Abstract
Tetrabromobisphenol A (TBBPA) is a widely used flame retardant in printed circuit boards, paper, and textiles. In a two-year study, TBBPA showed evidence of uterine tumors in female Wistar-Han rats and liver and colon tumors in B6C3F1 mice. In order to gain further insight into early gene and pathway changes leading to cancer, we exposed female Wistar Han rats to TBBPA at 0, 25, 250, or 1000 mg/kg (oral gavage in corn oil, 5×/week) for 13 weeks. Because at the end of the TBBPA exposure period, there were no treatment-related effects on body weights, liver or uterus lesions, and liver and uterine organ weights were within 10% of controls, only the high dose animals were analyzed. Analysis of the hepatic and uterine transcriptomes showed TBBPA-induced changes primarily in the liver (1000 mg/kg), with 159 transcripts corresponding to 132 genes differentially expressed compared to controls (FDR = 0.05). Pathway analysis showed activation of interferon (IFN) and metabolic networks. TBBPA induced few molecular changes in the uterus. Activation of the interferon pathway in the liver occurred after 13-weeks of TBBPA exposure, and with longer term TBBPA exposure this may lead to immunomodulatory changes that contribute to carcinogenic processes.
Keywords: Tetrabromobisphenol A, Toxicogenomics, Microarray, Interferon response transcripts, Pathway analysis
1. Introduction
Tetrabromobisphenol A (TBBPA) is a high production volume brominated flame retardant (Malkoske et al., 2016), used in printed circuit boards, paper, and textiles (U. S. EPA, 2015; Zhou et al., 2014). TBBPA exposure occurs from breast milk (Carignan et al., 2012; Harrad and Abdallah, 2015; Nakao et al., 2015), food ingestion (e.g. fish (Svihlikova et al., 2015)), industrial exposures (Zhou et al., 2014), dust in the home (Di Napoli-Davis and Owens, 2013), and at waste sites (Liu et al., 2016). TBBPA also accumulates in marine life and may be toxic to various fish species (He et al., 2015; Tang et al., 2015).
TBBPA caused clear evidence of uterine adenocarcinomas in female Wistar Han rats [Crl:WI(Han)] in a 2-year study (Dunnick et al., 2015; National Toxicology Program, 2014). These TBBPA-induced uterine tumors were highly malignant with metastases to the liver, pancreas, kidney, thyroid and other organ systems. Other TBBPA carcinogenic findings occurred in the liver, lower intestine, and vascular systems of male mice (National Toxicology Program, 2014). TBBPA has been classified as probably carcinogenic to humans (Group 2a) by the Interagency for Research on Cancer (IARC) based on sufficient evidence for carcinogenicity found in the 2-year rodent studies and mechanistic information reported in the literature (Grosse et al., 2016).
In this study we looked for TBBPA-induced transcriptomic changes in the liver because this organ is a primary site for metabolism of hormones and other chemicals (Tsuchiya et al., 2005), and in the uterus, a target site for TBBPA-induced tumors in rats (National Toxicology Program, 2014). The TBBPA carcinogenic effect in the uterus is of concern because endometrial tumors are a common malignancy in women with an estimated 50,000 new cases per year in the U.S. (Siegel et al., 2013), and one million new cases per year worldwide (Webb, 2015). Uterine cancer is predicted to be one of the three leading cancers in women by 2030 (Rahib et al., 2014). The majority of human uterine tumors are endometrial carcinomas (George et al., 2015); the same type of uterine tumors seen in rats after TBBPA exposure (National Toxicology Program, 2014). Environmental factors are thought to play a role in the development of uterine cancer (Lichtenstein et al., 2000), including chemical and hormone effects (e.g. tamoxifen and estrogen) (IARC, 2012).
TBBPA is a nongenotoxic chemical, and toxicokinetic studies of TBBPA in the female rat did not reveal any specific accumulation of the parent compound or metabolites in the uterus (compared to that in other organ systems) (Knudsen et al., 2014). Thus, these 13-week TBBPA studies were undertaken to identify molecular alterations in the liver and/or uterus to help characterize early changes along the pathway to cancer.
2. Materials and methods
2.1. Experimental design
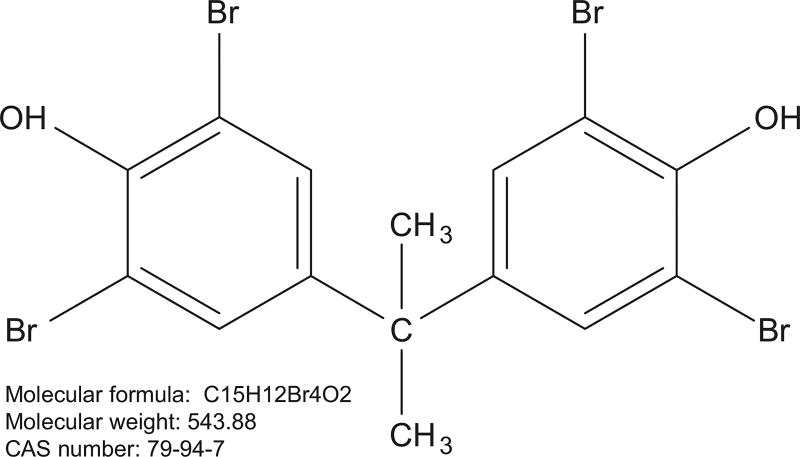
Tetrabromobisphenol A (CAS No. 79-94-7; Albemarle Corporation (Baton Rouge, LA), lot M032607 K) (Fig. 1) was prepared for oral gavage administration in corn oil to deliver TBBPA at doses of 0, 25, 250, or 1000 mg/kg body weight in a volume of 5 mL/kg body weight. Female Wistar Han IGS rats (Crl:WI(Han)) (25 animals/dose level) were obtained from Charles River (Raleigh, NC). 1000 mg/kg was a dose at which TBBPA induced uterine tumors in the female rat (National Toxicology Program, 2014), and transcriptomic patterns of liver and uterus were examined at this dose level. Two lower doses were added (25 and 250 mg/kg) to examine the uterus for transcriptomic changes, because this was the target organ in the 2-year cancer study. At the start of the study the animals were 5–6 weeks of age. The animals were housed two per cage. Tap water and NTP-2000 diet (Zeigler Brothers, Inc. Gardners, PA) were made available for ad libitum consumption.
Fig. 1.
TBPPA structure.
Body weights were obtained at one day prior to dosing and weekly thereafter. The treatment schedule was five days per week, excluding weekends and holidays, for up to 13 weeks. Animals were treated for a minimum of two consecutive days (within 24 h) prior to necropsy. Animals were not treated on the morning of scheduled sacrifice. At sacrifice, animals were euthanized with CO2. At necropsy uterine and liver organ weights were taken, and uterine and liver samples fixed in formalin for histopathologic evaluation at all dose levels. Sections were taken from the uterus (control and all dose levels) and liver (control and high dose) and flash frozen for the molecular studies.
The uterus with horns, vagina, and ovaries were harvested. The ovaries and vagina from each animal were fixed in formalin. The uterus was weighed then transected along the midline, including uterine body, such that one half of the uterus (including horn and body) was flash frozen in liquid nitrogen for RNA extraction. The other half of the uterus (including horn and body) was laid out, stretched with minimal tension, and pinned onto a piece of heavy card stock and placed in formalin. This was to prevent curling and shrinkage of the uterine body and horn so that it could be trimmed longitudinally without curling. This allowed appropriate microscopic interpretation of any changes.
The liver was removed and weighed, and then approximately a one-gram sample of the left lobe was collected and minced into approximately 3 mm cubes while on a weigh boat sitting in dry ice. The cubes were then frozen in liquid nitrogen in the weigh boat and, once frozen, transferred to three labeled, 2.0 mL cryotubes and flash frozen in liquid nitrogen. The liver samples were collected and frozen within 5 min of each animal’s sacrifice. An adjacent section (~3 mm) of the remaining left lobe of the liver was collected and placed in a labeled cassette and fixed in formalin for histopathology.
