi square = 3.691, P < .05 with degree of freedom = 1).
Fig. 2.
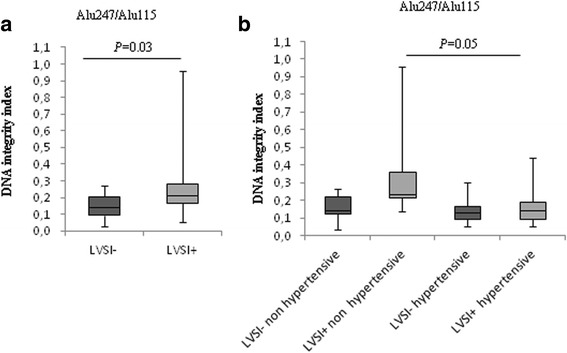
Hypertension affects serum DNA integrity indexes in LVSI positive ECs. Box-plots of DNA integrity index values in LVSI negative (LVSI-) and LVSI positive (LVSI+) serum samples (a), and association between LSVI status and blood pressure levels (b). P values, Mann–Whitney U test
Discussion
EC is the most common cancer of the female female reproductive organs and is the seventh most common cause of death from cancer in women in Western Europe. At diagnosis, about 75% of women have a cancer confined to the uterus (stage I) and the prognosis is good. However the prognosis for recurrent or metastatic EC remains poor, thus more sensitive methods to help clinical diagnosis and improve the stratification of EC patients are needed.
In our study, we analysed the quantities and degree of cfDNA integrity in EC serum samples in order to assess cfDNA content as a simple and inexpensive non-invasive complementary tool for EC stratification. We performed a quantification of cfDNA by qPCR for Alu repeats using as the template, serum without preeceding DNA purification, thus overcoming artifacts associated with DNA isolation, such as the prevailing short-comings of DNA extraction methods, [20]. We used two sets of Alu primers: the primer set for the 115 bp amplicon (Alu115) that amplifies both shorter (truncated by apoptosis) and longer DNA fragments, and a set for the 247 bp amplicon (Alu247) that amplifies only longer DNA fragments. It is generally accepted that the 180–200 base pairs or fragments length of multiples of this unit reflect apoptosis, the prevalent mechanism of cell death in physiologically conditions, whereas necrosis, producing much longer DNA fragments on account of an inefficient nuclease activity, seems to occur more frequently in tumor cells [22, 23]. Is has been suggested that DNA integrity, calculated as a ratio of longer to shorter DNA fragments, specifically represents the relative amount of non physiological non apoptotic cell death.
In our study, we observed a significant increase of both cfDNA content and DNA integrity index in high grade compared with G1 ECs, suggesting a role of cfDNA as potential prognostic biomarker in EC. Conversely, in a previous study Tanaka et al. did not find significant modulation in cfDNA content among both histological EC grade or stage [24]. We suggest that the difference in the results may be due to the different procedure applied and, in particular, to prior processing of samples and DNA extraction. However, ROC curve analysis revealed a poor diagnostic power of the cfDNA assay to differentiate low grade from high grad EC.
It has been shown that levels of inflammation are directly correlated with the amount of cfDNA release [25–28]. Levels of cfDNA concentration are also associated with pathophysiology of hypertension and vascular damage [29]. Several studies have demonstrated a correlation between hypertension and obesity and the relative risk of developing EC cancer [30]. We found an inverse relationship between the presence of these two inflammatory diseases and DNA integrity values in high grade EC. It is possible to hypothesise that the levels of inflammation status contribute to a larger cfDNA release of short fragments into the bloodstream. This mechanism may very likely lead to a relative increase in short cfDNA fragments, that may explain the decreased cfDNA integrity indexes observed.
It is worthwhile to note that high blood pressure and obesity affected specifically and exclusively the degree of cfDNA fragmentation in high grade EC sera, whereas not detectable differences in G1 EC samples were measured. It has been shown that the DNA integrity index varies between different cancers and also among individual cancers [31–33]. We hypothesise that cfDNA content in G1 EC is still under the clearance capacity of phagocytic cells and that nuclease activity is still efficient, thus effects may not be detected. Moreover, by qPCR-Alu115 it is not possible to amplify very small DNA fragments, and this may affect the detection level and the yield of this technique. In fact, we could not detect differences between cfDNA content in healthy control and G1 EC group. On the other hand, in higher grade EC, the rapid DNA release from a higher number of cancer cells may overwhelm phagocytosis resulting in the increased cfDNA accumulation of fragments detectable by qPCR into circulation due to both apoptosis ans necrosis.
