Abstract
Background
Cancer stem cells (CSCs), in choriocarcinoma and other carcinomas, possess the ability of self-renewal and multilineage differentiation potential. We previous isolated choriocarcinoma cancer stem-like cells (CSLCs), which hold the stemness characteristics of CSCs. Epigenetic modifications have emerged as drivers in tumorigenesis, but the mechanisms of CSCs are largely unknown, and new drug therapies are needed to break the persistence of CSCs.
Material/Methods
Quantitative real-time PCR (qRT-PCR) and Western blot analysis were performed to detect the expression of DNMTs, HDACs, and stemness-genes. DNMTs and HDACs silencing and overexpressing lentivirus were transfected into JEG-3 cells to investigate the epigenetic functions in CSLCs. In vivo expression of curcumol effects of CSLCs on DNMTs and HDACs were analyzed by immunohistochemistry.
Results
Expression of DNMT1, DNMT3b, HDAC1, and HDAC3 were increased in choriocarcinoma CSLCs. Consistent with the inhibitory effect of 5-AzaC and TSA on CSLCs, DNMT/HDAC knockdown displayed significant repression of self-renewal in CSLCs. Curcumol inhibited the stemness ability of CSLCs in vitro and in vivo, and the inhibitory effect we observed was mediated in part through repressing activity of DNMTs and HDACs. Importantly, curcumol showed a better effect than DNMT and HDAC inhibitors combined in eliminating CSLCs.
Conclusions
These findings indicate that DNMT- and HDAC-mediated epigenetic regulation plays an important role in the biology of choriocarcinoma CSLCs, and curcumol has the potential to be a new drug to fight CSLCs, warranting further investigation of epigenetic-based therapies.
MeSH Keywords: Choriocarcinoma, DNA Methylation, Histone Deacetylases, Neoplastic Stem Cells
Background
Choriocarcinoma is a highly malignant trophoblastic tumor characterized by abnormal trophoblastic hyperplasia, and is a type of gestational trophoblastic neoplasia. Despite well-established chemotherapy, about 25% of choriocarcinoma patients showed incomplete response, or relapsed due to tumor remission [1,2]. Loss of fertility resulting in hysterectomy is the most concern outcome for patients, and is the final problem remaining to be solved. Recently, the cancer stem cells (CSCs) subpopulation, which has a high tumorigenic character, has been discussed as an explanation for tumor development, chemoresistance, and relapse after initial treatment [3–5]. CSCs have 2 important characteristics: self-renewal and multipotency. Our previous study isolated the choriocarcinoma stem-like cells (CSLCs) from the human choriocarcinoma JEG-3 cell line, showing the characteristics of CSCs [6]. The driver mutations affect a wide range of epigenetic regulators in different cancers, which provides direct evidence for the importance of epigenetic dysregulation in the formation of CSCs [7,8].
Epigenetic modifications have emerged as drivers in tumorigenesis [9,10]. Aberrant DNA methylation and histone deacetylation are 2 principal factors in epigenetic phenomena, and have been verified to be related to tumorigenesis in many studies. As the key enzymes, DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) were recognized as antitumor drug targets in terms of epigenetic therapy [11,12]. DNMT1 maintains an established DNA methylation pattern, whereas DNMT3a and DNMT3b set up DNA methylation patterns in early development. Abnormal methylation by DNMTs overexpression induces tumor-suppressor genes silencing in human cancers. Given the reversibility of DNA methylation, DNMTs can be viable targets for the treatment of cancer. It was found that DNMT3a- and DNMT3b-mediated epigenetic regulation is pivotal for appropriate trophoblastic invasion [13]. Acetyl groups are removed on both histone and non-histone by HDACs, resulting in the alteration of protein functions [14]. HDACs have been shown to play crucial roles in the regulation of several proteins involved with cell cycle, proliferation, immunity, inflammation, and apoptosis [15,16]. Class I HDACs, especially HDAC1, HDAC2, and HDAC3, are found largely in the nucleus and account for the post-translational modification of histones into a deacetylated state, which cause transcription machinery alteration and alter nuclear signaling pathways relative to cell fate [17,18]. Especially in choriocarcinoma, HDAC1/2/3 inhibits the expression of multidrug resistance-associated protein 2 [19], and the HDAC inhibitor regulates the migration of JEG-3 cells [20]. Both DNMTs and HDACs induce tumor-genes silencing and cause the self-reinforcing nature of silencing mechanisms by interaction between them [21], and thus cause tumorigenesis by a bypass compensatory mechanism [22]. Interestingly, breast CSCs were recently reported to be affected by combining DNMT and HDAC inhibition [23].
Curcumol is one of the major active components of Curcuma zedoaria, like curcumin [24,25], which is traditionally used for the treatment of gynecological tumors in China. Curcumol has been reported to show anti-cancer effects in several cervical cancer cells and ovarian cancer cells [26,27]. Choriocarcinoma treatment using Chinese herbs uses Curcuma zedoaria for adjuvant therapy with chemotherapy. However, to the best of our knowledge, the effect of the single component, curcumol, is still largely unknown in choriocarcinoma. In the present study, we tested the effect of curcumol on choriocarcinoma CSLCs via regulating epigenetic machinery.
Material and Methods
Drug and cell
Curcumol (purity ≥96.7%), obtained from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, CN), was dissolved in DMSO and then diluted in PBS with 0.1% DMSO. DNMT inhibitor 5-azacytidine (5-AzaC) and HDAC inhibitor Trichostatin A (TSA) were procured from Selleck (Houston, USA). The human choriocarcinoma JEG-3 cell line was obtained from the American Type Culture Collection (ATCC, USA) [28]. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose (Gibco, USA), supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and antibiotics (50 U/ml penicillin, and 50 μg/mL of streptomycin) at 37°C in a humidified incubator with 5% CO2 atmosphere.
Sphere formation assay
JEG-3 CSLCs were isolated as described previously [6] for 7 days. The spheres were dissociated into single cells, then re-cultured for another 7 days. The first-generation spheres were treated with 5-zazcytidine (75 μM) or TSA (100 nM) for 7 days.