The care of animals on this study was according to NIH procedures as described in the “The U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals”, available from the Office of Laboratory Animal Welfare, National Institutes of Health, Department of Health and Human Services, RKLI, Suite 360, MSC 7982, 6705 Rockledge Drive, Bethesda, MD 20892-7982 or online at http://grants.nih.gov/grants/olaw/olaw.htm#pol. The protocol was approved by the laboratory where the animals were housed and dosed (Alion Science and Technology Animal Care and Use Committee).
2.2. Uterine and liver RNA preparation and microarray hybridization
A section of the frozen liver and one half of the frozen uterus (flash frozen and stored at −80° C) were placed into RNAlater® (Ambion, Inc., Austin, TX). RNA was extracted from control and treated liver and uterus to identify any molecular changes at a dose that caused uterine tumors (1000 mg/kg). RNA extraction was performed on tissues from 15 to 16 randomly selected animals per group.
RNA was extracted from the flash frozen uterine or liver tissues using the Invitrogen PureLink Mini kit (Invitrogen cat# 12183-018A, Carlsbad, CA) according to the manufacturer’s protocol. Frozen tissue samples were lysed and homogenized in TRIzol reagent (Invitrogen) using a rotor–stator homogenizer. Isolation of RNA was performed according to the mini kit protocol. On-column deoxyribonuclease (DNase) treatment was performed using the Invitrogen PureLink DNase kit (Invitrogen) to purify the RNA samples. RNA concentration and quality were measured on a Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples were aliquoted and stored at −80°C until they were analyzed.
Total RNA was used to synthesize double-stranded cDNA for each sample using Affymetrix GeneChip® Expression Analysis with 3’amplication two-cycle target labeling and control reagents (Affymetrix Inc. Santa Clara, CA). The cDNA served as a template to synthesize biotin-labeled antisense cRNA using an in vitro transcription (IVT) labeling kit. Labeled cRNA was fragmented and hybridized to the Affymetrix Rat Genome 230 2.0 Genechip® Array. Array hybridization, washing, and staining were performed according to the Affymetrix recommended protocol EuKGE_Ws2v5. The chips were scanned using an Affymetrix GeneChip® Scanner 3000. Quality control measurements were evaluated to determine if the data derived from the arrays were of sufficient quality prior to comparisons for differential expression.
2.3. Microarray data analysis
Gene expression analysis was conducted using Affymetrix Rat Genome 230 2.0 GeneChip® arrays (Affymetrix, Santa Clara, CA). Total RNA was amplified as directed in the Affymetrix 3′ IVT Plus kit protocol. 15 µg of amplified biotin-aRNAs were fragmented and 12.5 µg were hybridized to each array for 16 h at 45 °C in a rotating hybridization oven using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Array slides were stained with streptavidin/phycoerythrin utilizing a double-antibody staining procedure and then washed for antibody amplification according to the GeneChip® Hybridization, Wash and Stain Kit and user manual. Arrays were scanned in an Affymetrix Scanner 3000 and data was obtained using the GeneChip® Command Console and Expression Console Software (AGCC; Version 3.2 and Expression Console; Version 1.2).
2.4. Data normalization
Probe intensity data from all Rat Genome 230 version 2 Affymetrix GeneChip® arrays were read into the R software environment (http://www.R-project.org) directly from. CEL files using the R/affy package (Gautier et al., 2004). Probe-level data quality was assessed using image reconstruction, histograms of raw signal intensities and hierarchical clustering of samples. Normalization was carried out using the robust multi-array average (RMA) method using all probe intensity data sets together (Irizarry et al., 2003). The RMA method adjusts the background intensities of perfect match (PM) probes, applies quantile normalization, and calculates final expression measures using the Tukey median polish algorithm. RMA scatterplots were used as an additional quality control measure.
2.5. Statistical assessment of differential gene expression
Gene expression between control and treated liver and uterine samples was evaluated for each probe set using a bootstrap t-test approach. Pairwise tests were conducted while controlling the false discovery rate (FDR) at the 5% level. All statistical calculations were performed in the ORIOGEN software package using 10,000 bootstrap samples (Peddada et al., 2005).
Ingenuity Pathway Analysis database (www.Ingenuity.com) was used to compare experimental gene expression signatures to thousands of genomic signatures derived from published microarray data sets. This data mining approach is based on rank-based statistical procedures and is analogous to the Gene Set Enrichment method (Subramanian et al., 2005).
The upstream regulator analysis presented here is based on known relationships between transcriptional factor molecules and targets stored in the Ingenuity Knowledge Base. For each transcription factor, an overlap p-value is used to compare the overlap between the number of known transcription factor targets and the number of targets found in the TBBPA gene list. The activation z-score is used to compare the activation state of each transcription factor as described in the literature with the direction of change in TBBPA expression relative to control samples that are found in the current study. In addition, the interferome database (www.interferome.org) was used to identify significant transcripts in the interferon pathway (Rusinova et al., 2013).
2.6. Nanostring analysis
Microarray results were confirmed using the nCounter platform by NanoString© (www.nanostring.com) utilizing a Custom Code-Set consisting of 10 TBBPA responsive genes that were significantly altered by about 2-fold from control after microarray analysis. Genes evaluated by Nanostring were Agrn, Mx1, Mx2, Irf7, Usp18, Oas1a, Oas1b, Sng, Tsx, and Usp18, using the housekeeping genes Gapdh, Hprt1, Med15 and Rpl7 for normalization. For liver samples in Nanostring analysis, 100 ng of amplified cDNA, obtained utilizing the Nugen™ Ovation Pico WTA System (v2), was used. Gene expression was quantified on the nCounter Digital Analyzer™ and raw and normalized counts were generated with nSolver (v2.5)™ software. All data passed nSolver ‘s QA/QC.
2.7. Infection screening
Sentinel animals were randomly selected for parasite evaluation, gross observation for evidence of disease, and serum collection for serology at 4 weeks after receipt (5 rats) and at the end of the study (5 rats). Sentinels were from the vendor’s same animal room as the experimental rats and were subjected to the same environmental conditions. Serum samples were shipped to an independent laboratory (Idexx Bioresearch, Columbia, MO) for bacterial and viral titer testing. Serum was tested for antibodies to Mycoplasm pulmonis, RPV, RMV, KRV, H1, PMV, RCV/SDAV, RTV, and Sendai virus. All tests were negative.
3. Results
3.1. Body and organ weights and histopathologic findings
There was little or no treatment-related toxicity after 13 weeks of TBBPA exposure using conventional toxicity endpoints. This included no treatment-related effects on survival, body weight, or organ weights (Table 1). There were no treatment-related microscopic lesions in the liver or uterus.
Table 1.
Body weight, liver, and uterus weight in female Wistar Han rats after 13-weeks of TBBPA dosing.
| Dose (mg/kg) | Terminal Body wt. (g) | Liver wt. (g) | Relative Liver Weight | Uterus wt (g) | Relative Uterus Wt. | N |
|---|---|---|---|---|---|---|
| 0 | 241.7 ± 3.704 | 8.13 ± 0.190 | 33.55 ± 0.563 | 0.92 ± 0.081 | 3.87 ± 0.362 | 22–23 |
| 25 | 230.6 ± 3.435 | 7.74 ± 0.143 | 33.60 ± 0.427 | 1.07 ± 0.077 | 4.65 ± 0.31 | 25 |
| 250 | 236.1 ± 4.148 | 7.89 ± 0.1 | 33.45 ± 0.411 | 0.97 ± 0.070 | 4.14 ± 0.309 | 24 |
| 1000 | 244.6 ± 3.036 | 8.71 ± 0.11* | 35.63 ± 0.29** | 0.97 ± 0.06 | 3.99 ± 0.286 | 21 |
Relative organ weight = organ weight/body weight.
p < 0.05.
p < 0.01.