An important criterion for further therapy in cancer is represented by LSVI status. LVSI includes lymphatic vessel and blood vessel invasion involved in tumor spreading and metastasis by its ability to enhance dissemination of viable cancer cells and release of DNA from malignant tumor into the blood stream. LVSI had been suggested to be an important prognostic factor for relapse of disease and poor survival in patients with ovarian [34], vulva [35], cervical [36, 37], rectal [38], breast [39], ans lung cancers [40], and EC [41]. Therefore, we focused on cfDNA level related to LVSI status. Interestingly, we observed a significant increase of DNA integrity index in serum samples from EC with LVSI (LVSI+) compared to those from cancer without LVSI (LSVI-), further supporting the hypothesis that the relative higher content of longer cfDNA fragments in serum samples very likely derives from cancer cells. Interestingly, our data obtained from correlation analysis between cfDNA content and the inflammatory status due to hypertension or obesity in ECs presenting LVSI indicated that the higher DNA integrity index was specifically encountered in LSVI+ samples derived from non-hypertensive patients, further confirming a pivotal role of hypertension in affecting DNA integrity index values.
As a aperspective for EC patients, further analysis in large cohort are needed in order to better optimize the accuracy and reliability of cfDNA measurements and confirm its suitability for clinical use, taking into account a longer follow-up and specific markers of tumor origin [42–45].
Conclusions
Our pilot study provides reliable evidence on the utility of cfDNA analysis in EC for further validation studies, with some limitations represented by the short follow up that does not allow to reach a definitive endpoint in terms of clinical utility for prognosis. For this reason, we are currently collecting longer term follow up data.
We suggest that measurement of cfDNA content in serum samples by qPCR-Alu115 and qPCR-Alu247 may represent a potential simple and not expensive molecular tool to better surgical staging and help in EC stratification. In particular, on the basis of our result on the correlation between serum DNA integrity index and LVSI, we envisage the possibility that this approach might be useful to identify high grade EC with risk of metastasis. A longer follow up is necessary to validate this hypothesis.
Finally, from a molecular point of view, a better understanding of the source and clearance mechanisms of cfDNA, such as metabolic changes in the rate of cfDNA turnover, inefficient removal of dead cells or release of DNA from tumor microenviroment and blood cells, would improve the interpretation of our study.
Additional files
Table S1. CfDNA content and EC staging. Measurement of median cfDNA values obtained by qPCR-Alu115, qPCR-Alu247, and pPCR-Alu247/qPCR-Alu115 (DNA integrity index) in different EC stages. (DOCX 15 kb)
Table S2. Receiver operative characteristics (ROC) analysis and optimal cut-offs values for cfDNA evaluated by qPCR-Alu115, qPCR-Alu247 and qPCR-Alu247/qPCR-Alu115 in G2 and G3 EC versus G1 EC serum samples. (DOCX 11 kb)
Acknowledgements
This work has been founded by IRE Internal Projects to G.P. and E.V. Special thanks go to Silvia Bacchetti for editing the manuscript.
Funding
Not applicable.
Abbreviations
- BMI
Body mass index
- cfDNA
Cell-free DNA
- CI
Confidence interval
- EC
Endometrial cancer
- LVSI
Lymphovascular space invasion
- qPCR
Quantitative real time PCR
- ROC
Receiver operating characteristic
Authors’ contributions
LC, GC and EV conceived and designed the research project. LC and MDA performed the research. LC and GC wrote the manuscript. MC, EM, EB and BC collected clinical sample. AZ collected clinical data. GP and LP revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Experimental protocol was approved by the Ethics Committee of the Regina Elena National Cancer Institute (Rome, Italy), and performed in accordance with the relevant guidelines and regulations. Written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13046-018-0688-4) contains supplementary material, which is available to authorized users.
Contributor Information
Giacomo Corrado, Email: giacomo.corrado@alice.it.
