Abstract
Purpose
The decreased effectiveness of single-agent targeted therapies in advanced non–clear cell renal cell carcinoma (ncRCC) compared with clear cell renal cell carcinoma (RCC) supports the study of combination regimens. We evaluated the efficacy of everolimus plus bevacizumab in patients with metastatic ncRCC.
Patients and Methods
In this single-center phase II trial, treatment-naive patients received everolimus 10 mg oral once per day plus bevacizumab 10 mg/kg intravenously every 2 weeks. The primary end point was progression-free survival (PFS) at 6 months. Correlative analyses explored candidate tissue biomarkers through next-generation sequencing.
Results
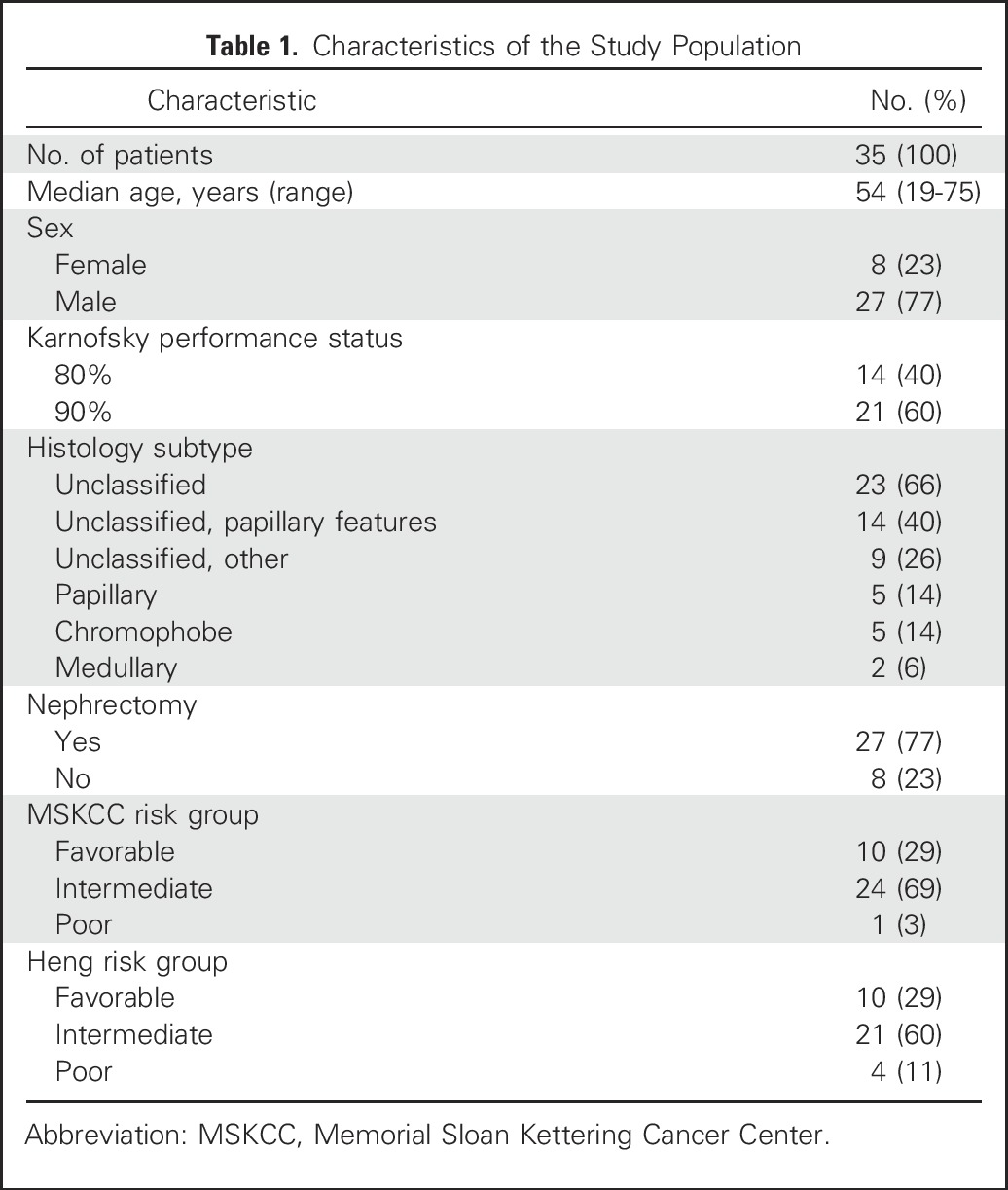
Thirty-five patients were enrolled with the following histologic subtypes: chromophobe (n = 5), papillary (n = 5), and medullary (n = 2) RCC and unclassified RCC (uRCC, n = 23). The majority of patients had papillary growth as a major component (n = 14). For 34 evaluable patients, median PFS, overall survival, and objective response rate (ORR) were 11.0 months, 18.5 months, and 29%, respectively. PFS varied by histology (P < .001), and ORR was higher in patients with significant papillary (seven of 18) or chromophobe (two of five) elements than for others (one of 11). Presence of papillary features were associated with benefit, including uRCC, where it correlated with ORR (43% v 11%), median PFS (12.9 v 1.9 months), and overall survival (28.2 v 9.3 months; P < .001). Several genetic alterations seemed to segregate by histology. In particular, somatic mutations in ARID1A were seen in five of 14 patients with papillary features but not in other RCC variants. All five patients achieved treatment benefit.
Conclusion
The study suggests efficacy for this combination in patients with ncRCC characterized by papillary features. Distinct mutational profiles among ncRCCs vary according to specific histology.
INTRODUCTION
Kidney cancer comprises several different malignancies that vary in pathobiology and sensitivity to approved systemic agents. Conventional clear cell renal cell carcinoma (RCC) comprises 60% to 80% of cases; these tumors are uniformly dependent on vascular endothelial growth factor (VEGF) signaling due to functional loss of the von Hippel Lindau protein.1 The remainder of subtypes are summarized as non–clear cell RCC (ncRCC), but constitute a diverse mixture of heterogeneous malignancies, including papillary, chromophobe, medullary, and collecting duct RCC. Cases that do not meet all criteria for these well-defined subtypes are categorized as unclassified RCC (uRCC).2
Although large randomized trials have standardized the therapeutic approach to metastatic clear cell RCC, phase III data to guide the management of metastatic ncRCC are limited to unplanned subgroup analyses.3 Phase II studies have reported some efficacy with VEGF- and mammalian target of rapamycin (mTOR)–directed agents across the majority of ncRCC variants.4-8 However, the antitumor effect observed has been more modest and less durable than what is seen in clear cell RCC. Likely, lower response rates to sunitinib and other antiangiogenic drugs reflect the lack of von Hippel-Lindau loss in ncRCC, with a more heterogeneous underlying molecular biology. Such a hypothesis would favor the use of combination regimens to treat ncRCC and underscores the need to better categorize patients, ideally by integrated histopathologic and molecular criteria, in the development of effective treatment approaches.