3.2. Liver transcriptomic alterations
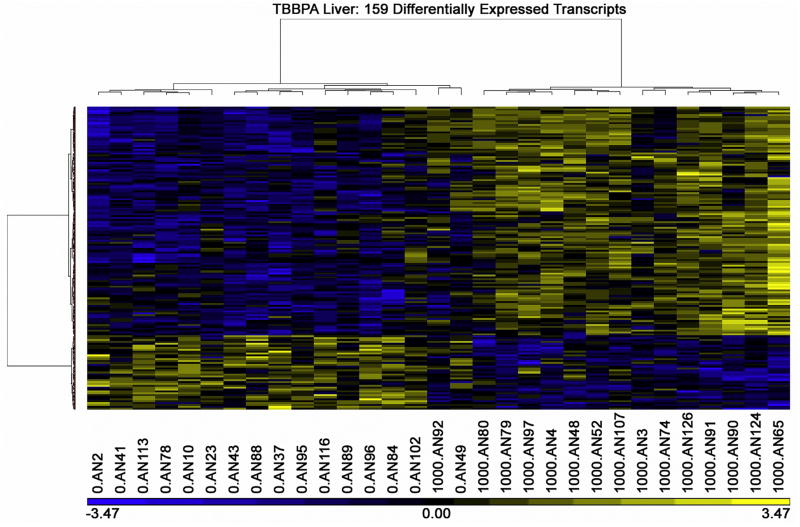
Using a false discovery rate threshold of 0.05, there were 159 differentially expressed liver transcript probes (Table 2). These 159 transcripts corresponded to expression level changes in 132 genes (6 genes were identified by two probes; Supplement 1). The heat map of these TBBPA liver transcript patterns showed a clear difference between control and TBBPA treated liver (Fig. 2).
Table 2.
TBBPA-induced liver transcripts after 13-weeks of TBBPA dosing (1000 mg/kg).
| Fold Change | ID | Symbol | Entrez Gene Name |
|---|---|---|---|
| 7.363 | 1367668_a_at | Scd2 | stearoyl-Coenzyme A desaturase 2 |
| 3.428 | 1387283_at | MX18 | MX dynamin-like GTPase 1 |
| 3.264 | 1389034_at | USP18* | ubiquitin specific peptidase 18 |
| 3.065 | 1368736_at | Tsx | testis specific X-linked gene |
| 2.773 | 1371152_a_at | OAS18 | 2′–5′-oligoadenylate synthetase 1, 40/46 kDa |
| 2.633 | 1382314_at | * | ISG15 ubiquitin-like modifier |
| 2.258 | 1371076_at | CYP2B6 | cytochrome P450, family 2, subfamily B, polypeptide 6 |
| 2.199 | 1394401_at | ELOVL6 | ELOVL fatty acid elongase 6 |
| 2.189 | 1382902_at | HERC6* | HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| 1.939 | 1379748_at | IFI44L* | interferon-induced protein 44-like |
| 1.904 | 1383564_at | IRF7* | interferon regulatory factor 7 |
| 1.844 | 1388056_at | Oas1b | 2′–5′ oligoadenylate synthetase 1B |
| 1.833 | 1381556_at | Ddx60* | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| 1.827 | 1369698_at | ABCC3* | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| 1.826 | 1371015_at | Mx1/Mx2* | MX dynamin-like GTPase 1 |
| 1.734 | 1387137_at | COMP | cartilage oligomeric matrix protein |
| 1.703 | 1377497_at | OASL* | 2′–5′-oligoadenylate synthetase-like |
| 1.666 | 1367707_at | FASN | fatty acid synthase |
| 1.658 | 1370913_at | RSAD2* | radical S-adenosyl methionine domain containing 2 |
| 1.627 | 1376908_at | IFIT3* | interferon-induced protein with tetratricopeptide repeats 3 |
| 1.565 | 1381960_at | ROPN1L | rhophilin associated tail protein 1-like |
| 1.543 | 1381206_at | PLCXD2 | phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| 1.527 | 1383424_at | CMPK2* | cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| 1.500 | 1390127_at | DIXDC1 | DIX domain containing 1 |
| 1.490 | 1391507_at | ZNF4678 | zinc finger protein 467 |
| 1.486 | 1370379_at | PRSS8 | protease, serine, 8 |
| 1.468 | 1376920_at | LOC500013 | similar to sterile alpha motif domain containing 9-like |
| 1.462 | 1388164_at | HLA-E | major histocompatibility complex, class I, E |
| 1.459 | 1391754_at | LOC100910735/Oas1i | 2 ' – 5 ' oligoadenylate synthetase 1I |
| 1.455 | 1379656_a_at | CORO2A | coronin, actin binding protein, 2A |
| 1.451 | 1373267_at | SH3YL1 | SH3 and SYLF domain containing 1 |
| 1.433 | 1383357_a_at | RELL1 | RELT-like 1 |
| 1.421 | 1383448_at | IRF98 | interferon regulatory factor 9 |
| 1.415 | 1367854_at | ACLY* | ATP citrate lyase |
| 1.408 | 1381014_at | IFI44* | interferon-induced protein 44 |
| 1.407 | 1367708_a_at | FASN* | fatty acid synthase |
| 1.378 | 1378581_at | CCDC62 | coiled-coil domain containing 62 |
| 1.350 | 1391702_at | ZNF446* | zinc finger protein 446 |
| 1.349 | 1371694_at | DPYSL2 | dihydropyrimidinase-like 2 |
| 1.346 | 1369716_s_at | Lgals5 | lectin, galactose binding, soluble 5 |
| 1.332 | 1390312_at | LOC684193 | similar to sterile alpha motif domain containing 9-like |
| 1.328 | 1385252_at | TRIM6–TRIM34 | TRIM6–TRIM34 readthrough |
| 1.320 | 1393044_at | CMPK2 | cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| 1.314 | 1380071_at | PARP12* | poly (ADP-ribose) polymerase family, member 12 |
| 1.299 | 1373514_at | RNF213* | ring finger protein 213 |
| 1.287 | 1388152_at | MAP2* | microtubule-associated protein 2 |
| 1.269 | 1388979_at | SMNDC1 | survival motor neuron domain containing 1 |
| 1.266 | 1373506_at | FAM102A | family with sequence similarity 102, member A |
| 1.247 | 1371901_at | DDHD2 | DDHD domain containing 2 |
| 1.247 | 1391463_at | Ddx588 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| 1.245 | 1390641_at | ZNF346 | zinc finger protein 346 |
| 1.237 | 1377878_at | FGFBP3 | fibroblast growth factor binding protein 3 |
| 1.236 | 1372705_at | CHERP | calcium homeostasis endoplasmic reticulum protein |
| 1.225 | 1374337_at | RNF2138 | ring finger protein 213 |
| 1.224 | 1368835_at | STAT1* | signal transducer and activator of transcription 1, 91 kDa |
| 1.222 | 1370803_at | ZWINT8 | ZW10 interacting kinetochore protein |
| 1.220 | 1373090_at | SSR1 | signal sequence receptor, alpha |
| 1.215 | 1397764_at | KCTD5 | potassium channel tetramerization domain containing 5 |
| 1.215 | 1389883_at | TMEM65 | transmembrane protein 65 |
| 1.212 | 1383621_at | MBTPS2 | membrane-bound transcription factor peptidase, site 2 |
| 1.211 | 1372930_at | SP110* | SP110 nuclear body protein |
| 1.210 | 1373288_at | ST5* | suppression of tumorigenicity 5 |
| 1.203 | 1376339_at | WWC3* | WWC family member 3 |
