ABSTRACT
Sensitive assays for HIV-1 neutralizing antibody detection are urgently needed for vaccine immunogen optimization and identification of protective immune response levels. In this study, we developed an easy-to-use HIV-1 pseudovirus neutralization assay based on a human CD4+ lymphoblastoid cell line A3R5 by employing a high efficient pseudovirus production system. Optimal conditions for cell counts, infection time, virus dose and concentration of DEAE-dextran were tested and identified. For T-cell line-adapted tier 1 virus strains, significantly higher inhibitory efficiency was observed for both monoclonal neutralizing antibody (4 fold) and plasma (2 fold) samples in A3R5 than those in TZM-bl assay. For circulating tier 2 strains, the A3R5 pseudovirus assay showed even much higher sensitivity for both neutralizing antibody (10 fold) and plasma (9 fold) samples. When sequential neutralizing antibody seroconverting samples were tested in both A3R5 and TZM-bl assays, the seroconverting points could be detected earlier for tier 1 (15.7 weeks) and tier 2 (68.3 weeks) strains in A3R5 assay respectively. The high sensitive pseudovirus assay using more physiological target cells could serve as an alternative to the TZM-bl assay for evaluation of vaccine-induced neutralizing antibodies and identification of the correlates of protection.
KEYWORDS: HIV-1, neutralizing antibody, pseudovirus, A3R5, TZM-bl
Introduction
It is well accepted that neutralizing antibody (NAb) plays a pivotal role in most successful vaccines against infectious agents, such as polio virus, rabies virus, measles, influenza virus, human papillomavirus.1 Since the identification of human immunodeficiency virus (HIV) as the pathogen of the acquired immunodeficiency syndrome (AIDS), development of the prophylactic HIV vaccine, which could induce robust and broad NAbs, has been the primary goal to fight against this epidemic. Up to now, a number of potent NAbs have been isolated from HIV-1 infected individuals2-4 and the protective potency of them has been conformed in animal models.5,6 Unfortunately, no candidate vaccine, however, has been reported to yield this kind of NAbs. The challenge to develop an effective HIV vaccine was not just the design of appropriate immunogens, but also the establishment of standardized assays to evaluate the protective immune responses to provide information relevant to the in vivo outcomes and guide further modification of the immunogens.
Great efforts have been invested in the development, standardization and implementation of in vitro assays for evaluating potency and breadth of NAbs against HIV-17-12. The early neutralization assays employed the T-cell line-adapted (TCLA) viruses to infect cell lines, which were lately proved poorly predictive of the in vivo outcomes. Subsequently, primary isolated viruses and peripheral blood mononuclear cells (PBMCs) were used for in vitro neutralization assays. However, due to the genetic polymorphisms of PBMCs from different donors and the variability of the primary virus isolates, the experiment variations of intra- or inter-laboratories were quite problematic, which largely restricted its wider applications. To circumvent variations of the tests, pseudoviruses and engeered cell lines were introduced to the neutralization assay. The pseudovirus neutralization assay based on TZM-bl cell was recommended as an optimized and validated approach to assess the sera from HIV-1 vaccines trials8, which showed a number of advantages including: high versatility of virus strains, high reproducibility, high throughput, ease and safety of operation, and facilitation of Good Laboratory Clinical Practices validation13. The greatest concern about the engineered cell-line based assays was physiologic relevance and representativeness of the value for the in vivo outcomes14. Disagreements have been reported between the results obtained between the PBMC-based and cell-line based assays15. These discrepancies were found to be attributed to the differences of the surface molecule concentration of target cells, especially the CC chemokine receptor 5 (CCR5) number16, which served as a coreceptor for HIV-1 entry. Cell lines with more physiological levels of CCR5 were introduced to the neutralization assay, such as the T-lymphoblastoid cell line A3R5 with similar surface CCR5 expression to PBMC, which employed infectious molecular clones17, 18. However, when A3R5 were used as target cells for pseudovirus infection, the luciferase signal-to-noise ratio was too weak to yield robust results (Montefiori, personal communication).
In this communication, we employed a highly efficient pseudovirus production system19 to develop a robust pseudovirus neutralization assay based on A3R5. With all the advantages of pseudoviurs, the A3R5 assay showed significantly higher sensitivity than the TZM-bl assay, especially for the detection of weak neutralization against tier 2 HIV-1 strains.
Results
Optimization of cell number for A3R5 neutralization assay
To determine the optimal cell density of A3R5 assay, two NAbs (PG9 and 2F5) and two HIV-1 positive plasma samples (HB118 and BJ170) were tested against one pseudovirus (11036 from subtype B) at different cell densities (from 1 × 104 to 3.2 × 105/well) (Fig. 1). The neutralization titer for the NAbs, showed as IC50, decreased sharply with the incremental input of cells (from 1 × 104 to 4 × 104/well), and then reached a plateau (from 4 × 104 to 3.2 × 105/well) (Fig. 1A). The similar performance was observed for the plasma samples, for which the ID50 values increased and then reached a plateau with the increasing of the target cell density (Fig. 1B). Data from the two types of samples demonstrated that the sensitivity of the assay increased with the incremental input of the target cells and reached a plateau when the cell number reached 4 × 104/well. It was of noted that goodness of fit (R2 value) for the neutralization curve reached the maximum point when the cell number reached 8 × 104/well, and then decreased slightly with the cell input increasing (Fig. 1C). Considering both the sensitivity and the variation, the optimal cell number was identified as 8 × 104/well for the A3R5 assay. When the cell number ranged from 4 × 104 to 3.2 × 105/well, little variation was found in the neutralization titers. The relatively wide range for the cell number demonstrated good robustness for the pseudovirus A3R5 assay.
Figure 1.