CD133+ cells isolation and flow cytometry analysis
The cells were labelled with a primary CD133 antibody (Miltenyi Biotec, GER), and the CD133+ and CD133− cells were subsequently magnetically isolated using the EasySep™ Human APC Positive Selection Kit (StemCell Technologies, CAN) following the manufacturer’s instructions. Trypan blue staining was used to assess the sorted cell viability, and higher than 90% viability was considered acceptable for further downstream experiments. The dissociated single cells from spheres were stained with anti-CD133/APC and analyzed using a FACSCanto II Flow Cytometer instrument (BD Biosciences, USA). Acquired data were analyzed with FlowJo software.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted with TRIzol reagent (Life Technologies, USA) following the manufacturer’s protocol. The RNA was reverse transcribed to complementary DNA (cDNA) using a Transcriptor First Strand cDNA Synthesis Kit (Life Technologies, USA). qRT-PCR was performed using SYBR PremixEx Taq II method (Life Technologies, USA) on the ABI 7500 real-time PCR system (Thermo Fisher, USA). The expression level was calculated using the 2−ΔΔCt method. Experiments were repeated at least 3 times. The following primers obtained from Sangon Biotech (Shanghai, China) were used:
DNMT1 forward: 5′-CAGGAAGAACGGCCGCAGCA-3′,
reverse: 5′-AGGCTTTGCCGGCTTCCACG-3′;
DNMT3a forward: 5′-CAGTGCAGGTGACGAACATT-3′,
reverse: 5′-TGTTCCACCACACCTGTTTTGA-3′;
DNMT3b forward: 5′-GGCAAGTTCTCCGAGGTCTCTG-3′,
reverse: 5′-TGGTACATGGCTTTTCGATAGGA-3′;
HDAC1 forward: 5′-GCCATCCTGGAACTGCTAAA-3′,
reverse: 5′-GGCTTGAAAATGGCCTCATA-3′;
HDAC2 forward: 5′-CCTGGAACAGGTGACATGTATGA-3′,
reverse: 5′-CGTAAGGGCACATTGAGACAATAG-3′;
NANOG forward: 5′-AGAACTCTCCAACATTCCTGAACCT-3′,
reverse: 5′-TGCCACCTCTTAGATTTCATTCTCT-3′;
OCT4 forward: 5′-CTTGCTGCAGAAGTGGGTGGAGGAA-3′,
reverse: 5′-CTGCAGTGTGGGTTTCGGGCA-3′;
SOX2 forward: 5′-GGGAAATGGGAGGGGTGCAAAAGA-3′,
reverse: 5′-TTGCGTGAGTGTGGATGGGATTGG-3′;
ABCG2 forward: 5′-GCAAGATGTACTGGCGAAGA-3′,
reverse: 5′-CAGGTAGGCAATTGTGAGGAA-3′.
Western blot analysis
The cells were harvested and lysed using ice-cold RIPA lysis buffer (Beyotime Biotechnology, CN). Following denaturation, equivalent amounts of protein from each sample (30–50 μg) were separated on 10% SDS-PAGE. For immunodetection, resolved proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Merck, Germany) in a semidry blotter (Bio-Rad) for 2 h using transfer buffer. The membranes were then blocked with TBST supplemented with 5% BSA for 1 h at room temperature, and then probed with indicated primary antibodies at 4°C overnight with gentle shaking. All the primary antibodies of DNMT1, DNMT3b, HDAC1, and HDAC3 were purchased from CST (Germany). The membranes were incubated with a secondary horseradish peroxidase (HRP)-conjugated antibody for 1 h. The bands of the target proteins were detected using enhanced chemiluminescence (ECL) reagent (Millipore, USA), and acquired by chemiluminescence system (Syngene, UK). The gray value of each band was measured by Image J software.
Global DNA methylation quantification
The MethylFlash Methylated DNA Quantification kit (Epigentek Inc., NY) was used to measure the levels of the DNA methylation mark 5-methylcytosine (5-mC). The genomic DNA was isolated first, then the methylation of DNA was detected by the antibody, and the absorbance was read using a microplate spectrophotometer (Thermo Fisher, USA). The percentage of 5-mC in total DNA was calculated.
HDAC activity analysis
The HDAC activity was analyzed using a colorimetric assay kit (Bio Vision, USA), according to the manufacturer’s protocol. Briefly, nuclear cell lysates were extracted using the Nuclear Extract Kit (Active Motif, Belgium) and were incubated with the HDAC colorimetric substrate, the density was read at 405 nm on a Multimode Reader (Promega, USA).
Cell transfection
Cells were seeded into 6-well plates before the day of transfection. At 30%–50% fusion, cells were transfected with different lentiviruses using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. After 12 h, the transfection medium was replaced with normal growth medium. The mRNA expression was assessed using qRT-PCR. The LV-RNAi of DNMT1 (CCGGCGACTACATCAAAGGCAGCAACTCGAGTTGCTG CCTTTGATGTAGTCGTTTTT), DNMT3b (CCGGCCTGTCATTGTT TGATGGCATCTCGAGATGCCATCAAACAATGACAGGTTTTTG), HDAC1 (CCGGGCTGCTCAACTATGGTCTCTACTCGAGTAGAGAC CATAGTTGAGCAGCTTTTT) and HDAC3 (CCGGCCTGCATTA TGGTCTCTATAACTCGAGTTATA GAGACCATAATGCAGGTTTTTG) and over-expression lentivirus (DNMT1 and HDAC1) were all provided by Genechem (Shanghai, China).
Xenograft tumorigenicity assay
All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of Central South University (ID: 201603115).
BALB/c-nude mice (female, 4–5 weeks of age, body weight 18–20 g) were purchased from the Experimental Animal Center of Central South University. The animals were acclimated to their new environment for 1 week; the cells were inoculated subcutaneously into the left flank of mice. Tumor formation was monitored after inoculation. Tumors measuring at least 5 mm in diameter were considered to demonstrate successful model establishment. Tumor volume was calculated every 4 days using the equation (L*W2)/2. After 3 weeks, mice were sacrificed after injection of cells, and the tumors were stripped and weighed. The overall survival analysis was performed and recorded as the whole survival time per mouse.
