ABSTRACT
The C-terminal domain (CTD) of the RNA polymerase II largest subunit consists of a unique repeated heptad sequence of the consensus Tyr1–Ser2–Pro3–Thr4–Ser5–Pro6–Ser7. An important function of the CTD is to couple transcription with RNA processing reactions that occur during the initiation, elongation, and termination phases of transcription. During this transcription cycle, the CTD is subject to extensive modification, primarily phosphorylation, on its non-proline residues. Reversible phosphorylation of Ser2 and Ser5 is well known to play important and general functions during transcription in all eukaryotes. More recent studies have enhanced our understanding of Tyr1, Thr4, and Ser7, and what have been previously characterized as unknown or specialized functions for these residues has changed to a more fine-detailed map of transcriptional regulation that highlights similarities as well as significant differences between organisms. Here, we review recent findings on the function and modification of these three residues, which further illustrate the importance of the CTD in precisely modulating gene expression.
KEYWORDS: kinases, phosphatases, phosphorylation, RNA processing, transcription
Introduction
RNA polymerase II (RNAP II) is responsible for transcription of all mRNAs as well as a large and seemingly growing number of non-coding (nc) transcripts. RNAP II is a highly conserved, multiprotein complex consisting of 12 subunits, the largest of which is Rpb1. The C-terminal domain (CTD) of Rpb1 is a unique protein domain consisting of a series of tandem heptad repeats of the consensus sequence Tyr–Ser–Pro–Thr–Ser–Pro–Ser (YSPTSPS), although the exact number of repeats and deviation from this sequence varies among organisms. All steps in the synthesis of a mature mRNA, or ncRNA, involve the CTD, as many different transcription and RNA processing factors associate with it in a dynamic manner throughout the transcription cycle. Modification of the CTD, principally phosphorylation, is important for many CTD functions. Phosphorylation occurs on all the non-proline residues, again in a dynamic manner with each residue/modification playing distinct roles. Two of these residues, Ser2 and Ser5, have garnered the most attention, and their functions are thus the best understood. Here, we highlight the residues Tyr1, Thr4, and Ser7, which also play significant but perhaps more specialized roles in RNAP II transcription and regulation. A number of excellent reviews have been published concerning CTD modification and function, and the reader is referred to these for a broader picture of this unusual and still in many ways enigmatic protein domain.1–5
The CTD has several features that allow for fine-tuned regulation of polymerase function. The length itself varies from 26 to 52 heptad repeats, with the exact number of repeats depending on the species; budding yeast has a 26 repeat CTD with little variation, while vertebrate species, including humans, have 52 repeats.6 These longer CTDs have more divergence from the consensus sequence in their more C-terminal repeats, whereas repeats closer to the N-terminus have less variation.1,7 Additionally, a 10-residue sequence is present at the C-terminus of vertebrate CTDs that helps stabilize it.8 The actual variation in CTD composition changes between species considerably, though. While CTDs from vertebrates show considerable similarity to each other, the CTD from Drosophila melanogaster displays much more variation, with only 3 of 45 repeats matching the consensus and with considerable divergence from a related organism, Aedes aegypti.2
The CTD can be modified in multiple ways, more so in metazoans than in yeasts. Besides phosphorylation, the threonine and serine residues can be glycosylated,9 and certain specific lysine residues can be ubiquitinated10 as well as methylated.11,12 These additional modifications occur primarily in metazoans, at least in part because Saccharomyces cerevisiae and Schizosaccharomyces pombe CTDs do not have the non-consensus lysine residues. Cis/trans isomerization of the prolines functions to alter binding of proteins associated with the CTD; the peptidylprolyl-cis/trans-isomerase Ess1 in yeast (Pin1 in mammals) performs this function by binding to a Ser5P CTD, and by changing CTD conformation it can alter CTD phosphorylation dynamics.13
Most early studies focused on the processes affected by Ser2 and Ser5. Two recent studies using mass spectrometry to analyze phosphorylation seem to vindicate this approach, as these residues were shown to be the most heavily modified, at least under the conditions analyzed.14,15 As “orphaned” residues, Tyr1, Thr4, and Ser7 have become more of a focus in recent years, with a growing body of work dedicated to determining their function. We review below studies on these thre residues and their place in modulating RNAP II function. Interesting themes that emerge are that these residues and their phosphorylation seem to affect more specialized genes and processes than Ser2/Ser5, and that significant differences in the function of these residues exist between species.
Serine-7
Ser7 was the first of the “orphan” residues to receive significant attention. A schematic highlighting important functions, and illustrating some of the major differences between mammalian cells and yeast, is presented in Fig. 1. Using the so-called “α-amanitin system,” in which endogenous RNAP II is inhibited by the drug so that exogenous α-amanitin-resistant mutant forms of Rpb1 can be analyzed,16 Egloff et al. found that a human Rpb1 derivative with all Ser7 residues mutated to Ala (S7A) was specifically defective in expression of certain small nuclear RNAs (snRNAs), in particular, the spliceosomal U1 and U2 snRNAs.17 Additionally, Ser7 phosphorylation was shown to facilitate interaction with Integrator, a 12 subunit complex that functions in 3′ cleavage of snRNAs.17,18 Further studies showed that before Integrator binds, Ser7P is necessary to recruit the Ser5 phosphatase RPAP2, which stably associates with Integrator after binding the CTD.19–21 Once RPAP2 is recruited, Ser5P is removed, leaving primarily a Ser7P/Ser2P CTD; Integrator subunit Int11 requires both Ser7P and Ser2P for efficient binding.19 The complexity of the proposed “CTD code”22 increased with better understanding of Ser7, as instead of general transcription mechanisms (as impacted by Ser2 and Ser5), the idea that a CTD residue can affect a specialized aspect of transcription, i.e., U snRNA 3′ end formation, came about.
Figure 1.