Everolimus, a rapalog-type inhibitor of the mTOR complex 1 (mTORC1), and bevacizumab, a recombinant humanized monoclonal antibody directed against VEGF-A, are both approved for the treatment of advanced RCC.9-11 The combination of everolimus and bevacizumab has been studied in clear cell RCC and was reported as tolerable when both drugs were given concurrently at standard doses.12 We conducted a single-center phase II study of everolimus plus bevacizumab in treatment-naive patients with advanced ncRCC. Correlative end points were included to refine our definitions of ncRCC variants and explore biomarkers that could enable rational patient selection for future studies.
PATIENTS AND METHODS
Eligibility
Patients age 18 years or older were required to have histologically confirmed, advanced ncRCC, including papillary, chromophobe, collecting duct, and medullary RCC and uRCC, per review at Memorial Sloan Kettering Cancer Center (MSKCC). Advanced disease was defined as unresectable, locally recurrent, or metastatic. Other inclusion criteria were no prior systemic therapy with a VEGF or mTORC1 inhibitor; measurable disease per Response Evaluation Criteria in Solid Tumors (RECIST) 1.113; Karnofsky performance status ≥ 70%; adequate renal, hepatic, and hematopoietic function at baseline; adequately controlled blood pressure; and absence of active brain metastases. The study was approved by the institutional review board at MSKCC; all patients provided written informed consent.
Study Design and Treatment
This was a single-institution, phase II study of everolimus plus bevacizumab in treatment-naive patients with advanced ncRCC sponsored by Novartis (Basel, Switzerland). Patients received concurrent therapy with everolimus (standard dose of 10 mg by mouth once per day) and bevacizumab (standard dose of 10 mg/kg intravenously every 14 days) until disease progression, intolerable toxicity, or withdrawal of consent. Archival tumor tissue and peripheral blood were collected for correlative analyses, including immunohistochemistry (IHC) and next-generation sequencing (NGS), from tumor and germline DNA.
End Points and Clinical Assessments
Cycle length was 28 days; cross-sectional imaging was repeated every two cycles for efficacy assessment per RECIST 1.1.13 Clinical and laboratory assessment were performed twice during cycle 1 and once during subsequent cycles. Urinalysis and fasting blood draw were conducted every two cycles. Toxicities were assessed after 14 days of study treatment then once per subsequent cycle and graded per Common Terminology Criteria for Adverse Events version 4.0.14 The protocol guided dose modifications for everolimus toxicity (5 mg once per day and 5 mg every other day). No dose reductions were recommended for bevacizumab, but dosing could be delayed or permanently discontinued if held > 8 weeks. In the event of bevacizumab discontinuation, everolimus treatment could be continued.
Correlative Analyses
Archival tumor tissue was not a requirement for eligibility, but efforts were maximized to obtain banked specimens for all patients. Samples were reviewed by a genitourinary pathologist (Y.-B.C.), who selected areas of high tumor content for analysis.
IHC staining for phosphorylated 4E-BP1 (p4E-BP1), a downstream effector of mTORC1,15 and ERG, a transcriptional regulator of angiogenesis and vascular homeostasis,16,17 were performed. Five-micrometer sections were stained with a p4E-BP1 (Thr37/46; 236B4) rabbit monoclonal antibody (Cell Signaling Technologies, Danvers, MA) or an ERG (EPR3864) rabbit monoclonal antibody (Ventana Medical Systems, Tucson, AZ) by using an automated platform (DISCOVERY XT; Ventana Medical Systems). A p4E-BP1 H score (range, 1 to 300) was calculated for each patient by multiplying the percentage of positive cells by the corresponding staining intensity (1 to 3). For ERG staining, an average count of positively stained endothelial cells per square millimeter (approximately five high-power fields) was documented.
For NGS analysis, we used the MSKCC IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) platform as previously described.18 In the version used here, the assay achieves pull-down capture with target-specific probes for exons from 341 cancer-related genes, including oncogenes, tumor suppressor genes, and components of pathways deemed actionable by targeted therapies (Appendix Table A1, online only, provides a full list). Deep-coverage NGS is then performed on an Illumina HiSeq system (San Diego, CA) across all coding sequences of these genes of interest. Coverage rate of targeted coding sequences and tumor-to-normal coverage ratios are used to investigate copy number alterations for individual genes.
Statistical Analysis
The primary end point was progression-free survival (PFS) after 6 months, per RECIST 1.1.13 Secondary end points were objective response rate (ORR), overall survival (OS), and treatment-emergent adverse events.
With a single-stage design for 34 patients, the regimen was to be considered promising if 22 or more patients were progression free at 6 months. Patients who left the study sooner without documented progression or as a result of death were conservatively treated as events for primary end point analysis. The design discriminates between true 6-month PFS rates ≤ 50% and ≥ 70%, with type I and II error rates of 6% and 19%, respectively. A 6-month PFS rate of 50% was chosen on the basis of phase II data for single-agent sunitinib in the same target population,4 the agent most frequently chosen for the treatment of advanced ncRCC outside of clinical trials.
PFS and OS were also calculated by using time-to-event methods to account for censoring. PFS was defined as time from treatment start to disease progression or death within 28 days of treatment end. Patients who did not progress or die within 28 days of treatment end were censored at the date of treatment end or last clinic visit. OS was defined as time from treatment start to death as a result of any cause. PFS and OS were calculated by using the Kaplan-Meier method, with the log-rank test used for subgroup comparisons. The ORR was calculated for the entire cohort and specific histologic subgroups. Correlation between IHC scores and achievement of 6-month PFS was investigated by Wilcoxon rank sum test. The NGS analysis was descriptive without formal statistical design. Statistical analysis was performed with SAS 9.4 (SAS Institute, Cary, NC) and R 3.1.0 (survival and Hmisc packages) software. Data cutoff was November 2015.