Immunohistochemistry analysis
Fresh tissues were immediately fixed with 4% paraformaldehyde and paraffin-embedded. All the tissue samples were sectioned into 4-μm sections. Slides were deparaffinized, rehydrated, and treated with 3% hydrogen peroxide. Antigen retrieval was performed by boiling the sections for 10 min in citrate buffer antigen retrieval solution (pH 6.0) at 100°C for 3 min. The sections were subsequently incubated overnight with primary antibody (DNMT1, 1: 200, DNMT3b, 1: 200, HDAC1, 1: 100, HDAC3, 1: 200), and then were washed with PBS and incubated with IgG anti-rabbit HRP-conjugated secondary antibodies IgG (CST, Darmstadt) for 30 min. The slides were stained with DAB (Solarbio, Beijing, China) and the stained cells were then analyzed. Five fields were randomly selected. The IHC scores were calculated according the method described previously [29].
Statistical analysis
All data are expressed as means ±SD. Differences were determined by one-way ANOVA test and chi-square test. Data not conforming to the homogeneity of variance were analyzed using the Kruskal-Wallis test. Survival curves were plotted by the Kaplan-Meier method and compared by log-rank test. A probability of p<0.05 was considered statistically significant. All statistical analyses were performed using SPSS 17.0 software.
Results
Increased expression of DNMTs and HDACs in choriocarcinoma CSLCs
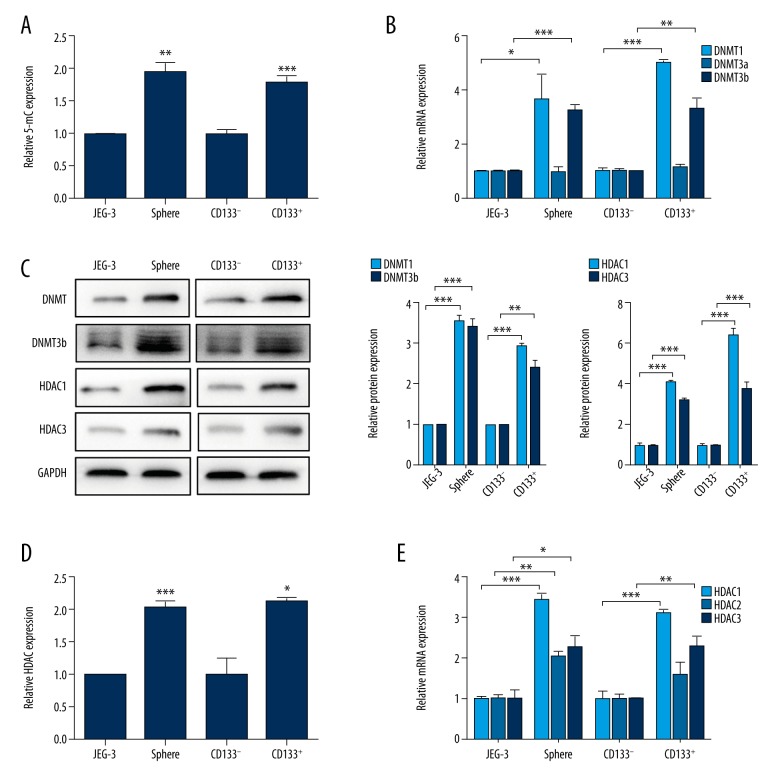
The choriocarcinoma CSLCs were isolated using 2 different methodologies. The first one, using free-serum spheres formation, was as we described earlier. In another, using the CSC genetic marker, we separated CD133+ cells from CD133− cells by FACS sorting. To determine the DNA methylation levels in CSLCs and non-CSLCs, we measured the levels of the DNA methylation markers 5-mC. The results showed that the CSLC population exhibited significantly higher expression of 5-mC, irrespective of the method used for isolating CSLCs (Figure 1A, JEG-3 vs. Sphere, p<0.01; CD133− vs. CD133+, p<0.001). Further, significantly higher mRNA and protein expression of DNMT1 and DNMT3b were observed in the CSLCs, regardless of the isolation method used (Figure 1B, 1C; p<0.05). Moreover, the HDAC activity was higher in CSLCs than in non-CSLCs (Figure 1D; p<0.05). In both of the 2 different isolation methods, significantly higher mRNA (Figure 1E, p<0.05) and protein (Figure 1C, p<0.001) expression of HDAC1 and HDAC3 were found in all CSLCs except for HDAC2. However, no significant difference was observed in the expression of HDAC2 mRNA in CD133+ cells as compared to CD133− cells. These findings indicate higher expression of DNMTs and HDACs in CSLCs.
Figure 1.
Higher DNMT and HDAC expression in choriocarcinoma CSLCs. (A) Quantification of 5-mC in non-CSLCs versus CSLCs (JEG-3 vs. sphere and CD133− vs. CD133+). Data are shown as relative expression compared with non-CSLCs (JEG-3 and CD133−). (B) RT-PCR analysis of DNMTs mRNA levels in non-CSLCs vs. CSLCs. (C) Western blot analysis of DNMT1, DNMT3b, HDAC1, and HDAC3 protein levels in non-CSLCs vs. CSLCs. GAPDH was used as the internal control (left). Respective relative changes in DNMTs (middle) and HDACs (right) protein levels. (D) Total HDAC activity of non-CSLCs vs. CSLCs. (E) RT-PCR analysis of HDACs mRNA levels in non-CSLCs vs. CSLCs. Data represent mean ±SD from at least 3 independent experiments, * p<0.05; ** p<0.01; *** p<0.001.
