Abstract
Although most reef-building corals live near the upper threshold of their thermotolerance, some scleractinians are resilient to temperature increases. For instance, Pocillopora acuta specimens from an upwelling habitat in Southern Taiwan survived a nine-month experimental exposure to 30°C, a temperature hypothesized to induce stress. To gain a greater understanding of the molecular pathways underlying such high-temperature acclimation, the protein profiles of experimental controls incubated at 27°C were compared to those of conspecific P. acuta specimens exposed to 30°C for two, four, or eight weeks, and differentially concentrated proteins (DCPs) were removed from the gels and sequenced with mass spectrometry. Sixty unique DCPs were uncovered across both eukaryotic compartments of the P. acuta-dinoflagellate (genus Symbiodinium) mutualism, and Symbiodinium were more responsive to high temperature at the protein-level than the coral hosts in which they resided at the two-week sampling time. Furthermore, proteins involved in the stress response were more likely to be documented at different cellular concentrations across temperature treatments in Symbiodinium, whereas the temperature-sensitive host coral proteome featured numerous proteins involved in cytoskeletal structure, immunity, and metabolism. These proteome-scale data suggest that the coral host and its intracellular dinoflagellates have differing strategies for acclimating to elevated temperatures.
Introduction
Given the significant anthropogenic pressures currently placed upon coral reef ecosystems [1], there is an urgent need to understand how corals will respond to the elevated temperatures that will characterize their habitats in the coming years [2–3]. Although corals tend to exist at the upper threshold of their thermotolerance [4], some can tolerate dramatic fluctuations [5–6] and increases [7] in temperature. Oliver and Palumbi [8] hypothesized that corals living in thermodynamically extreme environments may be more adept at surviving future increases in temperature, and such was confirmed in Taiwan [9]; Pocillopora acuta (misidentified as P. damicornis in the cited reference) specimens from a reef (“Houbihu”) characterized by highly variable temperatures due to episodic upwelling in Taiwan’s southernmost embayment, Nanwan, survived a nine-month exposure to 30°C, a temperature they experience for only several hours in a typical year and therefore one hypothesized to induce stress.
Since molecular biology-driven approaches have revolutionized our understanding of the fundamental biology of anthozoan-dinoflagellate endosymbioses (e.g., [10–18]), as well as how these environmentally sensitive mutualisms respond to environmental change (e.g., [19–28]), a next generation sequencing-based transcriptome profiling approach (RNA-Seq) was previously taken to attempt to uncover how the aforementioned pocilloporid corals from upwelling reefs of Southern Taiwan acclimated to such dramatic and prolonged increases in temperature [29]. The expression levels of numerous mRNAs in both compartments (coral host and dinoflagellate endosymbionts of the genus Symbiodinium) were affected by both short- (2-week) and long-term (36-week) elevated temperature exposure. From this transcriptome-scale dataset, the authors surmised that effective modulation of protein turnover might have enabled such high-temperature acclimation; indeed, protein turnover had already been hypothesized by other coral reef researchers to be a key cellular process underlying the capacity for certain coral species to acclimate to high temperatures [30–31].
Although protein homeostasis/turnover may ultimately be implicated in coral acclimation to high temperatures, it is possible that the concentrations of the proteins encoded by the differentially expressed genes (DEGs) uncovered in the aforementioned transcriptomic analysis did not actually differ between corals incubated at the control and high-temperature treatments. In fact, the degree of congruency between mRNA expression and protein concentration was markedly low in another pocilloporid, Seriatopora hystrix, exposed to either stable (26°C) or variable temperatures (23–29°C over a 6-hr period) for seven days [32]; only 2 of the 167 differentially concentrated proteins (DCPs) were associated with an mRNA that was also differentially expressed between treatments. This result highlights the role of post-transcriptional processes in S. hystrix, and alternative splicing, in fact, was found to vary dramatically across temperature treatments based on proteome profiling data from those same S. hystrix samples [32].
Given the need to better understand the molecular mechanisms underlying the ability of certain corals to acclimate to elevated temperatures, as well as the previously documented lack of congruency between mRNA expression and protein abundance in reef corals and their in hospite Symbiodinium populations, proteins co-extracted from the same P. acuta biopsies from which transcriptomes were previously sequenced [29] were electrophoresed across two dimensions (2D) herein. Proteins that were uniquely translated by samples of one treatment (i.e., “uniquely synthesized proteins” [USPs]), or, alternatively, documented at different concentrations between treatments (i.e., DCPs), were isolated and sequenced with mass spectrometry (MS). MS-based approaches have proven useful in identifying proteins responsive to extreme high temperature events over short-term time periods (1–2 days) in reef corals [33], and it was hypothesized herein that 1) a low degree of congruency would be documented between mRNA expression and protein concentration in samples of the two-week sampling time (the transcriptomes of the four-and eight-week samples were not sequenced.), and 2) a molecular mechanism underlying high-temperature acclimation could be uncovered for this widely distributed, model reef-builder using the proteome-scale data generated. Since the Symbiodinium compartment showed a more pronounced mRNA-level response upon incubation at elevated temperatures than the coral hosts in which they resided [29], it was also hypothesized that high temperature exposure would more dramatically affect their proteomes than those of their anthozoan hosts.
Materials and methods
The experiment
The details of this long-term temperature experiment can be found in another work [9]. Briefly, P. acuta nubbins created from colonies originally collected from Houbihu, Taiwan (21°56’18.01”N, 120°44’45.54”E) under Kenting National Park permit 0992900842 (to Dr. Tung-Yung Fan) were exposed to either a control temperature of 27°C or an elevated one of 30°C (n = 3 replicate coral reef mesocosms for each of the two treatments). Despite the fact that upwelling events routinely occur in the boreal summer at Houbihu, corals of Houbihu experience only ephemeral periods of 30°C in situ [34]; this temperature was therefore hypothesized to induce stress. Nubbins (1–2 per mesocosm x 3 mesocosms/treatment x 2 treatments) were collected after 0, 2, 4, 8, 24, or 36 weeks of treatment exposure, and the physiological [9] and mRNA expression (i.e., RNA-Seq) data [29] were described previously. Corals acclimated to the elevated temperature regime; however, the gastrodermal tissue layer became thicker in response to a long-term (36-week) exposure to 30°C [9], and Symbiodinium ubiquitin ligase mRNA expression increased dramatically upon a 2-week exposure to this temperature [29].
Protein extraction, 2D gel electrophoresis, and MS
Proteins were extracted with TRIzol® (Life Technologies) as described previously [9] from one randomly selected nubbin from each of three replicate tanks at each of the two temperatures at the two-, four-, and eight-week sampling times (n = 18 protein extractions). Additional details of the protein extraction protocol can be found in the S1 File.
Proteins (~150 μg) were electrophoresed across 2D as described previously [35]. For each sampling time, a control-temperature protein sample was electrophoresed at the same time as a high-temperature sample, and three replicate control vs. high-temperature pairs were analyzed for each treatment and sampling time (18 gels in total were run.). