SUMMARY
The flight muscles of Drosophila are highly enriched with mitochondria; but the mechanism by which mitochondrial complex I (CI) is assembled in this tissue has not been described. We report the mechanism of CI biogenesis in Drosophila flight muscles; and show that it proceeds via the formation of ~315-, ~550-, and ~815 kDa CI assembly intermediates. Additionally, we define specific roles for several CI subunits in the assembly process. In particular, we show that dNDUFS5 is required for converting an ~700 kDa transient CI assembly intermediate into the ~815 kDa assembly intermediate. Importantly, incorporation of dNDUFS5 into CI is necessary to stabilize or promote incorporation of dNDUFA10 into the complex. Our findings highlight the potential of studies of CI biogenesis in Drosophila to uncover the mechanism of CI assembly in vivo; and establish Drosophila as a suitable model organism and resource for addressing questions relevant to CI biogenesis in humans.
Graphical abstract
INTRODUCTION
Mitochondrial CI (NADH: ubiquinone oxidoreductase) is the first and largest of the electron transport chain complexes in the mitochondrion, and has a molecular mass approaching 1 MDa [reviewed in (Hirst, 2013)]. Human CI has 44 distinct subunits (Table S1); 14 of which are directly involved in transferring electrons from NADH to ubiquinone, or for generation of the membrane potential. Because these 14 subunits are conserved from bacteria to humans, and form the catalytic centers of the enzyme, they are referred to as the core or central subunits. The 30 remaining subunits, are referred to as accessory or supernumerary subunits; because they are not directly involved in catalysis, and are expressed to varying extents among eukaryotes (Table S1) [reviewed in (Hirst, 2013)]. A current hypothesis is that the accessory subunits may regulate ROS formation, complex assembly or stability, and cellular homeostasis in vivo. Of note, disease-causing mutations in several accessory subunits have been identified (Berger et al., 2008; Budde et al., 2000; Hoefs et al., 2008; Hoefs et al., 2011; Kirby et al., 2004; Ostergaard et al., 2011; Scacco et al., 2003); and genetic disruption of some accessory subunits in cell lines impair CI assembly (Guerrero-Castillo et al., 2017; Stroud et al., 2016). However, a definitive role for many of the accessory subunits in vivo remains to be established.
CI has two major arms: a hydrophobic membrane arm and a hydrophilic peripheral arm that juts into the mitochondrial matrix. The two arms are oriented almost perpendicularly to each other resulting in a characteristic boot or L-shaped structure (Clason et al., 2010; Efremov et al., 2010; Radermacher et al., 2006; Zickermann et al., 2015). Several cryo-electron microscopy density maps and higher resolution atomic structures of CI from various eukaryotes have recently been described (Fiedorczuk et al., 2016; Vinothkumar et al., 2014; Zhu et al., 2016; Zickermann et al., 2015). The accessory subunits were found to form a cage around the core subunits, and were particularly concentrated around the membrane domain. These observations lend further credence to the hypothesis that the accessory subunits may be involved in stabilizing the complex during or after biogenesis in vivo.
Surprisingly, despite the outstanding genetic capabilities of Drosophila, a systematic genetic analysis of CI assembly has not been described in this organism. Instead, previous in vivo genetic analyses of the regulation of eukaryotic CI assembly have been performed primarily in the aerobic fungus Neurospora crassa (Duarte et al., 1995). Although the N. crassa model of CI assembly is renowned for being the first system for which a model of CI assembly was described, there are notable deviations from the assembly pathway in mammalian systems (Nehls et al., 1992; Tuschen et al., 1990). Thus, it is important to develop additional genetically tractable CI assembly model systems that more closely resemble and recapitulate the human system. Importantly, Drosophila has a comparable number of CI subunits (similar to the human and bovine enzymes) and over a dozen putative assembly factors, all of which have clear human orthologs; making it a suitable model organism for studying CI assembly. Studying CI assembly in Drosophila has the added advantage of being in an in vivo context, where the effects of both developmental signals and environmental perturbations can be examined. Accordingly, we have analyzed the role of several nuclear-encoded CI subunits in CI assembly in Drosophila muscles.
We describe the mechanism of CI assembly in Drosophila flight muscles. Specifically, we show that many of the accessory subunits regulate specific stages of CI biogenesis in vivo, such that, when their level of expression is reduced, CI activity is diminished due to impaired CI assembly. We demonstrate that CI biogenesis in Drosophila involves the formation of ~315-, ~550-, and ~815 kDa assembly intermediates, and that RNAi-mediated knockdown of either dNDUFS2 or dNDUFS3 decreases the amount of the ~315 kDa assembly intermediate that is formed. Furthermore, we show that a specific accessory subunit – dNDUFA5 – is required for the formation and/or stabilization of the ~315 kDa assembly intermediate in vivo. Additionally, we define a specific role for another accessory subunit (dNDUFS5); and show that it is required for converting a transient CI assembly intermediate (an ~700 kDa assembly intermediate) into the ~815 kDa assembly intermediate, during one of the terminal steps of CI assembly. Four components of the Mitochondrial Complex I Assembly (MCIA) complex (dECSIT, dNDUFAF1, dACAD9 and dTIMMDC1), associated with the ~700 kDa assembly intermediate, further confirming that it is a true assembly intermediate in CI biogenesis. Importantly, incorporation of dNDUFS5 into CI is necessary to stabilize or promote incorporation of dNDUFA10 into the complex. We also identify several roles for many of the dNDUFB subunits. Altogether, our analyses reveal how studies of CI biogenesis in Drosophila can uncover mechanisms of CI assembly in vivo, and establish Drosophila as an important genetically pliable model organism for addressing questions relevant to mammalian CI biogenesis.
RESULTS
Drosophila Flight Muscles Are Suitable For Studying CI Assembly
CI consists of a hydrophilic matrix arm and a hydrophobic membrane arm that are oriented almost orthogonally to each other (Figure 1A). Subunits with the prefix NDUFA (NDUFA1-3 and NDUFA5-13) are thought to be part of the matrix arm, whereas the NDUFB subunits (NDUFB1-NDUFB11) are part of the membrane arm. In addition, subunits that are found in the vicinity of the 8 Fe-S clusters (NDUFS) or single Flavoprotein (NDUFV) are also localized in the matrix. All the NDUFA and NDUFB subunits are accessory subunits (Figure 1A). We used the Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool (DIOPT) to identify 42 putative orthologs of the 44 human CI subunits (Figure 1B and Table S1) (Hu et al., 2011). To facilitate comparison with their human orthologs, in this manuscript we refer to Drosophila orthologs of the human CI subunits as dNDUFS1, dNDUFS2, etc. Their designated gene nomenclature in Drosophila are shown in Table S1.
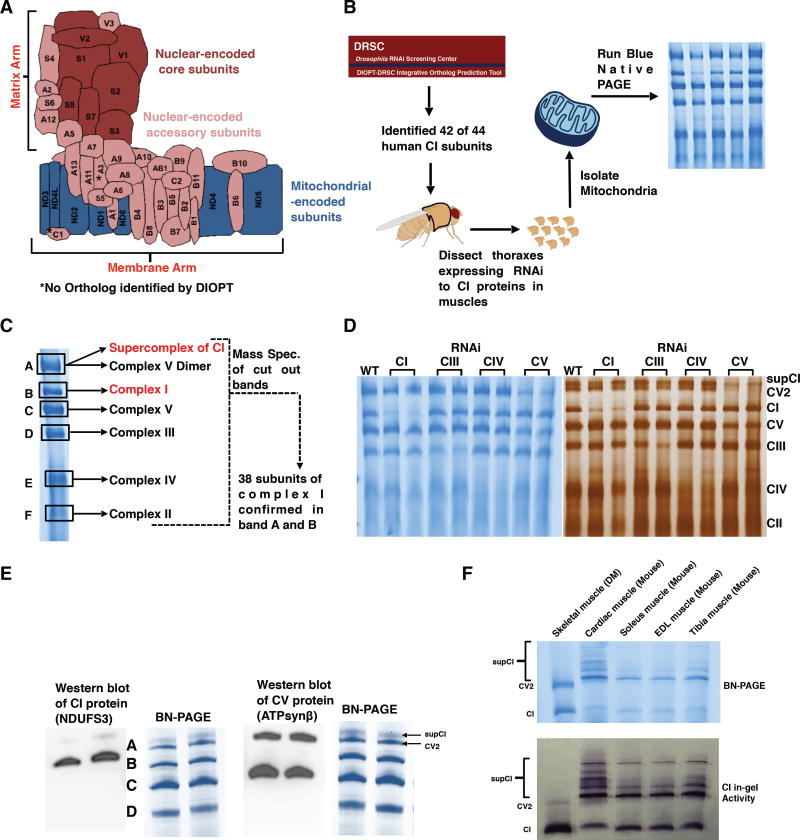
Figure 1. Drosophila Flight Muscles Are Suitable For Studying Complex I Assembly.
(A) Schematic representation of how the 44 different subunits of bovine CI are arranged to produce the L-shaped topology; adapted from (Guarani et al., 2014). The asterisk denotes subunits for which an ortholog was not identified in Drosophila by DIOPT.
(B) Summary of the experimental procedure for studying CI assembly in Drosophila. Transgenic RNAi constructs to the nuclear-encoded subunits were expressed specifically in thoracic muscles using the mhc-Gal4 driver. Mitochondria were isolated from thoraxes of 1 week-old flies, solubilized in 1% digitonin, and analyzed by blue native polyacrylamide gel electrophoresis (BN-PAGE).
