Abstract
Extensive prior research has focused on somatic copy-number alterations (SCNAs) affecting cancer genes, yet the extent to which recurrent SCNAs exert their influence through rearranging cis-regulatory elements remains unclear. Here, we present a framework for inferring cancer-related gene overexpression resulting from cis-regulatory element reorganization (e.g., enhancer hijacking), by integrating SCNAs, gene expression data, and information on chromatin interaction domains. Analysis of 7,416 cancer genomes uncovered several pan-cancer candidate genes, including IRS4, SMARCA1 and TERT. We demonstrate that IRS4 overexpression in lung cancer associates with recurrent deletions in cis, and present evidence supporting a tumor-promoting role. We additionally pursued cancer type-specific analyses, uncovering IGF2 as a target for enhancer hijacking in colorectal cancer. IGF2-containing tandem duplications result in the de novo formation of a 3D contact domain comprising IGF2 and a lineage-specific super-enhancer, which mediates high-level gene activation. Our framework enables systematic inference of cis-regulatory element rearrangements mediating dysregulation in cancer.
Introduction
Recent studies have provided numerous insights into the extent by which somatic DNA alterations affect protein-coding genes1–9. 98-99% of the genome, however, is made up of non-coding regions, a substantial fraction of which contain cis-regulatory elements (CREs)10–13. CREs, such as enhancers, can control gene expression over long distances – up to a megabase or more – accompanied by physical contacts of enhancers with the promoters of their target genes14–17. Several recent studies have uncovered somatic point mutations modulating gene regulation in cancer cells18,19,18–20, including those affecting CREs near TERT18,19, PAX521 and TAL122.
By comparison, much less focus has been placed on characterizing the effects SCNAs may have on CREs, in spite of the relevance of SCNAs in cancer4,23–31 and surveys suggesting that several common cancers are largely driven by SCNAs32. Individual studies focused on the pediatric cancer entities medulloblastoma28 and neuroblastoma29,30 as well as leukemia33,34, recently uncovered examples where recurrent SCNAs, including gains and losses, mediate gene overexpression by juxtaposing enhancers near cancer-related genes, a process termed enhancer hijacking. Importantly, the identification of enhancer hijacking events has challenged the previously widely followed principle that the type of SCNA can be used to define the function of putative cancer genes, with gains representing candidate oncogenic and losses tumor suppressor loci35. The extent to which enhancer hijacking occurs in different cancers yet remains unclear, since studies focused on identifying this process across different cancer types have been lacking. Relevant cancer driver genes acting in different cancers may thus far have been overlooked, since cancer genome analyses do not systematically search for this process.
Here we describe a computational framework denoted Cis Expression Structural Alteration Mapping (CESAM), which employs statistical concepts from expression quantitative trait locus (eQTL) mapping to integrate SCNAs, expression and chromatin interaction domain data36, to systematically identify SCNAs mediating gene dysregulation in cis. Using CESAM, we provide an estimate for the incidence of enhancer hijacking amongst the jumble of DNA rearrangements occurring in cancer genomes, report the first validated cases of enhancer hijacking in common solid tumors, and describe new mechanisms by which recurrent SCNAs mediate gene dysregulation.
Results
CESAM: inference of SCNA breakpoints associating with expression alteration in cis
CESAM integrates SCNA breakpoint data with donor-matched transcriptome (mRNA-Seq) data to identify candidate genes in cis, the altered expression of which is associated with SCNA-mediated rearrangements (Fig. 1a). This is achieved by employing linear regression of the mRNA-seq data on donor-matched SCNA breakpoint occurrence data (Methods). Thereby, CESAM relates gene expression values to binned SCNA breakpoints occurring in the vicinity of each gene. Breakpoint binning is achieved by making use of published data on topologically associating domains (TADs)36 (Fig. 1a), 3D chromosomal domains with a mean size of 830kb, which are largely invariant across cell types36–38. TADs can confine physical and regulatory interactions between enhancers and their target promoters38–42 and if disrupted can result in ectopic expression34,42.
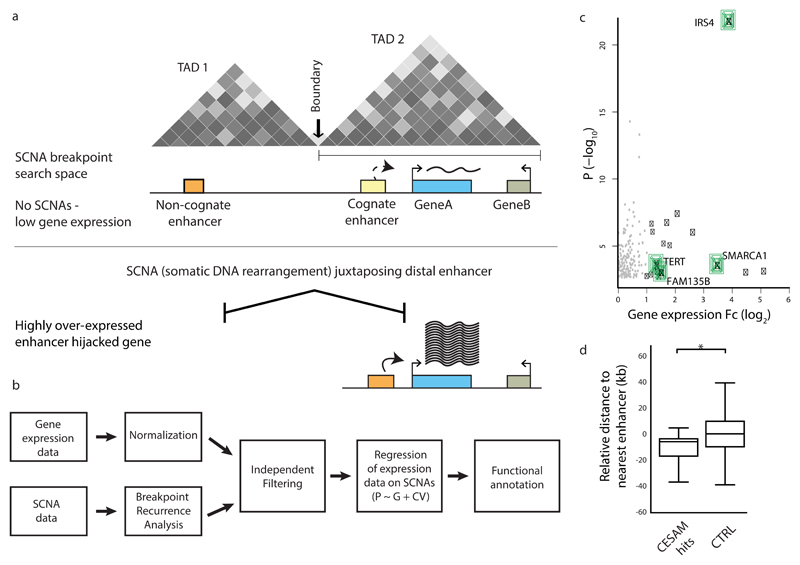
Figure 1. CESAM: framework for uncovering SCNAs driving gene dysregulation in cis.
(a) Principle behind CESAM. TADs are depicted as Hi-C-based contact maps36 with grey shading indicating locus interactions (darker shading indicates stronger interactions as measured by Hi-C). SCNA breakpoints are binned within each TAD (referred to as SCNA breakpoint search space). (b) Detailed analysis workflow of CESAM. (c) Volcano plot of CESAM hits in a pan-cancer setting, with nominal P-values plotted versus expression fold-change (Fc). Candidate genes identified by CESAM are shown as black dots (genes discussed in the text highlighted in green). Grey dots denote loci removed based on CESAM’s filtering criteria (which includes removal of expression alterations driven by gene dosage change). (d) Relative distance to nearest annotated enhancers at distal breakpoints of SCNAs identified by CESAM (‘CESAM hits’) versus SCNAs not implicated by CESAM, which are here used as control (CTRL) (P=0.001; based on 1000 permutations using the standard deviation of the observed proximity). Negative values refer to closer proximity to genomic feature relative to background.
For TADs that are recurrently affected by SCNAs, expression association is tested independently for each gene located within the given TAD (Fig. 1a and Supplementary Fig. 1). CESAM further pursues independent filtering43 to avoid testing genes that are (i) lowly expressed, (ii) display minor levels of expression variance, or (iii) are recurrently deleted or amplified to a copy-number ≥4 (Fig. 1b and Methods). To adjust for multiple-testing CESAM controls the false discovery rate (FDR) at 5%. Finally, CESAM summarizes functional annotations to facilitate inspection of proximal CREs.
Pan-cancer analysis of SCNAs affecting gene expression in cis
We employed CESAM to analyze 7,416 previously published cancer genomes involving 26 tumor types from The Cancer Genome Atlas (TCGA) data portal (http://cancergenome.nih.gov/). In this resource, SCNAs were defined based on SNP6 arrays. While these exhibit lower resolution than whole genome sequencing (WGS) for SCNA inference, presently the number of available specimens profiled both using mRNA-Seq and SNP6 arrays markedly exceeds published WGS datasets with matched expression data (http://icgc.org). We first performed a pan-cancer analysis with CESAM, identifying 18 gene loci with marked expression upregulation (fold-change ≥2) in conjunction with cis SCNAs (Fig. 1c, Supplementary Fig. 1, Table 1 and Supplementary Table 1). These encompassed several genes previously implicated in cancer, including the FAM135B gene (found altered in oesophageal cancer44), SMARCA145, a member of the SWI/SNF family of chromatin remodeling proteins, and TERT, which encodes a catalytic subunit of telomerase. A relatively high expression fold change when measured at the pan-cancer level (>25-fold) was observed for clustered deletions associated with upregulation of the insulin receptor substrate 4 (IRS4) gene. Simulations demonstrated enrichment of annotated enhancers, clustered enhancers also referred to as super-enhancers46, as well as promoters but not fragile sites at the distal end of SCNAs implicated by CESAM (Fig. 1d and Supplementary Fig. 1), in supporting of CRE-mediated activation mechanisms.
Table 1. Ranked list of CESAM pan-cancer candidate genes.