| 1.200 | 1398435_at | SLC22A15* | solute carrier family 22, member 15 |
| 1.199 | 1371996_at | AEBP2 | AE binding protein 2 |
| 1.198 | 1372779_at | B3GNT2* | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| 1.198 | 1383034_at | Rybp* | RING1 and YY1 binding protein |
| 1.195 | 1367693_at | YWHAH | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| 1.194 | 1395081_at | NCOA7* | nuclear receptor coactivator 7 |
| 1.187 | 1399157_at | URI1 | URI1, prefoldin-like chaperone |
| 1.184 | 1369559_a_at | CD47* | CD47 molecule |
| 1.180 | 1369978_at | PRPSAP2 | phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| 1.177 | 1382540_at | PRPF40A | PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| 1.174 | 1394761_at | ARHGAP42 | Rho GTPase activating protein 42 |
| 1.172 | 1374482_at | CTPS2 | CTP synthase 2 |
| 1.172 | 1382177_at | PML* | promyelocytic leukemia |
| 1.169 | 1383158_at | AGFG1 | ArfGAP with FG repeats 1 |
| 1.168 | 1380960_at | CORO2A | coronin, actin binding protein, 2A |
| 1.167 | 1388997_at | ARF3 | ADP-ribosylation factor 3 |
| 1.166 | 1387770_at | Ifi27* | interferon, alpha-inducible protein 27 |
| 1.163 | 1384362_at | ZBTB24 | zinc finger and BTB domain containing 24 |
| 1.159 | 1375431_at | C2orf69 | chromosome 2 open reading frame 69 |
| 1.157 | 1370244_at | CTSV* | cathepsin V |
| 1.156 | 1374815_at | STARD3NL | STARD3 N-terminal like |
| 1.155 | 1389686_at | PRKX | protein kinase, X-linked |
| 1.152 | 1373563_at | SNRNP27 | small nuclear ribonucleoprotein 27 kDa (U4/U6.U5) |
| 1.151 | 1376492_at | UNKL | unkempt family zinc finger-like |
| 1.150 | 1392916_at | MAP7 | microtubule-associated protein 7 |
| 1.148 | 1370351_at | TDRD78 | tudor domain containing 7 |
| 1.147 | 1379669_at | ARHGAP42 | Rho GTPase activating protein 42 |
| 1.147 | 1373670_at | STAT2* | signal transducer and activator of transcription 2, 113 kDa |
| 1.146 | 1389646_at | CDC23 | cell division cycle 23 |
| 1.142 | 1388930_at | TMEM123 | transmembrane protein 123 |
| 1.131 | 1372360_at | ABI1 | abl-interactor 1 |
| 1.128 | 1390387_at | SH3D19 | SH3 domain containing 19 |
| 1.119 | 1374637_at | TMCO6 | transmembrane and coiled-coil domains 6 |
| 1.115 | 1374707_at | CCAR2 | cell cycle and apoptosis regulator 2 |
| 1.113 | 1374406_at | KLHDC2* | kelch domain containing 2 |
| 1.112 | 1384022_at | AGFG1 | ArfGAP with FG repeats 1 |
| 1.104 | 1368217_at | RALBP1* | ralA binding protein 1 |
| 1.101 | 1373974_at | OSBP | oxysterol binding protein |
| 1.099 | 1375639_at | E2F6 | E2F transcription factor 6 |
| 1.080 | 1384971_at | DEPDC7 | DEP domain containing 7 |
| −1.102 | 1383244_at | IQCE* | IQ motif containing E |
| −1.122 | 1393123_at | C8G | complement component 8, gamma polypeptide |
| −1.125 | 1369898_a_at | GIP | gastric inhibitory polypeptide |
| −1.137 | 1371916_at | MSRB* | methionine sulfoxide reductase B1 |
| −1.137 | 1380624_at | VANGL1 | VANGL planar cell polarity protein 1 |
| −1.145 | 1370682_at | LILRA6* | leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 |
| −1.146 | 1376761_at | HDAC4 | histone deacetylase 4 |
| −1.152 | 1370202_at | PLA2G16* | phospholipase A2, group XVI |
| −1.156 | 1368399_a_at | CPQ* | carboxypeptidase Q |
| −1.160 | 1370853_at | CAMK2N1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| −1.160 | 1370267_at | GSK3B | glycogen synthase kinase 3 beta |
| −1.168 | 1376410_at | MMP17 | matrix metallopeptidase 17 (membrane-inserted) |
| −1.170 | 1398351_at | USP7 | ubiquitin specific peptidase 7 (herpes virus-associated) |
| −1.174 | 1367586_at | Ldha/RGD1562690 | lactate dehydrogenase A |
| −1.176 | 1377112_at | CDA* | cytidine deaminase |
| −1.194 | 1399070_at | SETD5 | SET domain containing 5 |
| −1.199 | 1372490_at | GPD1L* | glycerol-3-phosphate dehydrogenase 1-like |
| −1.207 | 1368190_at | REN | renin |
| −1.220 | 1383989_at | SOX4* | SRY (sex determining region Y)-box 4 |
| −1.222 | 1372195_at | TNNC2* | troponin C type 2 (fast) |
| −1.245 | 1396188_at | RHOJ | ras homolog family member J |
| −1.253 | 1381193_at | LPGAT1* | lysophosphatidylglycerol acyltransferase 1 |
| −1.253 | 1374048_at | NRTN | neurturin |
| −1.263 | 1370375_at | GLS2 | glutaminase 2 (liver, mitochondrial) |
| −1.292 | 1393949_at | HYAL3 | hyaluronoglucosaminidase 3 |
| −1.293 | 1382981_at | AHI1 | Abelson helper integration site 1 |
| −1.317 | 1374073_at | SLC46A1 | solute carrier family 46 (folate transporter), member 1 |
| −1.349 | 1387307_at | HAL* | histidine ammonia-lyase |
| −1.445 | 1397552_at | EML4 | echinoderm microtubule associated protein like 4 |
| −1.451 | 1387656_at | SLC4A1 | solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| −1.503 | 1393971_at | LOC102549203 | uncharacterized LOC102549203 |
| −1.658 | 1393910_at | Fam13a* | family with sequence similarity 13, member A |
| −1.736 | 1387052_at | GPT | glutamic-pyruvate transaminase (alanine aminotransferase) |
| −1.801 | 1368520_at | APOA4 | apolipoprotein A-IV |
| −1.809 | 1387391_at | CDKN1A* | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
Interferon pathway transcript.
Fig. 2.
TBBPA (1000 mg/kg) liver transcript heat map.