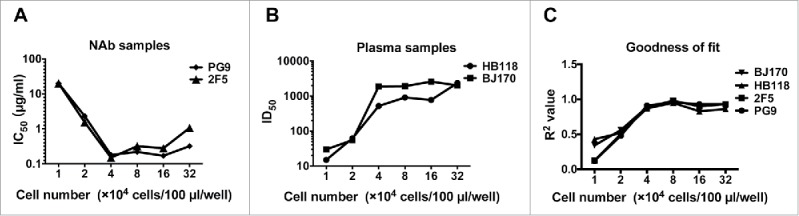
Optimization of cell numbers for the A3R5 pseudovirus assay. (A) Influence of cell numbers on neutralization results when tested against monoclonal neutralizing antibody samples (PG9 and 2F5) with pseudovirus 11036. (B) Influence of cell numbers on neutralization results when tested against serum samples (HB118 and BJ170) with 11036. (C) Effect of the cell density on the fitness of the neutralization curve.
Optimization of pseudovirus inoculum for neutralization
We next investigated the optimal inoculum of pseudovirus for A3R5 assay. The amount of pseudovirus inoculum was defined as TCID50. The relationship between the pseudovirus input and the sensitivity of the assay was analyzed by detecting and calculating the ID50 with different virus dose (starting from 200 to 16,000 TCID50/well) (Fig. 2). A subtype B tier 2 pseudovirus 11036 and a plasma sample from a subtype B HIV-1 infected individuals were employed. Although the input dose of 200 showed the highest sensitivity (Fig. 2A), the neutralization curve was non-smooth (R2 = 0.83) (Fig. 2B), which usually brought great variation. When the pseudovirus inoculum reached 800 TCID50/well or more, the acceptable R2 (not less than 0.98) values could be obtained for the neutralization curve (Fig. 2B). While the sensitivity would decrease dramatically with the virus input not less than 16,000 (Fig. 2A), the validated pseudovirus dose was identified as a range from 800 to 16,000 TCID50/well. The relatively wide range for the virus input also demonstrated good robustness for the pseudovirus A3R5 assay.
Figure 2.
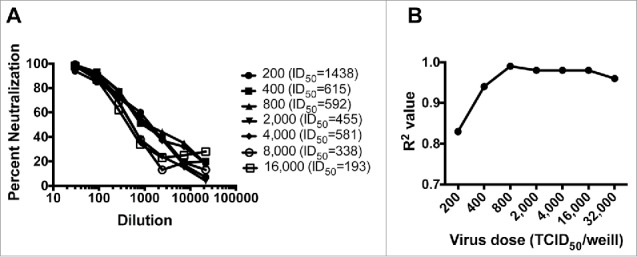
Optimization of the pseudovirus input for the A3R5 pseudovirus assay. (A) The neutralization curve under different pseudovirus inoculum. (B) Effect of the pseudovirus input amount on the fitness of the neutralization curve.
Optimization of diethylaminoethyl (DEAE)-dextran concentration for pseudovirus infection
Next, we optimized the DEAE-dextran concentration ranging from 1.7 to 144 μg/ml. RLU values were found to increase with the increasing amounts of DEAE-dextran between 1.7 to 9.9 μg/ml; the RLU readings decreased sharply with the concentration of DEAE-dextran reaching over 24.2 μg/ml (Fig. 3A). The decreased RLU values were found to be due to cellular toxicity induced by DEAE-dextran, especially when DEAE-dextran concentration reached 15.5 μg/ml or higher (Fig. 3B). The optimal concentration of DEAE-Dextran was identified as 10 μg/ml to yield satisfactory signal-to-noise ratio without obvious effects on cell viability.
Figure 3.
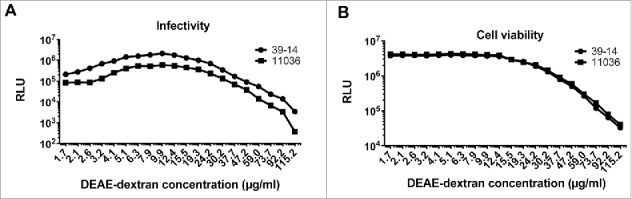
Optimization of DEAE-dextran concentration for the A3R5 pseudovirus assay. Effect of the DEAE-dextran concentration on infectivity of the pseudovirus (A) and the viability of the A3R5 cells (B).
Determination of incubation time for the pseudovirus A3R5 assay
As part of optimization process, we investigated the proper incubation time for pseudovirus infection assay. A3R5 cells were infected with two-fold serial diluted pseudovirus (39–14 and 11036 from subtype B) and read with luminometer at day 1, 2, 3, 4, 5, 6 and 7 post infection respectively. RLU values increased with prolonging incubation time and reached a maximum point at day 3, and then decreased (Fig. 4). So the incubation time was identified as 3 days.
Figure 4.
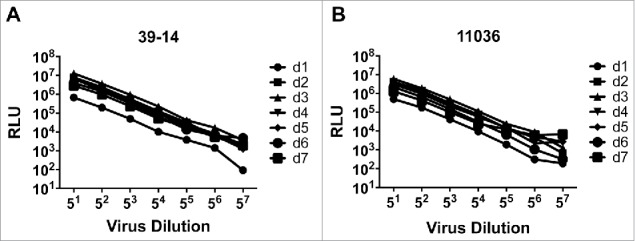
Determination of incubation time for the pseudovirus A3R5 assay. Relationship between the RLU and the incubation time for pseudovirus 39–14 (A) and 11306 (B).
Pseudovirus infection to A3R5 cell
To further investigate the infectivity of the pseudovirus to A3R5, a total of 30 pseudoviruses (10 from CRF01_AE, 10 from B/B’ and 10 from CRF07/08_BC/C) were screened for their ability to infect the target cells. The average relative luminescence unit (RLU) for the 30 pseudoviruses (5 fold dilution) reached 947041 (the RLU for cell control was 52), which made the luciferase signal-to-noise ratio (18212) high enough for the neutralization assay. When the infectivity of the pseudoviruses derived from different subtypes was compared, no significant difference (p > 0.05, one-way ANOVA) were found (Fig. 5A). Similarly, neither the infection phase nor the neutralization sensitivity showed significant effects (p > 0.05, unpaired t-test) on the infectivity of the pseudoviruses (Fig. 5B and C). Pseudoviruses generated in this high-titer system could efficiently infect the A3R5 cells to meet the requirement of the signal-to-noise for neutralization assay.