The list is ranked by FDR-corrected P-value, and is continued as Supplementary Table 1. For each candidate gene, tumor types with at least 3 samples exhibiting upregulation are depicted. FC, dosage-adjusted expression fold-change for SCNA-carrier samples vs. non-carrier controls. Padj, adjusted p-values, according to the Benjamini and Hochberg procedure.
| Gene | Chromosome location of TAD | FC | Padj |
|---|---|---|---|
| IRS4 | chrX: 107,720,001-108,600,000 | 15.0 | 2.47E-18 |
| TBL1X | chrX: 8,640,000-10,080,000 | 4.4 | 1.15E-05 |
| MTHFD1L | chr6: 151,200,000-151,760,000 | 3.4 | 4.01E-05 |
| LIPA | chr10: 91,000,000-91,520,000 | 2.3 | 4.63E-05 |
| PPP3CA | chr4: 101,080,000-103,480,000 | 2.4 | 0.000142 |
| MLLT4 | chr6: 167,400,000-169,000,000 | 6.2 | 0.000150 |
| NCOR1 | chr17: 15,880,000-16,440,000 | 3.1 | 0.000703 |
| GIGYF2 | chr2: 233,160,000-233,840,000 | 3.6 | 0.000822 |
| BTD | chr3: 15,600,000-16,280,000 | 2.6 | 0.00862 |
| SMARCA1 | chrX: 128,400,000-129,200,000 | 11.5 | 0.0127 |
| TERT | chr5: 40,000-1,720,000 | 2.7 | 0.0128 |
| OSGIN1 | chr16: 83,680,000-84,240,000 | 34.5 | 0.0250 |
| TSC22D3 | chrX: 105,560,000-107,240,000 | 2.7 | 0.0260 |
| STUB1 | chr16: 680,000-1,280,000 | 22.6 | 0.0275 |
| FAM135B | chr8: 138,840,001-139,800,000 | 2.9 | 0.0298 |
| KCNQ1 | chr11: 2,160,000-3,600,000 | 2.3 | 0.0353 |
| INSC | chr11: 16,760,001-17,520,000 | 2.1 | 0.0406 |
| PTCHD1 | chrX: 21,720,000-23,560,000 | 2.8 | 0.0463 |
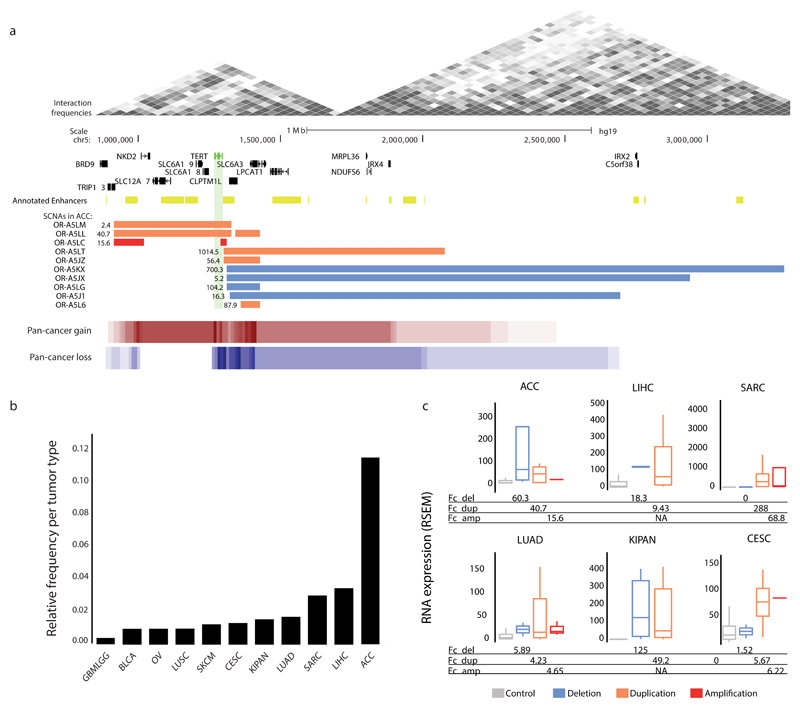
We first characterized the TERT locus, which CESAM identified in the highest number of cancers including kidney cancer, sarcoma and adrenocortical carcinoma (ACC). We observed the highest frequency in relation to cohort size (11.8%) for ACC (Fig. 2ab and Supplementary Table 1). Pronounced clustering of SCNAs became evident at the TERT promoter, where overexpression-associated cis SCNAs were previously described in chromophobe kidney cancer47. TERT-overlapping SCNAs (i.e. gains), however, occurred more rarely in ACC (Fig. 2a). Across cancers we observed TERT expression fold-changes of 2.7-fold in SCNA carrier vs. pan-cancer non-carrier samples. Within individual cancer types (i.e., compared to non-carrier samples from the same tumor cohort), however, we frequently observed much higher fold changes, e.g. >50-fold in ACC, kidney cancer and sarcoma. Both losses and gains contributed to overexpression, with deletions in cis occasionally resulting in even higher fold-changes than high-level (copy-number ≥4) TERT amplicons (Fig. 2c, Supplementary Fig. 2). Recent studies have implicated similar mechanisms of TERT upregulation in neuroblastoma29 and chromophobe kidney cancer47, lending support towards common mechanisms of TERT activation involving cis SCNAs in different cancer types.
Figure 2. Analysis of the TERT locus: a CESAM pan-cancer hit.
(a) Depiction of the TERT locus, the abnormal expression of which CESAM inferred to be mediated by cis SCNAs, for adrenocortical carcinoma (ACC) and summarized across cancer types (pan-cancer copy-number gains and losses). Gene expression values reflecting fold changes versus non-carrier ACC samples are indicated adjacent to each SCNA. (b) Fraction of donors per tumor type for which CESAM inferred TERT dysregulation along with SCNAs in cis in at least 3 donors (c) TERT expression values (unadjusted RSEM gene expression values) for different cancer types broken down by SCNA class (see Supplementary Table 1 for tumor-type abbreviations).
TAD boundary intersecting deletions associate with IRS4 dysregulation in sarcoma and squamous cancers
We next turned our focus to IRS4, a locus that CESAM identified in diverse cancer types. SCNAs in cis of IRS4, a gene located on chromosome X, were most commonly seen in lung squamous carcinoma (LUSC; N=22; see Fig. 3a), sarcoma (N=7), and cervical squamous carcinoma (N=3), although overall 48 samples from 10 tumor types exhibited IRS4 overexpression (Supplementary Fig. 3 and Supplementary Table 2). While not yet implicated in these cancer types, IRS4 was previously shown to have cell cycle promoting capabilities in vitro, i.e. IRS4 overexpression has been shown to enhance insulin-like growth factor-1 (IGF1) induced cell proliferation in the 3T3 cell line48 and to mediate proliferation and cell migration in hepatoblastoma cells49. The gene is presumed to act via the PI3K/AKT pathway49–52 with IRS4 overexpression inducing phosphatidylinositol 3,4,5-trisphosphate and AKT activation49–53, and AKT inhibitors blocking the growth promoting effect of IRS4 in vitro49. In spite of these prior findings, it is presently unclear whether IRS4 has any tumor-promoting role in vivo – and the relatively high recurrence level (e.g. 4.4% in LUSC; Supplementary Table 1 and Supplementary Fig. 4) of cis alterations associated with IRS4 overexpression prompted us to investigate this locus in further detail.
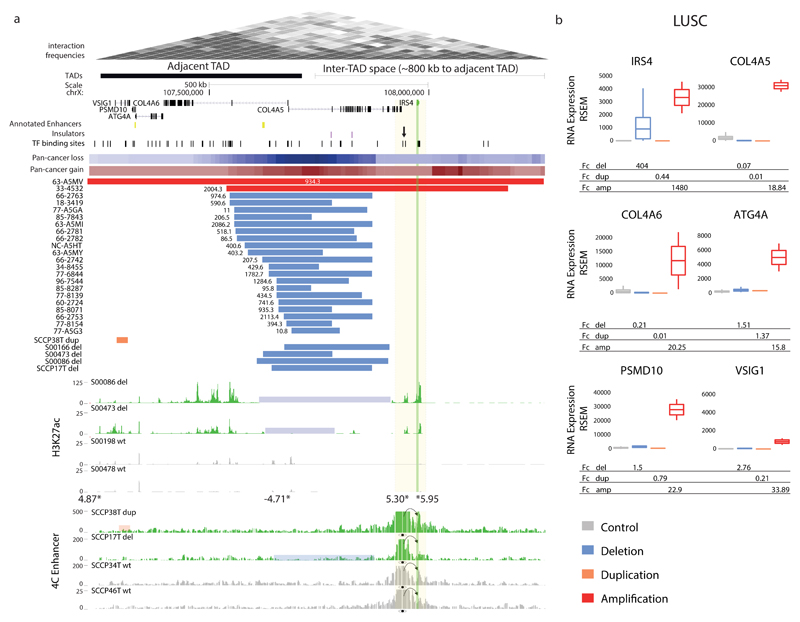
Figure 3. Recurrent SCNAs in cis associate with marked IRS4 expression increase.
(a) Recurrent deletions at a TAD boundary near IRS4, and IRS4 amplifications, associate with IRS4 dysregulation in LUSC. A region near IRS4 exhibiting clustered transcription factor (TF) binding sites (candidate CRE) is highlighted with an arrow. The recurrent deletions were evident both in male and female samples (indicating that both hemizygous and complete losses result in IRS4 overexpression). Summarized SCNAs across cancer types (pan-cancer copy-number gains and losses) shown as heatmaps. The full list of pan-cancer SCNAs at the locus is in Supplementary Table 2. Deletion-carrier samples (del, highlighted in blue) exhibited marked H3K27ac62 at the IRS4 promoter and adjacent candidate CRE. SCNA carrier samples in which chromatin analyses were performed were confirmed to exhibit outlier expression using semi-quantitative RT-PCR and qPCR (Supplementary Fig. 3, Supplementary Table 3, and data not shown). Asterisks depict differentially occupied peaks identified by genome-wide H3K27ac analysis (values adjacent to asterisks show Fc in H3K27ac signal for deletion-carriers vs. non-carriers). Lastly, 4C-Seq experiments using the candidate CRE as a viewpoint in carrier versus non-carrier samples are depicted. dup, duplication; WT, wild-type locus. (b) LUSC expression measurements (unadjusted RSEM gene expression values) for carriers versus non-carriers, revealing IRS4 as the most plausible target. IRS4 expression analyses revealed ~400-fold upregulation in deletion-carriers and >1000-fold for gene amplification carriers (number of control=470; del=24; dup=1; amp=2).