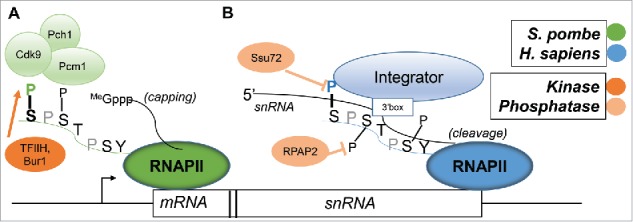
Ser7 facilitates mRNA capping in S. pombe and snRNA processing in H. sapiens. (A) In S. pombe, Cdk9 (with cyclin partner Pcm1) binds to CTD Ser7P, and enables the recruitment of 5′ cap methyltransferase Pcm1 to a Ser5P CTD. Ser7 is phosphorylated by TFIIH subunit Cdk7/Mcs6, as well as by Bur1. (B) In H. sapiens, the 12-subunit Integrator complex preferentially recognizes a Ser7P-Ser2P CTD at the 3′ end of snRNA genes. Ser7P recruits RPAP2 to the CTD, which dephosphorylates Ser5P, and Integrator is able to recognize both the Ser7P-Ser2P CTD as well as the 3′ box of the snRNA, enabling cleavage and processing of the snRNA. TFIIH subunit Cdk7 phosphorylates Ser7, while phosphatase Ssu72 has been shown to dephosphorylate Ser7P.
More recent studies have complicated somewhat the view of Ser7 in snRNA gene expression. Analysis of a chicken DT40 cell line expressing as the only source of Rpb1 a tetracycline (tet)-repressible derivative (tet-off) revealed that full viability in the presence of tet was restored by expression of an S7A derivative.23 Unexpectedly, U1 and U2 transcription and 3′ processing were completely normal in these cells, which contrasts with the results described above.17 While it is possible that the discrepancy reflects differences between chicken and humans, this seems unlikely due to the high conservation of all factors involved. Instead, the results of Egloff et al. may involve a synthetic effect between S7A and α-amanitin, as the inhibitor can lead to degradation of several proteins, including the elongation factor DSIF,24 known to be important for snRNA expression.25 However, other results of Hsin et al.23 do support, albeit indirectly, a role for Ser7 in snRNA 3′ processing. While DT40 cells expressing an Rpb1 variant with only repeats 1–26 was entirely viable and snRNA processing unaffected, a derivative with repeats 27–52 was inviable and showed a dramatic and specific decrease in snRNA 3′ processing. A parsimonious explanation for these results is that 20 of the 26 position 7 residues in the C-terminal half of the CTD are not Ser. Since many of these are non-conservative changes, e.g., Lys, perhaps this is sufficient to disrupt Integrator interaction and hence U snRNA 3′ processing.
The idea that CTD codes might be different for different organisms also arose from studies of Ser 7. For example, snRNA 3′ ends in S. cerevisiae are not formed by Integrator, which does not exist in yeast, but instead by the Nrd1–Nab3–Sen1 (NNS) complex, which primarily recognizes CTD Ser5P,26 and conversely the NNS does not appear to exist in vertebrate cells.27,28 Even within yeasts, there appear to be differences in Ser7 function. In S. pombe, CTD Ser7 has been associated with mRNA capping (traditionally associated with Ser5), as Ser7P is one of the components that enables efficient binding of 5′-cap methyltransferase Pcm1 to the CTD.29 Additionally, S7A mutation in S. pombe leads to reduced histone H3K4 and H3K36 trimethylation, and also exacerbates the effects of elongation factor Spt5 mutants.30 The same S7A mutation also led to de-repression of the PHO1 gene, while the phospho-mimetic S7E caused hyper-repression, which points to a role for Ser7 in phosphate homeostasis in fission yeast.31 However, in S. cerevisiae, no concrete function has yet been assigned to Ser7.
Despite the lack of a clear function, Ser7 is indeed phosphorylated in S. cerevisiae. Ser7P was found globally on transcribing RNAP II,32 and the TFIIH kinase Kin28/Cdk7 was shown to phosphorylate Ser7 in vitro and to be required in vivo, and across organisms.33,34 As this kinase has already been associated with not only Ser5 phosphorylation but also mRNA capping,35,36 that similar associations with Ser7 phosphorylation exist is perhaps not surprising. Additionally, the CTD kinase Bur1 also shows Ser7 phosphorylation activity distal to promoters of highly transcribed genes, consistent with the role of Bur1 in elongation.37 How Ser7 is dephosphorylated is less clear. Ser7P is not a target of Rtr1 (a Ser5 phosphatase, though there is some question about this; see38), but Ssu72 (another Ser5 phosphatase; see39) has been shown to have Ser7 dephosphorylation activity.40,41
In human cells, RNAP II is known to be the target of a different modification, glycosylation.42 The enzyme O-GlcNAc transferase (OGT) glycosylates both Ser5 and Ser7, and has been suggested to assist in proper preinitiation complex formation.43 Despite the ubiquitous nature of Ser7 phosphorylation across eukaryotes, glycosylation does not exist in yeast, and the primary findings from work on Ser7 highlight that the variation between organisms is significant and can have important implications for the function of these residues across species. Going forward, it will therefore be wise to consider carefully differences between organisms, and how some mechanisms in one species may be repurposed or removed in another.
