Abstract
Background
Differences in the concentrations of circulating 25-hydroxyvitamin D [25(OH)D] are associated with a wide range of health outcomes; however, most studies on genetic variants that impact 25(OH)D levels have been conducted in European populations. Here we aimed to identify common genetic variants that affect vitamin D concentrations in individuals of self-reported Arab ethnicity.
Methods
The study included 1151 Arab subjects living in Kuwait. Common variants of single-nucleotide polymorphisms and genes previously associated with vitamin D levels, such as GC, PDE3B, CYP2R1, and NADSYN1, were genotyped. Raw vitamin D level data were corrected for age, body mass index, and sex and then normalized. Regression tree analyses were performed to identify the impact of genetic variants on vitamin D levels.
Results
Compared with other gene variants, the GC gene variants exhibited the greatest impact on vitamin D levels in our study population, of which rs2298850 had the lowest p value (0.003). Individuals homozygous for the derived allele C had lower vitamin D levels. Analyses of the interaction between the number of years for which the subjects had lived in Kuwait and genetic variation in the GC gene showed that those with the CC genotype of rs2298850 who had lived in Kuwait for < 51 years had a mean 25(OH)D level of 10 ng/ml, whereas those who were homozygous for the ancestral allele had a mean 25(OH)D level of 17 ng/ml. Furthermore, subjects who had lived in Kuwait for > 51 years had higher vitamin D levels (mean 28 ng/ml) regardless of the genotype of their GC gene.
Conclusions
The GC gene may play a major role in determining vitamin D levels in Arab populations.
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1396-8) contains supplementary material, which is available to authorized users.
Keywords: Vitamin D, Arab population, GC gene, Recursive partitioning analysis
Background
Vitamin D deficiency occurs worldwide and is a common public health problem. Previous studies have shown that vitamin D deficiency is associated with the risk of developing osteoporosis, type 1 diabetes, cardiovascular diseases, asthma, and even certain types of cancer [1–6].
Vitamin D levels are dependent on skin pigmentation. Consequently, African Americans are more susceptible to vitamin D deficiency because their darker skin tone limits the penetration of ultraviolet light, in turn reducing the cutaneous synthesis of vitamin D. Ethnic differences in the prevalence of common genetic polymorphisms are another likely explanation for the low vitamin D levels found in African Americans [7–9]. Socio-cultural factors can also affect vitamin D levels. For example, most women in Arab Gulf countries wear veils and are rarely exposed to sunlight, which reduces vitamin D synthesis in the body [10]. Despite the emergence of new data on the high prevalence of vitamin D deficiency, most studies on the genetic and environmental factors affecting vitamin D status have focused on Western populations, and their conclusions do not necessarily apply to populations in different geographic regions. Given the lack of substantial data on genetic and environmental risk factors in populations of Middle Eastern origin in which the prevalence of vitamin D deficiency is high, we sought to determine the genetic and lifestyle risk factors underlying vitamin D deficiency in Arabs.
Recent genetic studies have associated vitamin D deficiency with several candidate genes, including the cytochrome P450, family 2, R (CYP2R1) gene; the group-specific component (GC) gene, and the 7-dehydrocholesterol reductase/NAD synthetase 1 (DHCR7/NADSYN1) gene. These genes are involved in hydroxylation, vitamin D transport, and cholesterol synthesis, respectively. The best indicator of vitamin D levels in humans is the serum concentration of its main circulating metabolite, 25-hydroxyvitamin D [25(OH)D]. The association between polymorphisms in these genes and 25(OH)D levels has been previously studied in African Americans and found that a CYP2R1 SNP, rs12794714, exhibited the strongest signal of association [11], and also among 30,000 individuals of European descent [12] where variants near genes DHCR7 (CYP2R1, CYP24A1), and GC identified to influence vitamin D status.
This study aimed to examine the effect each of the genes listed above has on vitamin D levels in a group of Arabs living in Kuwait for various lengths of time. Our main objective was to examine the influence of lifestyle factors and genetic variation on vitamin D status.
Methods
Study subjects
This cross-sectional study included 1151 Arab adults (age > 18 years) living in Kuwait. These subjects were part of a large multi-ethnic cohort that was randomly selected as previously described [13]. Our subjects were randomly selected from the computerized registry of the Public Authority of Civil Information. The study conformed to the principles outlined in the Declaration of Helsinki and was approved by the Scientific Advisory Board and Ethical Review Committee at Dasman Diabetes Institute. Informed written consent was obtained from all subjects before their enrolment in the study.