(C) The constituents of each of the six major bands observed during BN-PAGE was analyzed by mass spectrometry. 38 subunits of Drosophila CI were confirmed by mass spectrometry. The 38 subunits correspond to 37 different orthologs of human CI. Two paralogs of human NDUFV1 were confirmed by mass spectrometry (see Table S1). See Table S2 for all the peptides identified in the six major bands shown.
(D) BN-PAGE (left panel) and Silver staining (right panel) of samples from thoraxes following RNAimediated knockdown of complex I (CI), complex III (CIII), complex IV (CIV) and complex V (CV) proteins to confirm the identities of the bands. SupCI and CV2 denote a supercomplex of CI and a dimer of CV respectively. The exact RNAi constructs expressed starting from left to right were to the white gene (wildtype, WT), dNDUFV1 (CI), dNDUFS1 (CI), dUQCRC-2 (CIII), dUQCRC-Q (CIII), dCox5A (CIV), cyclope (CIV), dATPsyn-β (CV), and ATPsyn-b (CV).
(E) Immunoblotting with anti-NDUFS3 and anti-ATPsynβ antibodies of native gels to detect CI and CV respectively. Note that band A is a doublet consisting predominantly of a dimer of CV, and a supercomplex of CI.
(F) BN-PAGE (top panel) and CI in-gel enzyme activity (lower panel) indicate that most of CI exists as the holoenzyme in Drosophila melanogaster (DM) skeletal muscles, in contrast to cardiac, soleus, EDL and tibia muscles from mice where a significant portion of CI exists as a supercomplex.
To confirm whether the putative CI orthologs identified by DIOPT were bona fide CI subunits in Drosophila flight muscles, we isolated mitochondria from thoraxes of wild-type flies, solubilized their membranes in 1% digitonin, and resolved their oxidative phosphorylation (OXPHOS) complexes into various bands using blue native polyacrylamide gel electrophoresis (BN-PAGE) (Rera et al., 2011; Wittig et al., 2006). We solubilized the mitochondrial membranes in 1% digitonin because we found that 1% digitonin was the optimal detergent concentration for isolating and resolving OXPHOS complexes in their native state in Drosophila (Figure S1), as has been reported previously (Rera et al., 2011; Wittig et al., 2006). Subsequently, we cut out each of the bands detected by coomassie staining of the gel, and identified their composition by mass spectrometry (Figure 1C). We confirmed the existence of 37 of the 42 putative CI orthologs based on their presence in the band corresponding to the CI holoenzyme (Band B) and/or supercomplex (Band A) (Figure 1C, Tables S1 and S2). Notably, the Drosophila ortholog of NDUFA4 (ND-MNLL) – a protein that was previously considered a CI subunit, but has been reassigned as a complex IV (CIV) subunit (Balsa et al., 2012) – co-migrated with the CIV band (Band E) (Figure 1C and Table S2). In addition, 4 of the subunits we were unable to detect are highly hydrophobic membrane-embedded core subunits encoded in the mitochondrion (ND2, ND3, ND4L and ND6); thus they may have escaped detection due to their highly hydrophobic nature. Interestingly, these subunits were not identified in a previous proteomic analysis of CI in mouse cell lines (Balsa et al., 2012).
Coomassie- or silver-stained native gels containing mitochondrial protein complexes from flies expressing RNAi to CI, complex III (CIII), CIV, and complex V (CV) proteins further confirmed the identities of the bands cut for mass spectrometry (Figure 1D). Because our mass spectrometry data suggested that a portion of CI might be co-migrating with CV and possibly CIII, we tested whether this co-migration was the result of supercomplex formation. We were unable to find antibodies that cross-react with any of the Drosophila CIII proteins, but antibodies that cross-react with dNDUFS3 (a CI protein) and dATPsynß (a CV protein) were commercially available, and were used to examine the identity of “band A” via western blotting. As is evident in the silver staining gel (Figure 1D), immunoblotting revealed that “band A” was actually a doublet; and the lower band in the doublet corresponds to a dimer of CV, as has been observed in other contexts (Figure 1E) (Rera et al., 2011; Wittig et al., 2006). In addition, CI in flight muscles was found to exist predominantly as the holoenzyme, with a relatively small portion involved in CI-CIII supercomplex formation, which migrates as an upper band in the doublet (Figure 1E). Notably, the observation that CI in Drosophila flight/skeletal muscles occurs predominantly as the holoenzyme (i.e. free CI, not involved in supercomplex formation), contrasts markedly with CI in cardiac or skeletal muscles from mice, where a significant portion of CI is trapped in supercomplex formation (Figure 1F). Thus, in addition to the genetic capabilities of Drosophila, and the fact that it has a comparable number of CI subunits as the human enzyme; it is a suitable model for studying CI assembly because most of CI in flight muscles exists as the holoenzyme. Accordingly, a defect in CI biogenesis can easily be scored and quantified. Consequently, we decided to examine the role of the nuclear-encoded CI subunits in CI assembly.
Disruption Of Several CI Subunits In Flight Muscles Impair CI Assembly
We found that loss-of-function alleles for many Drosophila CI genes are lethal (not shown). Therefore, to ascertain which CI subunits are required for CI biogenesis in Drosophila, we used the Gal4/UAS system to express transgenic RNAi constructs (henceforth referred to as UAS-RNAi lines) to both core and accessory CI subunits (Brand and Perrimon, 1993). We examined the effect of knocking down the subunits specifically in muscles (using either Dmef2-Gal4 or mhc-Gal4). Transgenic expression of many of the UAS-RNAi constructs using Dmef2-Gal4 – a muscle-restricted Gal4 driver that is expressed strongly throughout development – caused lethality (not shown). However, a genetic cross between each of the UAS-RNAi lines and mhc-Gal4 produced viable flies, as the mhc-Gal4 driver has a weaker expression relative to Dmef2-Gal4 during the initial larval stages (Figure S2). Accordingly, we decided to analyze CI assembly in mitochondria isolated from thoraxes of mhc-Gal4/UAS-CIRNAi flies (henceforth referred to as mhc>CIRNAi flies) using BN-PAGE.
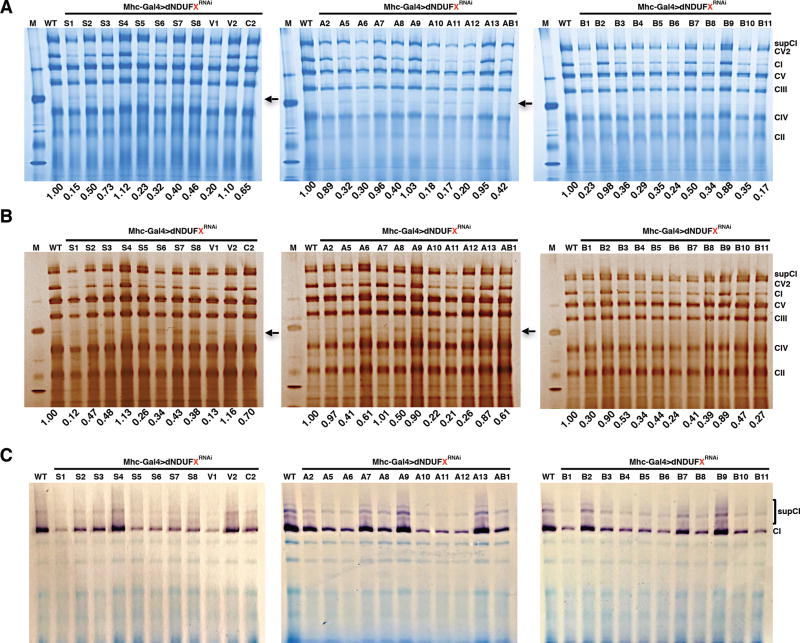
We observed that, in general, both core and accessory subunits, produced CI assembly defects whenever the extent of transcript knockdown was more than 50% (Figure 2A). To further assess the extent of the CI assembly deficit for each subunit, we quantified the amount of CI relative to the amount of CV in each lane, and normalized it to the corresponding value in the wild-type lane. Interestingly, this revealed that some of the most robust CI assembly deficits were observed when accessory/supernumerary subunits (such as dNDUFA10-12 and dNDUFB4-6) were genetically impaired (Figures 2A and 2B). Similar results were obtained with silver staining of the protein complexes in the native gels (Figure 2B). Finally, in-gel CI enzyme activity assay revealed that the assembly deficits correlated with a reduction in CI activity (Figure 2C). Altogether, these results indicate that many of the core and accessory subunits are essential for viability and biogenesis of the CI holoenzyme or supercomplex in flight muscles. Accordingly, we turned our attention towards elucidating the mechanism of CI assembly in Drosophila flight muscles.
Figure 2. Disruption of Several CI Core And Supernumerary Subunits Impair CI Assembly In Drosophila.
BN-PAGE (A), Silver staining (B), and CI in-gel enzyme activity (C) of mitochondria isolated from thoraxes following RNAi-mediated knockdown of the CI proteins indicated (mhc-Gal4>dNDUFXRNAi ). The values listed below each lane indicate the residual amount of CI normalized to the amount in the wildtype (mhc-Gal4>w1118) lane.