Figure 5.
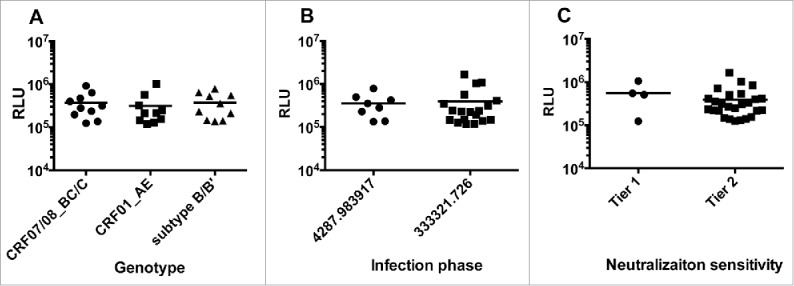
Infectivity of pseudoviruses to A3R5 cells. Pseudoviruses were diluted at 5 fold in triplicate and infected A3R5 cell. The average relative luminescent unit (RLU) values are indicated. (A) The infectivity of pseudoviruses from different subtypes. (B) The infectivity of pseudoviruses from different infection phases. (C) The infectivity of pseudoviruses with different neutralization sensitivities.
Validation of the A3R5 assay
Specificity
To test non-specific background of human plasma samples, twenty HIV-1 negative samples were used to establish a threshold or Limit of Blank (LOB) in A3R5 assay. These plasma samples were tested against a panel of seven pseudoviruses (SF162, Bal26 from tier 1 subtype B; the remaining from tier 2 strains, 11317 from subtype C; 11056, 11058 from subtype B; BJ5.11, GX24.8 from CRF01_AE) (Fig. 6A). The ID50 value 43 was established as the negativity threshold (non-parametric calculation)18 in A3R5 assay based on the 140 plasma/virus combinations data. Therefore, we defined the LOB as 50 for the ID50 value. When 50 for ID50 was used as the cutoff value, only 1 of 140 plasma/virus was considered as positive (false positive rate 0.7%) for A3R5, which is acceptable.
Figure 6.
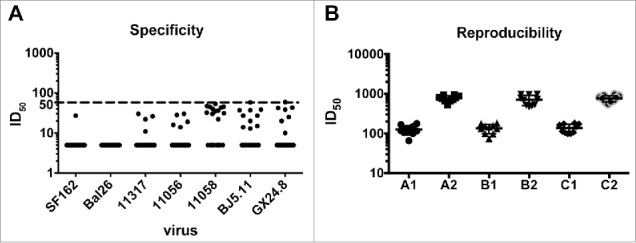
Validation of the pseudovirus A3R5 assay. (A) specificity: 20 HIV-1-negative plasma samples were tested against a panel of seven pseudoviruses (SF162, Bal26 from tier 1 subtype B; the remaining from tier 2 strains, 11317 from subtype C; 11056, 11058 from subtype B; BJ5.11, GX24.8 from CRF01_AE). (B) Reproducibility: two plasma samples (HJ182 and HB188) were employed to test against pseudovirus 11036. Each plasma sample was tested ten times in three independent runs by different operators.
Reproducibility
To determine the reproducibility of theA3R5 assay, two plasma samples (HJ182 and HB188) were employed to test against pseudovirus 11036. Each plasma sample was tested ten times in three independent runs by different operators (Fig. 6B). When the titers of plasma samples were calculated, the intra-assay and inter-assay coefficients of variation were 16.9–26.6% and 21.2–25.6%, respectively, indicating good reproducibility.
Linear range and detection limits
The linear range for the A3R5 assay is the interval between the upper and lower concentration of an inhibitor for which it has been proved that the analytical procedure has a suitable level of linearity. Neutralization curves generated with plasma samples give a consistent pattern of linearity over a range of 20–80% reductions in RLU within a determined virus dose range (Fig. 2A). Values in this linear range were proportional to the concentration of neutralizing antibodies in the sample.
Limit of quantitation (LOQ) is the lowest concentration of tested sample that can be quantified. The LOQ of the A3R5 assay was defined as lowest concentration of neutralizing antibody or the highest dilution of plasma that reduces RLU by 50%. The 50% point was chosen because it lies in the middle of neutralization curves (20–80% reductions in RLU). Failure to score at least 50% reduction of RLU at the determined dilution of concentration was considered as negative.
Comparison of the pseudovirus A3R5 assay with the TZM-bl assay
Firstly, we compared performances of two subtype B tier 1 pseudoviruses (SF162 and Bal.26) in the two kinds of target cells, for which we employed five NAbs (2F5, 4E10, PG16, 2G12 and b12) and five subtype B HIV-1 positive plasma samples (HB4, HB118, HB120, BJ182 and TJ208) (Fig. 7). For two tier 1 pseudoviruses, both of the NAb and plasma samples showed higher inhibitory efficiency in the A3R5 assays (Wilcoxon matched pairs test, p<0.05 for NAbs, and p < 0.01 for plasma samples. Compared to the TZM-bl assay, the sensitivity of A3R5 assay enhanced 4 and 2 fold for NAb (Fig. 7E) and plasma samples (Fig. 7F) respectively.
Figure 7.