Threonine-4
Differences between organisms’ use of the CTD became increasingly apparent as Thr4 was explored on the heels of Ser7. (Fig. 2 highlights the functions of Thr4 across species.) For example, in chicken DT40 cells, using the same approach as described above for analysis of Ser7, Hsin et al. found that Thr4 was both essential for viability and necessary for efficient 3′ end processing of non-polyadenylated replication-dependent histone transcripts, as processing (but not transcription) of these mRNAs, and recruitment of histone 3′ processing factors, was reduced in cells expressing a T4V CTD derivative.44 This immediately suggested a difference between yeast and vertebrates, as yeast histone mRNA 3′ ends are polyadenylated like all other mRNAs.45 Hsin et al.44 also compared levels of poly-(A)+ transcripts as well as U1 snRNA levels and found no changes (setting Thr4 apart from Ser2 or Ser7 function), bolstering the case for specialization. Extending this to mammalian cells, ChIPseq analyses showed an increase in Thr4P signal in the 3′ region of genes, and a T4A mutant showed a lethal phenotype (analogous to T4V in DT40 cells) as well as an apparent genome-wide elongation defect, as determined by elevated levels of promoter-proximal RNAP II and lower levels at 3′ ends of genes.46 It should be noted that Hsin et al.44 also observed small increases in promoter-proximal RNAP II on several genes. Whether the greater effects on elongation observed by Hintermair et al.46 were due to differences between chicken and human cells or to the use of the α-amanitin system as described above is not clear. Recently, a cell-cycle link was extended to Thr4, as Thr4-phosphorylated RNAP II was found to associate with centrosomes (across all cell-cycle phases save interphase) and the midbody in M phase HeLa cells.47
Figure 2.
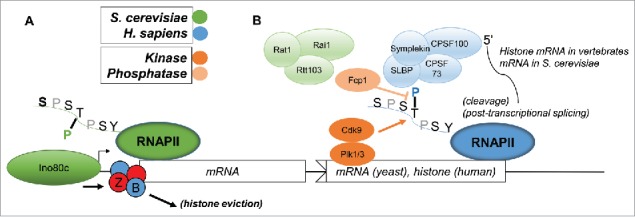
Divergent functions of Thr4 between S. cerevisiae and H. sapiens. (A) In S. cerevisiae, Thr4P has been shown to recruit the multi-subunit Ino80 chromatin remodeling complex to certain classes of promoters (see text), which evicts the promoter-proximal H2A.Z (red)/H2B (blue) histone dimers. Upon eviction of these dimers and replacement with H2A/H2B dimers, transcription of these genes is enabled. (B) In H. sapiens, histone mRNA synthesis requires Thr4 for efficient 3′ processing. Thr4 is required for recruitment of stem-loop binding protein (SLBP) and CPSF100, required along with other canonical 3′ processing factors such as Symplekin and CPSF73, to replication-dependent histone genes. In S. cerevisiae, histone mRNA 3′ ends are formed identically to all other mRNAs, and evidence shows that instead Thr4P plays a role in post-transcriptional splicing and Rtt103 (and Rat1/Rai1) recruitment during termination. In H. sapiens, Cdk9 and Plk1/Plk3 have been shown to phosphorylate Thr4, the later perhaps specifically in M phase, and Thr4P is dephosphorylated by Fcp1. Neither the kinase(s) nor phosphatase that acts on Thr4 is known in S. cerevisiae.
While Thr4 is essential for viability in vertebrates, this is not the case in S. cerevisiae. Substitutions with Ala are well-tolerated by yeast, even when paired with an S7A derivative.48 RNAP II with Thr4P was found globally on chromatin, suggesting a role during transcription, while distribution after the poly-(A) site was lower.49 Consistent with this, in a subsequent study Thr4 phosphorylated RNAP II was detected exclusively in the chromatin fraction, and properties of a T4V derivative linked Thr4 to chromatin remodeling and histone eviction from promoters of genes required for survival under low-phosphate conditions (PHO genes) and for galactose metabolism.50 Normally, the repressive histone dimer H2A.Z/H2B is evicted from promoters for such genes by chromatin remodeler INO80 during activation. In the T4V cells, this process was defective, and as a result the cells, which are fully viable in rich media, are inviable in phosphate-depleted or galactose-containing media. Schwer et al. found that expression of PHO genes was also dysregulated in S. pombe.51 These results together are again consistent with Thr4 being involved in relatively specific functions, which differ between yeast and mammalian cells.
More recent studies have however suggested broader roles for Thr4 in yeast, specifically in transcription termination and post-transcriptional splicing. Comparing Ser5P and Thr4P RNAP II interactomes, Harlen et al. found Thr4P devoid of spliceosomal components (associated with co-transcriptional splicing).52 Additional RNA analysis found that the T4V mutant cells partially retained introns in 90% of post-transcriptionally spliced genes, and a reanalysis of RNA-seq data of Rosonina et al.50 was consistent with a modest splicing defect. A proteomics analysis found that the known CTD-interacting protein Rtt103 (but not the exonuclease complex Rat1/Rai1, which function together in transcription termination) was found to associate with the Thr4P CTD. NET-seq analysis, which precisely maps the positions of elongating RNAP II, revealed an increase in RNAP II occupancy at poly(A) sites, where Rtt103 occupancy would be the highest, in a T4V mutant strain, similar to rtt103Δ cells. RNA-seq analysis replicated a global downstream shift of poly(A) site selection observed by Rosonina et al.,50 consistent with a reduction of 3′ end cleavage. More recent NET-seq experiments extended these findings to human cells, as Thr4P was found to correlate with termination in regions of protein-coding genes.53
The termination defect itself comes about when Rtt103 cannot effectively bind the CTD (such as in the T4V mutant), which prevents effective RNAP II disengagement from chromatin and continued transcription past the poly(A) site. An additional study using a T4A mutant found that this termination defect also affects a subset of snoRNA genes, and this was distinct from a known Ser2P requirement.54 Additionally, NMR studies revealed that the Rtt103 CTD interacting domain (CID) binds to a Thr4P CTD as well as a Ser2P CTD, but the presence of both together inhibits binding. 55 While no study suggests that Thr4 is necessary globally for transcription, there is some question as to how prevalent Thr4P is in yeast, with one estimate at only 2% of Ser2P levels14 and another suggesting it is as abundant as Ser2P.15
There are currently two candidates, not mutually exclusive, for the Thr4 kinase. Initially, it was reported that CDK9 was necessary for Thr4 phosphorylation in DT40 cells, as inhibiting CDK9/P-TEFb with DRB or flavopiridol inhibited Thr4 phosphorylation.44 Importantly, this did not reflect a requirement for Ser2P, as Thr4P was detected on an S2A derivative. Additionally, siRNA-mediated knockdown (KD) of CDK9 also decreased Thr4P in human 293 cells, and purified Cdk9 phosphorylated Thr4 in vitro.23 A role for CDK9 in Thr4 phosphorylation is consistent with earlier studies on histone 3′ end processing, since CDK9 KD resulted in increased RNAP II read-through of the natural 3′ end site to a downstream polyadenylation signal, resulting in an increase in poly-(A)+ histone mRNA.56 Other studies indicate that Polo-like kinases Plk3 and Plk1 are involved with Thr4 phosphorylation. Plk3, active throughout the cell cycle and playing additional roles in hypoxic stress response, although principally nucleolar,57 was found to phosphorylate Thr4 in vitro and KD in HeLa cells reduced Thr4P by 50%.46 Plk1, an M-phase specific kinase associated with RNAP II in centrosomes and the midbody, was also found to phosphorylate Thr4 in vitro, and mutation of Thr4 (T4A) inhibited M phase progression.47 It is certainly possible that both Cdk9 and Plk1/3 target Thr4 under different conditions, as multiple kinases seem to target all other CTD residues. Indeed, Ser5 is also targeted in M phase, with the help of Pin1, by Cdc2/CylinB.58 The identity of the Thr4 kinase(s) in S. cerevisiae, and whether Cdk9 or Plk homologues are involved, is currently unknown.