Proteomic Analyses And Immunoblotting Identify Assembly Intermediates Of CI
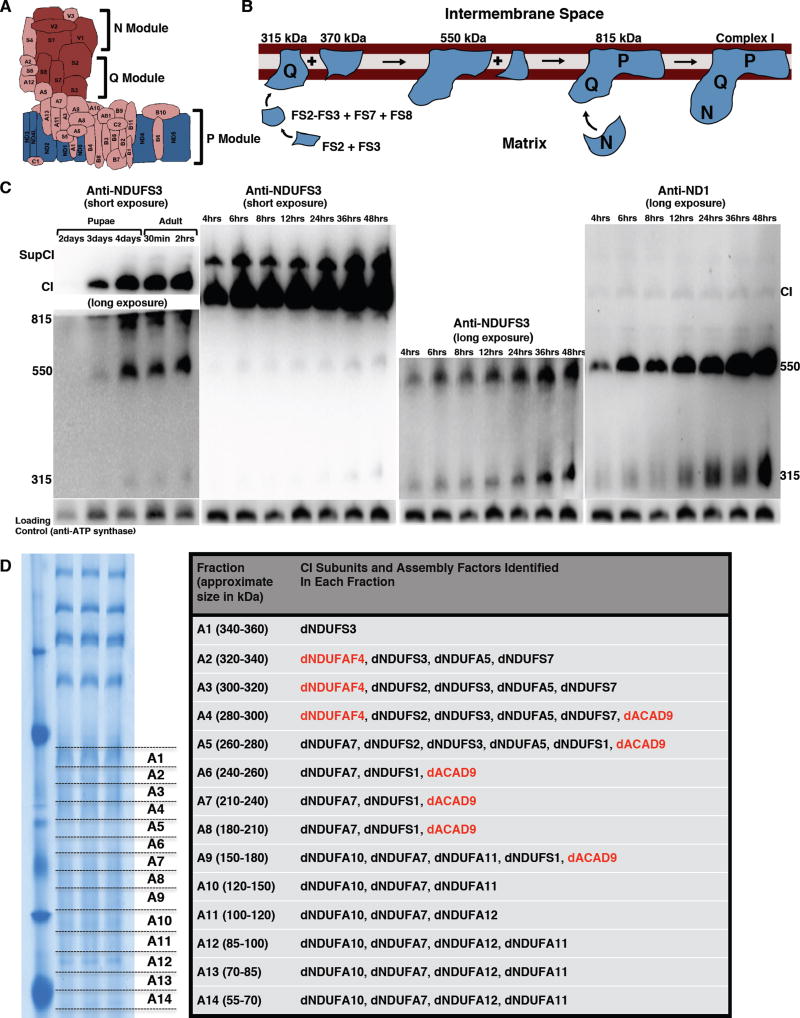
Studies from some mammalian cell lines have shown that CI biogenesis proceeds via a series of assembly intermediates that combine with each other, or other subunits, to form the ~950 kDa boot-shaped holoenzyme. The assembly intermediates generally correspond to partial or complete domains of the 3 functional modules of CI. The NADH Dehydrogenase module (N module) is located at the tip of the matrix arm, and is the site of NADH oxidation. Situated between the N module and the membrane, is the Q module, which is responsible for Ubiquinone reduction. The proton-conducting P module in the membrane arm can be subdivided into a proximal PP module (roughly corresponding to the first half of the P module that connects with the Q module) and a distal PD module (Figure 3A).
Figure 3. Proteomic Analyses And Immunoblotting Identify Assembly Intermediates Of CI.
(A) Schematic of CI showing the three modules of the enzyme. The NADH Dehydrogenase module (N module) is located at the tip of the matrix arm, and is the site of NADH oxidation. Situated between the N module and the membrane arm, is the Q module, which is responsible for Ubiquinone reduction. The proton-conducting P module is in the membrane arm.
(B) The current model of CI assembly in mammalian systems (reviewed in (Vartak et al., 2014). The assembly process begins with the formation of an assembly intermediate containing NDUFS2 and NDUFS3, which combines with NDUFS7 and NDUFS8. The subcomplex of NDUFS2, NDUFS3, NDUFS7 and NDUFS8 ultimately combines with ND1 to form the ~315 kDa assembly intermediate that is anchored to the membrane. The ~315 kDa subcomplex (also called the Q module) combines with an independently-formed ~370 kDa assembly intermediate to form an ~550 kDa assembly intermediate. This assembly intermediate which consists of the Q module and part of the P module grows by the addition of more subunits to form the ~815 kDa assembly intermediate, via mechanisms that are very poorly defined. The ~815 kDa assembly intermediate now consists of the complete Q and P modules. Finally, the N module is added to produce the 950kDa fully-assembled complex. Assembly factors or chaperones that assist in this process, but are not present in the fully assembled complex, have been omitted for clarity.
(C) Western blot of samples obtained from thoraxes from pupae aged between 2 and 4 days after pupariation, and of flies from 0.5 hours to 48 hours post-eclosure to detect the assembly intermediates, fully assembled CI, and a supercomplex containing complex I (supCI) after BN-PAGE. The anti-NDUFS3 antibody strongly detects CI and supCI; and weakly detects the ~315 kDa, ~550 kDa and ~815 kDa assembly intermediates after a short exposure. However, after a longer exposure, the ~315 and ~550 kDa assembly intermediates can clearly be seen. In the right panel, the membrane was stripped and re-probed with anti-NDI. Anti-ND1 detects the ~315 kDa and ~550 kDa assembly intermediates, and a very faint band corresponding to CI. Note that in all 4 panels there is a general increase in accumulation of assembly intermediates, holoenzyme and CI-containing supercomplex with time. The CV antibody (anti- ATPsynß) was used as a loading control because commercially-available anti-CII antibodies we tested did not cross-react with Drosophila CII.
(D) Proteomic analyses of assembly intermediates that form in the native gel sized between ~50 kDa and ~350 kDa. See Table S3 for all the peptides identified.
The current model posits that CI assembly in mammalian systems begins with the formation of a small assembly intermediate containing NDUFS2 and NDUFS3, that combines with NDUFS7 and NDUFS8 (Figure 3B). This assembly intermediate is the primary component of the Q module; and ultimately combines with ND1 to form an ~315 kDa assembly intermediate that is anchored to the mitochondrial inner membrane. The ~315 kDa assembly intermediate combines with an independently-formed ~370 kDa assembly intermediate to form an ~550 kDa assembly intermediate (Figure 3B). The ~550 kDa assembly intermediate, which consists of the complete Q module and a portion of the P module, grows by the addition of more subunits to form the ~815 kDa assembly intermediate, via mechanisms that are very poorly defined. At this point, the ~815 kDa assembly intermediate is generally considered to be composed of the complete Q and P modules. Finally, an independently-formed assembly intermediate consisting of NDUFS1, NDUFV1, NDUFV2, NDUFV3, NDUFS4, NDUFS6 and NDUFA12 which together form the N module, is added as a “cap” to the ~815 kDa assembly intermediate to produce the ~950 kDa holoenzyme [Figure 3B; the ~315-, ~370-, ~550-, and ~815 kDa assembly intermediates were previously estimated as ~400-, ~460-, ~650- and ~830 kDa subcomplexes respectively (Andrews et al., 2013; Vartak et al., 2014)].
As some flight muscles are formed by 24 hours after pupal formation (Roy and VijayRaghavan, 1999), we decided to ascertain the extent of CI biogenesis starting at 48 hours (i.e. 2 days) post-pupariation. Specifically, we isolated mitochondria at various time points, and examined CI assembly via western blotting of the native complexes. Because current models of mammalian CI assembly postulate that NDUFS3 and ND1 are both part of the ~815 kDa, ~550 kDa, and ~315 kDa assembly intermediates, western blot with anti-NDUFS3 or anti-ND1 antibodies will be expected to detect these 3 assembly intermediates, and possibly lower molecular weight assembly intermediates (if the respective epitopes are not masked when the assembly intermediate is formed). In addition, the fully assembled CI and CIcontaining supercomplexes will be expected to be detected as well. Indeed, immunoblotting with anti-NDUFS3 revealed that a portion of CI is assembled during pupal development and continues during the first 48 hours after flies eclose (emerge as adults from pupae) (Figure 3C). Notably, there is a time-dependent increase in the accumulation of the ~315 kDa, ~550 kDa and ~815 kDa assembly intermediates during the first 48 hours after eclosure, that correlates with an increase in the amount of fully-assembled CI in flight muscles (Figure 3C). Although we were able to reproduce the time-dependent increase in accumulation of the ~315 kDa and ~550 kDa assembly intermediates with the anti-ND1 antibody (Figure 3C), the higher molecular weight bands were only weakly detectable, conceivably because the epitope to which this antibody was raised for this hydrophobic subunit becomes less exposed to the aqueous environment during the final stages of CI biogenesis (Figure 3C). Moreover, while we were able to detect subcomplexes of CV that migrate with an apparent mass of about 100 kDa at this stage of development (Figure S3), we were unable to detect dNDUFS3-containing assembly intermediates with an apparent mass of less than 200 kDa. There are at least two possible explanations for this result: (i) the smaller NDUFS3-containing assembly intermediates may not be present at this stage; or (ii) the epitope of dNDUFS3 in the smaller assembly intermediates was inaccessible to the antibody, perhaps as a result of being masked by bound assembly factors and/or other interactors. Therefore, we used proteomic analyses to distinguish between these two possibilities.