We focused our analysis on LUSC, where CESAM identified a set of recurrent deletions (N=20) clustering 103kb downstream of IRS4 within a region demarcated by chrX: 107,549,609-107,872,288 (hg19) (Fig. 3a). IRS4 expression was increased by on average 400-fold when comparing LUSC deletion carriers to non-carrier control LUSC samples, and 25-fold when specifically comparing pan-cancer deletion carriers to pan-cancer non-carrier controls, whereas other genes in cis, by comparison, exhibited only modest expression alteration (Fig. 3b, Supplementary Figs. 5, 6). We also observed focal high-level IRS4 gene amplifications in two LUSC samples as well as in several samples from other tumor types exhibiting massive overexpression, supporting IRS4 as the most plausible target of recurrent SCNAs at this genomic locus (Fig. 3 and Supplementary Fig. 6). The cis deletions, notably, intersected with a TAD boundary downstream of IRS4, which also coincides with CTCF binding sites at an inferred insulator region54 (Fig. 3a). In sarcomas, and to a lesser extent cervical squamous carcinoma, CESAM identified recurrent deletions at the exact same genomic interval in association with IRS4 overexpression (Supplementary Figs. 3, 6), an interval in which clustered deletions and IRS4 expression are also seen in benign uterine leiomyoma55.
Amongst TCGA lung cancer cohorts, SCNAs associated with IRS4 overexpression are confined to LUSC, with none of the TCGA lung adenocarcinoma samples exhibiting such events. This is noteworthy since altered PI3K/AKT pathway signaling has been found to be particularly abundant in LUSC56,57. We also observed inversely correlated expression of IRS4 and its paralog IRS2 (r=-0.11; P=0.008, Pearson product-moment correlation; Supplementary Fig. 4) in LUSC. The mutual exclusivity pattern suggests complementary roles in activating the PI3K/AKT pathway signaling50,53,58. Additionally, we observed a significant co-occurrence of deletions in cis of IRS4 and amplifications of the FGFR1 cancer census gene on chromosome 8 (Pearson’s chi-square test, X2=7.6; P=0.006, Supplementary Fig. 4). This is notable since IRS4 associates with FGFR1 and can promote FGFR1 signaling59, and since FGFR1 can also activate PI3K/AKT pathway signaling60. Collectively these data implicate IRS4 as a candidate genetic target in LUSC.
To investigate the tumor growth-promoting effects of IRS4 in vivo, we subcutaneously injected a lung squamous cancer cell line, HCC-15, with and without an IRS4 overexpressing vector into athymic nude mice, performing two independent experimental replicates (with N=8 and N=12 mice, respectively; see Supplementary Note). This was achieved by introducing a transgenic IRS4 and an empty control lentivirus vector into HCC-15 cells. We observed palpable tumor formation in mice receiving transgenic IRS4 overexpression plasmids as well as the empty control, albeit with a significantly increased tumor growth in tumors harboring the IRS4 overexpression plasmids in both experimental replicates (P=0.046 and P=0.03, respectively; two-tailed t-test; Supplementary Fig. 7 and Supplementary Table 3). Resected tumors maintained IRS4 overexpression, shown by immunohistochemistry, quantitative reverse transcription PCR (RT-qPCR) and flow cytometry (Supplementary Fig. 7 and Supplementary Table 3), which strongly suggests a tumor promoting effect of IRS4 overexpression.
Based on the pronounced clustering of deletions downstream of IRS4 we hypothesized that alterations in chromatin structure or landscape may underlie IRS4 dysregulation. To investigate this hypothesis we performed experiments in primary LUSC specimens (Methods). Expression analyses in 94 primary LUSCs demonstrated IRS4 overexpression greater than 10-fold in 11 (12%) samples based on RT-qPCR (Supplementary Table 4). We performed rearrangement screens in several samples using long-range paired-end sequencing61, and identified IRS4 proximal rearrangements in nine out of ten IRS4 overexpressing specimens (Supplementary Fig. 8 and Supplementary Table 4). To investigate the chromatin landscape in these samples, we performed chromatin immunoprecipitation followed by sequencing (ChIP-Seq) in three deletion carriers versus two controls (non-carrier LUSC samples both lacking the cis deletion and lacking IRS4 overexpression). Several observations emerged from these experiments. First, we identified an accumulation of the active chromatin mark H3K27ac62 on both sides of the commonly deleted region. Second, when comparing the deletion carriers to the controls, we observed four regions with differential H3K27ac marks (Fig. 3a and Supplementary Note). The strongest differential H3K27ac peaks within the respective wider genomic region of interest corresponded to IRS4, followed by a region 26kb downstream of the gene exhibiting clustered transcription factor binding sites (Fig. 3a and Supplementary Fig. 3). None of the non-carriers exhibited measurable H3K27ac marks at this putative CRE, suggesting that its activity is confined to samples with IRS4 upregulation. In addition, an H3K27ac peak at the bidirectional promoter of two nearby genes, COL4A5 and COL4A6 encoding collagen type IV subunits, showed significant loss of signal consistent with deletion of these genes’ promoter. Furthermore, we also observed modest differential H3K27ac signals near VSIG1, an immunoglobulin-domain containing gene that is only lowly expressed in LUSC (and similarly in other cancers), and which exhibits no, or only modest, expression changes in conjunction with cis SCNAs (Supplementary Figs. 5, 6).
To investigate whether the differences we observed in the chromatin landscape of deletion carriers versus non-carriers are accompanied by differences in 3D chromosome conformation, we additionally performed 4C-Seq (chromosome conformation capture sequencing63) experiments employing the putative CRE downstream of IRS4 as a viewpoint. These experiments revealed tight physical proximity between the putative CRE and IRS4, indicating that this genomic region indeed interacts with and hence represents a candidate IRS4 enhancer. Interestingly, the physical contacts between this CRE and the IRS4 promoter were also present in tumor specimens without the cis SCNA (Fig. 3a and Supplementary Fig. 3), an observation verified with 4C-Seq experiments using the IRS4 promoter as the viewpoint (Supplementary Fig. 3). These results suggest that TAD boundary or insulator loss-mediated spreading of active chromatin in the context of already established promoter-enhancer interactions is resulting in IRS4 overexpression (see our model in Supplementary Fig. 9).
IGF2: a CESAM hit in colorectal cancers exhibiting IGF2 locus tandem duplication
We next performed individual tumor type-focused analyses with CESAM, pursuing independent assessment across 26 cancer types. We identified between 1 and 14 candidates per cancer type, with 98 genes implicated by CESAM altogether in these tumor type-focused analyses (see Supplementary Table 1 and Supplementary Fig. 10). A CESAM candidate catching our attention was the IGF2 locus on chromosome 11, which CESAM implicated in colorectal cancer (CRC). IGF2 was >250-fold overexpressed in CRCs harboring nearby SCNAs compared to CRC non-carrier controls, whereas other genes nearby showed no or only modest expression alterations (Fig. 4abc). 22 out of 378 (6%) CRCs from the TCGA resource exhibited IGF2 upregulation in conjunction with cis SCNAs (Fig. 4a). Previously, IGF2 high-level overexpression in CRC was thought to result from recurrent focal locus amplification3,64,65, i.e. elevated gene dosage of a locus encompassing both IGF2 and the miR-483 microRNA gene3,47,48. The microRNA gene, which is embedded within intron 8 of IGF2, was recently implicated as a driver oncogene64,65. Given the joint upregulation of IGF2 and miR-483 in CRC3,64,65, and since both have been implicated in dysplasia and tumorigenicity64,65, we herein refer to this locus as the ‘IGF2 locus’ for simplicity.
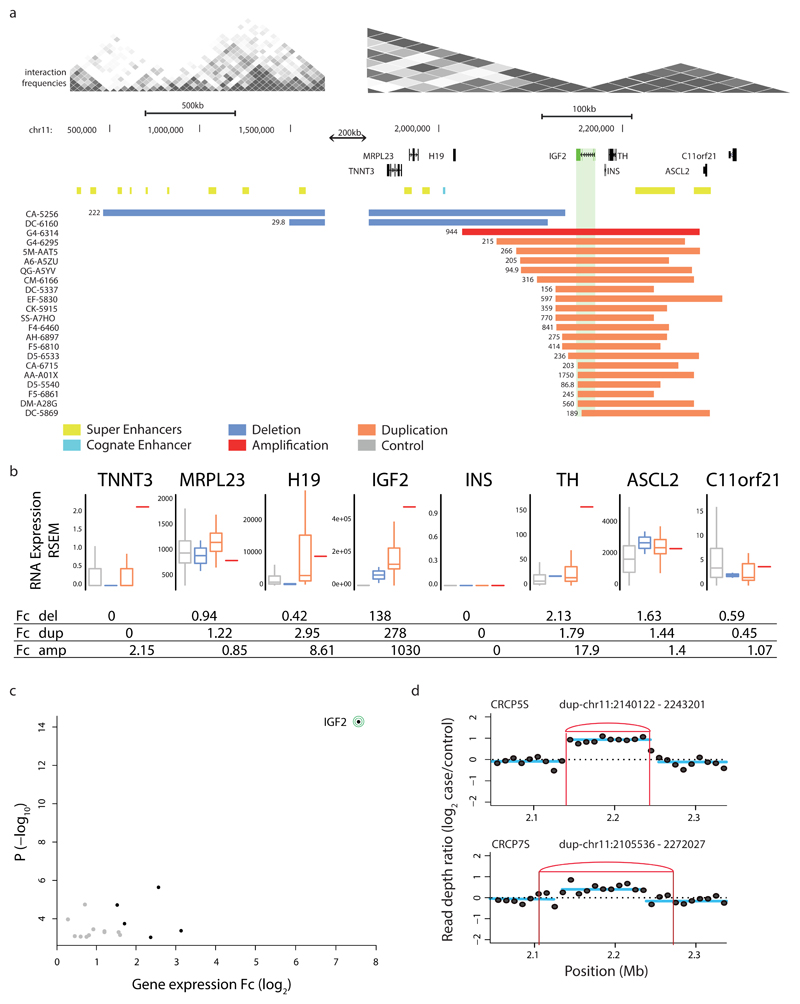
Figure 4. SCNAs associating with marked IGF2 locus overexpression in cis in CRC.