Fcp1, well known to dephosphorylate Ser2 at gene 3′ ends,59 also appears to be a Thr4 phosphatase. The enzyme was shown to dephosphorylate Thr4 in vitro, and Fcp1 KD in vivo increased Thr4P levels in 293 cells.23
Tyrosine-1
Determining the roles of Tyr1 phosphorylation has connected early successes with ongoing mystery. Much like Ser7 and Thr4, it was apparent very early on that significant differences between metazoan and yeast exist concerning Tyr1 (as illustrated in Fig. 3). Tyr1 was discovered to be phosphorylated in mammalian cells relatively quickly, but by a kinase, c-Abl, that has no known yeast equivalent.60,61 As to function, Tyr1P was found many years later to be present on RNAP II at transcriptional enhancers and promoters in human cells, albeit in the antisense direction.62 These findings are not limited to mammalian cells; upstream antisense RNAs (uaRNAs) were found to accumulate in DT40 cells when an Rpb1 containing Y1F substitutions was expressed.63 This increase in uaRNA accumulation was not due to increased transcription (RNAP II levels were in fact decreased) and thus appears to be due to increased stability of these naturally unstable RNAs. As in human cells, Tyr1P levels on upstream regions affected by the Y1F CTD, but not on the corresponding downstream sense genes, were elevated. From these studies, it appears that “antisense” RNAP II is excessively Tyr1 phosphorylated, and this in some way facilitates turnover of uaRNAs. Both of these studies, which used either the α-amanitin62 or tet-off63 system, found that Y1F substitutions were lethal following inhibition of the endogenous Rpb1.
Figure 3.
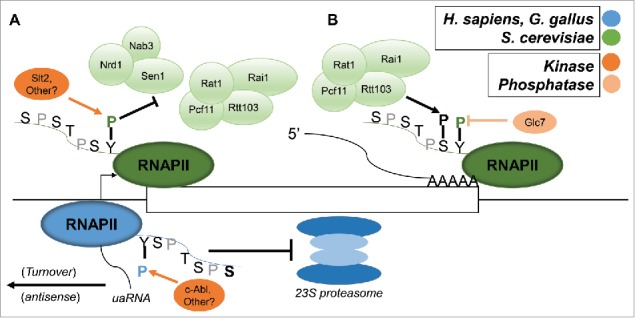
Tyr1 has multiple functions across species. (A) At the 5′ ends of genes, Tyr1 in vertebrates is phosphorylated, likely by c-Abl, and Tyr1P is important for efficient turnover of 5′ upstream antisense RNAs (uaRNAs). Tyr1 phosphorylation also enhances stability and prevents degradation of the unstructured CTD by the 23S proteasome in vertebrate cells. In S. cerevisiae, this function has not been described, and Tyr1 phosphorylation (by Slt2 or other unidentified kinases) is associated with antitermination, as it prevents efficient binding of Nrd1 (in yeast) or Rtt103 (yeast and humans) to a Ser5P or Ser2P CTD, respectively. (B) At 3′ ends of genes, CTD Tyr1P is dephosphorylated by Glc7, a subunit of cleavage and polyadenylation factor (CPF). This leads to Ser2P-CTD recognition by Pcf11 and Rtt103/Rat1/Rai1, which facilitates efficient termination. This function appears conserved from yeast to human.
Unexpectedly, Tyr1P also plays an important role in maintaining CTD stability in vertebrate cells. A Y1F-derivative expressed in DT40 cells was shown to be present at lower levels compared with a WT derivative, and a low molecular weight Rpb1 form lacking the CTD was observed.63 Remarkably, CTD stability (but not cell viability) could be completely restored by a single Tyr residue in the final heptad. Furthermore, in vitro assays showed that the CTD of a GST-CTD derivative could be completely degraded by purified 23S proteasomes, but a derivative Tyr1 phosphorylated by c-Abl was stable, implicating Tyr1 phosphorylation in CTD stability. Perhaps in keeping with this function Tyr1P was the only CTD phosphorylation found on cytoplasmic/nucleoplasmic (as opposed to chromatin-associated) RNAP II. Related results were observed in human cells, as a truncated Rpb1 apparently lacking the CTD was observed in cells expressing a Y1F derivative.62
The hunt for Tyr1 function in yeast cells has been more challenging. A truncated Y1F Rpb1 derivative (12 repeats) was shown to be lethal in S. cerevisiae,64 but more recent experiments found that a similar truncated derivative was viable with only minor growth defects in S. pombe.65 In contrast, in S. cerevisiae, a full-length Y1F Rpb1 was viable, but displayed a severe slow growth phenotype.50,66 The reason(s) for these discrepancies is unclear, but suggest differences not only between yeast and vertebrates but also between budding and fission yeasts themselves. Also at play may be differences in the lengths/composition of the CTDs analyzed, which could lead to synthetic effects.