Mitochondria were isolated from thoraxes of wild-type flies that had been aged for 24 hours after eclosure, and subjected to BN-PAGE. Subsequently, the region of the gel between ~50 kDa and ~350 kDa, was excised and divided into 14 slices (labeled fraction A1 to A14) for in-gel digestion and subsequent proteomics analyses (Figure 3D). We observed that dNDUFS2, dNDUFS3 and dNDUFS7 co-migrated in fractions corresponding to a mass of approximately 280–320 kDa (Figure 3D and Table S3). Interestingly, the CI assembly factor, dNDUFAF4, was also found in these fractions (Figure 3D and Table S3). In addition, dNDUFA5 co-migrated with dNDUFS2, dNDUFS3 and dNDUFS7 (Figure 3D), confirming that it is a component of the ~315 kDa assembly intermediate in vivo. Importantly, although several other CI subunits migrated in fractions corresponding to a mass of approximately 50–250 kDa, neither dNDUFS2 nor dNDUFS3 were found in these fractions. Combining these results with the observation that there is an increase in accumulation of the ~315 kDa assembly intermediate between 4 and 48 hours after eclosion, it appears that in an in vivo context, in Drosophila flight muscles, the constituents of the ~315 kDa assembly intermediate are combined almost synchronously.
Specific Subunits Regulate The Biogenesis Or Stability Of Specific Assembly Intermediates Of CI
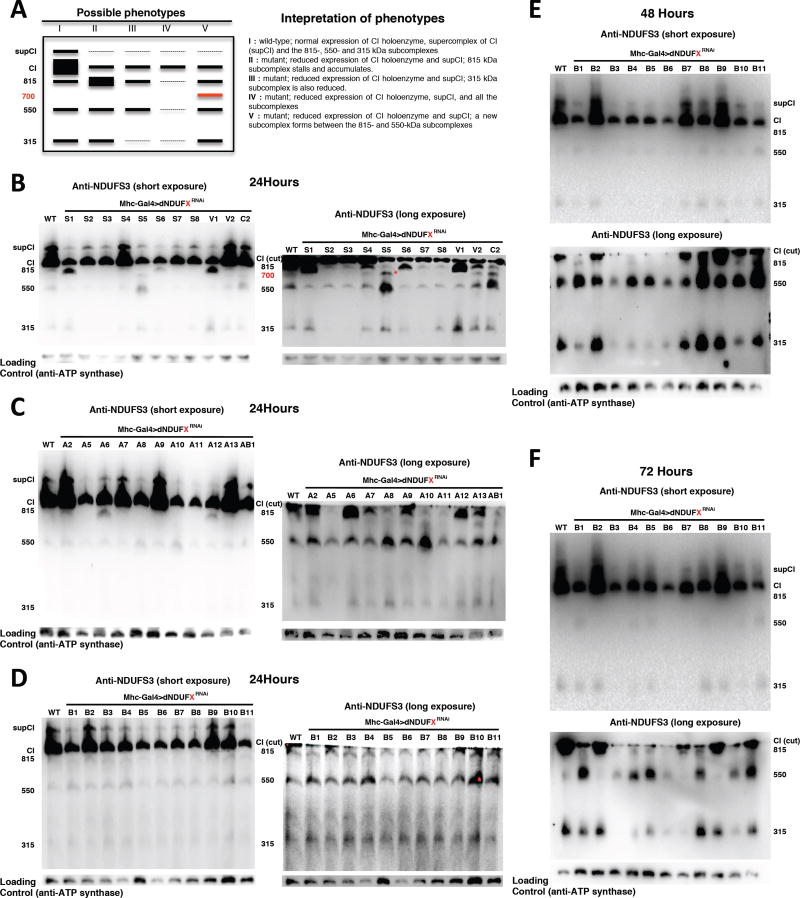
If the assembly intermediates observed are bona fide intermediates in the pathway of CI assembly in Drosophila, then at least some of these assembly intermediates will stall and accumulate, or they may disintegrate when specific CI subunits that are required for CI assembly are disrupted (Figure 4A). To test this hypothesis, we analyzed the CI assembly intermediates from thoraxes of Mhc>CIRNAi flies, 24 hours after eclosure using an anti-NDUFS3 antibody. As expected, the various subunits that produced CI assembly deficits in Figure 2 also resulted in a reduction of the level of the holoenzyme or the CI-containing supercomplex (Figures 4B–F).
Figure 4. Specific Subunits Regulate the Biogenesis or Stability of Specific Assembly Intermediates Of CI.
(A) The left panel depicts a schematic of the distribution of assembly intermediates on immunoblots as a result of RNAi-mediated disruption of various CI subunits. The right panel describes how various results can be interpreted.
(B–D) Distribution of assembly intermediates in thoraxes dissected 24 hours after eclosion with transgenic RNAi expression of the CI subunits shown. The ~815 kDa assembly intermediate accumulates in thoraxes expressing transgenic RNAi to dNDUFS1, dNDUFV1, dNDUFA6 and dNDUFA12; the ~315 kDa assembly intermediate is decreased in thoraxes expressing transgenic RNAi of dNDUFS2, dNDUFS3, dNDUFS7 and dNDUFA5. In addition, another assembly intermediate accumulates in thoraxes expressing RNAi to dNDUFS5 and dNDUFC2 (denoted by * in B). In panels labeled long exposure, the region of the membrane just at or below CI was cut and imaged.
(E and F) Distribution of assembly intermediates in thoraxes dissected 48 hours (E) and 72 hours (F) after eclosion with transgenic RNAi expression of the NDUFB subunits shown. RNAi-mediated knockdown of the expression of dNDUFB3 decreased the extent of accumulation of all the assembly intermediates; and the 550 kDa assembly intermediate accumulated when the expression of dNDUFB1, dNDUFB8 and dNDUFB11 were reduced. In addition, the extent of accumulation of the 315 kDa assembly intermediate was diminished following RNAi-mediated disruption of dNDUFB1, dNDUFB4, dNDUFB5, dNDUFB6 and dNDUFB10.
Disruption of dNDUFS1, dNDUFV1 and dNDUFA12, which are components of the N module of CI, and are thus expected to be added as part of the “cap” during the final step in CI assembly, resulted in a stalling and accumulation of the ~815 kDa assembly intermediate (Figures 4B and C). However, unexpectedly, disruption of dNDUFA6, not known to be part of the N module, also stalled the ~815 kDa subcomplex (Figure 4C). RNAi-mediated knockdown of dNDUFS2, dNDUFS3, dNDUFS5, dNDUFS7, and dNDUFS8 led to a reduction in the amount of the ~815 kDa assembly intermediate (relative to wild type), as they impaired some of the initial steps of CI biogenesis (Figures 4B and C). In addition, the amount of the ~315 kDa assembly intermediate was drastically reduced when the expression of dNDUFS2, dNDUFS3, or dNDUFS7 was impaired (Figure 4B); in line with our proteomic results in Figure 3D and current mammalian CI assembly models that show that the first step in CI biogenesis involves the formation of an assembly intermediate consisting of NDUFS2 and NDUFS3 (Figure 3B) [reviewed in (Vartak et al., 2014)]. Notably, we found that RNAi-mediated knockdown of dNDUFA5 depleted the ~315 kDa assembly intermediate (Figure 4C). Combining this result, with our proteomic data showing that dNDUFA5 co-migrates with dNDUFS2, dNDUFS3 and dNDUFS7 (Figure 3D), we conclude that although dNDUFA5 is an accessory subunit, it is a critical component of, and required for formation or stabilization of the ~315 kDa assembly intermediate (i.e. the Q module) in vivo.
Disruption of most of the dNDUFB subunits did not markedly alter the stability or extent of accretion of the CI assembly intermediates 24 hours after eclosion (Figure 4D), but by 48 and 72 hours after eclosion some notable and consistent phenotypes between the two time points were observed (Figure 4E and 4F). For instance, RNAi-mediated disruption of dNDUFB3 decreased the extent of accumulation of all the assembly intermediates; and the 550 kDa assembly intermediate accumulated when dNDUFB1, dNDUFB8 and dNDUFB11 were impaired at both time points (i.e. 48 and 72 hours post-eclosion). Surprisingly, although none of the NDUFB subunits are known to be part of the 315 kDa assembly intermediate, the extent of accumulation of the 315 kDa assembly intermediate was diminished when the expression of dNDUFB1, dNDUFB4, dNDUFB5, dNDUFB6 and dNDUFB10 were reduced (Figure 4E and 4F). Taken together, these results indicate that specific subunits regulate the biogenesis or stability of specific CI assembly intermediates during CI assembly in Drosophila thoraxes.