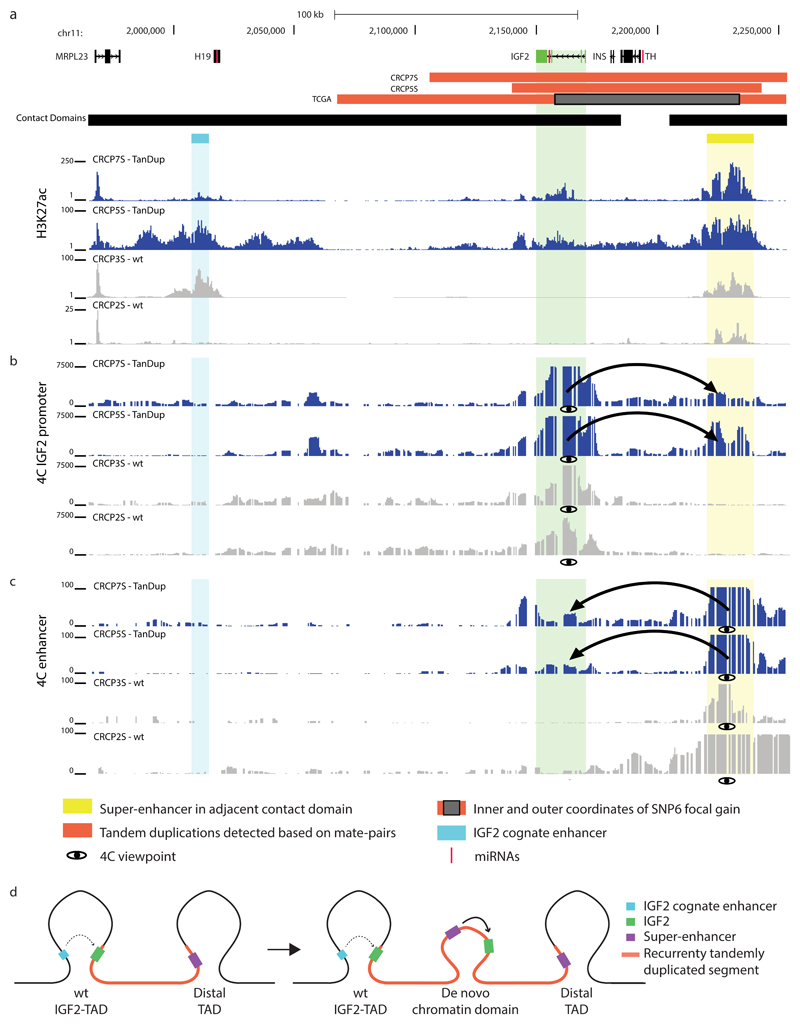
(a) Recurrent somatic duplications at the IGF2 locus (green) associating with IGF2 overexpression encompass a TAD boundary and a super enhancer (yellow) in the adjacent TAD, but do not encompass the known IGF2 cognate enhancer (light blue). Somatic deletions in cis extend over additional TAD boundaries. (b) Boxplots depict expression-SCNAs relationships for all protein-coding genes within the respective TAD, with IGF2 showing the by far most marked relationship making it the most likely target of these recurrent SCNAs in cis (boxplots separating into del carriers, dup carriers, amplification (amp; >4 copies) carriers, and control samples lacking SCNAs in cis). (c) Volcano plot of CESAM hits in CRC, with nominal P-values plotted versus log2-expression change based on all samples with SCNAs in TAD (CESAM hits are depicted in black; IGF2 is highlighted). (d) Structural variant detection by long-insert size paired-end sequencing4, followed by DELLY276 analysis, identified presence of TAD-spanning IGF2 locus tandem duplication in spheroid samples CRCP5S and CRCP7S (IGF2 outlier expression was verified in both samples by qPCR; see Supplementary Fig. 13, Supplementary Table 4).
Amongst the CRC samples exhibiting IGF2 dysregulation, 20 harbored gains and two harbored focal deletions in cis (Fig. 4a). Detailed examination of the SNP6 data showed that the corresponding gains at this locus typically underlie single-copy duplications (copy-number ratio of 1.25–1.75; see Fig. 4a and Supplementary Fig. 11), whereas only a single sample of the TCGA cohort showed higher-level locus amplification (copy-number ‘6’). The unusually high and consistent upregulation (>250-fold) in this context suggests that rather than gene dosage increase, specific locus rearrangements may drive IGF2 dysregulation.
To further characterize the mechanism of IGF2 activation we next performed experiments with spheroid cultures derived from primary CRC samples (Fig. 4d, Supplementary Fig. 12 and Methods). Expression profiling using RT-qPCR identified two CRC-derived spheroids overexpressing IGF2, denoted CRCP5S and CRCP7S (Supplementary Fig. 12 and Supplementary Table 5). Using long-range paired-end sequencing61 we uncovered single-copy tandem duplications both in CRCP5S and CRCP7S respectively, which seamlessly overlapped with the SNP6-based single-copy duplications in terms of size and position (Fig. 4ad). These data indicate that the recurrent gain at the IGF2 locus results from single-copy tandem duplications.
IGF2 activation through a super-enhancer mediated by de novo contact domain formation
We next performed ChIP-Seq at the IGF2 locus, detecting an accumulation of the active chromatin mark H3K27ac at a previously identified IGF2 enhancer66 herein referred to as IGF2 cognate enhancer (Fig. 5a, Supplementary Fig. 11). An even more pronounced H3K27ac peak, however, intersected with an element previously inferred to represent a lineage-specific super-enhancer in CRC cell lines (VACO-400 and VACO-9M)46. To verify enhancer function, we performed luciferase assays revealing enhancer activity of cloned fragments of this previously inferred super-enhancer46 in the HCT116 colon cancer but not in a control (HeLa) cancer cell line (Supplementary Fig. 12 and Supplementary Table 6). Notably, to our knowledge this super-enhancer has not previously been reported to physically interact with or regulate IGF2, and it indeed may not normally have the capacity to do so since it resides in an adjacent TAD (Fig. 5a and Supplementary Fig. 11).
Figure 5. Verification of IGF2 enhancer hijacking and model for mechanism involving de novo contact domain formation.
(a) ChIP-seq for H3K27ac62 yielding signals consistent with the activity of a previously annotated77 lineage-specific super-enhancer in the TAD adjacent to the IGF2 locus, but within the region accompanied by the recurrent somatic tandem duplication (TanDup). (b) 4C-Seq experiments using IGF2 promoter region as viewpoint demonstrate physical interaction between IGF2 and the super-enhancer in TanDup-carrier samples, but not in non-carrier samples (WT). (c) 4C-Seq experiments using the super-enhancer as viewpoint verify the highly specific physical interaction with IGF2 in TanDup-carriers (but not in WT samples). Further control data, for an additional WT sample, are in Supplementary Figure 11. (d) New model for high-level gene overexpression at the IGF2 locus in CRC, which involves TanDup-mediated de novo contact domain formation resulting in the hijacking of a lineage-specific super-enhancer.
Interestingly, the IGF2 locus tandem duplications extend over the intervening TAD boundary and also encompass this super-enhancer (Fig. 5a and Supplementary Fig. 11). We hence used 4C-Seq to investigate whether IGF2 dysregulation could be driven by topological or contact domain reorganization. And indeed, these data revealed the lineage-specific super-enhancer as the strongest interaction partner of IGF2 in CRCP5S and CRCP7S with complete absence of this interaction in control spheroids (Fig. 5b and Supplementary Fig. 11), an interaction that we verified in a reciprocal 4C-Seq experiment using the super-enhancer as viewpoint (Fig. 5c and Supplementary Fig. 11). By comparison, 4C-Seq reads connecting IGF2 with its cognate enhancer were absent, indicating that IGF2 is not activated by its cognate CRE in this context (Fig. 5b and Supplementary Fig. 11).
Our observations can be summarized in a model whereby a de novo 3D contact domain comprising a gene locus relevant to cancer (IGF2) and a super-enhancer forms in between preexisting TADs, resulting in oncogenic locus dysregulation (Fig. 5d). Indeed, the tandem duplications are inferred to result in copies of IGF2 and the super-enhancer being positioned in a head-to-tail orientation, with both now being able to contact each other via chromatin looping (see our model in Fig. 5d). We further examined the potential of the tandemly duplicated sequence to form a new contact domain by pursuing ChIP-Seq of CTCF, a DNA-binding protein that resides at contact domain boundaries36,67, and observed increased CTCF binding consistent with boundary use (Supplementary Fig. 11). Lastly, we also identified three larger somatic duplications of IGF2 in the TCGA data, which based on their size and location with respect to TAD boundaries are inferred to not lead to the formation of a 3D contact domain comprising IGF2 and this super-enhancer (Supplementary Fig. 13). Notably, none of these three IGF2 duplication carriers exhibited appreciable levels of IGF2 overexpression (exhibiting significantly lower IGF2 expression compared to tandem duplications with the potential to lead to 3D contact domain formation; P=0.01; Wilcoxon rank-sum test), lending additional support to our new model. Taken together, we show that rather than by gene dosage increase a hitherto undescribed mechanism – tandem duplication-mediated de novo contact domain formation resulting in physical interaction between the IGF2 promoter and a normally hidden super-enhancer – drives overexpression of IGF2 in CRC (Fig. 5d).