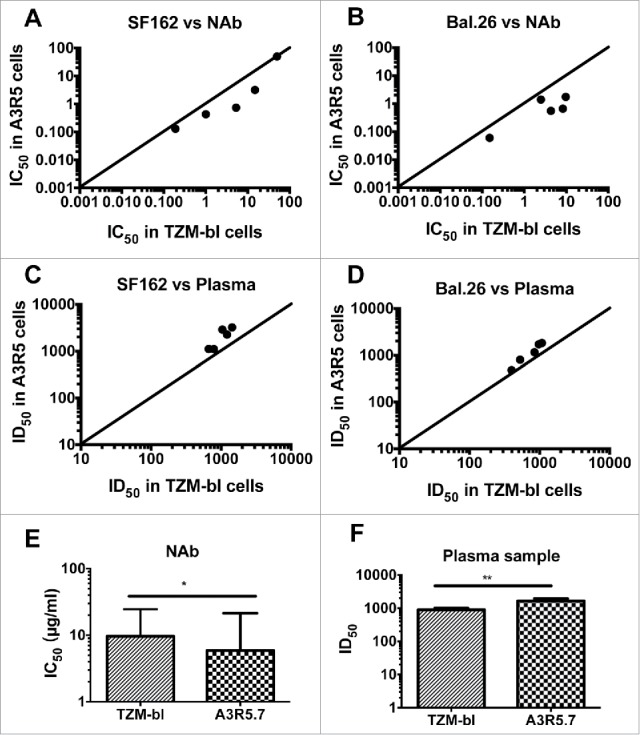
Comparison of the pseudovirus A3R5 assay with the TZM-bl assay tested against tier 1 pseudoviruses. Five NAbs (2F5, 4E10, PG16, 2G12 and b12) were tested against two pseudoviruses SF162 (A) and Bal.26 (B). Five plasma samples (HB4, HB118, HB120, BJ182 and TJ208) were tested against the same two pseudoviruses SF162 (C) and Bal.26 (D). The diagonal line depicts x = y (TZM-bl ID50/IC50 = A3R5 ID50/IC50 values). Statistical analyses for NAb (E) and plasma (F) samples were conducted respectively (Wilcoxon matched pairs test, * for p<0.05, and ** for p < 0.01).
Then, we investigated the sensitivity of the two assays, using the above NAbs and plasma samples for subtype B tier 2 pseudovirus (11036 and 11058), and another five CRF07/08_BC plasma samples (GZ48, HN60, GX66, GX90 and GX95) for subtype C tier 2 pseudoviruses (11317 and 11506) (Fig. 8). For the tier 2 pseudoviruses, the A3R5 assay gave much higher sensitivity against NAbs (10 fold higher) and plasma samples (9 fold higher) (Wilcoxon matched pairs test, p<0.001 for both NAbs and plasma samples) (Fig. 8I and J).
Figure 8.
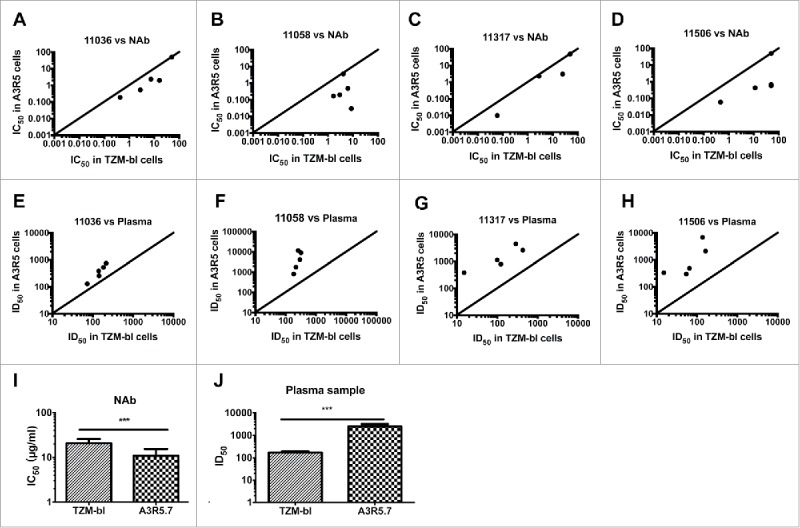
Comparison of the pseudovirus A3R5 assay with the TZM-bl assay tested against tier 2 pseudoviruses. Five NAbs were tested against four tier 2 pseudoviruses 11036 (A), 11058 (B) and 11317 (C), and 11506 (D). Five plasma samples were tested against the same four pseudoviruses 11036 (E), 11058 (F) and 11317 (G), and 11506 (H). The diagonal line depicts x = y (TZM-bl ID50/IC50 = A3R5 ID50/IC50 values). Statistical analyses for NAb (I) and plasma (J) samples were conducted respectively (Wilcoxon matched pairs test, *** for p<0.001=.
Besides NAb and chronic HIV-1 positive samples, we tested serial NAb seroconverting plasma samples against the pseudoviruses in both A3R5 and TZM-bl cells. Serial NAb seroconverting plasma samples were collected from 8 CRF01_AE-infected individuals (YA087, YA093, YA99, YA141, YA155, YA165, YA218 and YA233) from Beijing You'an Hospital (Table S1). Similarly, we detected the neutralizing antibody titers for samples from each blood collecting point against a tier 1 pseudovirus SF162 in both A3R5 and TZM-bl assays (Fig. 9). For most of the samples, higher ID50 values were obtained in the A3R5 assay. The average seroconverting point detecting in A3R5 was 15.7 weeks earlier than that in TZM-bl assay.
Figure 9.
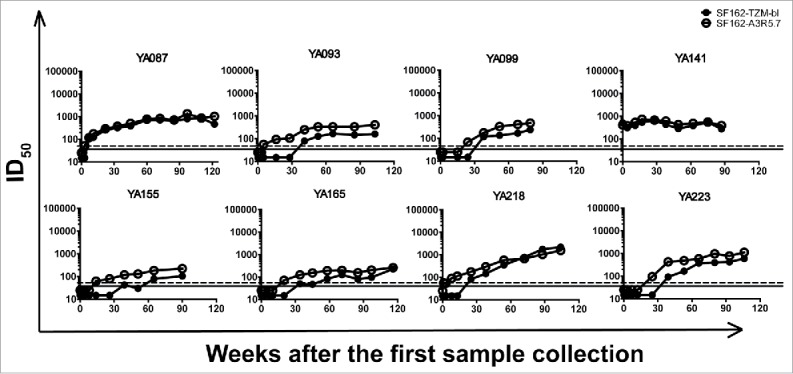
Comparison of A3R5 and TZM-bl assay using NAb seroconverting samples against tier 1 pseudoviurs. Serial NAb seroconverting plasma samples collected from 8 CRF01_AE-infected individuals were tested against pseudovirus SF162 in both A3R5 and TZM-bl assays. The dash and full lines indicate the cutoff values for A3R5 and TZM-bl assay respectively.