RESULTS
Baseline Characteristics
Thirty-five patients were enrolled and treated in the trial; clinical and pathologic features are summarized in Table 1. The majority of patients were categorized as favorable (29%) or intermediate (69%) risk according to MSKCC criteria.19 Histologic subtypes were uRCC (n = 23) and papillary (n = 5), chromophobe (n = 5), and medullary (n = 2) RCC. Patients with papillary RCC did not meet sufficient criteria to be further divided into type 1 or 2 subgroups. Within the large uRCC subgroup, several tumors were noted to have prominent papillary architectural features yet did not fulfill other criteria needed to establish a diagnosis of papillary RCC. During the course of the study, treatment benefit was noted for several patients within this subgroup. Consequently, after accrual was complete, a designated committee of genitourinary pathologists reviewed the specimens again for all 23 patients with uRCC. Blinded to treatment outcome, the committee divided cases of uRCC into two categories: uRCC with multinodular, intracystic papillary growth as a major component (uRCC with papillary features) or uRCC with various growth patterns without these specific papillary features specifically (uRCC without papillary features). Papillary features were seen in 14 of 23 patients with uRCC, and dedicated analyses for this subgroup were performed.
Table 1.
Characteristics of the Study Population
Efficacy
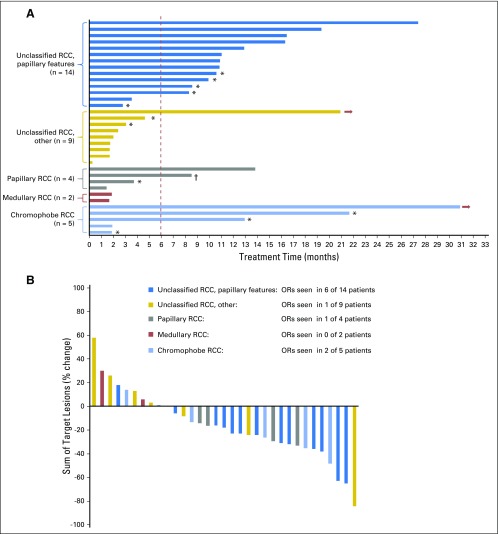
One patient was taken off the trial after the first dose of bevacizumab because of the need for a surgical procedure unrelated to his cancer or the study treatment. He was not evaluable and thus replaced with another subject. Efficacy outcomes for the 34 evaluable subjects are summarized in Table 2. The median follow-up for patients who remained alive was 23.6 months. Eighteen (53%) subjects were alive and progression free after 6 months and 10 (29%) after 12 months; two were still receiving active study treatment at the time of report (30.4 and 20.2 months on treatment, respectively; Fig 1A).
Table 2.
Summary Efficacy Analysis
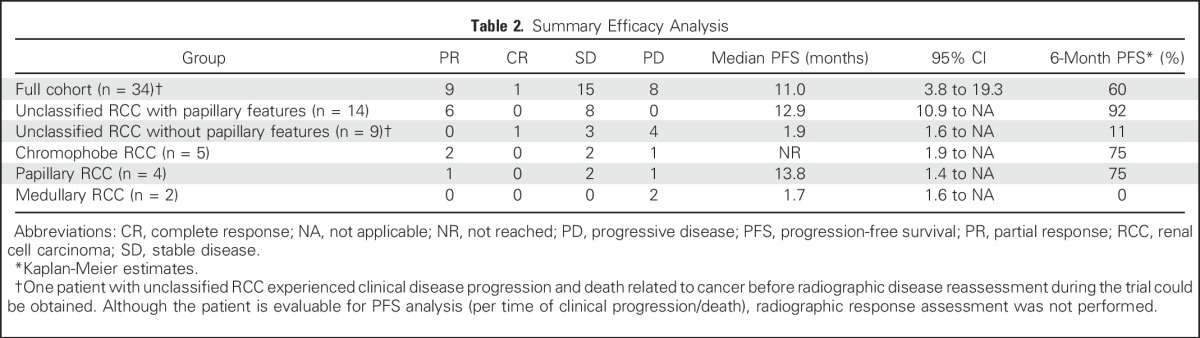
Fig 1.
Efficacy assessment by subject (colors encode renal cell carcinoma [RCC] variants). (A) Swimmer plot depicts individual patients as lines. Arrows indicate patients who remained on therapy. The primary end point for the trial was progression-free survival at 6 months (vertical line). (B) The best radiographic response for 33 evaluable patients treated. Best response was assessed per Response Evaluation Criteria in Solid Tumors 1.1. OR, objective response. (*) Discontinuation due to toxicity. (†) Voluntary withdrawal from study.
PFS varied significantly by histology (log-rank P < .001), as did the rate of patients who achieved a PFS ≥ 6 months (three of five with chromophobe RCC, zero of two with medullary RCC, two of four with papillary RCC, 12 of 14 with uRCC with papillary features, and one of nine with uRCC without papillary features). Objective responses were observed in a sizable proportion of subjects with significant papillary (seven of 18) or chromophobe (two of five) tumor components but rarely in patients with uRCC without papillary features (one of nine) or those with medullary RCC (zero of two; Table 2; Fig 1B).