Phenotypic data
Physical and anthropometric measurements, including body weight, height, and waist circumference, of all subjects were recorded. Blood samples were obtained after at least 10 h of overnight fasting and were analyzed for fasting blood glucose, hemoglobin A1c, and fasting insulin levels as well as for lipid profiles that included triglycerides, total cholesterol, and low- and high-density lipoprotein levels. Glucose levels and lipid profiles were measured using a Siemens Dimension RXL chemistry analyzer (Diamond Diagnostics, Holliston, MA, USA). Serum 25(OH)D levels were measured with a chemiluminescent competitive immunoassay using a DiaSorin LIAISON analyzer (DiaSorin Inc., MN, USA) following the manufacturer’s protocol. The intra-assay coefficients of variation were 5.5 and 4.0% at 10 and 25 ng/ml, respectively, and the inter-assay coefficients of variation were 8 and 6% at 15 and 40 ng/ml, respectively.
DNA isolation and genotyping
From each subject, 4-ml blood was drawn into ethylenediaminetetraacetic acid tubes. DNA was isolated using commercial kits following the manufacturer’s recommendations as previously reported [13]. DNA quantity and quality were checked using an Epoch Microplate Spectrophotometer before genotyping.
Genotyping was performed with TaqMan on an Applied Biosystems 7500 Real-Time PCR System as previously reported [13]. After polymerase chain reaction amplification, an endpoint plate read was performed. Fluorescence measurements obtained during the plate read were used in Sequence Detection System software to plot fluorescence values based on the signals from each single plate well. The results were plotted on a two-dimensional scatter plot of the major versus minor allele. Genotyping calls were assessed based on the allele discrimination plots and manually reviewed by examining the single amplification plots.
Statistical analyses
The effects of various parameters were investigated using linear regression analyses as implemented in the R environment. Vitamin D levels of all subjects were adjusted for sex, age, number of years in Kuwait, triglyceride levels, total cholesterol levels, and hip circumference. The residuals were normalized using the rntransform function implemented in the GenABEL package [14] that performs quantile normalization of residuals from a generalized linear model analysis.
In addition, conditional inference-based recursive partitioning (implemented in the R “party” package) [15] was used to determine the influence of the genetic variables on normalized vitamin D levels (as a quantitative trait). This approach searched predictor variables having main effects and higher-order interactions. Stepwise modeling and splitting was applied to produce a classification tree that showed how each genotype affected vitamin D levels. This approach was divided into two phases: in the first phase, we analyzed each gene separately, highlighting which single-nucleotide polymorphism (SNP) had the largest effect on vitamin D levels. In the second phase, we constructed a regression tree using only the SNPs identified as significant in the phase 1 analyses after Bonferroni correction. The final tree is based on the splitting variables of each node with the highest statistical significance. Regression trees allows to construct an hierarchy of the variables with the first node corresponding to the variable with the strongest effect, the second and the third nodes correspond to variables with significant impact of the phenotype. Recursive partitioning is widely used in medicine and genetics to describe interaction between variables.
The genotype distributions and derived allele frequencies of the SNPs of interest were compared with similar distributions from the reference dataset of 1000 Genomes Project. The distribution of the combined annotation-dependent depletion (CADD) score for each variant was obtained [16].
Results
The descriptive characteristics of all subjects are summarized in Table 1. In total, 1151 subjects (702 men and 449 women), with mean age of 46 (5th percentile: 27, 95th percentile: 64) years and a mean of 35 (5th: 5, 95th: 61) years of living in Kuwait, were analyzed. The mean 25(OH)D level in our cohort was 12.24 (5th: 4.2, 95th: 44.5) ng/ml.
Table 1.