Finally, the CRF01_AE seroconverting samples were tested against three tier 2 pseudovirruse strains derived from the same subtype (BJ5.11, GX24.8 and GX71.18)20. The neutralizing titers detected in A3R5 were much higher than those in TZM-bl cells. And the average seroconverting points was 68.3 weeks earlier in A3R5 than that in TZM-bl assay (Fig. 10).
Figure 10.
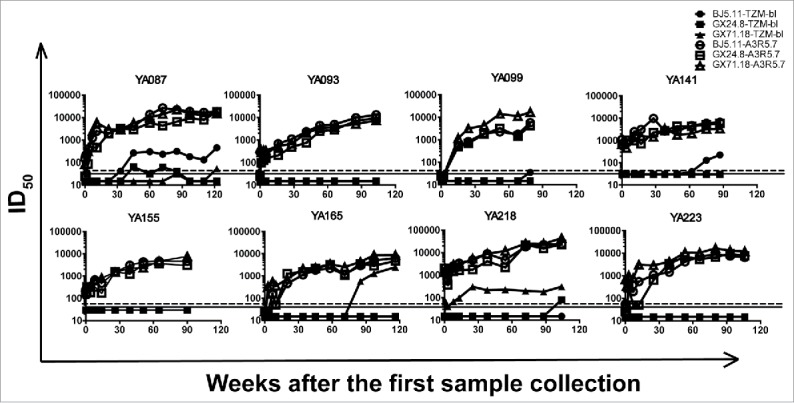
Comparison of A3R5 and TZM-bl assay using NAb seroconverting samples against tier 2 pseudoviurses. Serial NAb seroconverting plasma samples were tested against 3 tier 2 pseudoviruses in both A3R5 and TZM-bl assays. The dash and full lines indicate the cutoff values for A3R5 and TZM-bl assay respectively.
Discussion
Here we developed a high-sensitive pseudovirus neutralization assay based on a T-lymphoblastoid cell line A3R5 by using a high-efficient pseudovirus production system19. According to our knowledge, HIV-1 pseudovirus has not been successfully applied for neutralization detection in the primary PBMC or T-lymphocyte derived cell lines due to its relatively weak signals. Almost 100 times more RLUs could be generated in our new system than the traditional one to yield robust signal-to-noise ratio to meet the requirements for A3R5 and PBMC (data not shown) assays.
Compared with the traditional pseudovirus neutralization based on TZM-bl cells, the A3R5 assay showed significantly higher sensitivity for both quality and quantity assessment, which was similar to the comparison of pseudovirus TZM-bl assay and the IMC (infectious molecular clone) A3R5 assay17. The enhanced sensitivity was observed for both tier 1 and tier 2 pseudoviruses in A3R5 assay. The enhanced fold change of neutralization titers for tier 2 pseudoviruses was greater than that for tier 1 pseudoviruses, which was also observed in the post-vaccinated samples21. In that study, samples from the two phase III clinical trials were tested using the pseudovirus TZM-bl and IMC A3R5 assay. Neutralization of tier 1 viruses was detected in both RV14422 and Vax00323. Sporadic weak neutralization of tier 2 viruses was only detected in Vax003 using A3R5 approach21. It was reported that the ultra-sensitivity of the A3R5 assay was attributed to the relatively low level of surface CCR5 molecule compared to TZM-bl17. Similar CCR5 reporter density was also observed in our study (data not shown). However, the CCR5 was not the only cause. The surface CCR5 numbers of the A3R5 was higher than the CD3.8 stimulated PBMCs, while the A3R5 assay showed higher sensitivity than PBMC assay for most tests [17]. Another high sensitive method, TCLA/cell line assay, have been proved to be poorly predictive of the in vivo protection, mainly due to virus strains employed in this assay. The TCLA strains altered their neutralization sensitivity and envelope host cell protein composite during the passage in different cell types14, 24. Although the Envs of the pseudoviruses are identical to the primary ones, more comprehensive studies will be required to investigate the mechanism behind the super sensitivity, and the correlation between the potential values of certain Abs or vaccine immunogenicity and the neutralization efficiency identified in the A3R5 pseudovirus assay.
Although HIV-1 prophylactic vaccine still remains elusive, cross-reactive neutralizing antibodies have been found in about 50% HIV-1 chronically infected individuals, on average 2.5 years after infection25. Recently, this kind of broadly neutralizing antibodies has also been identified in some patients in the first year26-28 or even six months29 of infection in some rare conditions. Deciphering the mechanisms behind the development of broadly neutralizing antibodies, especially for the early infections, may facilitate the design of better immunogens and vaccination strategies. Correlation of neutralizing antibody induction and virus or host factors has not been fully identified, partially attributed to the limited numbers of neutralizers developed in the first months after infection29. Most of these studies employed the TZM-bl assay to determine the neutralization status of the infected subjects26-29, which may miss some modest or weak neutralizers due to the relative low sensitivity of the method. The newly developed pseudovirus neutralization assay based on A3R5 cells showed significantly higher sensitivity compared to the widely-used TZM-bl assay and could detect seroconverting points of neutralizing antibodies 68.3 weeks earlier, which could serve as an alternative to the TZM-bl assay, especially for the conditions requiring high sensitivity. For the immunogen design and modifications, high-sensitive evaluating assay could monitor minute advancements over the original one. Combining these small progresses may yield breakthroughs in the HIV-1 vaccine development. For HIV-1 vaccine clinical trials, high sensitive and easy-to-use neutralization assay are also urgently needed to establish the limit protection level for neutralizing antibodies.