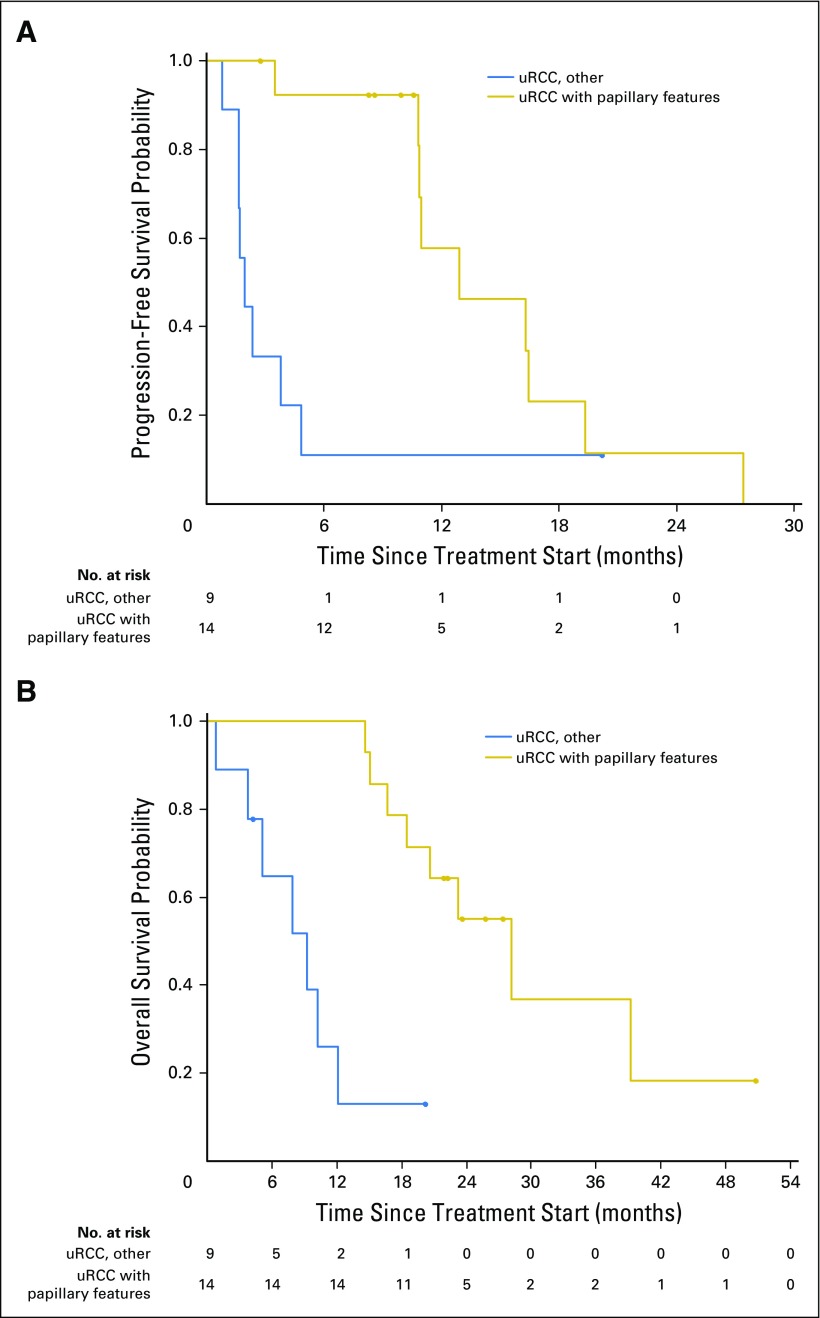
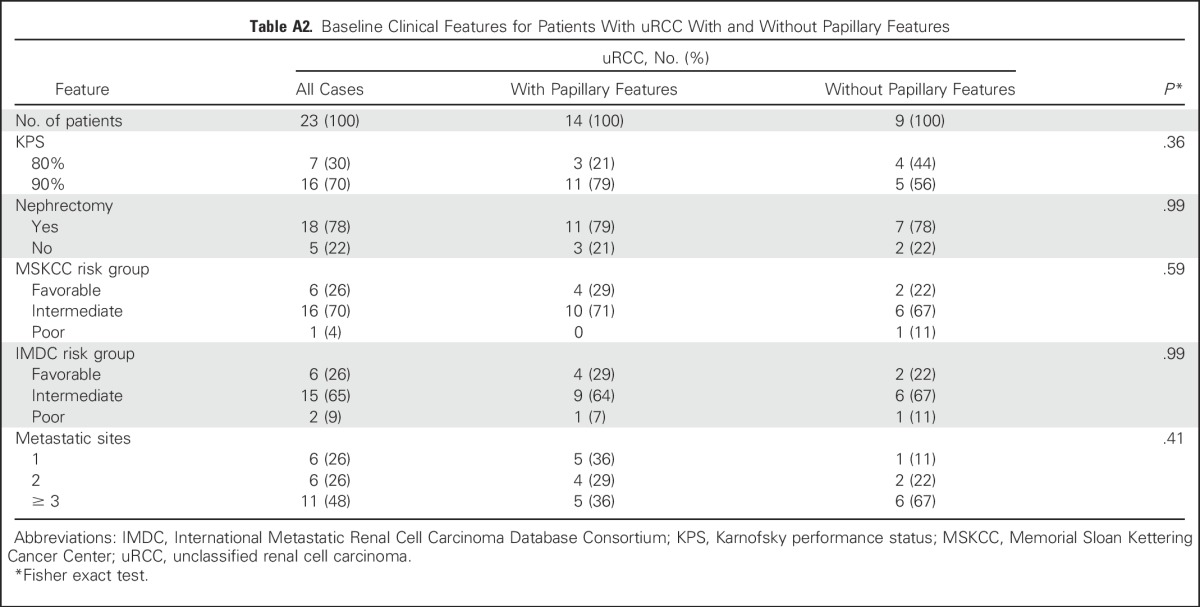
For patients with uRCC, the presence (n = 14) or absence (n = 9) of a major papillary component correlated strongly with ORR (43% v 11%), median PFS (12.9 v 1.9 months), and median OS (28.2 v 9.3 months; each log-rank for curves, P < .001; Fig 2). We compared baseline clinical features that could account for such discrepancies in outcome, including extent of disease, nephrectomy status, and MSKCC and International Metastatic Renal Cell Carcinoma Database Consortium risk status, but found no significant difference between the two subgroups (Appendix Table A2, online only). Only one subject with uRCC without papillary features remained on treatment > 6 months. In this subject, bevacizumab was discontinued after 95 days on study due to proteinuria. At the time, radiographic assessment was consistent with a partial response (PR). She then continued study treatment with everolimus monotherapy and ultimately achieved a complete response (CR). At the time of this report (20.2 months) this patient was still receiving treatment. As noted below, this subject’s tumor analysis revealed the presence of a somatic mutation in the kinase domain region of MTOR.
Fig 2.
Kaplan-Meier curves that summarize outcomes for patients with unclassified renal cell carcinoma (uRCC). Separate curves depict the comparison between subjects with uRCC with (n = 14; red line) and without (n = 9; black line) significant papillary features. (A) Median progression-free survival differed significantly between groups (12.9 v 1.9 months; log-rank P = .01). (B) Median overall survival was significantly longer for uRCC with papillary features than for other uRCC variants (28.2 v 9.3 months; log-rank P < .001).