Identification Of An ~700 kDa Assembly Intermediate Of CI In Drosophila
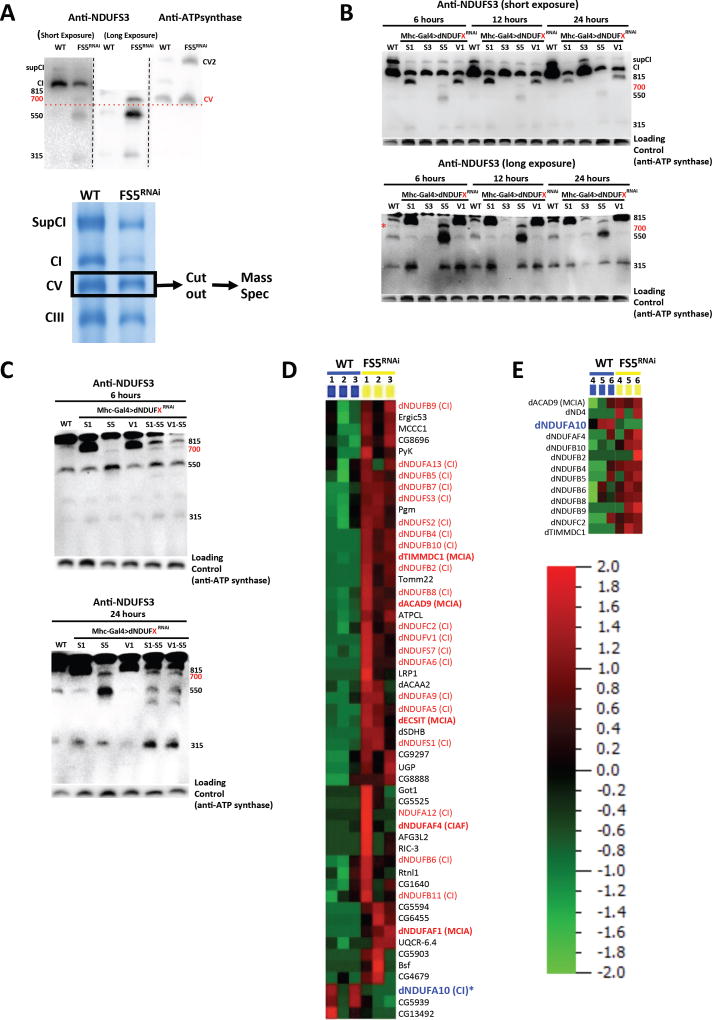
An assembly intermediate that accumulates between the ~550 and ~815 kDa assembly intermediates was detected on immunoblots of samples from mhc>dNDUFS5RNAi and mhc>dNDUFC2RNAi thoraxes (Figure 4B). We estimate its size to be ~700 kDa because it co-migrates with CV, previously estimated to be ~700 kDa in blue native gels (Figure 5A) (Abdrakhmanova et al., 2006). The accumulation of the ~700 kDa assembly intermediate in samples from mhc>dNDUFS5RNAi thoraxes was notable, because it suggested that this could be the point of entry of dNDUFS5 during CI assembly. NDUFS5 is a membrane-associated accessory subunit that extends into the intermembrane space; it is currently unclear at what point it becomes incorporated into CI. In contrast to the ~315, ~550 and ~815 kDa assembly intermediates, the ~700 kDa assembly intermediate was not readily perceptible by anti-NDUFS3 immunoblotting in the wild-type sample or most of the other mutant samples isolated 24 hours after eclosure (Figure 4B). This raised the possibility that it could simply be a degradation product, perhaps emanating from the ~815 kDa assembly intermediate.
Figure 5. Identification Of An ~700 kDa Assembly Intermediate Of CI In Drosophila.
(A) Top Panel: Immunoblots of samples obtained from wildtype and mhc>dNDUFS5RNAi thoraxes of flies aged for 6 hours after eclosure depicting co-migration of the ~700 kDa intermediate and CV. In the left and middle panels, anti-NDUFS3 antibodies detect the fully assembled CI, the ~700 kDa subcomplex, as well as other assembly intermediates in dNDUFS5RNAi thoraxes. Note that in the middle panel, the region of the membrane just below CI was cut and imaged. In the right panel, anti-ATPsynβ detects the CV monomer (700kDa) and dimer as shown. Lower Panel: Mitochondrial protein complexes from wildtype and mhc>dNDUFS5RNAi thoraxes were resolved by BN-PAGE and the region corresponding to the ~700 kDa assembly intermediate (i.e. CV, demarcated) was cut out, subjected to tryptic digestion, and analyzed by label-free quantitative LC-MS/MS.
(B) Immunoblots from samples obtained after 6 hours, 12 hours and 24 hours post eclosure from thoraxes where NDUFS1, NDUFS3, NDUFS5 and NDUFV1 were knocked down as a result of transgenic RNAi exression. Note that the ~815 kDa assembly intermediate accumulates as a result of disruption of NDUFS1 and NDUFV1, and the ~700 kDa assembly intermediate stalls and accumulates in NDUFS5 mutants at all time points. Importantly, upon prolonged exposure of the immunoblot, a band corresponding to the ~700 kDa assembly intermediate can also be observed in wild-type samples (denoted with the * in the lower panel), which confirms that it is an authentic, albeit transient assembly intermediate.
(C) The accumulation of the ~815 kDa assembly intermediate was significantly attenuated in mhc>dNDUFS5RNAi,dNDUFS1RNAi thoraxes relative to mhc>dNDUFS1RNAi thoraxes; instead there is an accumulation of the ~700 kDa assembly intermediate. Similar results were obtained when samples from mhc>dNDUFS5RNAi,dNDUFV1RNAi thoraxes were compared to samples from mhc>dNDUFV1RNAi thoraxes.
(D) Proteomic changes in the gel slice sample from wildtype and mhc>dNDUFS5RNAi thoraxes corresponding to the ~700 kDa assembly intermediate. Relative protein abundance among biological samples is expressed by spectral counts on a log scale. Several CI subunits and CIAFs, most notably components of the MCIA complex are upregulated in the ~700 kDa assembly intermediate. However, the amount of dNDUFA10 (denoted with an asterisk) is reduced in mhc>dNDUFS5RNAi thoraxes relative to wild type. See Table S4 for all the peptides identified
To determine whether the ~700 kDa assembly intermediate is a true assembly intermediate, we decided to look at earlier time points (6 and 12 hours post-eclosion) to ascertain whether it ever appears in wild-type samples. Immunoblotting at these time points revealed that accumulation of the ~700 kDa assembly intermediate in mhc>dNDUFS5RNAi thoraxes is present by the 6-hour time point, and gradually tapers off afterwards (Figure 5B). Importantly, at the 6-hour time point a faint band corresponding to the ~700 kDa assembly intermediate can be observed in wild-type samples, indicating that the ~700 kDa assembly intermediate exists in wild-type samples; and rapidly matures to the ~815 kDa assembly intermediate. The stalling of the ~700 kDa assembly intermediate in mhc>dNDUFS5RNAi thoraxes occurred concurrently with an accumulation of both the ~550 kDa and ~315 kDa assembly intermediates, and a diminution of the ~815 kDa assembly intermediate relative to wild-type levels. Thus, dNDUFS5 may be required for converting the ~700 kDa assembly intermediate into the ~815 kDa assembly intermediate, such that when this fails, there is a backlog of the ~700, ~550 and ~315 kDa assembly intermediates. To test this hypothesis, we compared the assembly intermediates that accumulate in mhc>dNDUFS5RNAi,dNDUFS1RNAi and mhc>dNDUFS5RNAi,dNDUFV1RNAi thoraxes with that in mhc>dNDUFS1RNAi and mhc>dNDUFV1RNAi thoraxes respectively. We reasoned that because the ~815 kDa assembly intermediate accumulates in mhc>dNDUFS1RNAi and mhc>dNDUFV1RNAi thoraxes (Figure 4B), if dNDUFS5 is required for converting the ~700 kDa assembly intermediate into the ~815 kDa assembly intermediate, then the extent of accumulation of the ~815 kDa assembly intermediate in either mhc>dNDUFS5RNAi,dNDUFS1RNAi and/or mhc>dNDUFS5RNAi,dNDUFV1RNAi thoraxes should be reduced relative to mhc>dNDUFS1RNAi and mhc>dNDUFV1RNAi respectively. In agreement with this proposition, we observed that the accumulation of the ~815 kDa assembly intermediate was significantly attenuated in mhc>dNDUFS5RNAi, dNDUFS1RNAi thoraxes relative to mhc>dNDUFS1RNAi thoraxes (Figure 5C). This was also accompanied by an accumulation of the ~700 kDa assembly intermediate (Figure 5C). Similar results were obtained by comparing mhc>dNDUFS5RNAi,dNDUFV1RNAi and mhc>dNDUFV1RNAi thoraxes (Figure 5C). Accordingly, we deduce from these results that when dNDUFS5 expression levels are impaired, the transient ~700-kDa assembly intermediate stalls and accumulates, impeding progression of CI biogenesis and ultimately resulting in a bottleneck of the ~550 kDa and ~315 kDa assembly intermediates as well.
To gain further insight into the identity of the ~700 kDa assembly intermediate, a single gel slice encompassing the region shown in Figure 5A was excised from native gels containing samples from wildtype and mhc>dNDUFS5RNAi thoraxes. Proteins from the gel slice were digested and analyzed by LC mass spectrometry; and a label-free spectral counting approach was used to generate a heat map for some of the proteins that showed altered expression levels between the samples. In agreement with our results showing a stalling and accumulation of the ~700 kDa assembly intermediate in this portion of the gel, we observed that several CI subunits were upregulated in the mhc>dNDUFS5RNAi sample relative to wildtype (Figure 5D). However, in stark contrast to the other CI subunits, we consistently observed (in 6 biological replicates taken at different time points of the day to control for circadian regulation) that dNDUFA10 was downregulated in the mhc>dNDUFS5RNAi sample; indicating that incorporation of dNDUFS5 into CI is necessary to stabilize or promote incorporation of dNDUFA10 into the complex (Figure 5D). In mammalian systems, at least five CI assembly factors – ECSIT, TMEM126B, NDUFAF1, ACAD9 and TIMMDC1 – are typically found associated with CI assembly intermediates, and have been dubbed the Mitochondrial Complex I Assembly (MCIA) complex (Guarani et al., 2014; Heide et al., 2012; Nouws et al., 2010; Vogel et al., 2007). We found four of these assembly factors (dECSIT, dNDUFAF1, dACAD9 and dTIMMDC1), associated with the 700 kDa assembly intermediate that were upregulated in the mhc>dNDUFS5RNAi samples, further confirming that it is a true assembly intermediate in CI biogenesis (Figure 5D and Table S4).
