ABSTRACT
We recently reported that bacteria can be found within pancreatic ductal adenocarcinoma (PDAC) tumors. Some of these bacteria can metabolize and thereby inactivate the nucleoside analog gemcitabine. We demonstrated that the long isoform of the bacterial enzyme cytidine deaminase (CDD) mediates the metabolism of gemcitabine. The clinical effect of overcoming this potential mechanism of drug resistance has yet to be studied.
KEYWORDS: chemoresistance, drug metabolism, gemcitabine, mycoplasma, novel therapeutic targets, pancreatic ductal adenocarcinoma, PDAC, mechanisms of resistance to therapy, tumor microbiome
Resistance to therapy is a major challenge in the treatment of cancer patients. Components of the tumor microenvironment such as vasculature, cancer-associated fibroblasts, inflammatory cells, and extracellular matrix, have all been implicated in resistance to various therapies.1 In a recently published study,2 we characterized the bacterial species present in the tumor microenvironment of pancreatic ductal adenocarcinoma (PDAC) tumors. Bacteria were detected in the majority of tumors tested and some of these bacteria were able to metabolize and inactivate gemcitabine, a commonly used therapy in the treatment of pancreatic cancer.
Our interest in intratumoral bacteria and their effect on treatment against cancer was instigated by a serendipitous finding. We previously reported that human dermal fibroblast (HDF) cells, when co-cultured with multiple colorectal and pancreatic cancer cell lines, could render these cancer cells resistant to gemcitabine.3 Conditioned medium from these HDF cells was by itself sufficient to generate gemcitabine resistance. Filtering this medium through a 0.45μm filter, as well as treating the HDF cells with antibiotics, abolished gemcitabine resistance, suggesting that microorganisms could be responsible. Follow-up studies indeed showed that the HDF cells were contaminated with Mycoplasma hyorhinis (M. hyorhinis) and that these bacteria deaminated gemcitabine into its inactive metabolite 2′2-difluorodeoxyuridine (dFdU) as has been previously reported.4 An additional study has demonstrated that gemcitabine efficacy can also be altered by Escherichia coli identified in human tumors.5
To determine whether other types of bacteria could mediate resistance to gemcitabine, we tested a total of 27 bacterial species. Thirteen of these bacteria, including M. hyorhinis, rendered RKO human colorectal cancer cells resistant to gemcitabine. In search of the mechanism behind this bacteria-mediated resistance, we considered the bacterial enzyme cytidine deaminase (CDD), as it was previously shown to be responsible for deamination of gemcitabine by Mycoplasma.4 Exploration of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database6 revealed that the cdd gene sequence was absent in 44.2% of all bacteria listed in the KEGG database, whereas the remainder of the bacteria had either a short (44.4%) or a long (11.2%) isoform of cdd. Interestingly, with the exception of M. hyorhinis, all of the bacterial species that mediated resistance to gemcitabine express the long isoform of the CDD enzyme (CDDL). Bacteria that either lack CDD or express the short isoform of the enzyme (CDDS) were unable to mediate resistance to gemcitabine. We do not yet understand the basis of M. hyorhinis-mediated resistance to gemcitabine, given that M. hyorhinis contains CDDS. The presence of CDD alone is not the only determinant in the ability of bacteria to metabolize gemcitabine. Loss of the nucleoside transporter, NupC, in CDDL-expressing bacteria partially abolished the bacteria-mediated resistance, suggesting that NupC plays a role in bacterial internalization of gemcitabine (Fig. 1). The effect of CDDL-expressing bacteria on gemcitabine efficacy was then ascertained in vivo. Mice containing CDDL-expressing bacteria in their tumors did not respond to gemcitabine. Treating these mice with antibiotics restored gemcitabine efficacy. Additionally, inoculation of bacteria-free tumors with bacteria in which the cddL gene was knocked out did not confer any resistance towards gemcitabine treatment.
Figure 1.
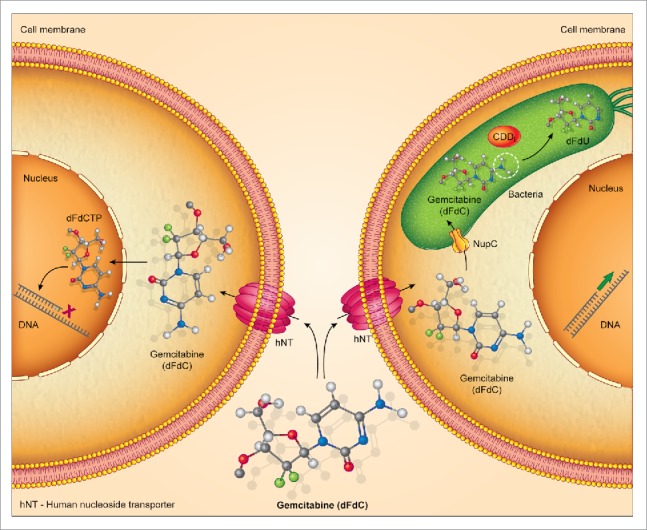
Intratumoral bacteria can metabolize gemcitabine. Bacteria expressing the long isoform of cytidine deaminase (CDDL) can metabolize gemcitabine into its inactive metabolite 2′2-difluorodeoxyuridine (dFdU). This prevents gemcitabine from inhibiting DNA synthesis in cancer cells. The bacterial nucleoside transporter, NupC, may be involved in bacterial internalization of gemcitabine.