Inhibition of DNMTs and HDACs suppresses CSLCs self-renewal
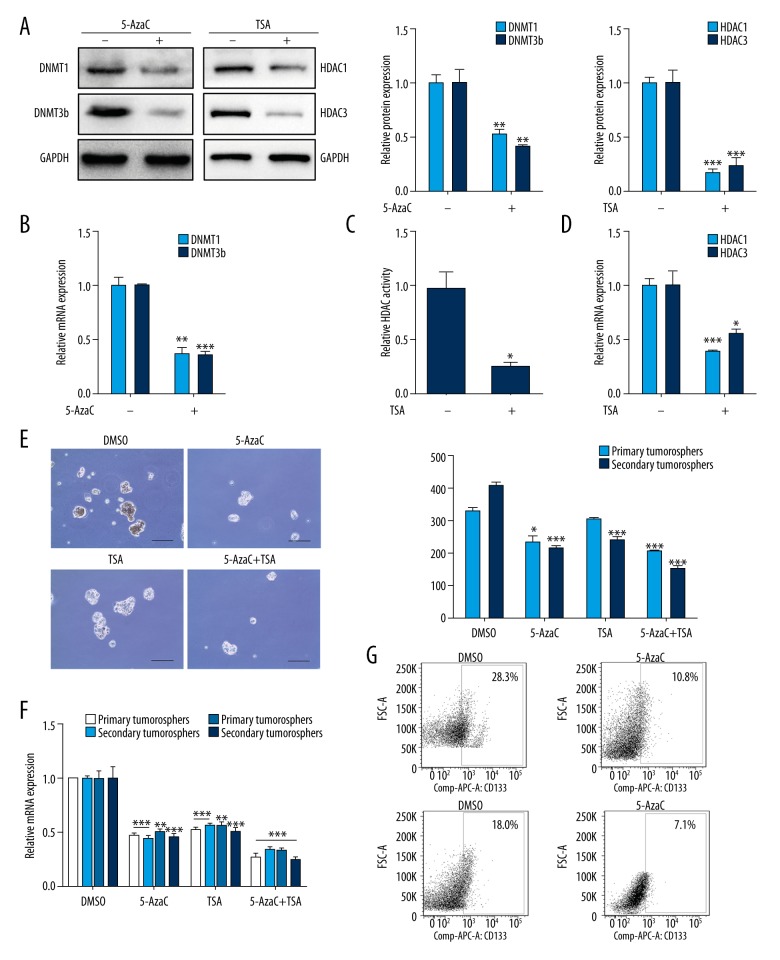
To further confirm the results above, DNMTs inhibitor 5-AzaC and HDACs inhibitor TSA were used to assess whether DNMT and HDAC inhibition could ablate choriocarcinoma CSLCs self-renewal. JEG-3 cells were treated with 5-AzaC or TSA in the process of sphere formation every other day for 7 days. DNMT1 and DNMT3b expression were reduced by 5-AzaC in spheres at both the mRNA and protein levels (Figure 2A, 2B, p<0.01). Similar suppression in the HDAC activity in spheres was observed with the treatment of TSA (Figure 2C, p<0.05); however, a significant decrease in HDAC1 and HDAC3 mRNA (Figure 2D, p<0.05) and protein (Figure 2A, p<0.01) expression was observed. For the self-renewal, 5-AzaC and TSA were significantly abrogated by CSLCs in both first- and second-generation (Figure 2E, p<0.05) and reduced the expression of stemness-associated genes, including NANOG, OCT4, SOX2, and ABCG2 (Figure 2F, p<0.01). The CD133 cell surface expression levels were significantly suppressed in spheres treated with 5-AzaC and TSA (Figure 2G, p<0.05). These data indicated that DNMTs and HDACs play significant roles in preserving the stemness state of CSLCs.
Figure 2.
DNMT and HDAC inhibition decrease CSLCs self-renewal. (A) Western blot was used to analyze DNMT1/3b and HDAC1/3 protein levels following 5-AzaC or TSA treatment (left). Respective changes in DNMTs (middle) and HDACs (right) protein levels. (B) RT-PCR analysis of DNMT1 and DNMT3b mRNA levels with 5-AzaC treatment. (C) Total HDAC activity of CSLCs with TSA treatment. (D) RT-PCR analysis of HDAC1 and HDAC3 mRNA levels with TSA treatment. (E) Representative images of spheres (10×, left) and sphere counts (right) in first- and second-generation in cells with 5-AzaC or TSA treatment. (F) RT-PCR analysis of stemness-associated genes in sphere cells with 5-AzaC or TSA treatment (groups of 5-AzaC, TSA and 5-AzaC+TSA vs. DMSO group). Results were normalized to expression of GAPDH. (G) Flow cytometry analysis of CD133 cell surface expression in sphere cells with 5-AzaC or TSA treatment. Data represent mean ±SD from at least 3 independent experiments, * p<0.05; ** p<0.01; ***p<0.001.
Inhibition of CSLCs self-renewal with RNAi of DNMT1/3b and HDAC1/3
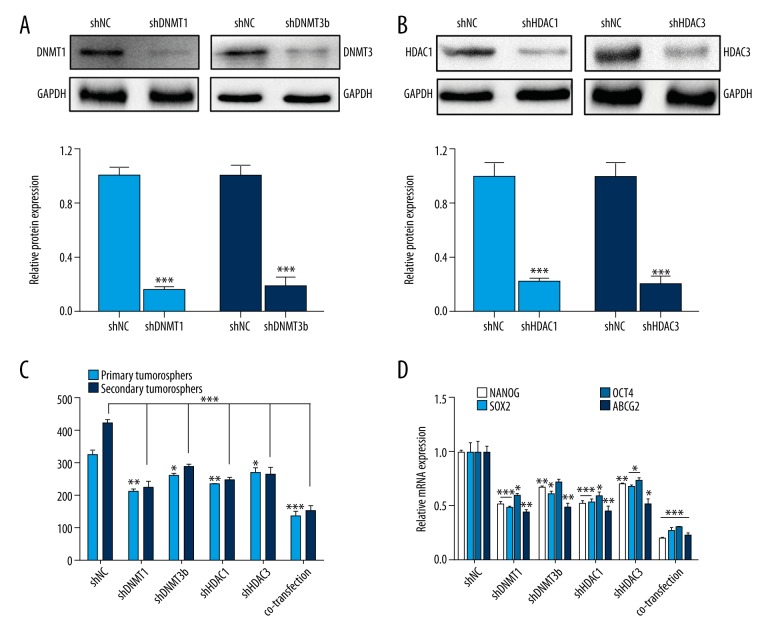
To further verify our hypothesis that DNMT and HDAC were indeed crucial for maintaining the stemness of CSLCs, we established transfected cells with LV-shRNA of DNMT1/3b and HDAC1/3 alone or combined. The expression of DNMT1/3b and HDAC1/3 protein decreased significantly in the transfected cells (Figure 3A, 3B, p<0.001). Further, the RNAi of DNMTs and HDACs inhibited the sphere formation (Figure 3C, p<0.05) and expression of stemness-associated genes (Figure 3D, p<0.05). Taken together, the results further support the vital role of DNMTs and HDACs in choriocarcinoma CSLCs.
Figure 3.