Several studies have begun to provide insight into the precise functions of Tyr1 and its phosphorylation in yeast. The pattern of Tyr1P globally was shown to resemble that of Ser2P, with the minor difference of a reduction in Tyr1P before the decrease in Ser2P at gene 3′ ends.49 By measuring binding affinities of several termination factor CIDs to CTD diheptad phosphopeptides, it was suggested that Tyr1P impairs termination factor recruitment to the CTD, specifically preventing Nrd1 association after promoter clearance and enabling Pcf11 and Rtt103 to bind to Ser2P-CTD after the drop in Tyr1P at 3′ ends.49 CIDs from all three of these proteins were shown to bind Ser 2P (or Ser5P, for Nrd1) CTD peptides but not Tyr1P-containing versions. Rtt103 in fact requires an intact and unmodified Tyr residue, since it can only bind efficiently to Ser2P-CTD peptides if a specific Asn residue can form a hydrogen bond with the hydroxyl group of Tyr1.67 Extending these results, Y1F mutation was found to disrupt Rtt103 binding to the CTD and impair Nrd1 recruitment to chromatin.66 Aside from the Nrd1/NNS complex, the presence of Tyr1P during elongation and near gene 3′ ends also promotes association of elongation factor Spt6, which has an SH2 domain that specifically recognizes Tyr1P and serves as a histone chaperone, with the CTD, thus linking the Tyr1 with elongation-promoting histone modifications.68 While not all of these findings can extend to vertebrate cells, since the NNS complex does not appear to exist outside of yeast, some overlap with Rtt103 (in vertebrates, Kub5-Hera) and Spt6 exists, though exactly how much is still in question.
The kinase(s) and phosphatase(s) responsible for modifying Tyr1 differ between yeast and vertebrate organisms. The protein kinase c-Abl was identified many years ago as a likely Tyr1 kinase in mammalian cells.60,61 c-Abl, a known nuclear tyrosine kinase, was shown to phosphorylate RNAP II Tyr1 efficiently in vitro. It contains both an SH2 domain that precedes its catalytic domain and is required for catalytic domain activation and efficient, near stoichiometric phosphorylation, and also a CTD-interacting domain specific for RNAP II. A c-Abl-related kinase, Arg, was also shown capable of phosphorylating Tyr1 in vitro.69 The precise roles of these kinases in vivo, and whether other Tyr1 kinases exist in mammalian cells, remains to be determined.
Determining the identity of the Tyr1 kinase(s) in yeast has been more challenging. This reflects in part the fact that that Tyr phosphorylation in budding yeast is very rare, constituting <0.1% total phosphorylation.70,71 Furthermore, there is no yeast homolog of c-Abl, and the only known SH2 domain-containing protein is, interestingly, the Tyr1P-interacting protein Spt6.72 Recent work though has identified the MAP kinase Slt2/Mpk173 as a Tyr1 kinase in S. cerevisiae.66 Slt2 phosphorylates Tyr1 in vitro, and in vivo Slt2 is necessary for full Tyr1 phosphorylation and also modulates Tyr1P levels during stress responses, which were found to rise following for example cell wall stress and heat shock. Interestingly, Slt2 also regulates other aspects of the transcription machinery, such as the Mediator kinase/cyclin pair Srb10/11 (by phosphorylating Srb11),74 which is retained on chromatin in cells expressing the Y1F derivative.66
Glc7 appears to be the principal Tyr1 phosphatase in yeast. Glc7 was initially identified as a Ser–Thr phosphatase and plays several roles, for example, as a subunit of the cleavage and polyadenylation factor (CPF) complex75 and as a required factor in the cell wall integrity pathway.76 Schreieck et al. found that Glc7 dephosphorylates Tyr1P in vitro and, as a component of CPF, is required for Tyr1P dephosphorylation at the polyadenylation site, for recruitment of termination factors Pcf11 and Rtt103, and for termination.77 These findings are consistent with other results that link both NNS (Nrd1) and Rtt103-dependent termination with control of Tyr1 phosphorylation.49,66 Another potential candidate for Tyr1 dephosphorylation, Ssu72, is also a subunit of the CPF complex,78 and has structural homology with a class of protein tyrosine phosphatases termed low molecular weight PTPs.79 However, Ssu72 does not show Tyr phosphatase activity in vitro, preferring to dephosphorylate Ser5.80
Concluding remarks
With the advent of new tools and strategies to explore the different functions of the CTD, more of the gaps in our knowledge surrounding how the CTD is modified over the course of transcription and how it functions are being filled in. But differences between metazoans, or even between similar species such as budding and fission yeasts, have become apparent and raise more questions. For example, what is the significance of the involvement of the INO80 complex/Htz1 and Rtt103 for Thr4 phosphorylation beyond budding yeast? The INO80 complex is evolutionarily conserved81 and the Rtt103 homolog Kub5-Hera is well-studied,82 but how their functions impact RNAP II activity vary, and not all of these functions are conserved between organisms. A stronger example of this divergence is evident with Tyr1. Beyond affecting termination factor recruitment, what aspects of Tyr1 phosphorylation and Slt2 function are transferable to mammalian cells? Evidence that Slt2 is present with RNAP II on transcribed genes and homologs Erk1/2 are recruited to genes in a manner similar to Slt283,84 has been reported, but might one of these kinases, or a related one, have Tyr1P activity? Future studies will continue to describe the many intricacies of the CTD and discover new aspects of regulation, but now that the “orphans” are orphans no longer, continued comparison across organisms will prove enlightening.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Chong Han Ng for critical reading of this manuscript.
Funding
Work from the authors’ laboratory is supported by NIH grant R35 GM118136 to JLM, and NMY was partially supported by an NIH training grant 5T32GM008798.