Clinical and biochemical profiles for study subjects stratified by gender
| Parameters | Overall (n = 1151) |
Male (n = 702) |
Female (n = 449) |
|---|---|---|---|
| Age, years | 36 (11.6) | 45.6 (11.6) | 46.3 (11.6) |
| Height (cm) | 166.7 (9.1) | 172 (6.6) | 158.5 (5.67) |
| Weight (kg) | 87.4 (18) | 90.8 (17.6) | 82 (17.2) |
| BMI (kg/m2) | 31.4 (6) | 30.7 (5.5) | 32.6 (6.54) |
| Waist-circumference (cm) | 101.2 (13.17) | 102.53 (12.9) | 99.1 (13.34) |
| Hip | 110.2 (11.23) | 108.4 (10.8) | 113.18 (12.7) |
| FBG (mmol/l) | 6.24 (2.63) | 6.4 (2.8) | 5.9 (2.3) |
| HbA1c (DCCCT%) | 6.1 (1.63) | 6.3 (1.72) | 5.9 (1.4) |
| Triglycerides (mmol/l) | 1.6 (0.94) | 1.75 (1.04) | 1.37 (0.72) |
| Total cholesterol (mmol/l) | 5.12 (1.03) | 5.1 (1) | 5.12 (1.01) |
| LDL cholesterol (mmol/l) | 3.3 (0.95) | 3.4 (0.97) | 3.23 (0.94) |
| HDL cholesterol (mmol/l) | 1.1 (0.32) | 1.01 (0.25) | 1.29 (0.34) |
| 25(OH)D (ng/l) | 17.7 (14.4) | 15 (12.3) | 22 (16) |
| Duration in Kuwait (years) | 32.9 (17.3) | 30 (17.4) | 36.6 (16.7) |
Mean and standard deviation are reported
Single-gene tree regression analyses showed that only two genes (GC and CYP2R1) were significantly associated with vitamin D levels in our cohort (Fig. 1a, b). The associated SNP rs12794714 in CYP2R1 had a CADD score of > 10, whereas all other variants had low CADD scores of < 7.9 (Additional file 1: Table S1). Partitioning tree analyses revealed that this association occurred in the GC gene. The analyses showed that only the variants in GC gene had a significant effect after Bonferroni correction.
Fig. 1.
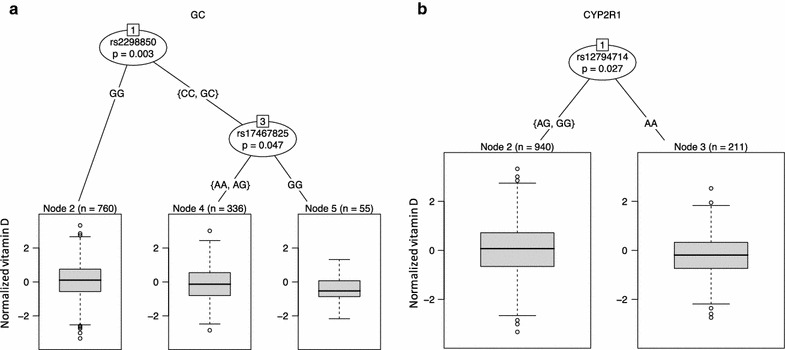
Regression tree for each gene in Arabs. a Regression tree analyses for variation in GC gene, b regression tree analyses for variation in CYP2R1. The p values are corrected for Bonferroni
Multi-regression analysis showed that among lifestyle and environmental factors, the number of years the subjects had lived in Kuwait had a strong impact on their vitamin D levels (p = 3.98e−06). Combining the genetic information with this parameter in a partitioning tree analysis, we discovered that subjects who had lived in Kuwait for > 51 years had vitamin D levels [mean (SD) raw level, 28 ± 15 ng/ml] higher than those in subjects who had lived in Kuwait for < 51 (16 ± 13) years. Vitamin D levels (10 ± 6 ng/ml) were lowest in subjects who had a homozygous CC genotype of rs2298850 compared with subjects who were heterozygous (15 ± 13 ng/ml) or had a homozygous GG genotype (17 ± 13 ng/ml; Fig. 2).
Fig. 2.
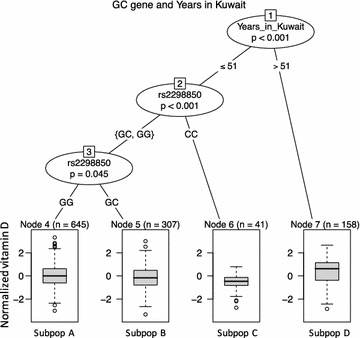
Partitioning tree analyses of GC gene variation and number of years living in Kuwait in Arabs individuals. The p values are corrected for Bonferroni
The tree analyses revealed four diverse groups (designated subpopulations A–D) corresponding to subpopulations with various percentages of subjects who could be considered vitamin D deficient according to cut-offs of 12 and 20 ng/ml. The vitamin D level classifications are summarized in Table 2.
Table 2.
