ABSTRACT
In a recent study published in Nature Communications, we showed that intron-encoded microRNA genes (miRNA) are frequent partners of fusion genes in the cancer genome. Analyzed from a functional rather than structural perspective, these rearrangements represent a new class of fusions we called “miRNA-convergent fusions”.
KEYWORDS: microRNA, gene fusions, cancer, genome aberrations
Detailed description of chromosomal aberrations, predominantly balanced chromosomal translocations, leading to fusion genes has been important for the understanding of the tumorigenic process, as well as for cancer diagnosis and therapeutic purposes. Originally, balanced translocations were studied by light-microscopy and chromosome-banding techniques. This was followed by use of in situ hybridization for more precise molecular mapping of breakpoints, and in then array-based analysis further increased the power of resolution. For the final identification of fusions genes, polymerase chain reaction using reverse transcribed messenger RNA and primers targeting the candidate fusion gene partners was the most commonly used approach.
Today, the advent of next-generation sequencing of all expressed RNA (RNA-seq) molecules allows unbiased, ab initio global identification of fusion genes. Because of the massive data produced by RNA-seq the expertise has moved from experimental biology to computer programming and statistical analyses. To handle the massive information the complexity of the data is usually reduced using a number of general assumptions, e.g., signals that are a priori considered spurious are filtered out and only data that fulfill certain structural constrains at the protein coding level are retained. From the resulting files, the frequency and statistical significance of each gene fusion is then calculated. Commonly used criteria to identify biologically/clinically relevant fusion genes are that the resulting gene should retain the capacity to code for a protein and should be recurrent. However, in parallel with the technical advances, our understanding of the functional genome has been changing. Consequently, our view of gene fusions has to be updated. Algorithms for fusion gene detection still regard introns as merely pieces of short-lived junk and focus almost exclusively on potential chimeric proteins. This reasoning follows a successful old tradition. However, small regulatory non-coding RNAs are frequently encoded by introns of protein coding genes, and are expressed together with the host gene transcript. As a result, regulatory non-coding RNAs included in introns of fusion genes may be fully functional even if the protein encoded by the fusion gene is not.
More than ten years ago, Croce and co-workers showed that microRNA genes (miRNAs) are often located in unstable cancer associated genomic regions.1 In our recent publication2 we focused on miRNA genes encoded by introns. We show that fusion genes detected by RNA-seq, including both out of frame and non-recurrent fusions, were enriched for miRNAs in their introns and we also showed that, as a consequence of the fusion, exchange of the upstream regions that normally regulate the miRNA may upregulate miRNA expression, irrespectively if the protein coding parts will produce a functional protein or not. To our knowledge only incidental findings have reported rearrangements leading to deregulation of intronic miRNAs in fusion genes. For instance, in B-cell lymphoblastic leukemia, a t(11;14)(q24.1;q32) translocation leads to upregulation of mir-125b-1,3 in B-cell lymphoma with t(3;7)(q27;q32) producing the B-cell lymphoma 6 protein (BCL6)-Fragile Site, Aphidicolin Type, Common, Fra(7)(Q32.3) (FRA7H) fusion gene (BCL6-FRA7H) leads to down-regulation of mir-29,4 and a t(6;7)(p25;q32) translocation anaplastic large cell lymphoma producing the Dual Specificity Phosphatase 22 (DUSP22)-FRA7H fusion gene (DUSP22-FRA7H) also upregulates mir-29.5 The lack of attention on intronic miRNAs can be exemplified by fusions involving the vacuole membrane protein 1 (VIMP1) gene, the host gene of oncomiR mir-21. Deletions in chromosome 17q23 may bring together the neighboring clathrin heavy chain (CLTC) and VMP1 genes. This is a recurrent, out of frame fusion that occurs in multiple cancer types including glioblastoma, neuroblastoma, lung cancer, breast cancer, bladder cancer, thyroid cancer, melanoma, and leukemia.6 Also out of frame are the fusions of VMP1 with the ribosomal protein S6 kinase beta-1 (RPS6KB1) in breast cancer (RPS6KB1–VMP1). Since the presence of the oncogenic mir-21 was obviated from further analysis, the functional consequence of these fusions was suggested to be associated with a tumor suppressive function.7 However, Inaki et al8 observed that VMP1 fusions lead to upregulation of mir-21 but this observation was however still ignored by other invetigators.9
We believe that our perception of functional relevant fusions in the context of miRNA genes has to be revised. As mentioned, most observed fusions upstream of miRNA-encoding introns are out of frame and, therefore, discarded form further analysis by most pipelines for RNA-seq data analyses. If, however, we include all fusions, as well as data on intronic miRNAs maintained in the fusions genes, a new pattern emerges, the recurrent inclusion of specific miRNAs in fusion genes downstream different 5’ partners. What makes these fusions special is the fact that the only requirement is the replacement of upstream regulatory regions that alter expression of the given miRNA. Consequently, it is the activity of the promoter that matters not the identity of the pair or the coding part of the gene. By focusing on functional outcome rather than on predefined structural features of the fusion, we rephrase the concept of recurrence and suggest that these types of promiscuous associations converge into a common function forming a specialized class of gene fusion that we called as “miRNA-convergent fusions” (Fig. 1). This structural freedom increases the number of potential oncogenic fusions considerably.