The Distal Portion Of The Membrane Arm Of CI Is Assembled Independently Of The Matrix Arm
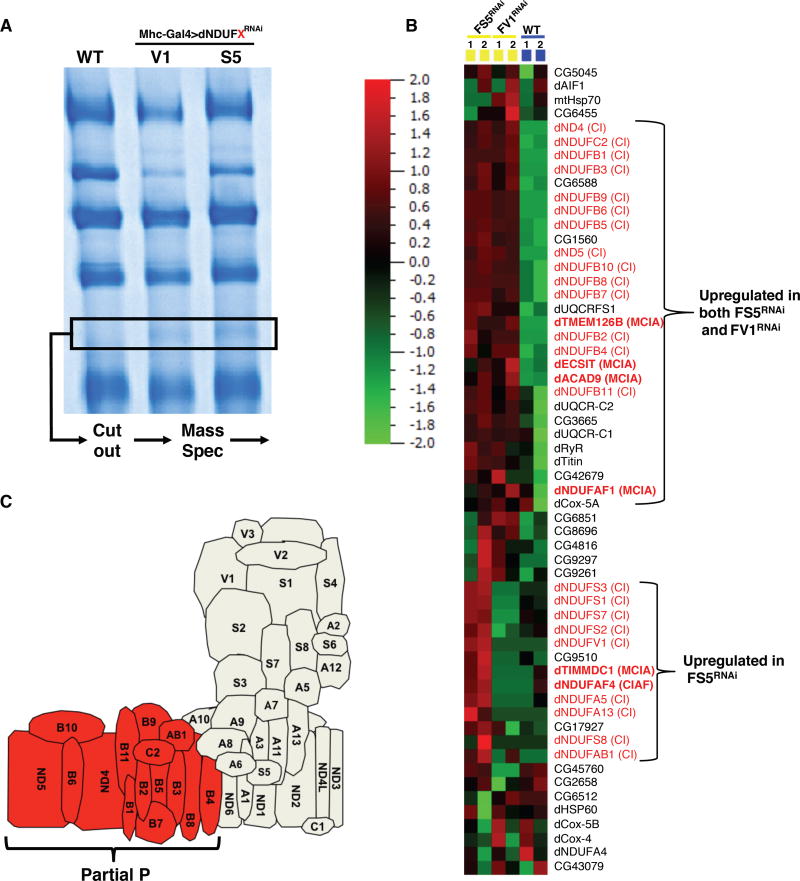
We noticed that in some instances where CI assembly was impaired, an additional band accumulated between the CIII and CIV bands in both the coomassie- and silver-stained gels (arrows in Figures 2A and B). A closer examination revealed that the accumulation of this intermediate was more readily evident in samples where subunits localized to the hydrophilic matrix domain were disrupted (i.e. the dNDUFS, dNDUFV and dNDUFA subunits) (Figure 1A). In line with our observations described in Figures 3, 4 and 5, we hypothesized that this band was likely another CI assembly intermediate that had stalled and accumulated as a result of a block in CI biogenesis. We decided to identify the constituents of this putative assembly intermediate via mass spectrometry.
We cut out the region of the gel corresponding to the stalled assembly intermediate in the wildtype, mhc>dNDUFS5RNAi and mhc>dNDUFV1RNAi thoraxes (Figure 6A), and used label-free quantification of peptides to ascertain which subunits and possibly assembly factors were altered between the two samples. Several components of the ETC machinery were downregulated; but there was a dramatic increase in CI subunits that are part of the distal membrane domain (i.e. all the dNDUFB subunits as well as dNDUFAB1, dNDUFC2, ND4 and ND5) (Figures 6B and 6C; Table S5). We note that there was no obvious accumulation of this assembly intermediate in blue native or silver-stained gels when any of these subunits (i.e. the dNDUFB subunits, or NDUFAB1 and NDUFC2 subunits) were disrupted (Figures 2A and B). Notably, many of these membrane-associated subunits were present in the corresponding gel slice from the wild-type samples (although at lower levels). All the components of the MCIA complex (i.e. dECSIT, dNDUFAF1, dACAD9, dTMEM126B and dTIMMDC1) were also found associated with this sub-complex.
Figure 6. CI Assembly In Drosophila Involves An Assembly Intermediate Containing Several Membrane-Associated Accessory Subunits.
(A) Mitochondrial protein complexes from wildtype, mhc>dNDUFS5RNAi and mhc>dNDUFV1RNAi thoraxes were separated by BN-PAGE and the region corresponding to the accumulated assembly intermediate (demarcated) was cut out, subjected to tryptic digestion, and analyzed by label-free quantitative LC-MS/ MS.
(B) Proteomic changes in the gel slice samples from wildtype, mhc>dNDUFS5RNAi and mhc>dNDUFV1RNAi thoraxes. Relative protein abundance among biological samples is expressed by spectral counts on a log scale. The color scale bar indicates the range of protein expression levels. See additional information in Table S5.
(C) Schematic representation highlighting the membrane subunits that are upregulated in the gel slice from the mhc>dNDUFS5RNAi and mhc>dNDUFV1RNAi thoraxes.
Proposed Model Of CI Assembly In Drosophila Muscle
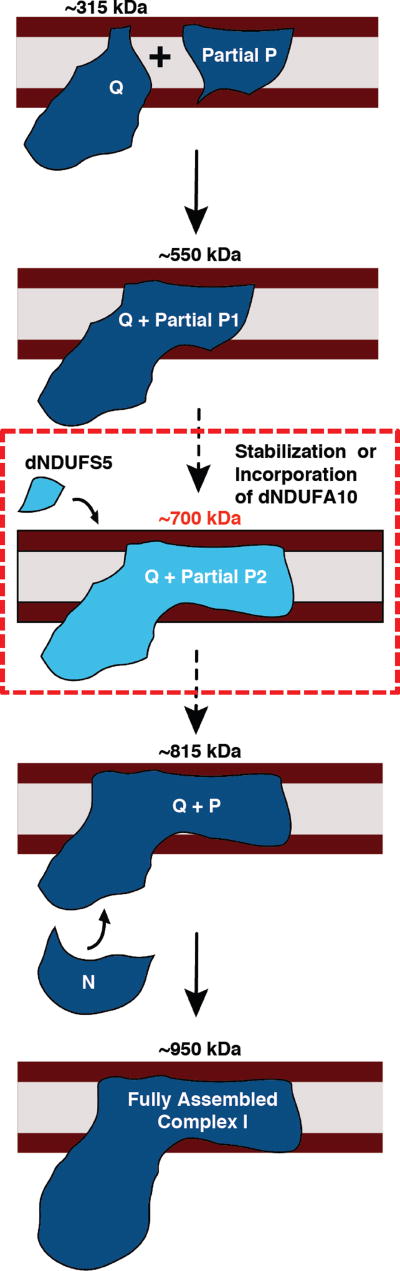
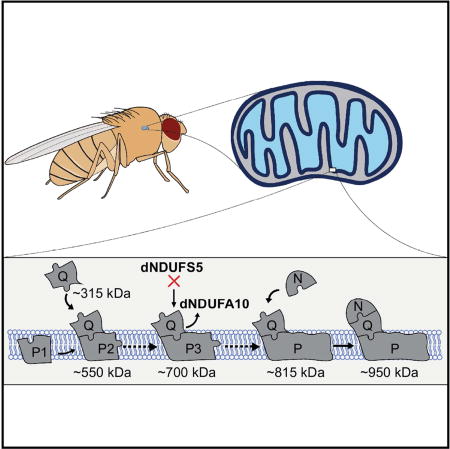
We propose a model for CI assembly in Drosophila flight muscles where dNDUFS2, dNDUFS3, dNDUFS7, dNDUFS8 and dNDUFA5 are combined in essentially one step to form the Q module, which is anchored to the membrane by dND1 (Figure 7). This assembly intermediate corresponds to the assembly intermediate in mammalian systems that was previously referred to as the ~400 kDa subcomplex, but has recently been re-estimated as the ~315 kDa subcomplex (Andrews et al., 2013; Vartak et al., 2014). This is consistent with the observation that assembly intermediates containing dNDUFS2, dNDUFS3, dNDUFS7, dNDUFS8 and dNDUFA5 co-migrate in blue native gels (Table S2); and that immunoblotting with both anti-ND1 and anti-NDUFS3 detect the ~315 kDa assembly intermediate (Figure 3C).
Figure 7.
An assembly intermediate consisting of dNDUFS2, dNDUS3, dNDUFS7, dNDUFS8 and dNDUFA5 are combined in essentially one step to form the Q module, which is anchored to the membrane by ND1. Subsequently, an independently-formed subcomplex comprising of membrane-associated subunits (Partial P1) is conjugated to the Q module, and possibly other subunits, to form an assembly intermediate comprised of the Q module and part of the P module (Q + Partial P2). This grows by the addition of more subunits to form a transient assembly intermediate of ~700kDa (Q + Partial P3). We propose that dNDUFS5 is then incorporated at this step, to promote incorporation or stabilization of dNDUFA10. Subsequently, the transient ~700 kDa assembly intermediate is rapidly converted to the ~815 kDa assembly intermediate, consisting of the complete P and Q modules (Q + P). Finally, the N module is added to produce the CI holoenzyme.