References
- [1].Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 2012; 26:2119–2137; PMID:23028141; https://doi.org/10.1101/gad.200303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Corden JL. RNA polymerase II C-Terminal domain: tethering transcription to transcript and template. Chem Rev 2013; 113:8423–8455; PMID:24040939; https://doi.org/10.1021/cr400158h [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 2013; 1829:55–62; PMID:22982363; https://doi.org/10.1016/j.bbagrm.2012.08.013 [DOI] [PubMed] [Google Scholar]
- [4].Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 2014; 15:163–175; PMID:24514444; https://doi.org/10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Conaway RC, Conaway JW. Orchestrating transcription with the pol II CTD. Nat Rev Mol Cell Biol 2015; 16:128; PMID:25693123; https://doi.org/10.1038/nrm3956 [DOI] [PubMed] [Google Scholar]
- [6].Corden JL. Tails of RNA polymerase II. Trends Biochem Sci 1990; 15:383–387; PMID:2251729; https://doi.org/10.1016/0968-0004(90)90236-5 [DOI] [PubMed] [Google Scholar]
- [7].Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol Biol Evol 2010; 27:2628–2641; PMID:20558594; https://doi.org/10.1093/molbev/msq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chapman RD, Palancade B, Lang A, Bendaude O, Eick D. The last CTD repeat of the mammalian RNA polymerase II large subunit is important for its stability. Nucleic Acids Res 2004; 32:35–44; PMID:14704341; https://doi.org/10.1093/nar/gkh172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J Cell Sci 2010; 123:13–22; PMID:20016062; https://doi.org/10.1242/jcs.053678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li H, Zhang Z, Wang B, Zhang J, Zhao Y, Jin Y. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol Cell Bio 2007; 27:5296–5305; https://doi.org/10.1128/MCB.01667-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sims RJ 3rd, Rojas LA, Beck D, Bonasio R, Schüller R, Drury WJ 3rd, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 2011; 332:99–103; PMID:21454787; https://doi.org/10.1126/science.1202663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dias JD, Rito T, Triglia ET, Kukalev A, Ferrai C, Chotalia M, Brookes E, Kimura H, Pombo A. Methylation of RNA polymerase II non-consensus lysine residues marks early transcription in mammalian cells. eLife 2015; 4:e11215; PMID:26687004; https://doi.org/10.7554/eLife.11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hanes SD. Prolyl isomerases in gene transcription. Biochim Biophys Acta 2015; 1850:2017–2034; PMID:25450176; https://doi.org/10.1016/j.bbagen.2014.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Suh H, Ficarro SB, Kang U, Chun Y, Marto JA, Buratowski S. Direct analysis of phosphorylation sites on the Rpb1 C-terminal domain of RNA polymerase II. Mol Cell 2016; 61:297–304; PMID:26799764; https://doi.org/10.1016/j.molcel.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schüller R, Forné I, Straub T, Schreieck A, Texier Y, Shah N, Decker TM, Cramer P, Imhof A, Eick D. Heptad-specific phosphorylation of RNA polymerase II CTD. Mol Cell 2016; 61:305–314; PMID:26799765; https://doi.org/10.1016/j.molcel.2015.12.003 [DOI] [PubMed] [Google Scholar]
- [16].Gerber HP, Hagmann M, Seipel K, Georgiev O, West MAL, Litingtung Y, Schaffner W, Corden JL. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 1995; 374:660–662; PMID:7715709; https://doi.org/10.1038/374660a0 [DOI] [PubMed] [Google Scholar]
- [17].Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 2007; 318:1777–1779; PMID:18079403; https://doi.org/10.1126/science.1145989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005; 123:265–276; PMID:16239144; https://doi.org/10.1016/j.cell.2005.08.019 [DOI] [PubMed] [Google Scholar]
- [19].Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem 2010; 285:20564–20569; PMID:20457598; https://doi.org/10.1074/jbc.M110.132530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 2012; 45:111–122; PMID:22137580; https://doi.org/10.1016/j.molcel.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Egloff S. Role of Ser7 phosphorylation of the CTD during transcription of snRNA genes. RNA Biol 2012; 9:1033–1038; PMID:22858677; https://doi.org/10.4161/rna.21166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Buratowski S. The CTD code. Nat Struct Biol 2003; 10:679–680; PMID:12942140; https://doi.org/10.1038/nsb0903-679 [DOI] [PubMed] [Google Scholar]
- [23].Hsin JP, Xiang K, Manley JL. Function and control of RNA polymerase II C-terminal domain phosphorylation in vertebrate transcription and RNA processing. Mol Cell Bio 2014; 34:2488–2498; https://doi.org/10.1128/MCB.00181-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tsao DC, Park NJ, Nag A, Martinson HG. Prolonged α-amanitin treatment of cells for studying mutated polymerases causes degradation of DSIF160 and other proteins. RNA 2012; 18:222–229; PMID:22194310; https://doi.org/10.1261/rna.030452.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci USA 2004; 101:7572–7577; PMID:15136722; https://doi.org/10.1073/pnas.0401493101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 2008; 15:795–804; PMID:18660819; https://doi.org/10.1038/nsmb.1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet 2012; 28:333–341; PMID:22622228; https://doi.org/10.1016/j.tig.2012.03.007 [DOI] [PubMed] [Google Scholar]
- [28].O'Reilly D, Kuznetsova OV, Laitem C, Zaborowska J, Dienstbier M, Murphy S. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res 2014; 42:264–275; PMID:24097444; https://doi.org/10.1093/nar/gkt892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].St. Amour CV, Sansó M, Bösken CA, Lee KM, Larochelle S, Zhang C, Shokat KM, Geyer M, Fisher RP. Separate domains of fission yeast Cdk9 (P-TEFb) are required for capping enzyme recruitment and primed (Ser7-phosphorylated) Rpb1 carboxyl-terminal domain substrate recognition. Mol Cell Biol 2012; 32:2372–2383; PMID:22508988; https://doi.org/10.1128/MCB.06657-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mbogning J, Pagé V, Burston J, Schwenger E, Fisher RP, Schwer B, Shuman S, Tanny JC. Functional interaction of Rpb1 and Spt5 C-terminal domains in co-transcriptional histone modification. Nucleic Acids Res 2015; 43:9766–9775; PMID:26275777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schwer B, Sanchez AM, Shuman S. RNA polymerase II CTD phospho-sites Ser5 and Ser7 govern phosphate homeostasis in fission yeast. RNA 2015; 21:1770–1780; PMID:26264592; https://doi.org/10.1261/rna.052555.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 2007; 318:1780–1782; PMID:18079404; https://doi.org/10.1126/science.1145977 [DOI] [PubMed] [Google Scholar]