Giulia Piaggio, Email: giulia.piaggio@ifo.gov.it.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 3.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 4.Thierry AR, El Messaoud S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35(3):347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marzese DM, Hirose H, Hoon DS. Diagnostic and prognostic value of circulating tumor-related DNA in cancer patients. Expert Rev Mol Diagn. 2013;13(8):827–844. doi: 10.1586/14737159.2013.845088. [DOI] [PubMed] [Google Scholar]
- 6.Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 7.Mandel P, Metais P. Les acides nucleiques du plasma sanguine chez I’homme. C R Seances Soc Biol Fil. 1948;142(3-49):241–243. [PubMed] [Google Scholar]
- 8.Bendich A, Wilczok T, Borenfreund E. Circulating DNA as a possible factor in oncogenesis. Science. 1965;148(3668):374–376. doi: 10.1126/science.148.3668.374. [DOI] [PubMed] [Google Scholar]
- 9.Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46(5):318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 10.Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59(1):211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 11.Rumore P, Muralidhar B, Lin M, Lai C, Steinman CR. Haemodialysis as a model for studying endogenous plasma DNA: oligonucleosome-like structure and clearance. Clin Exp Immunol. 1992;90(1):56–62. doi: 10.1111/j.1365-2249.1992.tb05831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90. [DOI] [PMC free article] [PubMed]
- 13.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20(4):430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 14.Holdenrieder S, Stieber P, Bodenmüller H, Busch M, Fertig G, Fürst H, et al. Nucleosomes in serum of patients with benign and malignant diseases. Int J Cancer. 2001;95(2):114–120. doi: 10.1002/1097-0215(20010320)95:2<114::AID-IJC1020>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holdenrieder S, Stieber P. Clinical use of circulating nucleosomes. Crit Rev Clin Lab Sci. 2009;46(1):1–24. doi: 10.1080/10408360802485875. [DOI] [PubMed] [Google Scholar]
- 17.Wang BG, Huang HY, Chen YC, Bristow RE, Kassauei K, Cheng CC, et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63(14):3966–3968. [PubMed] [Google Scholar]
- 18.Cheng J, Tang Q, Cao X, Burwinkel B. Cell-free circulating DNA integrity based on peripheral blood as a biomarker for diagnosis of cancer: a systematic review. Cancer Epidemiol. Biomarkers Prev. 2017: pii: cebp.0502.2017; 10.1158/1055-9965.EPI-17-0502. [DOI] [PubMed]
- 19.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC MC, Baldwin J, Devon K, Dewar K, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Umetani N, Kim J, Hiramatsu S, Reber HA, Hines OJ, Bilchik AJ, et al. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clin Chem. 2006;2(6):1062–1069. doi: 10.1373/clinchem.2006.068577. [DOI] [PubMed] [Google Scholar]
- 21.Walker JA, Kilroy GE, Xing J, Shewale J, Sinha SK, Batzer MA. Human DNA quantitation using Alu element-based polymerase chain reaction. Anal Biochem. 2003;315(1):122–128. doi: 10.1016/S0003-2697(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 22.Mangano A, Mangano A, Lianos GD, Cassinotti E, Roukos DH, Dionigi G, et al. Circulating free DNA in plasma or serum as biomarkers of carcinogenesis in colon cancer. Future Oncol. 2015;11(10):1455–1458. doi: 10.2217/fon.15.63. [DOI] [PubMed] [Google Scholar]
- 23.van der Vaart M, Pretorius PJ. Characterization of circulating DNA in healthy human plasma. Clin Chim Acta. 2008;395(1–2):186. doi: 10.1016/j.cca.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka H, Tsuda H, Nishimura S, Nomura H, Kataoka F, Chiyoda T, et al. Role of circulating free alu DNA in endometrial cancer. Int J Gynecol Cancer. 2012;22(1):82–86. doi: 10.1097/IGC.0b013e3182328c94. [DOI] [PubMed] [Google Scholar]
- 25.van der Vaart M, Pretorius PJ. Circulating DNA. Its origin and fluctuation. Ann N Y Acad Sci. 2008;1137:18–26. doi: 10.1196/annals.1448.022. [DOI] [PubMed] [Google Scholar]
- 26.Peters DL, Pretorius PJ. Origin, translocation and destination of extracellular occurring DNA--a new paradigm in genetic behaviour. Clin Chim Acta. 2011;412(11-12):806–811. doi: 10.1016/j.cca.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 27.Hefeneider SH, Cornell KA, Brown LE, Bakke AC, McCoy SL, Bennett RM. Nucleosomes and DNA bind to specific cell-surface molecules on murine cells and induce cytokine production. Clin Immunol Immunopathol. 1992;63(3):245–251. doi: 10.1016/0090-1229(92)90229-H. [DOI] [PubMed] [Google Scholar]
- 28.Frank MO. Circulating cell-free DNA differentiates severity of inflammation. Biol Res Nurs. 2016;18(5):477–488. doi: 10.1177/1099800416642571. [DOI] [PubMed] [Google Scholar]