Categorization of individuals according to raw values of vitamin D and regression tree analyses
| Group | % Deficient (< 12) ng/ml |
% Insufficient (12–20) ng/ml |
% Sufficient (> 20) ng/ml |
|---|---|---|---|
| SubpopA | 47 | 25 | 28 |
| SubpopB | 57 | 23 | 22 |
| SubpopC | 70 | 25 | 5 |
| SubpopD | 22 | 11 | 67 |
Discussion
Vitamin D status is a complex trait, and to date, most studies investigating circulating 25(OH)D levels have been conducted on Caucasian and Western populations. A meta-analysis showed that compared with Caucasian, non-Caucasian have lower vitamin D levels [17]. Other studies have shown that compared with Caucasians, Gujarati Indians in West London [18] and African and Mexican Americans [19] have lower plasma levels of vitamin D.
In the present study, regression tree analyses of normalized 25(OH)D levels using SNPs single and multiple genes revealed that the GC gene is the major actor in Arabs. The GC gene, which belongs to the albumin family, helps vitamin D activity by carrying vitamin D metabolites [7]. A major role of the GC gene has also been observed in French populations [20]. In our Arab cohort, the variant with the lowest p value was rs2298850. Notably, this SNP is an intronic variant with a CADD score of < 10, which indicates a low likelihood of deleteriousness. In our dataset, only rs12794714 had a CADD score of > 10 [16], which can be considered a cut-off to discriminate between deleterious and non-deleterious variants. This variant is the most significant for the CYP2R1 gene in Arabs and is also an expression quantitative trail locus for the COPB1 gene, which has been associated with vitamin D levels in children [21]. The effects of the variants in other genes were not significant compared with those of rs2298850 in regression tree analyses.
The results of our study also showed that the number of years subjects had lived in Kuwait had a positive impact on vitamin D levels. We hypothesized that this outcome results from acclimation to sun exposure, i.e., subjects tend to avoid sunlight initially but less so in later years. This lifestyle parameter interacted with genetic background but only in subjects who had lived in Kuwait for < 51 years. In the regression tree analysis, this outcome indicated a dominant effect of the lifestyle parameter over the underlying genetic variation. However, genetic variation in the GC gene represents a risk factor for vitamin D deficiency in Arabs, particularly those who are homozygous for the C allele. While the CC genotype showed significance with vitamin D levels, but not the GC heterozygous genotype, this may be attributed to possible conformational changes within the encoded protein impacted on by the minor alleles (when present in homozygous state). GC gene codes for a vitamin D-binding protein, and hence functional polymorphisms in this gene are expected to affect the overall function of this protein. Additionally, vitamin-D receptor’s polymorphisms may contribute to the vitamin levels, which would be a good future study too.
We used cut-off serum 25(OH)D levels of > 20 ng/ml as sufficient, of < 12 ng/ml as deficient, and of 12–20 ng/ml as insufficient to analyze the regression tree values. This cut-off chosen based on previously published data [22, 23]. Notably, most subjects in each group were vitamin D deficient, and the proportion of subjects with vitamin D sufficiency did not reach 30%, with exception of the subgroup D in which the majority of individuals (67%) is sufficient.
Among subjects who had lived in Kuwait for > 51 years, 22% had deficient vitamin D levels and 67% had sufficient levels, which contrasted with rs2298850-CC subjects who had lived in Kuwait for < 51 years, 70% of whom were deficient and 5% were sufficient. However, among rs2298850-GG individuals who had lived in Kuwait for < 51 years, 47% were deficient and 28% were sufficient, suggesting that other population-specific alleles or genes are involved in regulating vitamin D levels in Arab populations. These other genetic factors may explain the variance in vitamin D levels in rs2298850-GG individuals who have lived in Kuwait for < 51 years compared with those who have lived there for > 51 years. The identification of these genes or alleles would help to assess the risk of vitamin D deficiency in patients.
Notably, our results differ from those of a recent study [24] in which no evidence of any effect of the GC gene on vitamin D levels was found in subjects in Uzbekistan and Kazakhstan. Instead, that study showed an association with CYP2R1-rs10766197 and DHCR7/NADSYN1, which indicates population-specific genetic differences in the genes involved in the vitamin D pathway.
Conclusions
Our analyses showed as the distribution of serum 25(OH)D levels in individuals with self-reported Arab ethnicity is skewed towards low values, highlighting a high prevalence of deficiency and insufficiency in vitamin D.
As demonstrated, common markers in GC and CYP2R1 gene affect the variation in vitamin D level in Arab individuals, nevertheless our conditional inference trees show that polymorphisms in GC can explain most of the variation in vitamin D level when compared with several other variants already reported associated to serum 25(OH)D levels, showing that this gene is the major player in Arab population, interacting with lifestyle features. One limitation is that only know variants were included in this study, future research effort should be focused in the sequencing of this gene in order to find population-specific rare deleterious variants with higher impact on serum 25(OH)D for a better assessment of vitamin D deficiency in Arab populations.