RNAi of DNMT1/3b and HDAC1/3 inhibit CSLCs self-renewal. (A) Western blot analysis of DNMT1 and DNMT3b protein expression with the shRNA interference (shDNMT1, shDNMT3b vs. shNC). (B) Western blot analysis of HDAC1 and HDAC3 protein expression with the shRNA interference (shHDAC1, shHDAC3 vs. shNC). (C) Sphere counts in first- and second-generation in cells with overexpressing transfection of DNMT1, DNMT3, HDAC1, and HDAC3 (co-expression for 4 lentivirus transfections together; groups of shRNA vs. shNC group). (D) After the transfection, stemness-associate genes in the sphere cells were detected by RT-PCR (groups of shRNA vs. shNC group). Data represent mean ±SD from at least 3 independent experiments, * p<0.05; ** p<0.01; *** p<0.001.
Curcumol suppresses CSLCs self-renewal and DNMT/HDAC activity in vitro and in vivo
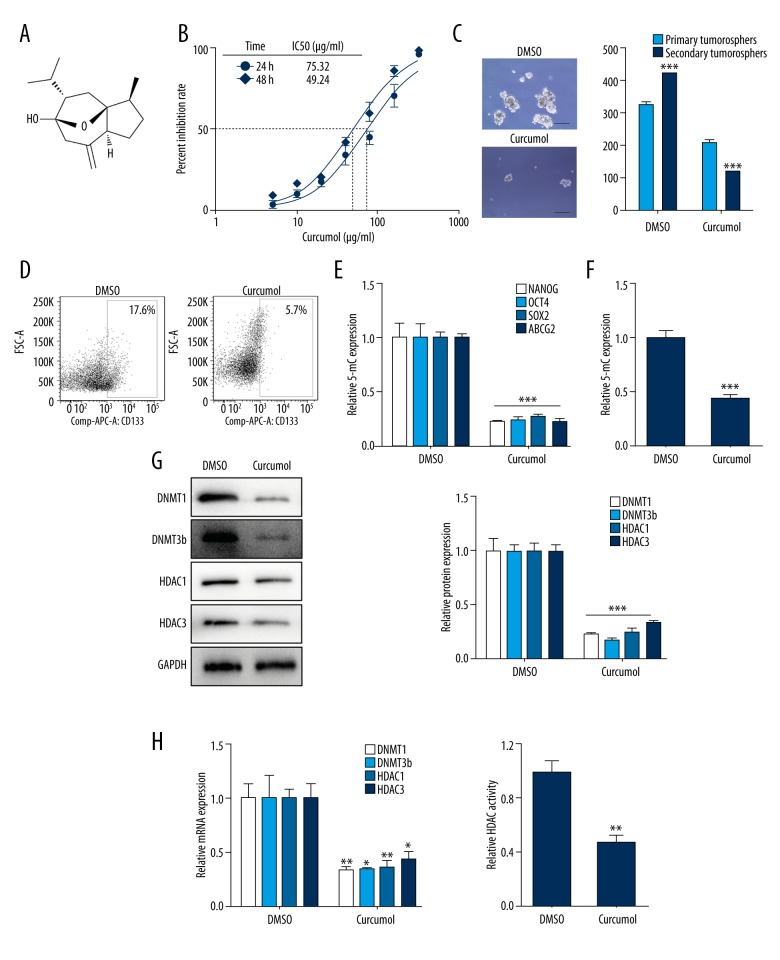
Next, to investigate the effect of curcumol (Figure 4A) on choriocarcinoma, 75 μg/ml curcumol was used in JEG-3 cells, and the mean 50% inhibitory concentrations (IC50) of cell growth (Figure 4B) was calculated. Primary JEG-3 cells were treated with 75 μg/ml curcumol every other day for 7 days for sphere formation. The treatment for functional level: (1) reduced the sphere formation rate (Figure 4C, p<0.001), (2) suppressed the CD133 surface expression percent (Figure 4D, p<0.01), and (3) decreased the expression of stemness-associated genes (Figure 4E, p<0.001). In terms of epigenetic profile, a higher expression of 5-mC indicated DNA methylation in CSLCs was abrogated by curcumol treatment (Figure 4F, p<0.001). Furthermore, the treatment led to a significant reduction in the expression of DNMT1 and DNMT3b (Figure 4G, 4H, p<0.05). Consistent with these findings, curcumol decreased HDAC activity in spheres (Figure 4I, p<0.01) and also significantly reduced the expression of the HDAC1/3 protein (Figure 4G, p<0.05) and mRNA (Figure 4H, p<0.001).
Figure 4.
Curcumol decreases CSLCs self-renewal and DNMT/HDAC activity in vitro. (A) Chemical structure of curcumol. (B) IC50 dose response curves of JEG-3 cells with different doses curcumol at 24 h and 48 h. (C) Representative tumorospheres images (10×) of DMSO and treated 75 μg/ml curcumol for 7 days (left) and sphere counts in first- and second-generation in cells (right). (D) Flow cytometry analysis of CD133 cell surface expression in sphere cells with curcumol treatment. (E) RT-PCR analysis of NANOG, OCT4, SOX2, and ABCG2 mRNA expression levels in sphere cells with curcumol treatment. (F) Quantification of 5-mC in sphere cells with curcumol treatment. (G) Western blot analysis of DNMT1/3b and HDAC1/3 in sphere cells with RA treatment. GAPDH was used as the internal control (left). Respective changes in DNMTs and HDACs (right) protein levels. (H) The mRNA expression levels of DNMT1/3b and HDAC1/3 in cells with curcumol treatment. (I) After curcumol treatment, HDAC activity was detected. Data represent mean ±SD from at least 3 independent experiments, * p<0.05; ** p<0.01; *** p<0.001.