Subsequently, another assembly intermediate consisting of most of the subunits in the membrane domain is formed. This assembly intermediate comprises part of the P module (i.e. Partial P1), and is conjugated to the Q module to form an assembly intermediate that corresponds to the ~550 kDa (formerly ~650 kDa) assembly intermediate previously described in mammalian systems (Figure 7). Although proteomic analyses of the assembly intermediate that accumulates in mhc>dNDUFS5RNAi and mhc>dNDUFV1RNAi thoraxes shows that all the dNDUFB subunits as well as dNDUFC1, dNDUFAB1, ND4 and ND5 subunits are present in the subcomplex (see Table S5), it is unlikely that all the membrane subunits are incorporated into the complex at this stage under normal (wild-type) conditions. We hypothesize that the accumulation of the membrane accessory subunits in response to genetic disruption of the matrix subunits may be a compensatory mitochondrial stress signaling mechanism impinging on the nucleus, and resulting in a system that is poised to rapidly resume CI biogenesis if and when the missing matrix subunit becomes available. The accretion of the Partial P module under conditions where other components of the CI assembly machinery are impaired provides further evidence that the various modules of the complex (i.e. the Q, P and N modules) are assembled largely independently of each other in vivo.
The ~550 kDa assembly intermediate grows by the addition of more subunits to form a transient assembly intermediate of ~700 kDa (Figure 7); we postulate that dNDUFS5 is then incorporated at or just prior to this stage together with possibly dNDUFA10 to rapidly convert the ~700 kDa assembly intermediate to the ~815 kDa assembly intermediate, consisting of the complete P and Q modules (Figure 7). Finally, the N module is added to produce the CI holoenzyme (Figure 7).
DISCUSSION
We have exploited the genetic capabilities of Drosophila to uncover the mechanism of CI assembly in vivo, in Drosophila flight muscles. Our immunoblotting and proteomic analyses reveal that during CI assembly in Drosophila, the first membrane-bound major assembly intermediate that forms contains at least the following six subunits: dND1, dNDUFS2, dNDUFS3, dNDUFS7, dNDUFS8 and dNDUFA5. Based on its constituents and migration pattern in native PAGE, we conclude that this assembly intermediate is the same assembly intermediate traditionally referred to as the ~315 kDa assembly intermediate from studies on mammalian CI assembly; and corresponds to the Q module of CI (Andrews et al., 2013; Vartak et al., 2014). Consistent with their roles in regulating formation of the Q module, we found that genetic disruption of dNDUFS2, dNDUFS3, dNDUFA5, and dNDUFS7 attenuated the amount of the ~315 kDa assembly intermediate formed.
Unexpectedly, we found an ~700 kDa assembly intermediate that is short-lived (at least relative to the ~315, ~550 and ~815 kDa assembly intermediates), as it is rapidly converted into the ~815 kDa assembly intermediate. Importantly, our proteomic analyses revealed that incorporation of dNDUFS5 into CI around this stage is necessary to stabilize or promote incorporation of dNDUFA10 into the complex. Similar to the ~315-, ~550-, and ~815 kDa assembly intermediates, the ~700 kDa sub-complex is a true assembly intermediate as it can be detected in wild-type muscles as well. Additionally, components of the MCIA complex are associated with the ~700 kDa assembly intermediate, as has been reported for other assembly intermediates observed in mammalian systems. RNAi-mediated disruption of dNDUFS5 led to a stalling and accumulation of this otherwise transient assembly intermediate, to a point where it is readily detectable by western blots; most likely because this is the stage at or around which dNDUFS5 is incorporated into the complex.
It is possible that mutations in some accessory subunits will have both primary and secondary effects. As a case in point, dNDUFS5 disruption may first impair conversion of the ~700 kDa assembly intermediate to the ~815 kDa assembly intermediate, and consequently, impair CI assembly (as we have shown); but ultimately, the accumulation of the ~700 kDa assembly intermediate can activate the mitochondrial unfolded protein response as well as other stress signaling cascades with far-reaching consequences (Haynes et al., 2013; Jensen and Jasper, 2014; Owusu-Ansah and Banerjee, 2009; Owusu-Ansah et al., 2013; Owusu-Ansah et al., 2008). As another example, when dNDUFB3 was disrupted no specific assembly intermediates were stalled or disintegrated. Instead, there was a general reduction in the level of expression of all assembly intermediates. It is possible that disruption of dNDUFB3 activates stress signaling pathways that induce apoptosis or culminate in a general reduction of protein synthesis, leading to a reduction in CI assembly.
We find that at least 42 of the 44 human CI proteins are conserved in Drosophila. The two human CI proteins for which a clear ortholog was not readily identified in Drosophila by DIOPT are NDUFA3 (9 kDa) and NDUFC1 (6 kDa), which are two of the smallest subunits of the complex. Interestingly, obvious orthologs of NDUFC1 are not found in C. elegans or Yarrowia lipolytica; and the orthologs in vertebrates such as Zebrafish and Xenopus have very weak homology (DIOPT score of 1) to the human protein.
Therefore it is possible that this subunit has significant sequence diversion in Drosophila, and although present, was not recognized by DIOPT. For most of the CI subunits where multiple paralogs were identified by DIOPT (i.e. NDUFS2, NDUFS7, NDUFV2, NDUFA7 and NDUFB2), only one of the paralogs was detected as a bona fide CI subunit in flight muscles. However, as an exception to this general rule, two of the three paralogs of NDUFV1 were detected as part of CI in skeletal muscles via mass spectrometry. ND-51 (CG9140) appears to be the authentic ortholog of human NDUFV1 as it is highly expressed in skeletal muscles relative to ND-51L (CG11423), and is comparable in size to the human ortholog (both are about 51 kDa). ND-51L is a 77 kDa protein with a stretch of about 200 amino acids at the N-terminus that is not present in either the Drosophila paralog (ND-51) or human ortholog (NDUFV1). It remains to be determined whether the expression of the subunits with multiple paralogs are regulated in a tissue-specific manner to generate mitochondria with varied CI activities; or whether they are regulated in the same tissue in response to different environmental conditions to fine-tune the activity of CI.
In summary, we have described the mechanism of CI assembly in Drosophila flight muscles, and defined specific roles for some of the accessory subunits in CI assembly. Importantly, although CI dysfunction has been implicated in a large number of pathologies, we find that knocking down the expression of various antioxidant enzymes or mitochondrial protein quality control genes does not solely impair CI assembly, indicating that destabilization of CI may not be the sole underlying factor in many mitochondrial disorders (Figure S4). In addition, our proteomic analyses established that incorporation of dNDUFS5 into CI is necessary to stabilize or promote incorporation of dNDUFA10 into the complex. We note that our analyses of CI assembly in an in vivo setting, where CI biogenesis is subject to both developmental and environmental cues, revealed that many of the accessory subunits are required for both assembly and viability. Moreover, several NDUFB subunits (dNDUFB1, dNDUFB4, dNDUFB5, dNDUFB6 and dNDUFB10) seem to regulate the stability of the 315 kDa assembly intermediate, in apparent deviation from what will be expected from current models of mammalian CI assembly. However, the mechanism of CI biogenesis in Drosophila flight muscles is remarkably similar to what has been described in mammalian systems; and the differences observed here may be due to the fact that we have analyzed CI assembly in an in vivo setting. Accordingly, Drosophila is a suitable organism for addressing questions relevant to mammalian CI biogenesis. We anticipate that future studies using the full repertoire of genetic tools and resources in Drosophila should foster the discovery of paradigms for regulating CI assembly in humans.
EXPERIMENTAL PROCEDURES
Drosophila Strains and Genetics
For a list of stocks used and detailed experimental procedures, see the Extended Experimental Procedures described in the supplementary file
Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
BN-PAGE was performed using NativePAGE gels from Life Technologies, following the manufacturer’s instructions.
Silver Staining
Silver staining of native gels was performed with the SilverXpress staining kit from Life Technologies, following the manufacturer’s protocol.
In-gel Complex I Activity
Complex I activity in native gels was performed by incubating the native gels in 0.1 mg/ml NADH, 2.5 mg/ml Nitrotetrazolium Blue Chloride, 5 mM Tris-HCl (pH 7.4) at room temperature.
Immunoblotting
For immunoblotting of samples in native gels, protein complexes from native gels were transferred to PVDF membranes (BIO-RAD), and probed with the relevant antibodies using standard procedures.
Mass Spectrometry Analyses
After mass spectrometry with a Thermo Fusion Tribrid mass spectrometer, tandem mass spectra from raw files were searched against a Drosophila protein database using the Proteome Discoverer 1.4 software (Thermo Finnigan, San Jose, CA). The Proteome Discoverer application extracts relevant MS/MS spectra from the .raw file and determines the precursor charge state and the quality of the fragmentation spectrum. The Proteome Discoverer probability-based scoring system rates the relevance of the best matches found by the SEQUEST algorithm. The Drosophila protein database was downloaded as FASTA-formatted sequences from Uniprot protein database (database released in May, 2015). The peptide mass search tolerance was set to 10ppm. A minimum sequence length of 7 amino acids residues was required. Only fully tryptic peptides were considered. To calculate confidence levels and false positive rates (FDR), Proteome Discoverer generates a decoy database containing reverse sequences of the non-decoy protein database and performs the search against this concatenated database (non-decoy + decoy). Scaffold (Proteome Software) was used to visualize searched results. The discriminant score was set at less than 1% FDR determined based on the number of accepted decoy database peptides to generate protein lists for this study. Spectral counts were used for estimation of relative protein abundance between samples.