Discussion
We developed CESAM to enable systematic discovery of enhancer hijacking events in cancer genomes, and inferred 18 candidate enhancer hijacking events in a pan-cancer analysis and 98 in tumor-type specific analyses. Earlier studies have provided comprehensive views of recurrent SCNAs in cancer, and the GISTIC algorithm23,27 has emerged as an important standard for identifying recurrent SCNAs in cancer. Our analyses using CESAM in pan-cancer and tumor-type specific settings, notably, identified sixteen cancer-related genes previously assigned to GISTIC peaks as CESAM hits (e.g., IRS4 and FAM135B). Our data collectively suggest that activation of cancer genes by juxtaposition of CREs is a fairly common process, which may be comparable to the number of recurrent in-frame gene fusions leading to 3’ target overexpression in cancer (e.g. recent work by Yoshihara et al. uncovered 39 such events to be recurrent in at least four cancer samples (a similar threshold as used in our study) in an analysis encompassing 4,300 TCGA donors68).
Hits uncovered by CESAM include, to our knowledge, the first validated cases of enhancer hijacking in adult solid cancers. We provide in vivo evidence for a tumor growth-promoting role of IRS4 – a gene dysregulated in conjunction with deletions in cis in several cancers. The identified upregulation of IRS4 (~400-fold overexpression in deletion carriers) in LUSC is associated with a marked gain in active chromatin marks at the gene’s promoter as well as at a candidate enhancer region. Notably, our observations of a stable promoter-enhancer chromatin looping state present both in an active and silent context shows resemblance with observations from gene regulation during metazoan development, where marked changes in expression typically do not involve alterations in enhancer-promoter contacts, but arise amongst pre-existing chromatin loops69. Our data are compatible with disruptions of CTCF insulators at TAD boundaries through recurring deletions34,42, the consequence of which appears to be the spreading of active chromatin marks in the context of IRS4 (see our model depicted in Supplementary Fig. 9). Consistent with our findings, CRISPR-mediated deletion of a CTCF insulator region at the Hox gene cluster has recently been shown to lead to spreading of active chromatin to neighbouring gene regions in embryonic stem cells70.
Furthermore, our tumor type-specific analyses uncovered that enhancer hijacking mediates gene dysregulation at the IGF2 locus in CRC. This involves a previously undescribed mechanism, whereby tandem duplication-mediated de novo formation of a contact domain accompanying a super-enhancer normally inaccessible to IGF2 results in >250-fold gene upregulation. IGF2, an imprinted gene66,71,72, is associated with aggressive and chemotherapy resistant cancer (reviewed in64), and our findings unexpectedly revealed enhancer-hijacking as the dominant mechanism of high-level overexpression at this well-known locus.
Since CESAM does not consider recurrent focal amplicons leading to locus copy-numbers of four or higher, our analysis did not include super-enhancer amplification events. These have recently been shown to lead to up to four fold overexpression of super-enhancer target genes in epithelial cancers73 – another remarkable mechanism by which tumors can exploit the regulatory genome. Since some candidates uncovered by CESAM present with expression fold-change of >100-fold it is tempting to speculate that enhancer hijacking may result in comparably more pronounced expression changes, possibly by providing access to otherwise inaccessible regulatory regions (Fig. 5d and Supplementary Fig. 9). Finally, we note that previously described examples of enhancer hijacking occasionally involved balanced translocations28,74, which are incompletely captured by SCNA profiling. In the future, similarly sized sets of whole genome sequenced cancer genomes with matched expression data, including those that will be provided by the Pan-Cancer Analysis of Whole Genomes initiative75, may enable incorporating such events into systematic CESAM searches. Given its potential to systematically uncover enhancer hijacking events, CESAM has repercussions for the design of analysis strategies to uncover genetic driver alterations in cancer genomes.
Online Methods
Cis expression structural alteration mapping (CESAM)
CESAM integrates SCNA-derived breakpoints with RNA-Seq data (RSEM, RNA-seq by expectation-maximization) to identify expression changes associated with breakpoints in cis. SCNA (N=10,320, SNP6-derived) and RNA-seq (N=9,999) data (representing 27 tumor types), embargo-free, were downloaded from the TCGA data portal (15.11.2015, hg19). In total, 7,416 donors having both SCNA and expression data, and involving 26 tumor types, were considered in our analysis (this excludes breast cancer; see below).
SCNA-derived, TAD-bound breakpoint occurrence matrix
CESAM performs linear regression of expression (molecular phenotype) on SCNA-derived breakpoint (somatic genotype) data. To identify breakpoints associated with cis expression, we used recently published TAD data from the IMR90 cell line36 (mean TAD size: 830kb). A somatic genotype matrix based on ‘TAD bins’ was constructed using BEDTools (v2.24.0)78 by annotating for every sample the presence/absence of breakpoints within a TAD. For the purpose of CESAM we defined as ‘TAD bins’ annotated TAD boundaries36 extended by 50 kb on either side allowing for flexibility in boundary precision. We then (somatically) genotyped every TAD bin (row) in every donor (column), and excluded TAD bins with fewer than 4 donors based on our independent filtering criteria. In extended genomic regions where adjacent TAD-bins exhibit similar somatic genotypes – i.e. where donors show similar patterns of presence/absence across neighboring TADs (for example, in the presence of recurrent SCNAs harboring their breakpoints in two neighboring TADs) CESAM performs neighbor 'TAD bin merging' combining adjacent TAD bins with similar somatic breakpoint genotypes into 'meta bins' based on PLINK79. We triggered ‘TAD bin merging’ if from the midpoint of a given TAD there was an adjacent TAD with its midpoint being ≤1000kb apart showing a somatic genotype similarity of R2≥0.2. In practice, this can avoid that similar somatic genotypes are tested repeatedly in adjacent TAD bins.
RNAseq-derived gene expression matrix
RNA-seq derived gene expression matrices comprising RSEM values (hg19) were scaled by log2-transformation, and independent filtering43 was employed to remove genes with low expression variance (i.e. genes with variance below the 20th percentile). To alleviate the effect of gene dosage, CESAM’s regression analysis adjusts for somatic gene copy-number alterations by dividing each gene’s expression (prior to log2-transformation) by the tumor/normal gene copy-number ratio. The relationship between signal and copy-number is known to be not linear in SNP microarrays, which are subject to saturation effects80 especially affecting regions with high copy-number status. As this may affect our ability to reliably identify enhancer hijacking events in such regions with CESAM’s dosage-adjusted regression analysis, our independent filtering criteria further conservatively removed genes recurrently deleted or amplified to a level of four or more copies in >0.4% of samples (“4 per-mille criterion”; in very small cohorts, we used a minimum of two amplicons to trigger filtering). The 4-per-mille criterion is generally applied by rounding up to the next integer. In practice, whereas genes such as KRAS that are frequently highly amplified become excluded from CESAM analysis through this criterion, IGF2 and IRS4 would not be filtered even if using a more stringent “2-per-mille criterion” (i.e. when filtering genes with high-level amplicons in >0.2% of samples).
Regression analysis
The regression analysis of CESAM involves a cis-eQTL search with the FastQTL (v2.1) algorithm81, which conservatively uses a relatively large (2 Megabase) cis-window centered on the TAD’s midpoint to relate TAD-binned SCNA breakpoints with expression changes. Although in practice gene expression changes were encouragingly nearly always most highly associated with SCNA breakpoints residing in the same TAD, the enlarged cis search window can facilitate identifying genes whose expression correlates best with breakpoint occurrence in cis. We performed 1000 permutations with FastQTL for statistical inference, using default parameters81. To minimize the effect of confounders, we used the following covariates in the regression: (i) the total number of SCNAs for each sample, to adjust for SCNA burden effects, (ii) principal components (PC), based on principal component analysis82 on the somatic SCNA-derived breakpoint matrix. An optimization step was executed whereby PCs were added sequentially until the genomic inflation factor lambda (calculated using chi-squared statistics83 was in the desired range of <2). Since we failed to reach genomic inflation factor <2 for breast cancer samples, we excluded this cancer type from our CESAM analysis.
Integrative analysis and filtering of CESAM hits
We employed an FDR of 5% using the Benjamini-Hochberg procedure, and required >2-fold expression upregulation relative to controls, for reporting CESAM candidate genes. Fold change was computed as the median expression in the group of SCNA “carriers” compared to the median of "non-carrier" control donors (median values were set to a minimum value of 1 RSEM in cases were a lower median expression level was seen). Candidate genes were then additionally filtered to adjust for gene fusion events as well as previously unaccountable ‘residual’ gene dosage effects. For fusion gene removal, CESAM identified candidate genes showing a predominance of SCNAs at the 5' end of the gene, which were then compared with the TCGA fusion database68 encompassing recurrent in-frame fusions with 3' partners leading to gene overexpression (in practice this step readily identifies known fusions, e.g. of ERG in prostate cancer). We also performed literature searches to remove previously described putative fusion genes. To recognize residual dosage effects CESAM applies finally ‘population-based dosage filtering’, by evaluating for each CESAM candidate gene whether expression in SCNA carriers vs. non-carriers is correlated linearly with the somatic gene copy-number status. Genes significantly correlated with somatic gene copy-number (linear least-squares regression, R2>0.2, and p<0.05) are removed by this population-based dosage filtering module. In practice, while CESAM’s regression analysis uses RNAseq expression values that are already adjusted for copy-number, we occasionally observe residual effects of gene copy-number not properly accounted for attributable to array saturation effects80, which are recognized by the population-based dosage filter. To identify SCNAs juxtaposing distal CREs13,46 for any given SCNA with two breakpoints b1 and b2 – with b1 being closest to the candidate gene – CESAM identifies the closest CRE proximal to b2.
Code availability
The code of CESAM is made available upon request.
Primary lung squamous cell cancer samples
Primary squamous cell lung cancer samples were obtained from Oslo University Hospital and from Cologne University Hospital, following informed consent obtained from each patient with appropriate approved by Review Boards.