Toxicity
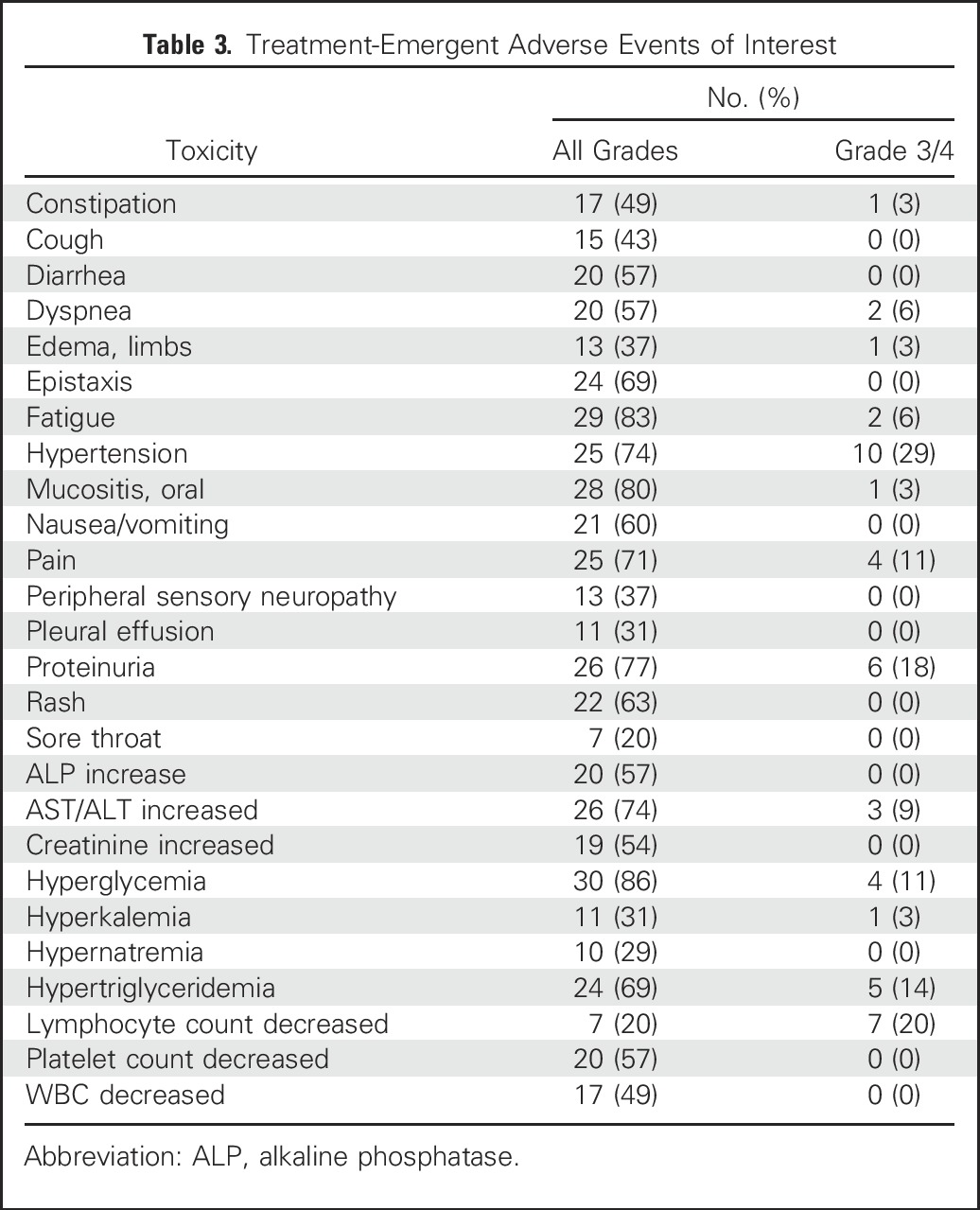
Treatment-emergent adverse events are summarized in Table 3. Treatment was generally well tolerated, although low-grade toxicities were commonly seen. High-grade (Common Terminology Criteria for Adverse Events grade 3 or greater) events were infrequent with the exception of hyperglycemia (11%), hypertriglyceridemia (14%), lymphopenia (20%), hypertension (29%), and proteinuria (18%), all established, class-specific events for mTORC1 or VEGF inhibitors. Eleven (32%) subjects required everolimus dose reductions, all for class-specific toxicities, including mucositis, cytopenias, and fatigue. Proteinuria developed in > 70% of patients (grade 2 or less for most). Nephrotic-range proteinuria developed in three cases, and eight (24%) had to discontinue bevacizumab permanently because of a persistent urine protein/creatinine ratio > 2. They continued everolimus as monotherapy until criteria for removal from trial were met. Two patients died while on study, both from a gastrointestinal hemorrhage. One death was related to progressive disease and the other possibly to bevacizumab.
Table 3.
Treatment-Emergent Adverse Events of Interest
Correlative Analysis
For 28 subjects, archival tissue was analyzed with deep targeted sequencing across 341 cancer-related genes; matched germline comparison from healthy cells was included for 17 of these. Average depth of coverage was 558 times. Findings are summarized in Figure 3. Although limited by small numbers, specific genetic alterations seemed to segregate by histology. Recurrent events included TP53 mutations in three of five patients with chromophobe RCC. Tumors from five of 14 subjects with a major papillary component (including papillary and uRCC variants) harbored mutations in ARID1A, whereas none were detected in the other RCC variants tested. All patients with ARID1A mutations achieved the primary end point of 6-month PFS, three of five with radiographic PR. Functional loss of fumarate hydratase was detected in tumors from four patients with uRCC with papillary features (all meeting the primary PFS end point) and one patient with uRCC without papillary features (who did not meet the PFS end point). As aforementioned, one patient with uRCC without papillary features achieved a durable CR and remains on study treatment; NGS analysis of her tumor tissue revealed the presence of an MTOR L2427R mutation that maps to the gene’s kinase domain. Functional study confirmed this mutation to be activating.
Fig 3.