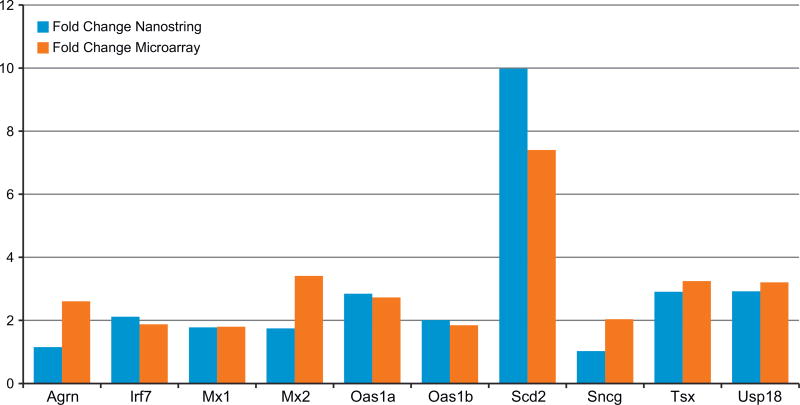
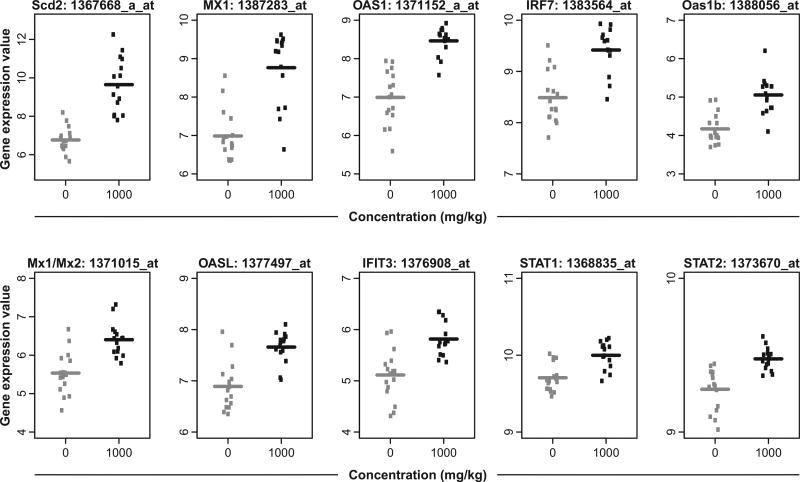
Nanostring analysis of selected transcripts confirmed the direction of the TBBPA-induced transcript change (Fig. 3), although Agrn and Sncg were minimally responsive to TBBPA by Nanostring analysis. Selected liver transcript signals were plotted by individual animals to display variance of control and TBBPA groups. Generally, transcript responses showed the same elevated direction of response of TBBPA treatment group compared to control (Fig. 4).
Fig. 3.
Selected Liver TBBPA (1000 mg/kg) transcripts confirmed by Nanostring analysis.
Fig. 4.
Plots of selected TBBPA (1000 mg/kg) induced-liver transcripts. The gene expression value shown refers to the RMA normalized gene expression measure described in the methods.
Ingenuity Pathway enrichment analysis provided evidence that the interferon (IFN) pathway was the most significant pathway affected by TBBPA treatment (Table 3). This led us to explore the IFN pathway more closely. There were 59 significantly altered TBBPA hepatic transcripts in the IFN pathway (www.interferome.org). Transcripts altered by TBBPA that are known to play critical roles in the interferon pathway included Irl-7, Mx1, Oas1, Isfg15, Ddx60, Stat1 and Stat2. Also upregulated were TBBPA transcripts involved in liver xenobiotic metabolism, including genes regulating fatty acid metabolism (Scd2, Cyp2b6, Elovl6, Herc6, Fasn) (Table 2).
Table 3.
Analysis of significant TBBPA liver transcripts and pathways (1000 mg/kg)*.
| Ingenuity Canonical Pathways | −log(p-value) | Ratio | z-score | Molecules |
|---|---|---|---|---|
| Interferon Signaling | 8.85E00 | 1.94E-01 | 2.449 | OAS1,IRF9,STAT1,IFIT3,STAT2,ISG15,MX1 |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | 4.47E00 | 7.94E-02 | 1.342 | IRF9,IRF7,STAT1,STAT2,ISG15 |
| Cell Cycle: G1/S Checkpoint Regulation | 3.27E00 | 6.25E-02 | −1.000 | E2F6,GSK3B,CDKN1A,HDAC4 |
| Cyclins and Cell Cycle Regulation | 2.94E00 | 5.13E-02 | NaN | E2F6,GSK3B,CDKN1A,HDAC4 |
| Pancreatic Adenocarcinoma Signaling | 2.46E00 | 3.77E-02 | NaN | E2F6,RALBP1,STAT1,CDKN1A |
| Acetyl-CoA Biosynthesis III (from Citrate) | 2.23E00 | 1E00 | NaN | ACLY |
| Role of JAK1, JAK2 and TYK2 in Interferon Signaling | 2.06E00 | 8.33E-02 | NaN | STAT1,STAT2 |
| Estrogen-mediated S-phase Entry | 2.06E00 | 8.33E-02 | NaN | E2F6,CDKN1A |
| JAK/Stat Signaling | 2.06E00 | 4.17E-02 | NaN | STAT1,STAT2,CDKN1A |
Analysis using ingenuity.com.
There were few TBBPA-induced changes in the uterine transcriptome. Using a false discovery rate threshold of 0.05, there were 4 differentially expressed transcripts at 1000 mg/kg, 8 at 250 mg/kg, and 5 at 25 mg/kg. All 17 transcripts at various dose levels were less than 1.5 fold changed versus control uterine transcript expression levels. None of the 17 uterine transcripts were common among the three TBBPA exposure levels examined (25, 250, and 1000 mg/kg), and none overlapped the 138 TBBPA liver transcripts found to be altered (1000 mg/kg). These transcript changes were not considered to alter organ function.
4. Discussion
In this study we found that TBBPA induced molecular changes in the liver after 13-weeks of exposure (1000 mg/kg), while there were few altered transcripts in the uterus. TBBPA caused liver expression changes in 159 significant Affymetrix probes (relative to controls [FDR ≤ 0.05]) which mapped to 132 genes. The TBBPA hepatic transcriptome included transcripts with functions in the IFN pathway (Noureddin et al., 2015; Sathish and Yuan, 2011; Schmeisser et al., 2010; Schneider et al., 2014) and in xenobiotic and fatty acid metabolism.
The TBBPA hepatic transcripts included upregulation of Scd2 (steraroly-coenzyme A desaturase 2), Elovl-6 (fatty acid elongase 6), and FasN (fatty acid synthase). Scd2 is a crucial enzyme in the synthesis of monounsaturated fatty acids which are required for maintaining a normal epidermal permeability barrier function, and are components of triglycerides (Miyazaki et al., 2005). Alterations in Scd2 levels may affect lipid content in non-hepatic organs (de Moura et al., 2016) where level and type of lipid is critical to function. Elovl-6 is one of six mammalian enzymes responsible for fatty acid elongation beyond 16 carbons to produce very long chain fatty acids. The FASN enzyme catalyzes de novo synthesis of fatty acids (Dorn et al., 2010). TBBPA also increased levels of the Cyp2b6, a transcript induced by phenobarbital and in xenobiotic metabolism (Liu et al., 2015). The expression of Cyp2b6 can have a 20–250 fold inter-individual variation (Wang and Tompkins, 2008). Because liver is a major site for chemical exposure (Hakk et al., 2000; Knudsen et al., 2014; Kuester et al., 2007) and is involved in estradiol metabolism (Gosavi et al., 2013; Raftogianis et al., 2000), these TBBPA-induced liver changes could affect hormone levels.
TBBPA induced IFN pathway transcripts (www.interferome.org) previously identified in liver cells (Rusinova et al., 2013). This included transcripts associated with IFN pathway regulation (e.g. Stat1, Stat2, Ilf7, Irf9, Pml) (Khodarev et al., 2012); antiviral activity (e.g. Mx1, Mx2, Ifit3, Isg15); and regulation of immune response (DDX58, Oas1) (Hertzog et al., 2011). Some of the TBBPA IFN pathway transcripts (e.g. Isg15) may be involved in hepatic cancer (Li et al., 2014), and could also play a role in the liver cancer that occurred in mice in the 2-year TBBPA study (National Toxicology Program, 2014). Ifl27 (induced by TBBPA) may promote cell proliferation (Hsieh et al., 2015), and is associated with tumorigenesis and invasion (Li et al., 2015).
Like TBBPA, tamoxifen causes uterine tumors (IARC, 2012), and induces interferon pathway transcripts (e.g. MX1 and interferon regulatory factors) in model systems (Dabydeen et al., 2015; Fawzy et al., 2012; Schild-Hay et al., 2009). Exposure of myeloid cells to a related chemical, bisphenol A, also stimulated interferon signaling (Panchanathan et al., 2015). These TBBPA findings are supported by recent in vitro studies which showed TBBPA alterations in IFN production in human cells (Almughamsi and Whalen, 2016).