Generation and culturing of tumor-initiating cell enriched primary CRC spheroid cultures
Primary human CRC samples or its derived metastases were obtained from Heidelberg University Hospital in accordance with the declaration of Helsinki. Informed consent on tissue collection was received from each patient, as approved by the University Ethics Review Board. The tumor tissue was minced and enzymatically digested using dispase (Stemcell Technologies). The digested tissue was filtered and the single cell suspension was cultured under serum free conditions in advanced DMEM/F-12 medium supplemented with glucose to 0.6% (Invitrogen), 2 mM L-glutamine (Invitrogen), 4 mg/ml BSA (Sigma-Aldrich), 5 mM HEPES (Sigma Aldrich), 4 μg/ml heparin (Sigma Aldrich), 1% penicillin/streptomycin (Invitrogen), in ultra-low attachment flasks with the addition of cytokines: 10 ng/ml FGF basic and 20 ng/ml EGF (R&D Systems) as previously described84. Cytokines were added twice a week. Depending on the patient culture, spheroids were dissociated manually by pipetting up and down 15-20 times or by treatment with accutase (PAA Laboratories GmbH) for 10-60 min. All spheroid cultures were authenticated and checked by Multiplexion (http://www.multiplexion.de/en/) for contamination against various species of bacteria, viruses, contaminating cells lines and murine cell contamination. CRC tumor initiating cells (TIC) were enriched in spheroid cultures from primary patient tumor tissue as previously described in Dieter et al.84.
Isolation of nucleic acids for DNA sequencing and RNA expression analysis
Following review by a pathologist, 30 μg of tumor tissue was used for the extraction of nucleic acids. In addition, patient derived TIC enriched spheroid cells were pelleted by centrifugation (800 rpm, 4°C, 5 min) and washed two times with PBS to get rid of the residual media. DNA and RNA of primary patient tissue and spheroids were isolated using DNeasy® Blood & Tissue Kit (Qiagen) and AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer’s instruction. RNA extracted in Oslo was isolated using Standard TRIZOL methods (Invitrogen, Carlsbad, CA), as specified by the manufacturer's instructions. The RNA was treated with on-column DNase I digestion protocol based on the manufacturer’s instruction (Qiagen) to get rid of any residual DNA. The DNA and RNA were quantified using Nanodrop and Qubit according to manufacturer’s instructions.
Chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-Seq)
105 to 107 cells were expanded, fixed with freshly prepared formaldehyde solution (11% Formaldehyde (Sigma Aldrich), 0.1M NaCl (Sigma Aldrich), 1mM EDTA (pH8) (Sigma Aldrich), 50mM HEPES (pH7.9) (Sigma Aldrich)) and agitated for 15 min at room temperature. The reaction was stopped by adding 1/20 volume Glycine Solution (Sigma Aldrich) and subsequently incubated for 5 min. The cells were washed with PBS to get rid of any media constituents and re-suspended in 10 ml chilled PBS-Igepal (0.5%) (Sigma Aldrich), and next centrifuged and resuspended in PBS-Igepal (0.5%) along with the addition of 100 μl PMSF (1mM) (Sigma Aldrich). The cells were then centrifuged, the supernatant was discarded and pellets were snap frozen on dry ice.
Samples were submitted to Active Motif (https://www.activemotif.com) for ChIP-Seq. Active Motif prepared chromatin and performed ChIP reactions. In brief, 3D cell cultures of pediatric tumors were fixed in PBS with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Chromatin was isolated using Active Motif’s proprietary buffer for low cell number ChIP-Seq. DNA was sheared to an average length of 300-500 bp with Active Motif’s EpiShear probe sonicator (53051) and cooled sonication platform (53080). Genomic DNA (Input) was prepared by treating aliquots of chromatin with RNase, proteinase K and heat for de-crosslinking, followed by ethanol precipitation. Pellets were resuspended and the resulting DNA was quantified on a NanoDrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield.
Chromatin was pre-cleared with protein A agarose beads (Life Technologies). Genomic DNA regions of interest were isolated using 4 ug of antibody against CTCF (Active Motif 61311, Lot. 2) and H3K27me3 (Millipore 07-449, Lot. 2475696). Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation.
Illumina sequencing libraries were prepared from the ChIP and input DNAs using the standard consecutive enzymatic steps of end-polishing, dA-addition, and adaptor ligation. After the final 15 cycle PCR amplification step, the resulting DNA libraries were quantified and sequenced (Illumina platform). Sequences (75 bp, single end) were aligned to the human genome (hg19) using BWA-mem (0.7.4)85. Duplicate reads were removed and only uniquely mapped reads (mapping quality >= 25) were used for further analysis. Alignments were extended in silico at their 3’-ends to a length of 200 bp, which is the average genomic fragment length in the size-selected library, and assigned to 32-nt bins along the genome. Filtering and peak calling was performed using HOMER (v4.7.2)86 with standard settings.
4C-Seq library preparation and sequencing
4C-Seq libraries were prepared following the protocol in Splinter et al.87 with some modifications. Briefly, 10M cells from each spheroid culture were dissociated and fixed with 2% formaldehyde. The fixed genomic DNA was digested using the NlaIII enzyme and subsequently self-ligated. A second digestion reaction was performed with DpnII, followed by ligation. After purification of the circularized DNA, inverse PCR was performed to obtain 4C-Seq libraries. 1.6 μg of template DNA was used for the amplification of the final libraries. For primary LUSC samples, cells were first dissociated with 0.0125% collagenase and nuclei isolated and subsequently fixed with 1% formaldehyde. Due to the low amount of tissue material, the 4C-Seq protocol was modified to use 1/3 of the volumes stated in the original protocol. For these libraries, 800 ng of template DNA was used for final library amplification. The reading primers (Supplementary Table 7) had 4–6 nucleotides of barcode sequences, to allow for de-multiplexing of pooled libraries. PCR products were purified, mixed altogether and sequenced on an Illumina HiSeq 2000 as well as an Illumina NextSeq platform in 100 and 75 bp paired-end read length mode, respectively. Alignment was performed using bwa-mem in single-end mode (v 0.7.4) to reference genome hg19. 4C interactions were identified using FourCSeq88.
Quantitative reverse transcription PCR (RT-qPCR) based expression measurements
RT-qPCR) was performed to identify samples with strong overexpression for IGF2 and IRS4. 35 CRC tumor sample RNAs for IGF2 were obtained from University Hospital Heidelberg (extracted with the AllPrep DNA/RNA Mini Kit (Qiagen)), whereas 94 Squamous cell carcinoma tumor sample RNAs for IRS4 were obtained from Oslo University (extracted with TRIzol (Invitrogen)). Only RNA samples with a RIN value > 3 and with tumor content >30% were used. Single-stranded cDNA was synthesized from 500 ng of total RNA using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen) according to the manufacturers’ protocol. qPCR primers were designed using the online Primer3 Plus program89 with the qPCR settings activated. Primer sequences were IRS4_F: CCCACACATGAGCAGAGAGA, IRS4_R: CTGACTGTCTGGGTTCAGCA, Globulin_F: TACATGTCTCGATCCCACTTAACTAT, Globulin_R: AGCGTACTCCAAAGATTCAGGTT, IGF2_F: TGGCATCGTTGAGGAGTGCTGT and IGF2_R: ACGGGGTATCTGGGGAAGTTGT. All primers were tested by running a standard curve and requiring the primer efficiency to be between 90-100% and as close as possible to that of the house keeping primer pair. The primer efficiency for globulin was 91.3%, 91.6% for IGF2 and 95.6% for IRS4. In addition, a single and discrete peak was detected in the melt curve analysis for all primers tested. The qPCR experiments were performed on a StepOnePlus 96 Fast machine (Applied Biosystems) in 20ul using a 96 well plate. The mastermix contained 10 μl 2 × SYBR Green PCR Master Mix (Applied Biosystems), 0.4 μl of each primer (10 μM), 2.5-5 ng of sample cDNA in 5 μl and 4.2 μl nuclease free H2O. The reaction program was run in default ramping speed mode and cycling conditions were 10 min at 95°C, 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by a melting curve stage. Non template controls were included in all experiments, replacing cDNA with H2O, and typically resulted in no detection at all. The results were analyzed using the StepOne analysis software v2.3 (Applied Biosystems). Relative expression levels for IGF2 and IRS4 were calculated relative to the house keeping gene globulin using the ΔΔ-Ct method. Each sample was measured in technical duplicates, and the relative fold expression difference was compared to the median expression value of all samples for IGF2, or the median of 7 representative samples with expression near the technical background for IRS4.
Massively-parallel DNA sequencing
Two types of Illumina next generation sequencing libraries, long-insert size paired-end mapping (mate-pair sequencing) as well as (regular) Illumina paired-end sequencing to 1x (low) coverage, were used to analyse somatic structural rearrangements in a locus-specific manner. In more detail, mate-pair DNA library preparation was performed using the Nextera Mate Pair Sample Preparation Kit (Illumina). In brief, 4 μg of high molecular weight genomic DNA was fragmented by the tagmentation reaction in 400 μl, followed by strand displacement. Samples were size-selected to 4–5 kb following the Gel-Plus path of the protocol. A total of 300–550 ng of size-selected DNA was circularized in 300 μl for 16 h at 30°C. After an exonuclease digestion step to get rid of remaining linear DNA, fragmentation to 300–700 bp with a Covaris S2 instrument (LGC Genomics), and binding to streptavidin beads, the libraries were completed via End Repair, A-Tailing, and Illumina Truseq adapter ligation. The final sequencing library was obtained after PCR for 1 min at 98°C, followed by nine cycles of 30 s at 98°C, 30 s at 60°C, 1 min at 72°C, and a final elongation step of 5 min at 72°C. Sequencing was carried out with Illumina HiSeq2000 (2x101 bp reads) instrument using v3 or v4 chemistry to reach an average spanning coverage of 20–30x. Short insert size library preparation was performed using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs). Briefly, 250ng of genomic DNA were fragmented with a Covaris S2 instrument (LGC Genomics) to 700-800 bp and then processed according to the manufacturers’ protocol and sequenced in 2x125 bp mode.4,90. We employed an in-house Illumina HiSeq 2000 platform for sequencing each library sequenced to average physical depths (spanning-coverage) of 35x for mate-pair sequencing and <2x for low-coverage/short-insert-size sequencing, using 100 bp paired-end reads.