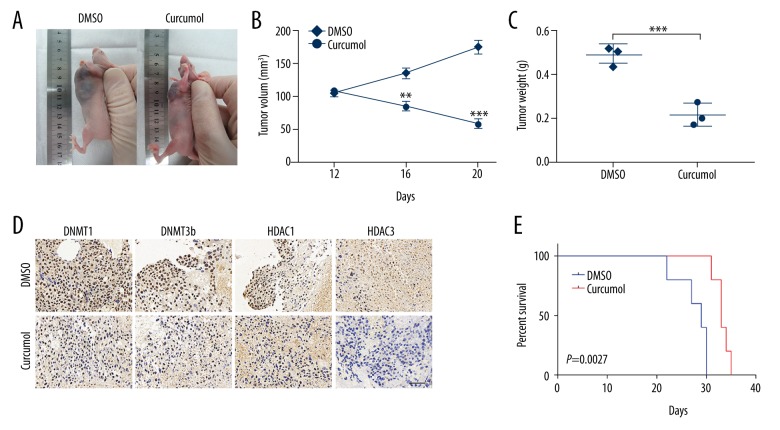
Further, for the in vivo studies, choriocarcinoma CSLCs tumor-bearing nude mice were orally treated with curcumol (200 mg/kg) daily for 10 days when the tumor grew to 100 mm3. Two weeks after the termination of treatment, changes in the tumors in mice were observed (Figure 5A). A significant shrinkage both in tumor volume and weight was observed after treatment with curcumol (Figure 5B, 5C, p<0.001). Further, IHC analysis showed that the curcumol-treated xenografts exhibited elevated DNMT1, DNMT3b, HDAC1, and HDAC3 expression (Figure 5D). The Kaplan-Meier analysis revealed that mice treated with curcumol survived longer than in the DMSO group (Figure 5E, p=0.0027). These data demonstrate that inhibition of curcumol overcomes the stemness potential of choriocarcinoma CSLCs.
Figure 5.
Curcumol decreases CSLCs self-renewal in vivo. (A) Tumor-bearing nude mice were treated with (200 mg/kg/day) orally for 10 days when the tumor grew to 100 mm3. Subcutaneous tumor in DMSO group vs. curcumol group of nude mice. (B) Tumor volumes were measured every 4 days for 3 weeks (DMSO vs. curcumol group). (C) Tumor weight in the indicated mice (DMSO vs. curcumol group). (D) IHC staining displaying the DNMT1, DNMT3b, HDAC1, and HDAC3 protein expression between DMSO and curcumol groups (IHC, 40×). (E) Kaplan-Meier curves of mice with curcumol treatment, compared with the DMSO group. Data represent mean ±SD from at least 3 independent experiments, ** p<0.01; *** p<0.001.
Curcumol affects CSLCs through regulation of DNMTs and HDACs
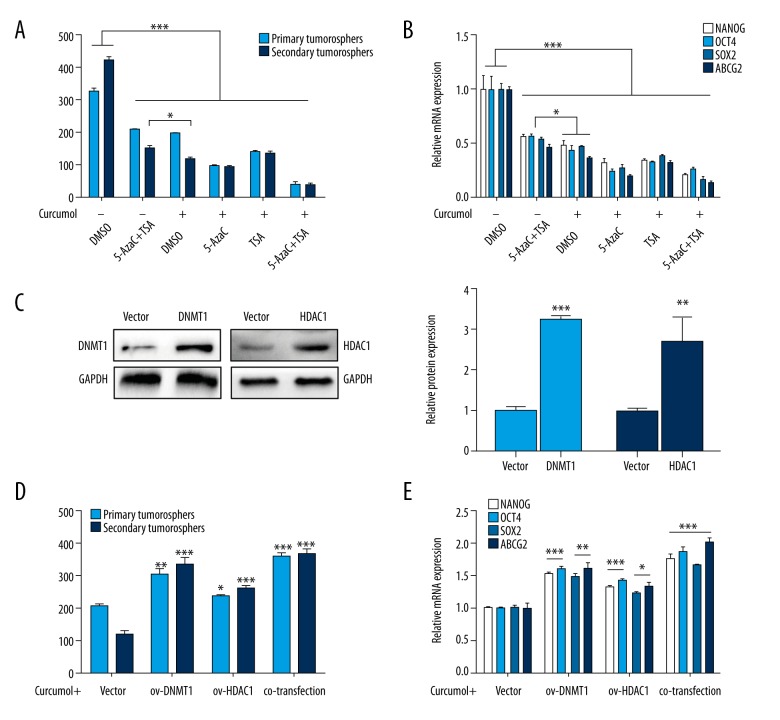
Next, we assessed whether the inhibition of curcumol in choriocarcinoma CSLCs is through the regulation of DNMTs and HDACs. The primary cells treated with curcumol had significantly fewer sphere counts as compared to the combination of 5-AzaC and TSA in the secondary generation (p<0.05), and the NANOG, OCT4, SOX2, and ABCG2 expression levels (p<0.05). Further, the treatments of curcumol combined with 5-AzaC or TSA were also evaluated. When curcumol was combined with 5-AzaC or TSA, both the combinations significantly enhanced the inhibition of CSLCs phenotypes, which was represented by decreased sphere formation rate and stemness-associated genes expression (Figure 6A, 6B, p<0.05). Nevertheless, given the exceptional role of DNMT1 and HDAC1 in CSLCs, we transfected their over-expression lentivirus into primary JEG-3 cells (Figure 6C, p<0.01). The curcumol-mediated reduction of CSLCs was reversed by DNMT1 and HDAC1 over-expression, including stemness-associated genes (Figure 6D, 6E, p<0.05). These results demonstrate that curcumol suppresses CSLCs through the significant regulation of DNMTs and HDACs in choriocarcinoma.
Figure 6.
Curcumol affects CSLCs via regulation of DNMT and HDAC. With the treatment of curcumol combined with 5-AzaC or TSA, (A) the numbers of spheres in first- and second-generation cultures from JEG-3 cells (all groups vs. DMSO group; 0 μM curcumol+5-AzaC+TSA vs. 75 μg/ml curcumol+DMSO) and (B) stemness-associated genes mRNA expression levels. With the transfection of over-expression lentivirus of DNMT1 or HDAC1, (C) Western blot analysis of DNMT1 and HDAC1 protein levels (left) and respective changes (right), (D) sphere counts in first- and second-generation in cells (over-expression groups vs. Vector group) and (E) respective stemness-associated genes mRNA expression levels. Data represent mean ±SD from at least 3 independent experiments, * p<0.05; ** p<0.01; *** p<0.001.
Discussion
Because cancer stem cells play pivotal roles in occurrence, development, recurrence, and metastasis of cancer, it is imperative to elucidate the underlying molecular mechanisms of CSCs. Recently, the CSC epigenetics was shown to explain the significance of epigenetic-modifying agents in the stem-like characteristics of tumor cells [30]. The present study demonstrated that curcumol could reduce the self-renewal of choriocarcinoma CSLCs with repression of the DNMT- and HDAC-mediated epigenetic regulation.