Authors’ contributions
Conceived and designed the experiments: NE. Performed the experiments: FA, MMA, MM, and OA. Analyzed the data: MMZ. Contributed reagents/materials/analysis tools: MA and AB. Contributed to the writing of the manuscript: MMZ, ST, and NE. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank our study team in the Department of Biostatistics and Epidemiology for their efforts and excellent work. We are grateful to the Clinical Laboratory and the Tissue Bank Core Facility at DDI for their contribution in performing the vitamin D profile analysis and handling samples, respectively. We are also indebted to the senior management, Scientific Advisory Board, and Ethical Review Committee of the Dasman Diabetes Institute for their unwavering support and valuable recommendations. Finally, we are grateful to Kuwait Foundation for the Advancement of Sciences for financial support of this Research Project (RA-2010-004).
Dr. Elkum, the study principal investigator, is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and analyzed in the present study are available from Dr. Naser Elkum upon reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study conformed to the principles outlined in the Declaration of Helsinki and was approved by the Scientific Advisory Board and Ethical Review Committee at Dasman Diabetes Institute. Informed written consent was obtained from all subjects before their enrollment in the study.
Funding
Kuwait Foundation for the Advancement of Sciences (KFAS) Award Number (RA-2010-004).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1: Table S1. Gene and markers description used in this study.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1396-8) contains supplementary material, which is available to authorized users.
Contributor Information
Massimo Mezzavilla, Email: massimo.mezzavilla@burlo.trieste.it.
Sara Tomei, Email: stomei@sidra.org.
Fadi Alkayal, Email: fadi.alkayal@dasmaninstitute.org.
Motasem Melhem, Email: motasem.melhem@dasmaninstitute.org.
Maisa M. Ali, Email: maisa.mahmoud@dasmaninstitute.org
Monira Al-Arouj, Email: monira.arouj@dasmaninstitute.org.
Abdullah Bennakhi, Email: abdullah.bennakhi@dasmaninstitute.org.
Osama Alsmadi, Email: Oalsmadi@gmail.com.
Naser Elkum, Email: nelkum@sidra.org.
References
- 1.Arem H, Yu K, Xiong X, Moy K, Freedman ND, Mayne ST, Albanes D, Amundadottir LT, Arslan AA, Austin M, et al. Correction: vitamin D metabolic pathway genes and pancreatic cancer risk. PLoS ONE. 2015;10:e0129983. doi: 10.1371/journal.pone.0129983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borkar VV, Devidayal, Verma S, Bhalla AK. Low levels of vitamin D in North Indian children with newly diagnosed type 1 diabetes. Pediatr Diabetes. 2010;11:345–350. doi: 10.1111/j.1399-5448.2009.00589.x. [DOI] [PubMed] [Google Scholar]
- 3.Bosse Y, Lemire M, Poon AH, Daley D, He JQ, Sandford A, White JH, James AL, Musk AW, Palmer LJ, et al. Asthma and genes encoding components of the vitamin D pathway. Respir Res. 2009;10:98. doi: 10.1186/1465-9921-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heist RS, Zhou W, Wang Z, Liu G, Neuberg D, Su L, Asomaning K, Hollis BW, Lynch TJ, Wain JC, et al. Circulating 25-hydroxyvitamin D, VDR polymorphisms, and survival in advanced non-small-cell lung cancer. J Clin Oncol. 2008;26:5596–5602. doi: 10.1200/JCO.2008.18.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilkkinen A, Knekt P, Aro A, Rissanen H, Marniemi J, Heliovaara M, Impivaara O, Reunanen A. Vitamin D status and the risk of cardiovascular disease death. Am J Epidemiol. 2009;170:1032–1039. doi: 10.1093/aje/kwp227. [DOI] [PubMed] [Google Scholar]
- 6.Simonelli C. The role of vitamin D deficiency in osteoporosis and fractures. Minn Med. 2005;88:34–36. [PubMed] [Google Scholar]