Structural variant calling was performed using the procedure described in91, by aligning reads to the to hg19 reference genome assembly with bwa-mem (v0.7.4), and using DELLY2 (v0.6.8)76 for structural variant discovery.
Identification of H3K27ac peaks with differential H3K27ac signal in candidate regions
Differential H3K27ac occupancy analysis was performed using Bioconductor, in particular, the DiffBind92 package. Briefly, LUSC (cis deletion-carriers n=3; non-carrier controls n=2) H3K27ac peaks, as well as CRC (tandem duplication-carriers n=2; non-carrier controls n=4) H3K27ac peaks called by Homer86 and corresponding H3K27ac ChIP-seq bam files were used as the input data for the analysis. Differentially bound peaks were identified with the modules “dba.count”, dba.contrast”, “dba.analyze”, and “dba.report” of the package, consecutively.
Specifically, we first performed an unbiased differential H3K27ac occupancy analysis comparing LUSC deletion-carriers to non-carrier controls at the IRS4 locus and its vicinity, and controlled the FDR at 5%. This analysis revealed only four peaks with differential H3K27ac signal within the relevant region shown in Fig. 3 (a nearly 1 Megabase long region, which includes a TAD as well as inter-TAD space). The two peaks exhibiting the most significant differential H3K27ac signal corresponded to the IRS4 gene itself as well as the inferred novel IRS4 enhancer, respectively (see asterisks in Fig. 3). In both cases H3K27ac signal was significantly higher in cis SCNA (deletion) carriers. The third most significant differential peak, which again exhibited more H3K27ac in SCNA carriers, localized ~20kb upstream of VSIG1. However, as opposed to IRS4, VSIG1 is barely expressed and its expression showed only slight increases in deletion-carriers (2.7-fold for VSIG1, vs. 400-fold for IRS4; see Fig. 3 and Supplementary Fig. 3), which strongly implicates IRS4 (rather than VSIG1) as the target of these recurrent cis SCNAs. The fourth peak localizing at the bidirectional promoter of the COL4A5/COL4A6 showed significantly less H3K27ac signal in deletion-carriers in line with promoter deletion in SCNA carriers and with the lower expression of COL4A5 and COL4A6 in LUSC deletion-carriers (Fig. 3). Together with the observation that occasional locus amplifications and duplications clearly drive IRS4 expression (Supplementary Fig. 6), these data nominate IRS4 as the most plausible candidate gene becoming aberrantly activated as a consequence of recurrent SCNAs in this genomic region.
Differential H3K27ac occupancy analysis for CRC samples showing IGF2 tandem duplication versus non-carrier controls did not reveal a single peak with differential H3K27ac signal on chromosome 11 when controlling the FDR at 5%. By comparison, when controlling the FDR at 20%, we identified only one large peak covering IGF2 itself and the respective TAD boundary (see H3K27ac occupied region in Fig. 5a) as differentially marked with H3K27ac on chromosome 11 (which is consistent with the massive activation of IGF2 as a consequence of recurrent locus rearrangements).
Cell line, vectors and virus preparation
HCC-15 cell line was purchased from DSMZ and cultured in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific) and Antibiotic-Antimycotic (Thermo Fisher Scientific). An IRS4 overexpressing vector, pLenti-IRS4-Myc-DDK, was purchased from OriGene. An IRES-eGFP sequence was cloned from a pIRES2-AcGFP1 vector (Takara-Clonetech) into the pLenti-IRS4-Myc-DDK vector using In-Fusion HD cloning kit (Takara-Clonetech) and it is referred as pLenti-IRS4. We used the following primers for this purpose: F-GGCCGCGGTCTGTACA-cttcgaattctgcagtcgacg; and R-GAATCCTACTTGTACAtcacttgtacagctcatccatgcc. The control vector was created by removing IRS4-Myc-DDK by restriction enzyme digest with EcoRI and it is referred as pLenti-empty. Plasmids used for lentivirus production were pMD2.G (VSV-G envelope) and psPAX2 (2nd generation lentiviral packaging plasmid); both gifts from Didier Trono - Addgene plasmids #12259 & #12260. Lentivirus production was conducted by transfection with Lipofectamine 3000 Reagent (Thermo Fisher Scientific) of equal amounts of pMD2.G, psPAX2 and pLenti-IRS4-Myc-DDK-IRES-GFP/pLenti-IRES-GFP, in 293FT cells (Thermo Fisher Scientific) according to the manufactures protocol. Cells were transduced with produced virus with the addition of 8ug/mL polybrene (Sigma-Aldrich) by spinfection (centrifuge 2000rpm for 2 hours) with the produced virus and were enriched by sorting according to eGFP intensity (see Flow cytometry methods). All cell lines were regularly checked for mycoplasma contamination.
Flow cytometry
Transduced HCC-15 cells were sorted for eGFP expression on a MoFloXDP cell sorter (Beckman Coulter Inc) equipped with a Coherent Innova 90C Argon ion laser (Coherent Inc.), tuned to 488nm at 200mW. Cells were sorted using a 100um Nozzle while running BD FACSFlow as sheath at 20psi/RT. Forward and side scatter height and area signals were used for gating of live cells and singlets. eGFP fluorescence was detected using a 530/40nm bandpass filter combined with a 488notch filter. eGFP positive cells were sorted in purity mode (1 drop envelope) into 6-well or 96-well dishes with culture media respectively. In order to measure eGFP intensity HCC-15 cells were run through LSR-Fortessa SORP instrument (BD Biosciences) with a 488 nm laser (530/30 BP). All post acquisition analysis was done with FlowJo 10.0.8 (Tree Star, Inc).
Immunohistochemistry
Immunohistochemistry was performed according to Sotillo et al93. Anti-IRS4 antibody used was purchase from Abcam (clone EP907Y - Product code ab52622, 1DegreeBio ID: 1DB-001-0001145254).
Mouse experiments
1 million transduced HCC15 cells were suspended in DMEM mixed 1:1, v/v with Matrigel (BD Biosciences) and subcutaneously implanted into both flanks of nude mice (Charles River Laboratories, NMRI-Foxn1nu /Foxn1nu (homozygous) male mice; 8 weeks old at time of injection). The total number of tumors N=8 for each group in the first experiment (i.e. two cell line injections in each of four mice, whereby we performed experiments in both flanks in each mouse), N=9 for control (5 mice) and N=12 (6 mice) for IRS4 overexpressing sample in the second experiment. While at this sample size effect sizes are not robustly estimated, differences in tumor growth became readily evident. Mice were randomly assigned into two groups and tumor sizes were measured twice weekly in two dimensions (length and width). Tumor volumes (V) were calculated as: V (cm3) = 0.5 X (length X width2). Mice were euthanized once the biggest tumor volume was ~2 cm3. Mice were housed and maintained according to animal use guidelines at EMBL Heidelberg. Both mouse grouping as well as tumor volume measurements were blinded.
Tissue Preparation for flow cytometry
Small parts of fresh tumors grown in nude mice were cut and digested in DMEM F-12 media (Lonza) with 25mM Hepes (Gibco), 100 I.U./ml Penicillin-Streptomycin, 150 U/ml Collagenase (Worthington Biochemical) and 20μg/ml Liberase (Roche) at 37°C for 3 hours. Supernatants were carefully removed after adding D-PBS (Gibco) and centrifuged at 1,000 rpm for 5 minutes at room temperature. Cell pellets were subsequently digested by 0.25% Trypsin (Gibco) for 45 minutes at 37°C and deactivated by DMEM F-12 with 25mM Hepes, 10% fetal bovine serum (FBS, Biowest) and DNAse I. After centrifuging and digesting with red blood cell lysis buffer (Sigma), cells were washed twice with D-PBS containing 2% FBS and filtered by 40μm mesh.
Luciferase enhancer assays
Enhancer regions were amplified by PCR and cloned into the luciferase reporter vector pGL4.24[luc2P/minP] (Promega) containing a multiple cloning site followed by a minimal promoter and the luciferase reporter gene. For amplification of several stretches of the CRC super-enhancer region we used the following primer sets:
E1 (Chr11:2213583-2218091):
forward primer 5’-AAGGTACCGAGGCTGAGAACACAGGCAA-3’
reverse primer 5’-AAGCTAGCCTTCCCGTCTCTGCGGATTT-3’
E2 (Chr11:2218986-2222624):
forward primer 5’-AAGAGCTCCAGGCCTGGGACATTACTCC-3’
reverse primer 5’-AACTCGAGCAACATGAGGTTGGGGGACA-3’
E3 (Chr11:2222436-2227610):
forward primer 5’-AACTCGAGGTTACTGGCCGTAGGGTCTTG-3’
reverse primer 5’-AAAAGCTTAGGACCAACTGAAAGGGTTCG-3’
E4 (Chr11:2227598-2233058):
forward primer 5’-AAGAGCTCCTGGGCCCTCAGACAATTAGA-3’
reverse primer 5’-AACTCGAGCGGCCAGTAACTGATAGGTCAA-3’
E5 (Chr11:2234046-2237062):
forward primer 5’-AAGCTAGCCCTCCGACACCAACAAGTCT-3’
reverse primer 5’-AACTCGAGAAGAGGCAAGGAACTTAGAGGC-3’
E6 (Chr11:2236031-2239626):
forward primer 5’-AAGAGCTCTTGTGGCTGTGTCGTACTCC-3’
reverse primer 5’-AACTCGAGGGCGGGTACCCTTGAGAAAA-3’
For testing enhancer region activity, HCT116 (CRC) and Hela (cervical cancer) cell lines were plated in 96-well plates in triplicates and transfected with 50 ng of enhancer region DNA using pGL4.24 reporter vectors and 10 ng of pRL-TK renilla luciferase control plasmid. 48 h post transfection, cells were lysed and luciferase activities were measured using the Dual-Luciferase® Reporter Assay System (Promega). Firefly luciferase signal of pGL4.24 vectors was normalized to renilla luciferase signal of pRL-TK vector and displayed as fold activity normalized to pGL4.24 empty vector control. Experiments were performed in triplicates for E5. For constructs E2, E4 and E6 triplicate experiments were performed for each of the 2 independent experiments.