Epigenetic regulation is a heritable mechanism during stem cell fate specification [31], which involves intrinsic and extrinsic mechanisms deciding cell fate [32]. CSCs exhibit remarkable functional differences from their progenies that share the same genetic information, indicating the epigenetic changes they regulate. Thus, epigenetics must be the potential driver of the inherent heterogeneity [33]. Chemotherapy is the first-line therapy in choriocarcinoma patients; however, due to the childbearing age and the intrinsic chemotherapeutic sensitivity, there is a serious shortage of grouped clinical specimens. Despite this limitation, the present in vitro study supports that hypermethylation and activated histone deacetylation are consistent differential factors in CSCs as compared to non-CSCs.
Embryonic stem cells that lack DNA methyltransferases remain viable; however, they die when induced to differentiate [34]. Numerous studies reported that DNMTs are essential for cancer stem cell maintenance and tumorigenesis [35,36], including DNMTs themselves and DNMT-mediated epigenetics [37,38]. In another choriocarcinoma cell line, BeWo cells, both DNMT3a and DNMT3b, were found to affect migration and invasion [13]. Furthermore, the epigenetic mechanism has also been found to play a dynamic role in oncogenes. For instance, DNA methylation of E-cadherin promotor helps to recruit HDACs to the site, leading to histone deacetylation and transcriptional silencing [39]. The combination of DNMT and HDAC inhibitors, one of the most common combination therapies, have been extensively evaluated in clinical trials for the treatment of a variety of cancers [40]. In the present study, DNMT1/3b and HDAC1/3 expression were consistently upregulated in CSLCs. The combination of 5-AzaC and TSA showed a noticeable effect on multipotential stemness, which indicated the critical role of DNMTs and HDACs in CSLCs.
Besides these known single-target compounds, many other multipotent-drugs are being developed and investigated in clinical trials. Thus, we sought to evaluate the effectiveness of the traditional drugs with new and pleiotropic functions in cancer therapy. As an active component of a traditional Chinese medicine used in gynecological therapy, curcumol attracted our attention due to its low toxicity and its anti-cancer effects on many tumors. Therefore, in this study we investigated the anti-cancer activities of curcumol against choriocarcinoma CSLCs. Numerous studies have reported the anti-cancer mechanisms of curcumol. For instance, it repressed colorectal cancer cells via regulating IGF-R and p38 MAPK signaling pathways [41] and Akt/GSK3β/cyclin D1 pathways [42]. Some traditional Chinese medicines, such as curcumin, have been shown to affect epigenetic regulation in cancer cells [43,44]. However, there are no reports on its epigenetic-regulatory effect in cancer cells, particularly in CSCs. Notably, in the present study, the stemness repression in choriocarcinoma CSLCs was observed in both in vitro and in vivo studies with treatment of curcumol. Moreover, curcumol enhanced the overall survival rate of tumor-bearing mice. We further investigated the underlying regulatory mechanisms of curcumol. As hypothesized, DNMTs and HDACs expressions were significantly decreased in CSLCs by curcumol, and the stemness repression of curcumol can be abrogated by DNMT/HDAC overexpression, remarkably. These findings indicated that DNMT/HDAC is the potential target of curcumol, which warrants further investigation. Moreover, we found that curcumol exhibited a significantly greater effect on stemness repression in CSLCs as compared to the combination of DNMT and HDAC inhibitors. Experimentation with various other types of inhibitors is required.
Conclusions
In conclusion, our results underlined the significance of epigenetic mechanism in choriocarcinoma CSLCs through regulation of DNMTs and HDACs. Our results also demonstrated the effectiveness of curcumol against CSLCs, indicating its potential therapeutic role in epigenetic cancer therapies.
Footnotes
Conflict of interest
None.
Source of support: This study was supported by the Graduate Independent Innovation Project Fund of Central South University (Grant number 2016zzts125) and the National Natural Science Foundation of China (Award number 81472434)
References
- 1.Abrão RA, de Andrade JM, Tiezzi DG, et al. Treatment for low-risk gestational trophoblastic disease: Comparison of single-agent methotrexate, dactinomycin and combination regimens. Gynecol Oncol. 2008;108:149–53. doi: 10.1016/j.ygyno.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Ma Y, Xiang Y, Wan XR, et al. The prognostic analysis of 123 postpartum choriocarcinoma cases. Int J Gynecol Cancer. 2008;18:1097–101. doi: 10.1111/j.1525-1438.2007.01132.x. [DOI] [PubMed] [Google Scholar]
- 3.Park CY, Tseng D, Weissman IL. Cancer stem cell-directed therapies: recent data from the laboratory and clinic. Mol Ther. 2009;17:219–30. doi: 10.1038/mt.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, Yi M, Chen S, et al. NOR1 suppresses cancer stem-like cells properties of tumor cells via the inhibition of the AKT-GSK-3β-Wnt/β-catenin-ALDH1A1 signal circuit. J Cell Physiol. 2017;232:2829–40. doi: 10.1002/jcp.25706. [DOI] [PubMed] [Google Scholar]
- 6.Cai J, Peng T, Wang J, et al. Isolation, culture and identification of choriocarcinoma stem-like cells from the human choriocarcinoma cell-line JEG-3. Cell Physiol Biochem. 2016;39:1421–32. doi: 10.1159/000447845. [DOI] [PubMed] [Google Scholar]
- 7.Wainwright EN, Scaffidi P. Epigenetics and cancer stem cells: Unleashing, hijacking, and restricting cellular plasticity. Trends Cancer. 2017;3:372–86. doi: 10.1016/j.trecan.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 10.Yi M, Yang J, Li W, et al. The NOR1/OSCP1 proteins in cancer: From epigenetic silencing to functional characterization of a novel tumor suppressor. J Cancer. 2017;8:626–35. doi: 10.7150/jca.17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway SJ, Woster PM, Greenlee WJ, et al. Epigenetics: Novel therapeutics targeting epigenetics. J Med Chem. 2016;59:1247–48. doi: 10.1021/acs.jmedchem.6b00098. [DOI] [PubMed] [Google Scholar]
- 12.Zhong T, Zhang Y, Guo Q, et al. Parental neuropathic pain influences emotion-related behavior in offspring through maternal feeding associated with DNA methylation of amygdale in rats. Neurochem Res. 2015;40:1179–87. doi: 10.1007/s11064-015-1578-1. [DOI] [PubMed] [Google Scholar]