While TBBPA has little activity as an estrogen receptor agonist or antagonist (Hamers et al., 2006), a feedback loop between estrogen signaling and IFN signaling has been reported (Panchanathan et al., 2015, 2010). Upregulation of interferon pathways by TBBPA and tamoxifen could affect estrogen signaling and ultimately the development of uterine cancer. The ability of TBBPA to interact with sulfotransferase (Gosavi et al., 2013) could disrupt estrogen homeostasis (Sanders et al., 2016). In addition, TBBPA can cause oxidative damage and disruption of thyroid hormone signaling (He et al., 2016; Iakovleva et al., 2016). Further work is needed to determine how these various TBBPA effects work together to cause cancer and how age affects biologic outcome (Hines, 2008).
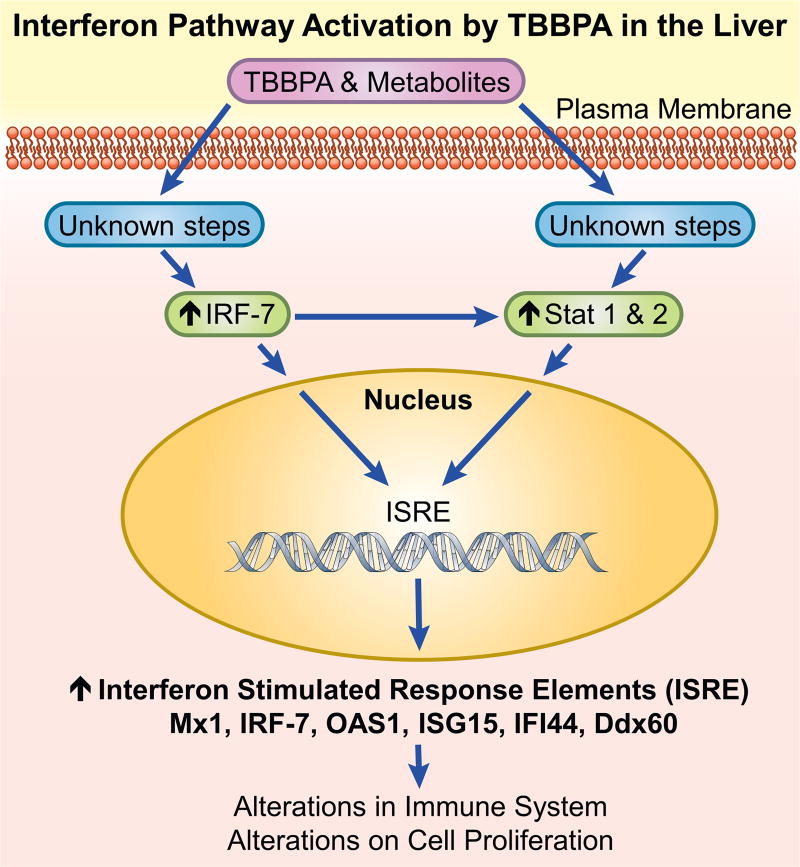
Decreases in immune function can facilitate cancer development (Yang and Rosenberg, 2016), and in several studies TBBPA was found to suppress the immune system. TBBPA was an immunosuppressant in in vitro cell systems including suppression of NK cell activity (Hurd and Whalen, 2011; Kibakaya et al., 2009). Mice treated with TBBPA were less able to mount a defense against viral infection (Watanabe et al., 2010). Interferon signaling modulates immune activity, and Irf7 when upregulated in the liver, as occurred in these TBBPA studies (Fig. 5), can regulate the immune system (Wang et al., 2013). Persistent IFN signaling can disrupt immune responsiveness potentially leading to immune suppression (Teijaro, 2016). This TBBPA immunosuppressent activity was thought to contribute to its carcinogenic properties in a recent review of TBBPA studies (Grosse et al., 2016).
Fig. 5.
Proposed Mechanism for TBBPA activation of interferon pathway.
One interpretation of the TBBPA-mediated IFN signature reported in our study might be as a general stress response to repeated chemical exposure. The expression of ‘IFN stimulated genes’ or ‘interferon signaling network genes’ (ISGs) in response to various viral, bacterial and chemical stimuli has been well characterized and described (de Veer et al., 2001; Schneider et al., 2014). In our studies animals were screened for a broad spectrum of infections and none were found, suggesting that the IFN pathway activation was not due to microbial or viral exposure. In addition, liver histopathology showed no overt signs of cell death or organ damage. Exactly, how TBBPA-mediated expression of ISGs in liver might influence or contribute to uterine carcinogenesis is unclear at this time. Although blood cytokine measurements were not performed in this study, it is possible that TBBPA could alter circulating or organ-localized production of interferon to affect ISGs leading to the IFN signature observed here in liver. For example, microrarray analysis in an independent study showed that acute pentachorophenol exposure in C57BL/6 mice increased expression of many interferon responsive transcripts in liver, including Stat1, Stat2, Irf7, Oas, Ifit as well as other IFN regulated genes (Kanno et al., 2013). These authors proposed a scheme where pentachorophenol produced metabolites and oxygenated radicals leading to DNA and protein damage that resulted in pathway activation of the Nrf2 Tir receptor and PRR (pattern response recognition) systems to initiate interferon signaling and expression of ISGs. Another study found in vitro TBBPA exposure disrupted IFN-γ secretion from various human immune cell preparations (Almughamsi and Whalen, 2016). Whether the uterus would respond similarly to TBBPA or other chemical exposures and engage IFN-dependent pathways is an untested hypothesis. IFN polymorphisms are associated with increased risk of cervical cancer (Sun et al., 2015) but the relationship of IFN-related mechanisms with uterine carcinogenesis requires further investigation.
In summary, conventional biochemical and toxicological measures and histologic lesions were not observed in either liver or uterus in female Wistar Han rats after 13 weeks of repeated TBBPA exposure from 25 to 1000 mg/kg. Toxicogenomic analysis showed no substantial changes in uterus but did reveal a robust gene expression in liver that involved activation of the interferon pathway. We speculate that long-term TBBPA exposures could lead to direct or indirect immunomodulatory changes that contribute to carcinogenic processes in the uterus.
Supplementary Material
HIGHLIGHTS.
Tetrabromobisphenol A (TBBPA) is a widely used flame retardant.
Oral administration of TBBPA to rats cause transcriptomic changes in the liver.
These transcriptomic changes indicated activation of the interferon pathway.
Acknowledgments
This research was supported [in part] by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. We thank M. Cesta, NIEHS, and G. Knudson, NCI for their review of this manuscript.
Footnotes
Conflict of interest
There is no conflict of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.toxlet.2016.11.019.