Supplementary Material
Acknowledgements
This research project has been funded, in part, through network of excellence funding by the European Commission (260791, to J.O.K and A.H.), grants from the German Ministry for Science and Education (BMBF) (01KU1505F to J.O.K., S.P.; 01ZX1303A to R.K.T. and M.P.; 01ZX1406 to M.P), and a European Research Council (ERC) Starting Grant to J.O.K. (336045), the Danish Medical Research Council (DFF - 4183-00233 to J.W.) the DFG (KFO 227/BA 4806/1-2 to C.R.B. and H.G.), the Baden-Württemberg Stiftung (P-LS-ASII/33) to C.R.B. and H.G., and iMed (Helmholtz Initiative on Personalized Oncology to H.G.). S.M.W. received funding through a SNSF Early Postdoc Mobility Fellowship (P2ELP3_155365) and an EMBO Long-Term Fellowship (ALTF 755-2014). T.D. was supported by a scholarship from the German Cancer Research Center and S.M.D. by the Heidelberg School of Oncology. The results reported here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov) and we acknowledge the specimen donors as well as the research groups involved in the sampling, sequencing and processing of these data. We are grateful to the NCT Tissue Bank for providing samples in accordance with the regulations of the tissue bank and the approval of the ethics committee of Heidelberg University. We are grateful to the GeneCore, IT, mouse facility and Flow Cytometry core facilities at EMBL for excellent assistance, and thank Eileen Furlong and Bernd Klaus for valuable discussions during early stages of this project, Nikos Sidiropoulos for assistance with Hi-C plots and Nina Habermann for assistance with manuscript formatting and proof reading.
Footnotes
Accession codes
EGAS00001002066
Data availability statement
Sequence data for ChIP, whole-genome mate-pair and 4C (bam files and processed bed files) has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001002066. Publically available TCGA Research Network data is available at http://cancergenome.nih.gov/.
Author Contributions:
Development of the CESAM methodology: J.W., J.O.K.; computational pan-cancer analysis of TCGA public resource dataset: J.W., A.P.D., S.M.W., T.Z., J.O.K.; design of experiments: J.W., A.P.D., B.R.M., T.D., C.R.B., H.G., J.O.K.; experiments in primary cancer samples: A.P.D., B.R.M., A.M.S., T.D., B.R., T.E., G.B., R.T., M.P., A.R.H., A.H., C.R.B., J.W., H.G., J.O.K.; experiments in cell lines: A.P.D., B.R., A.M.S., T.E., B.R.M. J.W., J.O.K., experiments in spheroid cultures: T.D., A.P.D., B.R.M., A.M.S., S.M.D., J.W., C.R.B., H.G., J.O.K.; enhancer luciferase experiments: C.S., S.D., C.R.B., H.G.; preparation of manuscript display items: J.W., A.P.D., B.R.M., Y.C., T.D., C.R.B., H.G., J.O.K., clinical information and human cancer tissue: M.S., A.H., M.P., H.B., W.W., O.T.B., P.N., S.P., I.P., S.K.S., E.T.; pathology analysis of spheroids W.W.; mouse experiments: Y.C., A.P.D., S.M.W.; design of mouse experiments: R.S., M.J., A.P.D., Y.C., J.W., J.O.K.
Competing Financial Interests Statement:
The authors declare no competing financial interests.
References
- 1.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rausch T, et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements with TP53 Mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones DT, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nature genetics. 2013;45:927–32. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–54. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–20. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roadmap Epigenomics, C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Encode-Project-Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–63. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 18.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46:1160–5. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46:1258–63. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 20.Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47:710–6. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 22.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–10. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–34. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peifer M, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–4. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valentijn LJ, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015 doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 31.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–8. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–33. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groschel S, et al. A Single Oncogenic Enhancer Rearrangement Causes Concomitant EVI1 and GATA2 Deregulation in Leukemia. Cell. 2014;157:369–81. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 34.Hnisz D, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–8. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Weiss WA. When deletions gain functions: commandeering epigenetic mechanisms. Cancer Cell. 2014;26:160–1. doi: 10.1016/j.ccr.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dekker J, Heard E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015;589:2877–84. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson E, Devenney PS, Hill RE, Lettice LA. Mapping the Shh long-range regulatory domain. Development. 2014;141:3934–43. doi: 10.1242/dev.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Symmons O, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waszak SM, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell. 2015;162:1039–50. doi: 10.1016/j.cell.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Lupianez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107:9546–51. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–5. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 45.Roy N, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015;29:658–71. doi: 10.1101/gad.256628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis CF, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–30. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu BH, Karas M, Koval A, LeRoith D. Insulin receptor substrate-4 enhances insulin-like growth factor-I-induced cell proliferation. J Biol Chem. 1999;274:31179–84. doi: 10.1074/jbc.274.44.31179. [DOI] [PubMed] [Google Scholar]
- 49.Xia Z, Zhang N, Ding D. Proliferation and migration of hepatoblastoma cells are mediated by IRS-4 via PI3K/Akt pathways. Int J Clin Exp Med. 2014;7:3763–9. [PMC free article] [PubMed] [Google Scholar]
- 50.Hoxhaj G, Dissanayake K, MacKintosh C. Effect of IRS4 levels on PI 3-kinase signalling. PLoS One. 2013;8:e73327. doi: 10.1371/journal.pone.0073327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Homma Y, et al. Insulin receptor substrate-4 binds to Slingshot-1 phosphatase and promotes cofilin dephosphorylation. J Biol Chem. 2014;289:26302–13. doi: 10.1074/jbc.M114.565945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimwell NJ, et al. Adenovirus 5 E1A is responsible for increased expression of insulin receptor substrate 4 in established adenovirus 5-transformed cell lines and interacts with IRS components activating the PI3 kinase/Akt signalling pathway. Oncogene. 2009;28:686–97. doi: 10.1038/onc.2008.417. [DOI] [PubMed] [Google Scholar]
- 53.Lingohr MK, et al. Decreasing IRS-2 expression in pancreatic beta-cells (INS-1) promotes apoptosis, which can be compensated for by introduction of IRS-4 expression. Mol Cell Endocrinol. 2003;209:17–31. doi: 10.1016/j.mce.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Consortium, E.P et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehine M, Makinen N, Heinonen HR, Aaltonen LA, Vahteristo P. Genomics of uterine leiomyomas: insights from high-throughput sequencing. Fertil Steril. 2014;102:621–9. doi: 10.1016/j.fertnstert.2014.06.050. [DOI] [PubMed] [Google Scholar]
- 56.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uchida T, Myers MG, Jr, White MF. IRS-4 mediates protein kinase B signaling during insulin stimulation without promoting antiapoptosis. Mol Cell Biol. 2000;20:126–38. doi: 10.1128/mcb.20.1.126-138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinsby AM, Olsen JV, Mann M. Tyrosine phosphoproteomics of fibroblast growth factor signaling: a role for insulin receptor substrate-4. J Biol Chem. 2004;279:46438–47. doi: 10.1074/jbc.M404537200. [DOI] [PubMed] [Google Scholar]
- 60.Ahmad I, Iwata T, Leung HY. Mechanisms of FGFR-mediated carcinogenesis. Biochim Biophys Acta. 2012;1823:850–60. doi: 10.1016/j.bbamcr.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–6. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van de Werken HJ, et al. 4C technology: protocols and data analysis. Methods Enzymol. 2012;513:89–112. doi: 10.1016/B978-0-12-391938-0.00004-5. [DOI] [PubMed] [Google Scholar]
- 64.Brouwer-Visser J, Huang GS. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 2015;26:371–7. doi: 10.1016/j.cytogfr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Li X, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20:769–77. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–89. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 67.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshihara K, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–54. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghavi-Helm Y, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 70.Narendra V, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–21. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venkatraman A, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–9. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 73.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–82. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nambiar M, Kari V, Raghavan SC. Chromosomal translocations in cancer. Biochim Biophys Acta. 2008;1786:139–52. doi: 10.1016/j.bbcan.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 75.Stein LD, Knoppers BM, Campbell P, Getz G, Korbel JO. Data analysis: Create a cloud commons. Nature. 2015;523:149–51. doi: 10.1038/523149a. [DOI] [PubMed] [Google Scholar]
- 76.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Attiyeh EF, et al. Genomic copy number determination in cancer cells from single nucleotide polymorphism microarrays based on quantitative genotyping corrected for aneuploidy. Genome Res. 2009;19:276–83. doi: 10.1101/gr.075671.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 83.Yang J, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–12. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dieter SM, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9:357–65. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 85.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, 1303.3997. 2013 [Google Scholar]
- 86.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Splinter E, de Wit E, van de Werken HJ, Klous P, de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 2012;58:221–30. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 88.Klein FA, et al. FourCSeq: analysis of 4C sequencing data. Bioinformatics. 2015;31:3085–91. doi: 10.1093/bioinformatics/btv335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Untergasser A, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mardin BR, et al. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weischenfeldt J, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23:159–70. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 92.S R, B G. DiffBind: differential binding analysis of ChIP-Seq peak data. Bioconductor. 2011 [Google Scholar]
- 93.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.