Oncogenomic changes detected by deep-sequence analysis from archival tumor specimens across 28 subjects. Columns represent individual subjects; rows represent selected genes of interest examined for each sample. Subjects who reached the primary efficacy goal of 6 months progression-free survival (PFS) are marked blue; those who did not are marked gold. Subjects censored within their first 6 months of the trial are marked gray. Colored squares mark the presence of somatic alterations detected by sequence analysis. ARID1A mutations were seen in five of 14 tumors with major papillary features; no ARID1A mutations were seen in all other renal cell carcinoma (RCC) variants. For patients with ARID1A mutations, PFS was > 6 months in five of five, and three of the five achieved a partial response.
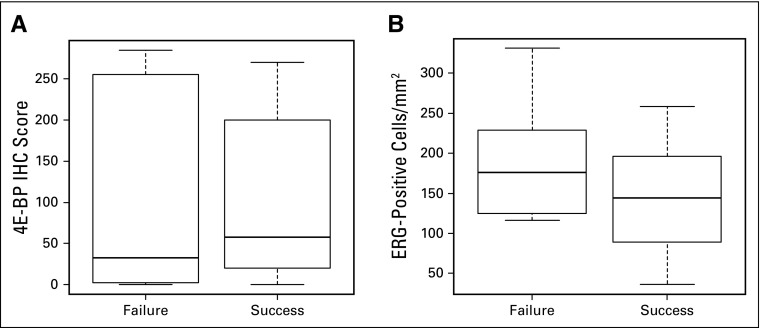
IHC testing for p4E-BP1 and ERG was performed on archival samples from 19 subjects. Six-month PFS correlated neither with p4E-BP1 IHC score (P = .96; Appendix Fig A1, online only) nor with microvessel density per number of ERG-positive cells per square millimeter (P = .38; Appendix Fig A1).
DISCUSSION
We tested everolimus plus bevacizumab in patients with advanced ncRCC, a heterogenous group of diseases with poorly defined standards of care.20 Although the trial did not meet its primary end point, a striking signal was observed for defined histologic subgroups, specifically those with a significant papillary tumor component. The ORR in this group was 39%, which is comparable with those reported for VEGF-targeted therapies in phase II trials of clear cell RCC.21,22 The majority of responses were durable, with 14 (78%) of 18 patients progression free at 6 months and the median PFS eclipsing 1 year (12.9 months). These outcomes compare favorably with those reported for first-line sunitinib in the largest efforts to date, the phase II ASPEN (Randomized Phase II Study of Afinitor [RAD001] vs. Sutent [Sunitinib] in Patients With Metastatic Non-Clear Renal Cell Carcinoma) and ESPN (Everolimus Versus Sunitinib Prospective Evaluation in Metastatic Non-Clear Cell Renal Cell Carcinoma) trials (median PFS, 8.3 and 4.1 months, respectively, among subjects with papillary histology).7,8,23
Activity was observed for patients with chromophobe RCC. Three of five remained on treatment for > 12 months, which adds to prior reports of benefit with rapalog therapies in this subgroup.6,7,23,24 Subjects with other variants (medullary RCC and uRCC without papillary features), achieved little or no benefit from everolimus plus bevacizumab. Novel treatment approaches beyond VEGF- and mTOR-directed therapy are needed for such patients.
We had conducted a phase I study of sunitinib plus everolimus in 20 treatment-naive patients, including seven with ncRCC.25 Three of the seven (papillary, chromophobe, chromophobe) achieved a PR, and all three remained on protocol for > 1 year. The current findings lend further support to combining mTOR- and VEGF-directed therapy in the papillary and chromophobe variants.
This prospective study is the first to our knowledge to report on defined subgroups of uRCC and to identify histologic hallmarks that seem to correlate with treatment benefit. Most uRCC tumors in this study (14 of 23) contained multinodular, intracystic papillary growth as a major component (uRCC with papillary features). These subjects were significantly more likely to benefit from treatment, as reflected by superior ORR, PFS, and OS (Fig 2). Of note, nine of 14 subjects with uRCC with papillary features were referred for management from outside institutions, where the majority had been classified as papillary RCC. Pathologic re-review at MSKCC determined that they did not meet sufficient criteria for a formal diagnosis of papillary RCC, and they were recategorized as having uRCC with papillary features. Others have investigated whether papillary RCC and uRCC with papillary morphology originate from a common cell of origin but were unable to demonstrate a unifying genomic background.26 Moreover, recent work suggested a number of oncogenomic variants within the spectrum of papillary RCC.26,27 The Southwest Oncology Group recently opened SWOG 1500 (NCT02761057), the largest prospective effort in papillary RCC to date, which incorporates not only a randomized comparison of various targeted treatment strategies, but also various correlative end points to better define the landscape of papillary RCC.
Our NGS analysis, although limited by sample size, yielded a number of thought-provoking findings. Acquired mutations in ARID1A were detected in five of 14 tumors with major papillary components (Fig 3), with all five subjects achieving benefit with a PFS > 6 months. No ARID1A mutations were detected in patients with PFS < 6 months, nor were any seen in tumors without papillary components. ARID1A is a member of the switch/sucrose nonfermentable chromatin-remodeling complex, which modulates DNA accessibility to other cellular machinery (repair, transcription, duplication) through regulation of nucleosome repositioning. Somatic mutations in ARID1A are frequently seen across human malignancies, and a haploinsufficient effect that promotes oncogenesis has been described.28,29 The exact mechanisms of its cancer-promoting effects remain largely unknown. Recent preclinical reports indicate that loss of ARID1A function and abnormal phosphatidylinositol 3-kinase (PI3K) pathway signaling can converge in tumorigenesis, which provides a rationale for targeting the PI3K/mTORC1 signaling axis in ARID1A mutant tumors.30 However, more recent data from in vitro breast cancer models suggest that loss of ARID1A through activated annexin A1 expression may confer resistance to mTOR inhibition.31 Although the current results support the former hypothesis, the small sample size prohibits conclusions from being drawn. Furthermore, concurrent bevacizumab may play an important role that is not accounted for in these models. NGS analysis of papillary RCC in The Cancer Genome Atlas project identified mutations in members of the switch/sucrose nonfermentable complex in 20% and 27% of papillary type 1 and 2 tumors, respectively.27 Altogether, the data suggest that ARID1A merits further study for its functional role in papillary RCC variants and as a candidate biomarker for future study of everolimus plus bevacizumab.