- [33].Kim M, Suh H, Cho EJ, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. JBC 2009; 284:26421–26426; https://doi.org/10.1074/jbc.M109.028993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Akhtar MS, Heidemann M, Tietjen J, Zhang D, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxyl-terminal domain of RNA polymerase II. Mol Cell 2009; 34:387–393; PMID:19450536; https://doi.org/10.1016/j.molcel.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 2000; 14:2452–2460; PMID:11018013; https://doi.org/10.1101/gad.824700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 2000; 14:2435–2440; PMID:11018011; https://doi.org/10.1101/gad.836300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tietjen JR, Zhang DW, Rodríguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. . Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 2010; 17:1154–1161; PMID:20802488; https://doi.org/10.1038/nsmb.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xiang K, Manley JL, Tong L. The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat Comm 2012; 3:946; https://doi.org/10.1038/ncomms1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 is an RNA polymerase II CTD phosphatase. Mol Cell 2004; 14:387–394; PMID:15125841; https://doi.org/10.1016/S1097-2765(04)00235-7 [DOI] [PubMed] [Google Scholar]
- [40].Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell 2009; 34:168–178; PMID:19394294; https://doi.org/10.1016/j.molcel.2009.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang DW, Mosley AL, Ramisetty SR, Rodríguez-Molina JB, Washburn MP, Ansari AZ. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem 2012; 287:8541–8551; PMID:22235117; https://doi.org/10.1074/jbc.M111.335687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem 1993; 268:10416–10424; PMID:8486697 [PubMed] [Google Scholar]
- [43].Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, Lewis BA. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J Biol Chem 2012; 287:23549–23561; PMID:22605332; https://doi.org/10.1074/jbc.M111.330910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 2011; 334:683–686; PMID:22053051; https://doi.org/10.1126/science.1206034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 2008; 9:843–854; PMID:18927579; https://doi.org/10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, et al. . Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J 2012; 31:2784–2797; PMID:22549466; https://doi.org/10.1038/emboj.2012.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hintermair C, Voss K, Forné I, Heidemann M, Flatley A, Kremmer E, Imhof A, Eick D. Specific threonine-4 phosphorylation and function of RNA polymerase II CTD during M phase progression. Sci Rep 2016; 6:27401; PMID:27264542; https://doi.org/10.1038/srep27401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stiller JW, Mcconaughy BL, Hall BD. Evolutionary complementation for polymerase II CTD function. Yeast 2000; 16:57–64; PMID:10620775; https://doi.org/10.1002/(SICI)1097-0061(20000115)16:1<57::AID-YEA509>3.0.CO;2-E [DOI] [PubMed] [Google Scholar]
- [49].Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012; 336:1723–1725; PMID:22745433; https://doi.org/10.1126/science.1219651 [DOI] [PubMed] [Google Scholar]
- [50].Rosonina E, Yurko N, Li W, Hoque M, Tian B, Manley JL. Threonine-4 of the budding yeast RNAP II CTD couples transcription with Htz1-mediated chromatin remodeling. Proc Natl Acad Sci USA 2014; 111:11924–11931; PMID:25071213; https://doi.org/10.1073/pnas.1412802111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Schwer B, Bitton DA, Sanchez AM, Bähler J, Shuman S. Individual letters of the RNA polymerase II CTD code govern distinct gene expression programs in fission yeast. Proc Natl Acad Sci USA 2014; 111:4185–4190; PMID:24591591; https://doi.org/10.1073/pnas.1321842111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Harlen KM, Trotta KL, Smith EE, Mosaheb MM, Fuchs SM, Churchman LS. Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. Cell Rep 2016; 15:2147–2158; PMID:27239037; https://doi.org/10.1016/j.celrep.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell 2017; 65:25–38; PMID:28017589; https://doi.org/10.1016/j.molcel.2016.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nemec CM, Yang F, Gilmore JM, Hintermair C, Ho YH, Tseng SC, Heidemann M, Zhang Y, Florens L, Gasch AP, et al. . Different phosphoisoforms of RNA polymerase II engage the Rtt103 termination factor in a structurally analogous manner. Proc Natl Acad Sci USA 2017; 114:E3944–E3953; PMID:28465432; https://doi.org/10.1073/pnas.1700128114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Jasnovidova O, Krejcikova M, Kubicek K, Stefl R. Structural insight into recognition of phosphorylated threonine-4 of RNA polymerase II C-terminal domain by Rtt103p. EMBO Rep 2017; 18(6):906–913; PMID:28468956; https://doi.org/10.15252/embr.201643723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep 2009; 10:894–900; PMID:19575011; https://doi.org/10.1038/embor.2009.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zimmerman WC, Erikson RL. Finding Plk3. Cell Cycle 2007; 6:1314–1318; PMID:17568195; https://doi.org/10.4161/cc.6.11.4275 [DOI] [PubMed] [Google Scholar]