References
- Almughamsi H, Whalen MM. Hexabromocyclododecane and tetrabromobisphenol A alter secretion of interferon gamma (IFN-gamma) from human immune cells. Arch. Toxicol. 2016;90(7):1695–1707. doi: 10.1007/s00204-015-1586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carignan CC, Abdallah MA, Wu N, Heiger-Bernays W, McClean MD, Harrad S, Webster TF. Predictors of tetrabromobisphenol-A (TBBP-A) and hexabromocyclododecanes (HBCD) in milk from Boston mothers. Environ. Sci. Technol. 2012;46(21):12146–12153. doi: 10.1021/es302638d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabydeen SA, Kang K, Diaz-Cruz ES, Alamri A, Axelrod ML, Bouker KB, Al-Kharboosh R, Clarke R, Hennighausen L, Furth PA. Comparison of tamoxifen and letrozole response in mammary preneoplasia of ER and aromatase overexpressing mice defines an immune-associated gene signature linked to tamoxifen resistance. Carcinogenesis. 2015;36(1):122–132. doi: 10.1093/carcin/bgu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Napoli-Davis G, Owens JE. Quantitation of tetrabromobisphenol-A from dust sampled on consumer electronics by dispersed liquid–liquid microextraction. Environ. Pollut. 2013;180:274–280. doi: 10.1016/j.envpol.2013.05.038. [DOI] [PubMed] [Google Scholar]
- Dorn C, Riener MO, Kirovski G, Saugspier M, Steib K, Weiss TS, Gabele E, Kristiansen G, Hartmann A, Hellerbrand C. Expression of fatty acid synthase in nonalcoholic fatty liver disease. Int. J. Clin. Exp. Pathol. 2010;3(5):505–514. [PMC free article] [PubMed] [Google Scholar]
- Dunnick JK, Sanders JM, Kissling GE, Johnson CL, Boyle MH, Elmore SA. Environmental chemical exposure may contribute to uterine cancer development: studies with tetrabromobisphenol A. Toxicol. Pathol. 2015;43(4):464–473. doi: 10.1177/0192623314557335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawzy IO, Negm M, Ahmed R, Esmat G, Hamdi N, Abdelaziz AI. Tamoxifen alleviates hepatitis C virus-induced inhibition of both toll-like receptor 7 and JAK-STAT signalling pathways in PBMCs of infected Egyptian females. J. Viral Hepat. 2012;19(12):854–861. doi: 10.1111/j.1365-2893.2012.01612.x. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- George SM, Ballard R, Shikany JM, Crane TE, Neuhouser ML. A prospective analysis of diet quality and endometrial cancer among 84,415 postmenopausal women in the women's health initiative. Ann. Epidemiol. 2015;25(10):788–793. doi: 10.1016/j.annepidem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosavi RA, Knudsen GA, Birnbaum LS, Pedersen LC. Mimicking of estradiol binding by flame retardants and their metabolites: a crystallographic analysis. Environ. Health Perspect. 2013;121(10):1194–1199. doi: 10.1289/ehp.1306902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse Y, Loomis D, Guyton KZ, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Mattock H, Straif K. Carcinogenicity of some industrial chemicals. Lancet Oncol. 2016;17(4):419–420. doi: 10.1016/S1470-2045(16)00137-6. [DOI] [PubMed] [Google Scholar]
- Hakk H, Larsen G, Bergman A, Orn U. Metabolism, excretion and distribution of the flame retardant tetrabromobisphenol-A in conventional and bile-duct cannulated rats. Xenobiotica. 2000;30(9):881–890. doi: 10.1080/004982500433309. [DOI] [PubMed] [Google Scholar]
- Hamers T, Kamstra JH, Sonneveld E, Murk AJ, Kester MH, Andersson PL, Legler J, Brouwer A. In vitro profiling of the endocrine-disrupting potency of brominated flame retardants. Toxicol. Sci. 2006;92(1):157–173. doi: 10.1093/toxsci/kfj187. [DOI] [PubMed] [Google Scholar]
- Harrad S, Abdallah MA. Concentrations of polybrominated diphenyl ethers, hexabromocyclododecanes and tetrabromobisphenol-A in Breast milk from United Kingdom women do not decrease over twelve months of lactation. Environ. Sci. Technol. 2015;49(23):13899–13903. doi: 10.1021/acs.est.5b00539. [DOI] [PubMed] [Google Scholar]
- He Q, Wang X, Sun P, Wang Z, Wang L. Acute and chronic toxicity of tetrabromobisphenol A to three aquatic species under different pH conditions. Aquat. Toxicol. 2015;164:145–154. doi: 10.1016/j.aquatox.2015.05.005. [DOI] [PubMed] [Google Scholar]
- He M, Li X, Zhang S, Sun J, Cao H, Wang W. Mechanistic and kinetic investigation on OH-initiated oxidation of tetrabromobisphenol A. Chemosphere. 2016;153:262–269. doi: 10.1016/j.chemosphere.2016.03.043. [DOI] [PubMed] [Google Scholar]
- Hertzog P, Forster S, Samarajiwa S. Systems biology of interferon responses. J. Interferon Cytokine Res. 2011;31(1):5–11. doi: 10.1089/jir.2010.0126. [DOI] [PubMed] [Google Scholar]
- Hines RN. The ontogeny of drug metabolism enzymes and implications for adverse drug events. Pharmacol. Ther. 2008;118(2):250–267. doi: 10.1016/j.pharmthera.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Hsieh WL, Huang YH, Wang TM, Ming YC, Tsai CN, Pang JH. IFI27, a novel epidermal growth factor-stabilized protein, is functionally involved in proliferation and cell cycling of human epidermal keratinocytes. Cell Prolif. 2015;48(2):187–197. doi: 10.1111/cpr.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd T, Whalen MM. Tetrabromobisphenol A decreases cell-surface proteins involved in human natural killer (NK) cell-dependent target cell lysis. J. Immunotoxicol. 2011;8(3):219–227. doi: 10.3109/1547691X.2011.580437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC) IARC mongraphs on the evaluation of carcinogenic risks to humans. Pharmaceuticals: A review of human carcinogens. 101A. IARC; Lyon, France: 2012. [Google Scholar]
- Iakovleva I, Begum A, Brannstrom K, Wijsekera A, Nilsson L, Zhang J, Andersson PL, Sauer-Eriksson AE, Olofsson A. Tetrabromobisphenol A is an efficient stabilizer of the transthyretin tetramer. PLoS One. 2016;11(4):e0153529. doi: 10.1371/journal.pone.0153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kanno J, Aisaki K, Igarashi K, Kitajima S, Matsuda N, Morita K, Tsuji M, Moriyama N, Furukawa Y, Otsuka M, Tachihara E, Nakatsu N, Kodama Y. Oral administration of pentachlorophenol induces interferon signaling mRNAs in C57BL/6 male mouse liver. J. Toxicol. Sci. 2013;38(4):643–654. doi: 10.2131/jts.38.643. [DOI] [PubMed] [Google Scholar]
- Khodarev NN, Roizman B, Weichselbaum RR. Molecular pathways: interferon/stat1 pathway: role in the tumor resistance to genotoxic stress and aggressive growth. Clin. Cancer Res. 2012;18(11):3015–3021. doi: 10.1158/1078-0432.CCR-11-3225. [DOI] [PubMed] [Google Scholar]
- Kibakaya EC, Stephen K, Whalen MM. Tetrabromobisphenol A has immunosuppressive effects on human natural killer cells. J. Immunotoxicol. 2009;6(4):285–292. doi: 10.3109/15476910903258260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen GA, Sanders JM, Sadik AM, Birnbaum LS. TITLE disposition and kinetics of tetrabromobisphenol A in female Wistar han rats. Toxicol. Rep. 2014;1:214–223. doi: 10.1016/j.toxrep.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuester RK, Solyom AM, Rodriguez VP, Sipes IG. The effects of dose, route, and repeated dosing on the disposition and kinetics of tetrabromobisphenol A in male F-344 rats. Toxicol. Sci. 2007;96(2):237–245. doi: 10.1093/toxsci/kfm006. [DOI] [PubMed] [Google Scholar]