Prior reports have linked alterations in PI3K pathway components, particularly TSC1, TSC2, and MTOR, to exceptional benefit in patients with RCC treated with rapalog monotherapy32 and mTORC1/VEGF-directed combination therapy.33 Detection of an MTOR kinase domain mutation (L2427R) in a subject with uRCC, who achieved a CR with single-agent everolimus (after early discontinuation of bevacizumab) adds to these data. A subject with an R1369W missense mutation in TSC2 continued to receive study treatment > 6 months, but a concurrent ARIDA1 mutation may have also accounted for his treatment response. One subject with uRCC who harbored a Y1151C missense mutation in MTOR did not benefit from the study treatment most likely because the alteration did not affect a functionally relevant portion of the gene.
In summary, this study demonstrates a benefit of everolimus plus bevacizumab in ncRCC, particularly in variants with significant papillary elements. Future studies are warranted and should narrow histologic entry criteria. The data also illustrate distinct mutational profiles among ncRCCs that vary according to specific histology and identify ARID1A as a potential marker of papillary architecture and as a candidate predictive biomarker in future trials of this promising regimen.
Appendix
Fig A1.
Analysis of archival tissue by immunohistochemistry (IHC) failed to demonstrate correlates of treatment benefit. (A) Box plot of median and range of 4E-BP1 IHC scores in patients who experienced disease progression or died within < 6 months of starting the study treatment (failure) versus those alive and free from disease progression at ≥ 6 months (success; median score, 32.5 v 57.5; Wilcoxon rank sum P = .96). (B) Box plot of median and range of ERG-positive cells/mm2 in patients who experienced disease progression or died within < 6 months of starting study treatment (failure) versus those alive and free from disease progression (success; median score, 176 v 144; Wilcoxon rank sum P = .38).
Table A1.
Target-Specific Probes for Exons From 341 Cancer-Related Genes and Components of Pathways Deemed Actionable by Targeted Therapies
Table A2.
Baseline Clinical Features for Patients With uRCC With and Without Papillary Features
Footnotes
Supported by Novartis International AG (Basel, Switzerland).
Presented at the 2015 Genitourinary Cancers Symposium, Orlando, FL, February 26-28, 2015, and 2015 ASCO Annual Meeting, Chicago, IL, May 29-June 2, 2015.
Authors’ disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
Clinical trial information: NCT01399918
See accompanying editorial on page 3825
AUTHOR CONTRIBUTIONS
Conception and design: Martin H. Voss, Ana M. Molina, Sujata Patil, Robert J. Motzer, Darren R. Feldman
Administrative support: Devyn T. Coskey
Provision of study materials or patients: Martin H. Voss, Ana M. Molina, Chung-Han Lee, James J. Hsieh, Robert J. Motzer
Collection and assembly of data: Martin H. Voss, Ana M. Molina, Ying-Bei Chen, Joshua L. Chaim, Devyn T. Coskey, Almedina Redzematovic, Patricia Wang, William Lee, S. Duygu Selcuklu, James J. Hsieh, Chung-Han Lee, Darren R. Feldman
Data analysis and interpretation: Martin H. Voss, Ying-Bei Chen, Kaitlin M. Woo, Devyn T. Coskey, Almedina Redzematovic, Michael F. Berger, Satish K. Tickoo, Victor E. Reuter, Sujata Patil, James J. Hsieh, Robert J. Motzer, Darren R. Feldman
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Phase II Trial and Correlative Genomic Analysis of Everolimus Plus Bevacizumab in Advanced Non–Clear Cell Renal Cell Carcinoma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Martin H. Voss
Honoraria: Novartis
Consulting or Advisory Role: Novartis, Calithera Biosciences, Natera, GlaxoSmithKline, Exelixis
Research Funding: Pfizer, Bristol-Myers Squibb, Genentech
Travel, Accommodations, Expenses: Novartis, Takeda Pharmaceuticals
Ana M. Molina
Honoraria: ASCO
Consulting or Advisory Role: Eisai, Novartis
Ying-Bei Chen
No relationship to disclose
Kaitlin M. Woo
No relationship to disclose
Joshua L. Chaim
No relationship to disclose
Devyn T. Coskey
No relationship to disclose
Almedina Redzematovic
No relationship to disclose
Patricia Wang
Employment: Helix OpCo
Travel, Accommodations, Expenses: Helix OpCo
William Lee
Employment: Helix OpCo
Leadership: Helix OpCo
S. Duygu Selcuklu
No relationship to disclose
Chung-Han Lee
Research Funding: Pfizer (Inst), Eisai (Inst)
Michael F. Berger
Consulting or Advisory Role: Cancer Genetics, Sequenom
Satish K. Tickoo
No relationship to disclose
Victor E. Reuter
No relationship to disclose
Sujata Patil
No relationship to disclose
James J. Hsieh
Honoraria: Chugai Pharmaceutical
Consulting or Advisory Role: Novartis, Chugai Pharmaceutical, Eisai
Research Funding: Novartis, Pfizer, CGI
Robert J. Motzer
Consulting or Advisory Role: Pfizer, Novartis, Eisai
Research Funding: Pfizer (Inst), GlaxoSmithKline (Inst), Bristol-Myers Squibb (Inst), Eisai (Inst), Novartis (Inst), Genentech (Inst)
Travel, Accommodations, Expenses: Bristol-Myers Squibb
Darren R. Feldman
Consulting or Advisory Role: Bayer AG, Gilead Sciences (I), Seattle Genetics
Research Funding: Novartis
REFERENCES
- 1.Linehan WM. Genetic basis of kidney cancer: Role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22:2089–2100. doi: 10.1101/gr.131110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovacs G, Akhtar M, Beckwith BJ, et al. The Heidelberg classification of renal cell tumours. J Pathol. 1997;183:131–133. doi: 10.1002/(SICI)1096-9896(199710)183:2<131::AID-PATH931>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 3.Dutcher JP, de Souza P, McDermott D, et al. Effect of temsirolimus versus interferon-alpha on outcome of patients with advanced renal cell carcinoma of different tumor histologies. Med Oncol. 2009;26:202–209. doi: 10.1007/s12032-009-9177-0. [DOI] [PubMed] [Google Scholar]
- 4. Molina AM, Feldman DR, Ginsberg MS, et al: Phase II trial of sunitinib in patients with metastatic non-clear cell renal cell carcinoma. Invest New Drugs 30:335-340, 2012. [DOI] [PMC free article] [PubMed]