Compared to the A3R5 assay using IMC, the new A3R5 pseudovirus assay could employ the same Env-expressing plasmids as the TZM-bl assay. Hundreds of plasmids have been constructed for the generation of pseudoviruses for TZM-bl assays, which could be transferred smoothly to produce pseudoviruses for A3R5 assay by cotransfecting 293T cells together with pSG3ΔEnv.fluc instead of pSG3ΔEnv. The A3R5 pseudovirus assay inherits all the advantages of the TZM-bl assay, such as high versatility of virus strains, high reproducibility, high throughput, low cost, ease and safety of operation.
Since it is unknown what level and breadth of neutralization in any assay will predict a significantly protective vaccine effect, the newly developed A3R5 pseudovirus assay could provide a complementary choice for the HIV-1 neutralizing antibody detection, especially for the samples with low neutralization potency against the circulating tier 2 strains.
In this communication, we developed and optimized a HIV-1 pseudovirus neutralization assay based on A3R5 cells, which showed significantly higher sensitivity over the traditional pseudovirus assay based on TZM-bl cells, especially when the circulating strains of pseudovirus were tested against. The easy-to-use and high sensitive method may facilitate the study of protection mechanism for HIV-1 vaccines and guide the optimization of the immnogen designs.
Material and methods
Cell lines, antibodies, plasma and plasmids
The following reagents were obtained from AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells and Env-deficient HIV-1 backbone plasmid pSG3△env (Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc.); A3R5 (Dr. Robert Mclinden, Henry M. Jackson Foundation for the Advancement of Military Medicine); broadly neutralizing antibodies (bNAbs) 2F5, 4E10, 2G12 (Dr. Hermann Katinger); PG9, PG16, G1b12 (Dr. Dennis Burton and Carlos Barbas). Plasmids SF162 (Drs. L. Stamatatos and C. Cheng-Mayer); BaL.26 (Dr. John Mascola); NIH Cat#11017, 11036 and 11058 from standard subtype B panel30; NIH Cat#11307, 11317, 11505 and 11506 from standard subtype C panel31.
Chronically HIV-1-infected plasma samples were selected from plasma pools deposited in our laboratory20, 32, 33, including 6 subtype B’ samples (HB4, HB118, HB120, BJ170, BJ182 and TJ208), 5 CRF07/08_BC samples (GZ48, HN60, GX66, GX90 and GX95). Serial NAb seroconverting plasma samples were collected from CRF01_AE-infected individuals (YA087, YA093, YA99, YA141, YA155, YA165, YA218 and YA233) from Beijing You'an Hospital (Table S1). HIV-1 negative plasma samples were selected from a blood bank (Shanghai RAAS Blood Products Co. Ltd, Shanghai, China)34 Written informed consent was obtained from all donors. All plasma samples were treat-naive and heated at 56°C 1 h before using.
Pseudoviruses derived from China have been described previously20, 33, including: SHX335.24, SH188.6, GX83.47, GZ187.10, GX91.2, YN192.16, GX28.31, BJX4.6, GX43-2, YN108R-4, GX75-20, YN148R-9, YN99R-5, BJ23-1, 176–1, 171–13, 121B-16, YA-192, 39–14, BJ5.11, GX24.8, GX71.18.
In the optimization of the A3R5 assay, two pseudoviruses were employed: 11306 (for cell number, virus dose, DEAE concentration and incubation time) and 39–14 (for DEAE concentration and incubation time). 11306 is a pseudovirus strain originated from US, which was firstly constructed in Dr. Montefiori's laboratory and served as a well-known standard subtype B strain [30]. 39–14 is a subtype B pseudovirus strain constructed in our laboratory33. A 30 pseudovirus panel was used to investigate the infectivity of pseudovirus to A3R5 cells and confirm the feasibility of this assay especially for strains from China.
293T and TZM-bl cells were maintained in a 5% CO2 environment at 37 °C in high glucose DMEM supplemented with 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml), and passaged every 2–3 days. A3R5 cells were cultured in RPMI-1640 supplemented with 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml) in 5% CO2 /37°C conditions.
Preparation and titration of pseudoviruses
293T cells were cotansfected with env-expressing plasmid and HIV-1 backbone plasmid (pSG3△env35 for TZM-bl assay, pSG3ΔEnv. fluc19 for A3R5 assay) at a mass ratio of 1:2 using Polyethylenimine (PEI) (Alfa Aesar, 43896) according to the manufacture's instruction. The supernatant containing pseudovirus was harvested 48 h after transfection and centrifuged at 1000 g for 10 min to remove cell debris. Then the supernatant was filtered with micropore filter (0.45 μm) and adjusted the FBS concentration to 20%, and stored at −80°C.
The 50% tissue culture infectious dose (TCID50) of pseudovirus for TZM-bl assay was determined as described previously12.
For A3R5 assay, a 5-fold serial dilution of pseudovirus was made with a beginning dilution of 5 fold in quadruplicate wells in a 96-well cell culture plate. Then 100 μl A3R5 cells were adjusted to 8 × 105/ml and added into each well of the above plate. Following 72 hours incubation at 37°C/5% CO2, the cell culture plate was centrifuged at 900 g for 10 min. Before the luciferase detection, the appropriate volume of supernatant was aspirated to leave 100 μl in each well without disturbing the cells, and then 100 μl substrate Bright-Glo (Promega, E2650) was added into each well and incubated for 2 min at room temperature. Then, a total of 150 μl of lysate was transferred to a solid white plate for measurement of luminescence using GLOMAX™ 96 microplate luminometer (Promega). The TCID50 value was calculated using the Reed-Muench method36.