Supplementary Material
The constituents of each of the six major bands observed during BN-PAGE was analyzed by mass spectrometry. Peptides identified in each of bands A to F are shown in the table.
The table shows all the peptides identified in the 14 fractions shown in Figure 3D
The table shows the total number and identity of peptides found in the 700 kDa assembly intermediate.
The table shows the CI subunits and other proteins identified as part of the membrane arm subcomplex.
Acknowledgments
We thank members of the Owusu-Ansah lab for general discussions; Eric Schon, Estela Area-Gomez, Henry Colecraft, Barbara Corneo, Wes Grueber, Laura Johnston, Richard Kitsis, Andrew Marks, Norbert Perrimon, Martin Picard, Elizabeth Pon, Mimi Shirasu-Hiza and David Walker for fly stocks, reagents and critical discussions; and Stavroula Kousteni for critical discussions, and for providing skeletal and cardiac muscle samples from mice. We acknowledge the Bloomington Drosophila Stock Center, the National Institute of Genetics (Japan) and the Vienna Drosophila Resource Center for various fly strains. We appreciate Steven Shikhel’s technical assistance in dissecting cardiac and skeletal muscles from mice; and the technical assistance obtained from the Proteomics Shared Resource at the Herbert Irving Comprehensive Cancer Center, Columbia University Medical Center. This work was supported by an Institutional Cardiovascular Research Training Grant (T32 HL120826) to CG and JK; and a NIH R21 grant (DK112074), a pilot grant from the Robert N. Butler Columbia Aging Center, and institutional start-up funds from the Department of Physiology and Cellular Biophysics, Columbia University Medical Center to EO-A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
EO-A conceived the project, designed experiments and secured funding for the work. CG, JK, ECo, and EO-A performed all experiments, except mass spectrometry. ECh performed mass spectrometry. EO-A, CG, JK, and ECh analyzed and discussed results. EO-A wrote the manuscript with feedback from ECh CG, and JK
References
- Abdrakhmanova A, Zwicker K, Kerscher S, Zickermann V, Brandt U. Tight binding of NADPH to the 39-kDa subunit of complex I is not required for catalytic activity but stabilizes the multiprotein complex. Biochim Biophys Acta. 2006;1757:1676–1682. doi: 10.1016/j.bbabio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Andrews B, Carroll J, Ding S, Fearnley IM, Walker JE. Assembly factors for the membrane arm of human complex I. Proc Natl Acad Sci U S A. 2013;110:18934–18939. doi: 10.1073/pnas.1319247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landazuri MO, Enriquez JA. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Berger I, Hershkovitz E, Shaag A, Edvardson S, Saada A, Elpeleg O. Mitochondrial complex I deficiency caused by a deleterious NDUFA11 mutation. Ann Neurol. 2008;63:405–408. doi: 10.1002/ana.21332. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Budde SM, van den Heuvel LP, Janssen AJ, Smeets RJ, Buskens CA, DeMeirleir L, Van Coster R, Baethmann M, Voit T, Trijbels JM, et al. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem Biophys Res Commun. 2000;275:63–68. doi: 10.1006/bbrc.2000.3257. [DOI] [PubMed] [Google Scholar]
- Clason T, Ruiz T, Schagger H, Peng G, Zickermann V, Brandt U, Michel H, Radermacher M. The structure of eukaryotic and prokaryotic complex I. J Struct Biol. 2010;169:81–88. doi: 10.1016/j.jsb.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte M, Sousa R, Videira A. Inactivation of genes encoding subunits of the peripheral and membrane arms of neurospora mitochondrial complex I and effects on enzyme assembly. Genetics. 1995;139:1211–1221. doi: 10.1093/genetics/139.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarani V, Paulo J, Zhai B, Huttlin EL, Gygi SP, Harper JW. TIMMDC1/C3orf1 functions as a membrane-embedded mitochondrial complex I assembly factor through association with the MCIA complex. Mol Cell Biol. 2014;34:847–861. doi: 10.1128/MCB.01551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Castillo S, Baertling F, Kownatzki D, Wessels HJ, Arnold S, Brandt U, Nijtmans L. The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell Metab. 2017;25:128–139. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Fiorese CJ, Lin YF. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 2013;23:311–318. doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide H, Bleier L, Steger M, Ackermann J, Drose S, Schwamb B, Zornig M, Reichert AS, Koch I, Wittig I, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16:538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Hirst J. Mitochondrial complex I. Annu Rev Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- Hoefs SJ, Dieteren CE, Distelmaier F, Janssen RJ, Epplen A, Swarts HG, Forkink M, Rodenburg RJ, Nijtmans LG, Willems PH, et al. NDUFA2 complex I mutation leads to Leigh disease. American journal of human genetics. 2008;82:1306–1315. doi: 10.1016/j.ajhg.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefs SJ, van Spronsen FJ, Lenssen EW, Nijtmans LG, Rodenburg RJ, Smeitink JA, van den Heuvel LP. NDUFA10 mutations cause complex I deficiency in a patient with Leigh disease. European journal of human genetics : EJHG. 2011;19:270–274. doi: 10.1038/ejhg.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MB, Jasper H. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 2014;20:214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby DM, Salemi R, Sugiana C, Ohtake A, Parry L, Bell KM, Kirk EP, Boneh A, Taylor RW, Dahl HH, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. The Journal of clinical investigation. 2004;114:837–845. doi: 10.1172/JCI20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls U, Friedrich T, Schmiede A, Ohnishi T, Weiss H. Characterization of assembly intermediates of NADH:ubiquinone oxidoreductase (complex I) accumulated in Neurospora mitochondria by gene disruption. Journal of molecular biology. 1992;227:1032–1042. doi: 10.1016/0022-2836(92)90519-p. [DOI] [PubMed] [Google Scholar]
- Nouws J, Nijtmans L, Houten SM, van den Brand M, Huynen M, Venselaar H, Hoefs S, Gloerich J, Kronick J, Hutchin T, et al. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 2010;12:283–294. doi: 10.1016/j.cmet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Ostergaard E, Rodenburg RJ, van den Brand M, Thomsen LL, Duno M, Batbayli M, Wibrand F, Nijtmans L. Respiratory chain complex I deficiency due to NDUFA12 mutations as a new cause of Leigh syndrome. J Med Genet. 2011;48:737–740. doi: 10.1136/jmg.2011.088856. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Ruiz T, Clason T, Benjamin S, Brandt U, Zickermann V. The three-dimensional structure of complex I from Yarrowia lipolytica: a highly dynamic enzyme. J Struct Biol. 2006;154:269–279. doi: 10.1016/j.jsb.2006.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, VijayRaghavan K. Muscle pattern diversification in Drosophila: the story of imaginal myogenesis. Bioessays. 1999;21:486–498. doi: 10.1002/(SICI)1521-1878(199906)21:6<486::AID-BIES5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Scacco S, Petruzzella V, Budde S, Vergari R, Tamborra R, Panelli D, van den Heuvel LP, Smeitink JA, Papa S. Pathological mutations of the human NDUFS4 gene of the 18-kDa (AQDQ) subunit of complex I affect the expression of the protein and the assembly and function of the complex. The Journal of biological chemistry. 2003;278:44161–44167. doi: 10.1074/jbc.M307615200. [DOI] [PubMed] [Google Scholar]
- Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, Osellame LD, Stait T, Beilharz TH, Thorburn DR, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- Tuschen G, Sackmann U, Nehls U, Haiker H, Buse G, Weiss H. Assembly of NADH: ubiquinone reductase (complex I) in Neurospora mitochondria. Independent pathways of nuclear-encoded and mitochondrially encoded subunits. Journal of molecular biology. 1990;213:845–857. doi: 10.1016/S0022-2836(05)80268-2. [DOI] [PubMed] [Google Scholar]
- Vartak RS, Semwal MK, Bai Y. An update on complex I assembly: the assembly of players. J Bioenerg Biomembr. 2014;46:323–328. doi: 10.1007/s10863-014-9564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel RO, Janssen RJ, van den Brand MA, Dieteren CE, Verkaart S, Koopman WJ, Willems PH, Pluk W, van den Heuvel LP, Smeitink JA, et al. Cytosolic signaling protein Ecsit also localizes to mitochondria where it interacts with chaperone NDUFAF1 and functions in complex I assembly. Genes Dev. 2007;21:615–624. doi: 10.1101/gad.408407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickermann V, Wirth C, Nasiri H, Siegmund K, Schwalbe H, Hunte C, Brandt U. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The constituents of each of the six major bands observed during BN-PAGE was analyzed by mass spectrometry. Peptides identified in each of bands A to F are shown in the table.
The table shows all the peptides identified in the 14 fractions shown in Figure 3D
The table shows the total number and identity of peptides found in the 700 kDa assembly intermediate.
The table shows the CI subunits and other proteins identified as part of the membrane arm subcomplex.