- 5.Tannir NM, Plimack E, Ng C, et al. A phase 2 trial of sunitinib in patients with advanced non-clear cell renal cell carcinoma. Eur Urol. 2012;62:1013–1019. doi: 10.1016/j.eururo.2012.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh Y, Lim HY, Ahn JH, et al. Phase II trial of everolimus for the treatment of nonclear-cell renal cell carcinoma. Ann Oncol. 2013;24:1026–1031. doi: 10.1093/annonc/mds582. [DOI] [PubMed] [Google Scholar]
- 7. doi: 10.1016/j.eururo.2015.10.049. Tannir NM, Jonasch E, Albiges L, et al: Everolimus Versus Sunitinib Prospective Evaluation in Metastatic Non-Clear Cell Renal Cell Carcinoma (ESPN): A randomized multicenter phase 2 trial. Eur Urol 69:866-874, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong AJ, Halabi S, Eisen T, et al. Everolimus versus sunitinib for patients with metastatic non-clear cell renal cell carcinoma (ASPEN): A multicentre, open-label, randomised phase 2 trial. Lancet Oncol. 2016;17:378–388. doi: 10.1016/S1470-2045(15)00515-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motzer RJ, Escudier B, Oudard S, et al. Efficacy of everolimus in advanced renal cell carcinoma: A double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 10.Escudier B, Pluzanska A, Koralewski P, et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: A randomised, double-blind phase III trial. Lancet. 2007;370:2103–2111. doi: 10.1016/S0140-6736(07)61904-7. [DOI] [PubMed] [Google Scholar]
- 11.Rini BI, Halabi S, Rosenberg JE, et al. Bevacizumab plus interferon alfa compared with interferon alfa monotherapy in patients with metastatic renal cell carcinoma: CALGB 90206. J Clin Oncol. 2008;26:5422–5428. doi: 10.1200/JCO.2008.16.9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hainsworth JD, Spigel DR, Burris HA, III, et al. Phase II trial of bevacizumab and everolimus in patients with advanced renal cell carcinoma. J Clin Oncol. 2010;28:2131–2136. doi: 10.1200/JCO.2009.26.3152. [DOI] [PubMed] [Google Scholar]
- 13.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 14. National Cancer Institute: Common Terminology Criteria for Adverse Events (CTCAE) Version 4.03. Washington, DC, US Department of Health and Human Services, 2010. [Google Scholar]
- 15.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birdsey GM, Dryden NH, Shah AV, et al. The transcription factor Erg regulates expression of histone deacetylase 6 and multiple pathways involved in endothelial cell migration and angiogenesis. Blood. 2012;119:894–903. doi: 10.1182/blood-2011-04-350025. [DOI] [PubMed] [Google Scholar]
- 17.Birdsey GM, Shah AV, Dufton N, et al. The endothelial transcription factor ERG promotes vascular stability and growth through Wnt/β-catenin signaling. Dev Cell. 2015;32:82–96. doi: 10.1016/j.devcel.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Motzer RJ, Bacik J, Murphy BA, et al: Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J Clin Oncol 20:289-296, 2002. [DOI] [PubMed]
- 20.Albiges L, Escudier B. Current and future strategies in nonclear-cell metastatic renal cell carcinoma. Curr Opin Urol. 2015;25:367–373. doi: 10.1097/MOU.0000000000000197. [DOI] [PubMed] [Google Scholar]
- 21.Motzer RJ, Rini BI, Bukowski RM, et al. Sunitinib in patients with metastatic renal cell carcinoma. JAMA. 2006;295:2516–2524. doi: 10.1001/jama.295.21.2516. [DOI] [PubMed] [Google Scholar]
- 22.Rixe O, Bukowski RM, Michaelson MD, et al. Axitinib treatment in patients with cytokine-refractory metastatic renal-cell cancer: A phase II study. Lancet Oncol. 2007;8:975–984. doi: 10.1016/S1470-2045(07)70285-1. [DOI] [PubMed] [Google Scholar]
- 23. Armstrong AJ, Broderick S, Eisen T, et al: Final clinical results of a randomized phase II international trial of everolimus vs. sunitinib in patients with metastatic non-clear cell renal cell carcinoma (ASPEN). J Clin Oncol 33, 2015 (suppl; 4507) [Google Scholar]
- 24.Voss MH, Bastos DA, Karlo CA, et al. Treatment outcome with mTOR inhibitors for metastatic renal cell carcinoma with nonclear and sarcomatoid histologies. Ann Oncol. 2014;25:663–668. doi: 10.1093/annonc/mdt578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molina AM, Feldman DR, Voss MH, et al. Phase 1 trial of everolimus plus sunitinib in patients with metastatic renal cell carcinoma. Cancer. 2012;118:1868–1876. doi: 10.1002/cncr.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsaud A, Dadone B, Ambrosetti D, et al. Dismantling papillary renal cell carcinoma classification: The heterogeneity of genetic profiles suggests several independent diseases. Genes Chromosomes Cancer. 2015;54:369–382. doi: 10.1002/gcc.22248. [DOI] [PubMed] [Google Scholar]
- 27. doi: 10.1056/NEJMoa1505917. Linehan WM, Spellman PT, Ricketts CJ, et al: Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med, 374:135-145, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu JN, Roberts CW. ARID1A mutations in cancer: Another epigenetic tumor suppressor. Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu RC, Wang TL, Shih IeM. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther. 2014;15:655–664. doi: 10.4161/cbt.28411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandler RL, Damrauer JS, Raab JR, et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat Commun. 2015;6:6118. doi: 10.1038/ncomms7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. doi: 10.1158/1078-0432.CCR-15-2996. Berns K, Sonnenblick A, Gennissen A, et al: Loss of ARID1A activates ANXA1, which serves as a predictive biomarker for trastuzumab resistance. Clin Cancer Res clincanres.2996.2015 [epub ahead of print on May 12, 2016] [DOI] [PubMed] [Google Scholar]
- 32.Voss MH, Hakimi AA, Pham CG, et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res. 2014;20:1955–1964. doi: 10.1158/1078-0432.CCR-13-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagle N, Grabiner BC, Van Allen EM, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4:546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]