Neutralization assay
Neutralization in the TZM-bl cells was performed as described previously12.
For neutralization assay based on A3R5, 3-fold serial dilutions of serum or monoclonal NAb samples were made in duplicate with the beginning dilution of 30 fold or 25 μg/ml in a total volume of 100 μl. Then 50 μl pseudoviruses were added into each well and incubated at 37°C for 60 min. Following the incubation, 100 μl A3R5 cells with a concentration of 8 × 105 cell/ml were added into the 96-plates to adjust the working concentration of DEAE-Dextran to 10 μg/ml. And then the luminescence was detected as described in the titration section.
Statistical analysis
All 50% inhibitory concentration/ 50% inhibitory dose (IC50/ID50) values were calculated using a formally validated Excel-based macro37. Neutralizing antibody titers were represented as the reciprocal of serum dilution required to reduce RLU by 50%. IC50/ID50 values were compared between A3R5 and TZM-bl assays for tier 1 and tier 2 pseudovirus respectively, using Wilcoxon matched pairs test. Statistical analyses were performed using GraphPad Prism Software 5.0 (GraphPad, San Diego, CA, USA).
Supplementary Materials: Table S1 Information of the serial NAb seroconverting plasma samples.
Disclosure of potential conflicts of interest
The authors declare no conflict of interest. The funding bodies had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We are grateful to Drs. John C. Kappes and Xiaoyun Wu for providing pSG3ΔEnv plasmid and TZM-bl cells; Dr. Robert Mclinden for A3R5; Dr. Hermann Katinger for bNAbs 2F5, 4E10, 2G12; Dr. Dennis Burton and Carlos Barbas for PG9, PG16, G1b12. Drs. L. Stamatatos and C. Cheng-Mayer for plasmids SF162; Dr. John Mascola for BaL.26; Dr. David Montefiori for plasmids 11017, 11036, 11058, 11307, 11317, 11505 and 11506.
Funding
This work was supported by the National Key Research and Development Plan of China under Grant 2016YFC1200904. The funding had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Author contributions
Qingqing Chen, Jianhui Nie, and Youchun Wang wrote the manuscript; Youchun Wang, Qingqing Chen, Jianhui Nie, Weijin Huang, and Hao Wu conceived and designed the experiments; Qingqing Chen, Jianhui Nie, Weijin Huang, Yanmei Jiao, Lan Li, Tong Zhang, and Juan Zhao conducted the experiments. All the authors have read and approved the final manuscript.
References
- [1].Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47(3):401-9. doi: 10.1086/589862. PMID:18558875 [DOI] [PubMed] [Google Scholar]
- [2].Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al.. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466-70. doi: 10.1038/nature10373. PMID:21849977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al.. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285-9. doi: 10.1126/science.1178746. PMID:19729618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15(8):866-70. PMID:19525964 [DOI] [PubMed] [Google Scholar]
- [5].Ferrantelli F, Rasmussen RA, Buckley KA, Li PL, Wang T, Montefiori DC, Katinger H, Stiegler G, Anderson DC, McClure HM, et al.. Complete protection of neonatal rhesus macaques against oral exposure to pathogenic simian-human immunodeficiency virus by human anti-HIV monoclonal antibodies. J Infect Dis. 2004;189(12):2167-73. doi: 10.1086/420833. PMID:15181562 [DOI] [PubMed] [Google Scholar]
- [6].Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, Nason MC, Montefiori D, Moldt B, Poignard P, et al.. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211(10):2061-74. doi: 10.1084/jem.20132494. PMID:25155019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fenyo EM, Heath A, Dispinseri S, Holmes H, Lusso P, Zolla-Pazner S, Donners H, Heyndrickx L, Alcami J, Bongertz V, et al.. International network for comparison of HIV neutralization assays: the NeutNet report. PLoS One. 2009;4(2):e4505. doi: 10.1371/journal.pone.0004505. PMID:19229336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mascola JR, D'Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol. 2005; 79(16):10103-7. doi: 10.1128/JVI.79.16.10103-10107.2005. PMID:16051803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Montefiori DC, Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol. 2009;485,395-405. doi: 10.1007/978-1-59745-170-3_26. PMID:19020839 [DOI] [PubMed] [Google Scholar]
- [10].Polonis VR, Brown BK, Rosa Borges A, Zolla-Pazner S, Dimitrov DS, Zhang MY, Barnett SW, Ruprecht RM, Scarlatti G, Fenyo EM, et al.. Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology. 2008;375(2):315-20. doi: 10.1016/j.virol.2008.02.007. PMID:18367229 [DOI] [PubMed] [Google Scholar]
- [11].Heyndrickx L, Heath A, Sheik-Khalil E, Alcami J, Bongertz V, Jansson M, Malnati M, Montefiori D, Moog C, Morris L, et al.. International network for comparison of HIV neutralization assays: the NeutNet report II. PLoS One. 2012;7(5):e36438. doi: 10.1371/journal.pone.0036438. PMID:22590544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nie J, Wang W, Wen Z, Song A, Hong K, Lu S, Zhong P, Xu J, Kong W, Li J, et al.. Optimization and proficiency testing of a pseudovirus-based assay for detection of HIV-1 neutralizing antibody in China. J Virol Methods. 2012;185(2):267-75. doi: 10.1016/j.jviromet.2012.07.011. PMID:22796285 [DOI] [PubMed] [Google Scholar]
- [13].Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, et al., Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131-46. doi: 10.1016/j.jim.2013.11.022. PMID:24291345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Polonis VR, Schuitemaker H, Bunnik EM, Brown BK, Scarlatti G, Impact of host cell variation on the neutralization of HIV-1 in vitro. Curr Opin HIV AIDS. 2009;4(5):400-7. doi: 10.1097/COH.0b013e32832edc50. PMID:20048704 [DOI] [PubMed] [Google Scholar]