- 7.Powe CE, Evans MK, Wenger J, Zonderman AB, Berg AH, Nalls M, Tamez H, Zhang D, Bhan I, Karumanchi SA, et al. Vitamin D-binding protein and vitamin D status of black Americans and white Americans. N Engl J Med. 2013;369:1991–2000. doi: 10.1056/NEJMoa1306357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aloia JF. African Americans, 25-hydroxyvitamin D, and osteoporosis: a paradox. Am J Clin Nutr. 2008;88:545S–550S. doi: 10.1093/ajcn/88.2.545S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aloia JF, Patel M, Dimaano R, Li-Ng M, Talwar SA, Mikhail M, Pollack S, Yeh JK. Vitamin D intake to attain a desired serum 25-hydroxyvitamin D concentration. Am J Clin Nutr. 2008;87:1952–1958. doi: 10.1093/ajcn/87.6.1952. [DOI] [PubMed] [Google Scholar]
- 10.Elsammak MY, Al-Wosaibi AA, Al-Howeish A, Alsaeed J. Vitamin D deficiency in Saudi Arabs. Horm Metab Res. 2010;42:364–368. doi: 10.1055/s-0030-1248296. [DOI] [PubMed] [Google Scholar]
- 11.Batai K, Murphy AB, Shah E, Ruden M, Newsome J, Agate S, Dixon MA, Chen HY, Deane LA, Hollowell CM, et al. Common vitamin D pathway gene variants reveal contrasting effects on serum vitamin D levels in African Americans and European Americans. Hum Genet. 2014;133:1395–1405. doi: 10.1007/s00439-014-1472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkum N, Alkayal F, Noronha F, Ali MM, Melhem M, Al-Arouj M, Bennakhi A, Behbehani K, Alsmadi O, Abubaker J. Vitamin D insufficiency in Arabs and South Asians positively associates with polymorphisms in GC and CYP2R1 genes. PLoS ONE. 2014;9:e113102. doi: 10.1371/journal.pone.0113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 15.Hothorn T, Zeileis A. Partykit: a modular toolkit for recursive partytioning in R. J Mach Learn Res. 2015;16:5. [Google Scholar]
- 16.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagenau T, Vest R, Gissel TN, Poulsen CS, Erlandsen M, Mosekilde L, Vestergaard P. Global vitamin D levels in relation to age, gender, skin pigmentation and latitude: an ecologic meta-regression analysis. Osteoporos Int. 2009;20:133–140. doi: 10.1007/s00198-008-0626-y. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson RJ, Llewelyn M, Toossi Z, Patel P, Pasvol G, Lalvani A, Wright D, Latif M, Davidson RN. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case–control study. Lancet. 2000;355:618–621. doi: 10.1016/S0140-6736(99)02301-6. [DOI] [PubMed] [Google Scholar]
- 19.Prentice A. Vitamin D deficiency: a global perspective. Nutr Rev. 2008;66:S153–S164. doi: 10.1111/j.1753-4887.2008.00100.x. [DOI] [PubMed] [Google Scholar]
- 20.Touvier M, Deschasaux M, Montourcy M, Sutton A, Charnaux N, Kesse-Guyot E, Assmann KE, Fezeu L, Latino-Martel P, Druesne-Pecollo N, et al. Determinants of vitamin D status in Caucasian adults: influence of sun exposure, dietary intake, sociodemographic, lifestyle, anthropometric, and genetic factors. J Investig Dermatol. 2015;135:378–388. doi: 10.1038/jid.2014.400. [DOI] [PubMed] [Google Scholar]
- 21.Anderson D, Holt BJ, Pennell CE, Holt PG, Hart PH, Blackwell JM. Genome-wide association study of vitamin D levels in children: replication in the Western Australian Pregnancy Cohort (Raine) study. Genes Immun. 2014;15:578–583. doi: 10.1038/gene.2014.52. [DOI] [PubMed] [Google Scholar]
- 22.Katrinaki M, Kampa M, Margioris A, Castanas E, Malliaraki N. Vitamin D levels in a large Mediterranean cohort: reconsidering normal cut-off values. Hormones. 2016;15:205–223. doi: 10.14310/horm.2002.1674. [DOI] [PubMed] [Google Scholar]
- 23.Shah S, Chiang C, Sikaris K, Lu Z, Bui M, Zebaze R, Seeman E. Serum 25-hydroxyvitamin D insufficiency in search of a bone disease. J Clin Endocrinol Metab. 2017;102:2321–2328. doi: 10.1210/jc.2016-3189. [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Mao J, Zhang M, Liu H, Li H, Lei H, Han L, Gao M. Vitamin D deficiency in Uygurs and Kazaks is associated with polymorphisms in CYP2R1 and DHCR7/NADSYN1 genes. Med Sci Monit. 2015;21:1960–1968. doi: 10.12659/MSM.894793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed in the present study are available from Dr. Naser Elkum upon reasonable request.