Figure 1.
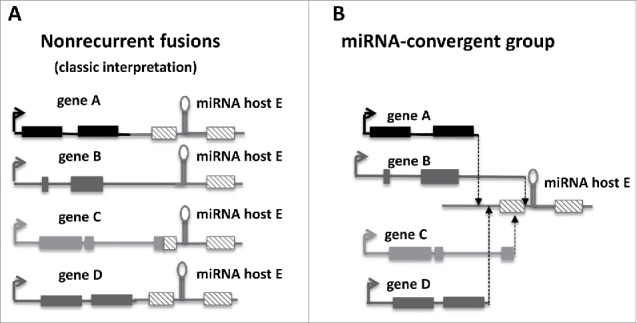
miRNA-convergent fusions. Different fusion pairs classified as nonrecurrent when considered individually (A) may functionally converge to regulate the transcriptional expression of a common miRNA forming a miRNA-convergent fusion group (B).
References
- 1.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al.. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004 doi: 10.1073/pnas.0307323101. PMID:14973191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Persson H, Søkilde R, Häkkinen J, Pirona AC, Vallon-Christersson J, Kvist A, Mertens F, Borg Å, Mitelman F, Höglund M, et al.. Frequent miRNA-convergent fusion gene events in breast cancer. Nat Commun. 2017;8:788. doi: 10.1038/s41467-017-01176-1. PMID:28983113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tassano E, Acquila M, Tavella E, Micalizzi C, Panarello C, Morerio C. MicroRNA-125b-1 and BLID upregulation resulting from a novel IGH translocation in childhood B-Cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49:682–687. doi: 10.1002/gcc.20776. PMID:20544842. [DOI] [PubMed] [Google Scholar]
- 4.Schneider B, Nagel S, Kaufmann M, Winkelmann S, Bode J, Drexler HG, MacLeod RA. T(3;7)(q27;q32) fuses BCL6 to a non-coding region at FRA7H near miR-29. Leukemia. 2008;22:1262–1266. doi: 10.1038/sj.leu.2405025. PMID:17989715. [DOI] [PubMed] [Google Scholar]
- 5.Feldman AL, Dogan A, Smith DI, Law ME, Ansell SM, Johnson SH, Porcher JC, Ozsan N, Wieben ED, Eckloff BW, et al.. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood. 2011;117:915–919. doi: 10.1182/blood-2010-08-303305. PMID:21030553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giacomini CP, Sun S, Varma S, Shain AH, Giacomini MM, Balagtas J, Sweeney RT, Lai E, Del Vecchio CA, Forster AD, et al.. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9:e1003464 doi: 10.1371/journal.pgen.1003464. PMID:23637631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, Rye IH, Nyberg S, Wolf M, Borresen-Dale AL, et al.. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12:R6. doi: 10.1186/gb-2011-12-1-r6. PMID:21247443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaki K, Menghi F, Woo XY, Wagner JP, Jacques PÉ, Lee YF, Shreckengast PT, Soon WW, Malhotra A, Teo AS, et al.. Systems consequences of amplicon formation in human breast cancer. Genome Res. 2014;24:1559–1571. doi: 10.1101/gr.164871.113. PMID:25186909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum AE, Venkitachalam S, Guo Y, Kieber-Emmons AM, Ravi L, Chandar AK, Iyer PG, Canto MI, Wang JS, Shaheen NJ, et al.. RNA sequencing identifies transcriptionally viable gene fusions in esophageal adenocarcinomas. Cancer Res. 2016;76:5628–5633. doi: 10.1158/0008-5472.CAN-16-0979. PMID:27503924. [DOI] [PMC free article] [PubMed] [Google Scholar]