- 13.Rahnama F, Shafiei F, Gluckman PD, et al. Epigenetic regulation of human trophoblastic cell migration and invasion. Endocrinology. 2006;147:5275–83. doi: 10.1210/en.2006-0288. [DOI] [PubMed] [Google Scholar]
- 14.Reilly CM, Regna N, Mishra N. HDAC inhibition in lupus models. Mol Med. 2011;17:417–25. doi: 10.2119/molmed.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemoine M, Younes A. Histone deacetylase inhibitors in the treatment of lymphoma. Discov Med. 2010;10:462–70. [PubMed] [Google Scholar]
- 16.Wang X, Guo H, Liu W, et al. Effects of siRNA-mediated knockdown of HDAC1 on the biological behavior of esophageal carcinoma cell lines. Med Sci Monit. 2016;22:1291–96. doi: 10.12659/MSM.895853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011;2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Ruijter AJ, van Gennip AH, Caron HN, et al. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem J. 2003;370:737–49. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan HY, Ma D, Zhou KY, et al. Effect of histone deacetylase inhibition on the expression of multidrug resistance-associated protein 2 in a human placental trophoblast cell line. Chin Med J (Engl) 2017;130:1352–60. doi: 10.4103/0366-6999.206352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma H, Q MY, Zhang X, et al. HSPC117 is regulated by epigenetic modification and is involved in the migration of JEG-3 cells. Int J Mol Sci. 2014;15:10936–49. doi: 10.3390/ijms150610936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson KD, Ait-Si-Ali S, Yokochi T, et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 22.Clements EG, Mohammad HP, Leadem BR, et al. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012;40:4334–46. doi: 10.1093/nar/gks031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathania R, Ramachandran S, Mariappan G, et al. Combined inhibition of DNMT and HDAC blocks the tumorigenicity of cancer stem-like cells and attenuates mammary tumor growth. Cancer Res. 2016;76:3224–35. doi: 10.1158/0008-5472.CAN-15-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng R, Deng Q, Liu Y, et al. Curcumin inhibits gastric carcinoma cell growth and induces apoptosis by suppressing the Wnt/β-catenin signaling pathway. Med Sci Monit. 2017;23:163–71. doi: 10.12659/MSM.902711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen D, Dai F, Chen Z, et al. Dimethoxy curcumin induces apoptosis by suppressing survivin and inhibits invasion by enhancing E-cadherin in colon cancer cells. Med Sci Monit. 2016;22:3215–22. doi: 10.12659/MSM.900802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu LC, Bian K, Liu ZM, et al. The inhibitory effect of the curcumol on women cancer cells and synthesis of RNA. Tumor. 2005;25:570–72. [Google Scholar]
- 27.Yane G, Jinzhu G, Hui H, et al. Effects of curcumol on proliferation and apoptosis in human cervical cancer CASKI cell line. Modern Oncology. 2009;17:1836–39. [Google Scholar]
- 28.Shi D, Tan Z, Lu R, et al. MicroRNA-218 inhibits the proliferation of human choriocarcinoma JEG-3 cell line by targeting Fbxw8. Biochem Biophys Res Commun. 2014;450:1241–46. doi: 10.1016/j.bbrc.2014.06.094. [DOI] [PubMed] [Google Scholar]
- 29.Wongtrakoongate P. Epigenetic therapy of cancer stem and progenitor cells by targeting DNA methylation machineries. World J Stem Cells. 2015;7:137–48. doi: 10.4252/wjsc.v7.i1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soslow RA, Dannenberg AJ, Rush D, et al. COX-2 is expressed in human pulmonary, colonic, and mammary tumors. Cancer. 2000;89:2637–45. doi: 10.1002/1097-0142(20001215)89:12<2637::aid-cncr17>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 31.Lunyak VV, Rosenfeld MG. Epigenetic regulation of stem cell fate. Hum Mol Genet. 2008;17:28–36. doi: 10.1093/hmg/ddn149. [DOI] [PubMed] [Google Scholar]
- 32.Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453:306–13. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
- 33.Zagorac S, Alcala S, Fernandez Bayon G, et al. DNMT1 inhibition reprograms pancreatic cancer stem cells via upregulation of the miR-17-92 cluster. Cancer Res. 2016;76:4546–58. doi: 10.1158/0008-5472.CAN-15-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 35.Pathania R, Ramachandran S, Elangovan S, et al. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat Commun. 2015;6:6910. doi: 10.1038/ncomms7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roscigno G, Quintavalle C, Donnarumma E, et al. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget. 2016;7:580–92. doi: 10.18632/oncotarget.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai Q, Xu YH, Chen Q, et al. The loss-of-function of DNA methyltransferase 1 by siRNA impairs the growth of non-small cell lung cancer with alleviated side effects via reactivation of RASSF1A and APC in vitro and vivo. Oncotarget. 2017;8:59301–11. doi: 10.18632/oncotarget.19573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilhelm T, Lipka DB, Witte T, et al. Epigenetic silencing of AKAP12 in juvenile myelomonocytic leukemia. Epigenetics. 2016;11:110–19. doi: 10.1080/15592294.2016.1145327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Shang Y. Epigenetic control of epithelial-to-mesenchymal transition and cancer metastasis. Exp Cell Res. 2013;319:160–69. doi: 10.1016/j.yexcr.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 40.Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Huang F, Bai Z, et al. Curcumol inhibits growth and induces apoptosis of colorectal cancer LoVo cell line via IGF-1R and p38 MAPK pathway. Int J Mol Sci. 2015;16:19851–67. doi: 10.3390/ijms160819851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Li XM, Bai Z, et al. Curcumol induces cell cycle arrest in colon cancer cells via reactive oxygen species and Akt/GSK3β/cyclin D1 pathway. J Ethnopharmacol. 2018;210:1–9. doi: 10.1016/j.jep.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 43.Boyanapalli SS Tony Kong ANL “Curcumin, the King of Spices”. Epigenetic regulatory mechanisms in the prevention of cancer, neurological, and inflammatory diseases. Curr Pharmacol Rep. 2015;1:129–39. doi: 10.1007/s40495-015-0018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Y, Shu L, Zhang C, et al. Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1. Biochem Pharmacol. 2015;94:69–78. doi: 10.1016/j.bcp.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]