- Li C, Wang J, Zhang H, Zhu M, Chen F, Hu Y, Liu H, Zhu H. Interferon-stimulated gene 15 (ISG15) is a trigger for tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget. 2014;5(18):8429–8441. doi: 10.18632/oncotarget.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xie Y, Zhang W, Gao J, Wang M, Zheng G, Yin X, Xia H, Tao X. Interferon alpha-inducible protein 27 promotes epithelial-mesenchymal transition and induces ovarian tumorigenicity and stemness. J. Surg. Res. 2015;193(1):255–264. doi: 10.1016/j.jss.2014.06.055. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li L, Wu H, Hu J, Ma J, Zhang QY, Ding X. Characterization of CYP2B6 in a CYP2B6-humanized mouse model: inducibility in the liver by phenobarbital and dexamethasone and role in nicotine metabolism in vivo. Drug Metab. Dispos. 2015;43(2):208–216. doi: 10.1124/dmd.114.061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Li J, Yan S, Zhang W, Li Y, Han D. A review of status of tetrabromobisphenol A (TBBPA) in China. Chemosphere. 2016;148:8–20. doi: 10.1016/j.chemosphere.2016.01.023. [DOI] [PubMed] [Google Scholar]
- Malkoske T, Tang Y, Xu W, Yu S, Wang H. A review of the environmental distribution, fate, and control of tetrabromobisphenol A released from sources. Sci. Total Environ. 2016 doi: 10.1016/j.scitotenv.2016.06.062. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Dobrzyn A, Elias PM, Ntambi JM. Stearoyl-CoA desaturase-2 gene expression is required for lipid synthesis during early skin and liver development. Proc. Natl. Acad. Sci. U. S. A. 2005;102(35):12501–12506. doi: 10.1073/pnas.0503132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao T, Akiyama E, Kakutani H, Mizuno A, Aozasa O, Akai Y, Ohta S. Levels of tetrabromobisphenol A, tribromobisphenol A, dibromobisphenol A, monobromobisphenol A, and bisphenol a in Japanese breast milk. Chem. Res. Toxicol. 2015;28(4):722–728. doi: 10.1021/tx500495j. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program. Toxicology studies of tetrabromobisphenol A (Cas no. 79–94-7) in F344/NTac rats and B6C3F1/N mice and toxicology and carcinogeogenesis studies of tetrabromobisphenol A in Wistar Han [Crl:WI (Han)] rats and B6C3F1/N mice. NTP Technical Report 587. 2014 doi: 10.22427/NTP-TR-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noureddin M, Rotman Y, Zhang F, Park H, Rehermann B, Thomas E, Liang TJ. Hepatic expression levels of interferons and interferon-stimulated genes in patients with chronic hepatitis C: A phenotype-genotype correlation study. Genes Immun. 2015;16(5):321–329. doi: 10.1038/gene.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R, Shen H, Zhang X, Ho SM, Choubey D. Mutually positive regulatory feedback loop between interferons and estrogen receptor-alpha in mice: implications for sex bias in autoimmunity. PLoS One. 2010;5(5):e10868. doi: 10.1371/journal.pone.0010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R, Liu H, Leung YK, Ho SM, Choubey D. Bisphenol A (BPA) stimulates the interferon signaling and activates the inflammasome activity in myeloid cells. Mol. Cell. Endocrinol. 2015;415:45–55. doi: 10.1016/j.mce.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddada S, Harris S, Zajd J, Harvey E. ORIOGEN: an order restricted inference for ordered gene expression data. Bioinform. 2005;21:3933–3934. doi: 10.1093/bioinformatics/bti637. [DOI] [PubMed] [Google Scholar]
- Raftogianis R, Creveling C, Weinshilboum R, Weisz J. Estrogen metabolism by conjugation. J. Natl. Cancer Inst. Monogr. 2000;27:113–124. doi: 10.1093/oxfordjournals.jncimonographs.a024234. [DOI] [PubMed] [Google Scholar]
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JM, Coulter SJ, Knudsen GA, Dunnick JK, Kissling GE, Birnbaum LS. Disruption of estrogen homeostasis as a mechanism for uterine toxicity in Wistar Han rats treated with tetrabromobisphenol A. Toxicol. Appl. Pharmacol. 2016;298:31–39. doi: 10.1016/j.taap.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathish N, Yuan Y. Evasion and subversion of interferon-mediated antiviral immunity by Kaposi's sarcoma-associated herpesvirus: an overview. J. Virol. 2011;85(21):10934–10944. doi: 10.1128/JVI.00687-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Hay LJ, Leil TA, Divi RL, Olivero OA, Weston A, Poirier MC. Tamoxifen induces expression of immune response-related genes in cultured normal human mammary epithelial cells. Cancer Res. 2009;69(3):1150–1155. doi: 10.1158/0008-5472.CAN-08-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser H, Mejido J, Balinsky CA, Morrow AN, Clark CR, Zhao T, Zoon KC. Identification of alpha interferon-induced genes associated with antiviral activity in Daudi cells and characterization of IFIT3 as a novel antiviral gene. J. Virol. 2010;84(20):10671–10680. doi: 10.1128/JVI.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA cancer. J. Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Lu Y, Pen Q, Li T, Xie L, Deng Y, Qin A. Interferon gamma +874T/A polymorphism increases the risk of cervical cancer: evidence from a meta-analysis. Tumour Biol. 2015;36(6):4555–4564. doi: 10.1007/s13277-015-3100-4. [DOI] [PubMed] [Google Scholar]
- Svihlikova V, Lankova D, Poustka J, Tomaniova M, Hajslova J, Pulkrabova J. Perfluoroalkyl substances (PFASs) and other halogenated compounds in fish from the upper Labe river basin. Chemosphere. 2015;129:170–178. doi: 10.1016/j.chemosphere.2014.09.096. [DOI] [PubMed] [Google Scholar]
- Tang B, Zeng YH, Luo XJ, Zheng XB, Mai BX. Bioaccumulative characteristics of tetrabromobisphenol A and hexabromocyclododecanes in multi-tissues of prey and predator fish from an e-waste site, South China. Environ. Sci. Pollut. Res. Int. 2015 doi: 10.1007/s11356-015-4463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR. Type I interferons in viral control and immune regulation. Curr. Opin. Virol. 2016;16:31–40. doi: 10.1016/j.coviro.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Yokoi T. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett. 2005;227(2):115–124. doi: 10.1016/j.canlet.2004.10.007. [DOI] [PubMed] [Google Scholar]
- U. S. EPA. Fame retardants in printed circuit boards. http://www.epa.gov OTS0574261. 2015 ( http://www2.epa.gov/saferchoice/design-environment-alternatives-assessments)
- Wang H, Tompkins LM. CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr. Drug Metab. 2008;9(7):598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XA, Zhang R, Zhang S, Deng S, Jiang D, Zhong J, Yang L, Wang T, Hong S, Guo S, She ZG, Zhang XD, Li H. Interferon regulatory factor 7 deficiency prevents diet-induced obesity and insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2013;305(4):E485–495. doi: 10.1152/ajpendo.00505.2012. [DOI] [PubMed] [Google Scholar]
- Watanabe W, Shimizu T, Sawamura R, Hino A, Konno K, Hirose A, Kurokawa M. Effects of tetrabromobisphenol A, a brominated flame retardant, on the immune response to respiratory syncytial virus infection in mice. Int. Immunopharmacol. 2010;10(4):393–397. doi: 10.1016/j.intimp.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Webb PM. Environmental (nongenetic) factors in gynecological cancers: update and future perspectives. Future Oncol. 2015;11(2):295–307. [PubMed] [Google Scholar]
- Yang JC, Rosenberg SA. Adoptive T-Cell therapy for cancer. Adv. Immunol. 2016;130:279–294. doi: 10.1016/bs.ai.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Guo J, Zhang W, Zhou P, Deng J, Lin K. Tetrabromobisphenol A contamination and emission in printed circuit board production and implications for human exposure. J. Hazard. Mater. 2014;273C:27–35. doi: 10.1016/j.jhazmat.2014.03.003. [DOI] [PubMed] [Google Scholar]
- de Moura RF, Nascimento LF, Ignacio-Souza LM, Morari J, Razolli DS, Solon C, de Souza GF, Festuccia WT, Velloso LA. Hypothalamic stearoyl-CoA desaturase-2 (SCD2) controls whole-body energy expenditure. Int. J. Obes. (Lond.) 2016;40(3):471–478. doi: 10.1038/ijo.2015.188. [DOI] [PubMed] [Google Scholar]
- de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001;69(6):912–920. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.