- [15].Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, et al.. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232-52. doi: 10.1128/JVI.78.23.13232-13252.2004. PMID:15542675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Choudhry V, Zhang MY, Harris I, Sidorov IA, Vu B, Dimitrov AS, Fouts T, Dimitrov DS. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentration. Biochem Biophys Res Commun. 2006;348(3):1107-15. doi: 10.1016/j.bbrc.2006.07.163. PMID:16904645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McLinden RJ, Labranche CC, Chenine AL, Polonis VR, Eller MA, Wieczorek L, Ochsenbauer C, Kappes JC, Perfetto S, Montefiori DC, et al.. Detection of HIV-1 neutralizing antibodies in a human CD4(+)/CXCR4(+)/CCR5(+) T-lymphoblastoid cell assay system. PLoS One. 2013;8(11):e77756. doi: 10.1371/journal.pone.0077756. PMID:24312168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sarzotti-Kelsoe M, Daniell X, Todd CA, Bilska M, Martelli A, LaBranche C, Perez LG, Ochsenbauer C, Kappes JC, Rountree W, et al.. Optimization and validation of a neutralizing antibody assay for HIV-1 in A3R5 cells. J Immunol Methods 2014;409:147-60. doi: 10.1016/j.jim.2014.02.013. PMID:24607608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nie J, Wu X, Ma J, Cao S, Huang W, Liu Q, Li X, Li Y, Wang Y. Development of in vitro and in vivo rabies virus neutralization assays based on a high-titer pseudovirus system. Sci Rep. 2017;7:42796. doi: 10.1038/srep42769. PMID:28202935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nie J, Zhang C, Liu W, Wu X, Li F, Wang S, Liang F, Song A, Wang Y. Genotypic and phenotypic characterization of HIV-1 CRF01_AE env molecular clones from infections in China. J Acquir Immune Defic Syndr. 2010;53(4):440-50. doi: 10.1097/QAI.0b013e3181cb8300. PMID:20090544 [DOI] [PubMed] [Google Scholar]
- [21].Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, et al.. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis. 2012;206(3):431-41. doi: 10.1093/infdis/jis367. PMID:22634875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al.. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209-20. doi: 10.1056/NEJMoa0908492. PMID:19843557 [DOI] [PubMed] [Google Scholar]
- [23].Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K. Bangkok Vaccine Evaluation, G., Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194(12):1661-71. doi: 10.1086/508748. PMID:17109337 [DOI] [PubMed] [Google Scholar]
- [24].Bastiani L, Laal S, Kim M, Zolla-Pazner S. Host cell-dependent alterations in envelope components of human immunodeficiency virus type 1 virions. J Virol. 1997;71(5):3444-50. PMID:9094615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28(2):163-9. doi: 10.1097/QAD.0000000000000106. PMID:24361678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 2011;7(1):e1001251. doi: 10.1371/journal.ppat.1001251. PMID:21249232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Euler Z, van den Kerkhof TL, van Gils MJ, Burger JA, Edo-Matas D, Phung P, Wrin T, Schuitemaker H. Longitudinal analysis of early HIV-1-specific neutralizing activity in an elite neutralizer and in five patients who developed cross-reactive neutralizing activity. J Virol. 2012;86(4):2045-55. doi: 10.1128/JVI.06091-11. PMID:22156522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van den Kerkhof TL, Euler Z, van Gils MJ, Boeser-Nunnink BD, Schuitemaker H, Sanders RW. Early development of broadly reactive HIV-1 neutralizing activity in elite neutralizers. AIDS. 2014;28(8):1237-40. doi: 10.1097/QAD.0000000000000228. PMID:24556870 [DOI] [PubMed] [Google Scholar]
- [29].Sanchez-Merino V, Fabra-Garcia A, Gonzalez N, Nicolas D, Merino-Mansilla A, Manzardo C, Ambrosioni J, Schultz A, Meyerhans A, Mascola JR, et al.. Detection of Broadly Neutralizing Activity within the First Months of HIV-1 Infection. J Virol. 2016;90(11):5231-45. doi: 10.1128/JVI.00049-16. PMID:26984721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al.. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108-25. doi: 10.1128/JVI.79.16.10108-10125.2005. PMID:16051804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, et al.. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80(23):11776-90. doi: 10.1128/JVI.01730-06. PMID:16971434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang S, Nie J, Wang Y, Comparisons of the genetic and neutralization properties of HIV-1 subtype C and CRF07/08_BC env molecular clones isolated from infections in China. Virus Res. 2011;155(1):137-46. doi: 10.1016/j.virusres.2010.09.012. PMID:20875470 [DOI] [PubMed] [Google Scholar]
- [33].Chong H, Hong K, Zhang C, Nie J, Song A, Kong W, Wang Y. Genetic and neutralization properties of HIV-1 env clones from subtype B/BC/AE infections in China. J Acquir Immune Defic Syndr. 2008;47(5):535-43. doi: 10.1097/QAI.0b013e3181663967. PMID:18209676 [DOI] [PubMed] [Google Scholar]
- [34].Nie J, Liu Y, Huang W, Wang Y. Development of a Triple-Color Pseudovirion-Based Assay to Detect Neutralizing Antibodies against Human Papillomavirus. Viruses. 2016;8(4):107. doi: 10.3390/v8040107. PMID:27120611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al.. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307-12. doi: 10.1038/nature01470. PMID:12646921 [DOI] [PubMed] [Google Scholar]
- [36].Matumoto M. A note on some points of calculation method of LD50 by Reed and Muench. Jpn J Exp Med. 1949;20(2):175-9. PMID:15396956 [PubMed] [Google Scholar]
- [37].Piehler B, Nelson EK, Eckels J, Ramsay S, Lum K, Wood B, Greene KM, Gao H, Seaman MS, Montefiori DC, et al.. LabKey Server NAb: a tool for analyzing, visualizing and sharing results from neutralizing antibody assays. BMC Immunol. 2011;12:33. doi: 10.1186/1471-2172-12-33. PMID:21619655 [DOI] [PMC free article] [PubMed] [Google Scholar]