The relevance of our finding in human tumors had still to be tested. To this end, human PDAC samples were chosen because gemcitabine is frequently used in the treatment of this cancer. Bacterial DNA was detected in 76% of tumors tested and only in 15% of normal pancreas tissue controls. The most common species identified belong to the Gammaproteobacteria class. Notably, the majority (98.4%) of bacteria expressing CDDL among those listed in the KEGG database belong to the Gammaproteobacteria class. Furthermore, bacteria cultured from 14 out of 15 PDAC tumors could render the RKO and HCT116 human colon carcinoma cell lines resistant to gemcitabine in vitro.
In summary, these findings indicate that some PDAC tumors contain bacteria that can potentially influence the efficacy of gemcitabine within the tumor. While co-treatment with gemcitabine and antibiotics restored gemcitabine efficacy in murine models, it is important to note that there are many other known resistance mechanisms to gemcitabine in PDAC7 and consequently the clinical impact of targeting these intratumoral bacteria has yet to be determined. Additionally, the larger amount of bacteria within tumors in our murine model compared to the numbers we found in patient samples indicates that more work is needed to understand the contribution of intratumoral bacteria to the efficacy of gemcitabine treatment in human tumors.
Treating patients with antibiotics in combination with gemcitabine may not be a straightforward approach, even if bacteria-mediated gemcitabine metabolism is found to be of clinical importance. Long-term treatment with antibiotics can lead to the emergence of antibiotic-resistant bacterial strains. Moreover, antibiotic treatment can affect bacterial communities throughout the body, including that of the gut. Several studies have demonstrated that bacteria present in the gut can profoundly affect the response to cancer treatments.8 Consequently, by using antibiotics, we might unknowingly affect the efficacy of treatment in other ways. An alternative approach to antibiotic treatment might be to utilize a small molecule to target the bacterial CDD enzyme. Such an approach may reduce the global effect on our microbial communities. Our preliminary studies demonstrate that the selective pressure imposed on bacteria by inhibiting CDD activity is negligible compared to that imposed by antibiotic treatment.
The study of the tumor microbiome is a growing field, and we are currently broadening our studies to characterize bacteria in additional tumor types. The presence of bacteria in tumors could have implications beyond gemcitabine metabolism that we, and others, are beginning to explore. We demonstrated, for example, that bacteria can mediate resistance to the drug Oxaliplatin, however, the mechanism driving this is still unknown. It has also been previously reported that Mycoplasma9 and other bacteria5,10 can alter the efficacy of multiple chemotherapies. Moreover, as bacteria are known to interact with their human hosts, the effects of intratumoral bacteria not only on the efficacy of other therapeutic agents, but also on various cancer traits such as angiogenesis, cell proliferation, invasion and metastasis, and tumor immunity, are important avenues for future research.
Funding
Israel Science Foundation (grant number 1877/14).
References
- 1.Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015;25:198–213. doi: 10.1016/j.tcb.2014.11.006. PMID:25540894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al.. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160 doi: 10.1126/science.aah5043. PMID:28912244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Straussman R. Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al.. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504 doi: 10.1038/nature11183. PMID:22763439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vande Voorde J, Sabuncuoğlu S, Noppen S, Hofer A, Ranjbarian F, Fieuws S, Balzarini J, Liekens S. Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J Biol Chem. 2014;289:13054–13065. doi: 10.1074/jbc.M114.558924. PMID:24668817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehouritis P, Cummins J, Stanton M, Murphy CT, McCarthy FO, Reid G, Urbaniak C, Byrne WL, Tangney M. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 2015;5:14554. doi: 10.1038/srep14554. PMID:26416623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. PMID:10592173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Sousa Cavalcante L, Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. PMID:25084222. [DOI] [PubMed] [Google Scholar]
- 8.Bashiardes S, Tuganbaev T, Federici S, Elinav E. The microbiome in anti-cancer therapy. Semin Immunol. 2017;32:74–81 doi: 10.1016/j.smim.2017.04.001. PMID:28431920. [DOI] [PubMed] [Google Scholar]
- 9.Bronckaers A, Balzarini J, Liekens S. The cytostatic activity of pyrimidine nucleosides is strongly modulated by Mycoplasma hyorhinis infection: Implications for cancer therapy. Biochem Pharmacol. 2008;76:188–197. doi: 10.1016/j.bcp.2008.04.019. PMID:18555978. [DOI] [PubMed] [Google Scholar]
- 10.Westman EL, Canova MJ, Radhi IJ, Koteva K, Kireeva I, Waglechner N, Wright GD. Bacterial inactivation of the anticancer drug doxorubicin. Chem Biol. 2012;19:1255–1264. doi: 10.1016/j.chembiol.2012.08.011. PMID:23102220. [DOI] [PubMed] [Google Scholar]