- 29.Jeong DW, Moon JY, Choi YW, Moon H, Kim K, Lee YH, et al. Effect of blood pressure and glycemic control on the plasma cell-free DNA.In hemodialysis patients. Kidney Res Clin Pract. 2015;34(4):201–206. doi: 10.1016/j.krcp.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aune D, Sen A, Vatten LJ. Hypertension and the risk of endometrial cancer: a systematic review and meta-analysis of case-control and cohort studies. Sci Rep. 2017;7:44808. doi: 10.1038/srep44808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitahara M, Hazama S, Tsunedomi R, Takenouchi H, Kanekiyo S, Inoue Y, et al. Prediction of the efficacy of immunotherapy by measuring the integrity of cell-free DNA in plasma in colorectal cancer. Cancer Sci. 2016;107(12):1825–1829. doi: 10.1111/cas.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holdenrieder S, Burges A, Reich O, Spelsberg FW, Stieber P. DNA integrity in plasma and serum of patients with malignant and benign diseases. Ann N Y Acad Sci. 2008;1137:162–170. doi: 10.1196/annals.1448.013. [DOI] [PubMed] [Google Scholar]
- 33.Ellinger J, Bastian PJ, Haan KI, Heukamp LC, Buettner R, Fimmers R, et al. Noncancerous PTGS2 DNA fragments of apoptotic origin in sera of prostate cancer patients qualify as diagnostic and prognostic indicators. Int J Cancer. 2008;122(1):138–143. doi: 10.1002/ijc.23057. [DOI] [PubMed] [Google Scholar]
- 34.O'Hanlan KA, Kargas S, Schreiber M, Burrs D, Mallipeddi P, Longacre T, et al. Ovarian carcinoma metastases to gastrointestinal tract appear to spread like colon carcinoma: implications for surgical resection. Gynecol Oncol. 1995;59(2):200–206. doi: 10.1006/gyno.1995.0008. [DOI] [PubMed] [Google Scholar]
- 35.Paladini D, Cross P, Lopes A, Monaghan JM. Prognostic significance of lymph node variables in squamous cell carcinoma of the vulva. Cancer. 1994;74(9):2491–2496. doi: 10.1002/1097-0142(19941101)74:9<2491::AID-CNCR2820740916>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 36.Roman LD, Felix JC, Muderspach LI, Varkey T, Burnett AF, Qian D, et al. Influence of quantity of lymph-vascular space invasion on the risk of nodal metastases in women with early-stage squamous cancer of the cervix. Gynecol Oncol. 1998;68(3):220–225. doi: 10.1006/gyno.1998.4943. [DOI] [PubMed] [Google Scholar]
- 37.Yu H, Zhang S, Zhang R, Zhang L. The role of VEGF-C/D and Flt-4 in the lymphatic metastasis of early-stage invasive cervical carcinoma. J Exp Clin Cancer Res. 2009;9(28):98. doi: 10.1186/1756-9966-28-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talbot IC, Ritchie S, Leighton MH, Hughes AO, Bussey HJ, Morson BC. Spread of rectal cancer within veins. Histologic features and clinical significance. Am J Surg. 1981;141(1):15–17. doi: 10.1016/0002-9610(81)90004-0. [DOI] [PubMed] [Google Scholar]
- 39.Katz A, Strom EA, Buchholz TA, Theriault R, Singletary SE, McNeese MD. The influence of pathologic tumor characteristics on locoregional recurrence rates following mastectomy. Int J Radiat Oncol Biol Phys. 2001;50(3):735–742. doi: 10.1016/S0360-3016(01)01500-0. [DOI] [PubMed] [Google Scholar]
- 40.Sun JG, Wang Y, Chen ZT, Zhuo WL, Zhu B, Liao RX, Zhang SX. Detection of lymphangiogenesis in non-small cell lung cancer and its prognostic value. J Exp Clin Cancer Res. 2009;16(28):21. doi: 10.1186/1756-9966-28-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mariani A, Webb MJ, Keeney GL, Aletti G, Podratz KC. Predictors of lymphatic failure in endometrial cancer. Gynecol Oncol. 2002;84(3):437–442. doi: 10.1006/gyno.2001.6550. [DOI] [PubMed] [Google Scholar]
- 42.Silvestris N, Ciliberto G, De Paoli P, Apolone G, Lavitrano ML, Pierotti MA, Stanta G; On the behalf of the “dynamic medicine OECI group”. Liquid dynamic medicine and N-of-1 clinical trials: a change of perspective in oncology research. J Exp Clin Cancer Res 2017;13;36(1):128. [DOI] [PMC free article] [PubMed]
- 43.Deng L, Gao Y, Li X, Cai M, Wang H, Zhuang H, et al. Expression and clinical significance of annexin A2 and human epididymis protein 4 in endometrial carcinoma. J Exp Clin Cancer Res. 2015;34:96. doi: 10.1186/s13046-015-0208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cicchillitti L, Corrado G, Carosi M, Dabrowska ME, Loria R, Falcioni R, Cutillo G, Piaggio G, Vizza E. Prognostic role of NF-YA splicing isoforms and Lamin a status in low grade endometrial cancer. Oncotarget. 2017;8(5):7935–7945. doi: 10.18632/oncotarget.13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. CfDNA content and EC staging. Measurement of median cfDNA values obtained by qPCR-Alu115, qPCR-Alu247, and pPCR-Alu247/qPCR-Alu115 (DNA integrity index) in different EC stages. (DOCX 15 kb)
Table S2. Receiver operative characteristics (ROC) analysis and optimal cut-offs values for cfDNA evaluated by qPCR-Alu115, qPCR-Alu247 and qPCR-Alu247/qPCR-Alu115 in G2 and G3 EC versus G1 EC serum samples. (DOCX 11 kb)