- [58].Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev 2003; 17:2765–2776; PMID:14600023; https://doi.org/10.1101/gad.1135503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 2001; 15:3319–3329; PMID:11751637; https://doi.org/10.1101/gad.935901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Baskaran R, Dahmus ME, Wang JY. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci USA 1993; 90:11167–11171; PMID:7504297; https://doi.org/10.1073/pnas.90.23.11167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Baskaran R, Escobar SR, Wang JY. Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine (CTD)-tyrosine kinase-specific for the mammalian RNA polymerase II: possible role in transcription elongation. Cell Growth Differ 1999; 10:387–396; PMID:10392900 [PubMed] [Google Scholar]
- [62].Descostes N, Heidemann M, Spinelli L, Schüller R, Maqbool MA, Fenouil R, Koch F, Innocenti C, Gut M, Gut I, et al. . Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. eLife 2014; 3:e02105; PMID:24842994; https://doi.org/10.7554/eLife.02105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hsin JP, Li W, Hoque M, Tian B, Manley JL. RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. eLife 2014; 3:e02112; PMID:24842995; https://doi.org/10.7554/eLife.02112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 1995; 140:1223–1233; PMID:7498765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell 2011; 43:311–318; PMID:21684186; https://doi.org/10.1016/j.molcel.2011.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yurko NY, Liu X, Hoque M, Tian B, Manley JL. MPK1/SLT2 links multiple stress responses with gene expression in budding yeast by phosphorylating Tyr1 of the RNAP II CTD. Submitted for publication 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol 2010; 17:1195–1201; PMID:20818393; https://doi.org/10.1038/nsmb.1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Burugula BB, Jeronimo C, Pathak R, Jones JW, Robert F, Govind CK. Histone deacetylases and phosphorylated polymerase II C-terminal domain recruit Spt6 for cotranscriptional histone reassembly. Mol Cell Biol 2014; 34:4115–4129; PMID:25182531; https://doi.org/10.1128/MCB.00695-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Baskaran R, Chiang GG, Mysliwiec T, Kruch GD, Wang JYJ. Tyrosine phosphorylation of RNA polymerase II carboxyl-terminal domain by the Abl-related gene product. J Biol Chem 1997; 272:18905–18909; PMID:9228069; https://doi.org/10.1074/jbc.272.30.18905 [DOI] [PubMed] [Google Scholar]
- [70].Modesti A, Bini L, Carraresi L, Magherini F, Liberatori S, Pallini V, Manao G, Pinna LA, Raugei G, Ramponi G. Expression of the small tyrosine phosphatase (Stp1) in Saccharomyces cerevisiae: a study on protein tyrosine phosphorylation. Electrophoresis 2001; 22:576–585; PMID:11258771; https://doi.org/10.1002/1522-2683(200102)22:3<576::AID-ELPS576>3.0.CO;2-P [DOI] [PubMed] [Google Scholar]
- [71].Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA 2007; 104:2193–2198; PMID:17287358; https://doi.org/10.1073/pnas.0607084104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Diebold ML, Loeliger E, Koch M, Winston F, Cavarelli J, Romier C. Noncanonical Tandem SH2 enables interaction of elongation factor Spt6 with RNA polymerase II. J Biol Chem 2010; 285:38389–38398; PMID:20926373; https://doi.org/10.1074/jbc.M110.146696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pearson G, Robinson F, Gibson TB, Xu B, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 2011; 22:153–183; PMID:11294822; https://doi.org/10.1210/edrv.22.2.0428 [DOI] [PubMed] [Google Scholar]
- [74].Strich R, Cooper KF. The dual role of cyclin C connects stress regulated gene expression to mitochondrial dynamics. Microb Cell 2014; 1:318–324; PMID:28357211; https://doi.org/10.15698/mic2014.10.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem 2003; 278:33000–33010; PMID:12819204; https://doi.org/10.1074/jbc.M304454200 [DOI] [PubMed] [Google Scholar]
- [76].Andrews PD, Stark MJ. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J Cell Sci 2000; 113:507–520. PMID:10639337 [DOI] [PubMed] [Google Scholar]
- [77].Schreieck A, Easter AD, Etzold S, Wiederhold K, Lidschreiber M, Cramer P, Passmore LA. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat Struct Mol Biol 2014; 21:175–179; PMID:24413056; https://doi.org/10.1038/nsmb.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].He X, Khan AU, Cheng H, Pappas DL Jr, Hampsey M, Moore CL. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev 2003; 17:1030–1042; PMID:12704082; https://doi.org/10.1101/gad.1075203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Meinhart A, Silberzahn T, Cramer P. The mRNA transcription/processing factor Ssu72 is a potential tyrosine phosphatase. J Biol Chem 2003; 278:15917–15921; PMID:12606538; https://doi.org/10.1074/jbc.M301643200 [DOI] [PubMed] [Google Scholar]
- [80].Xiang K, Manley JL, Tong L. An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev 2012; 26:2265–2270; PMID:23070812; https://doi.org/10.1101/gad.198853.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chen L, Cai Y, Jin J, Florens L, Swanson SK, Washburn MP, Conaway JW, Conaway RC. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J Biol Chem 2011; 286:11283–11289; PMID:21303910; https://doi.org/10.1074/jbc.M111.222505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Morales JC, Richard P, Rommel A, Fattah FJ, Motea EA, Patidar PL, Xiao L, Leskov K, Wu SY, Hittelman WN, et al. . Kub5-Hera, the human Rtt103 homolog, plays dual functional roles in transcription termination and DNA repair. Nucleic Acids Res 2014; 42:4996–5006; PMID:24589584; https://doi.org/10.1093/nar/gku160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kim KY, Levin DE. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell 2011; 144:745–756; PMID:21376235; https://doi.org/10.1016/j.cell.2011.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mikula M, Skrzypczak M, Goryca K, Paczkowska K, Ledwon JK, Statkiewicz M, Kulecka M, Grzelak M, Dabrowska M, Kuklinska U, et al. . Genome-wide co-localization of active EGFR and downstream ERK pathway kinases mirrors mitogen-inducible RNA polymerase 2 genomic occupancy. Nucleic Acids Res 2016; 44:10150–10164; PMID:27587583; https://doi.org/10.1093/nar/gkw763 [DOI] [PMC free article] [PubMed] [Google Scholar]


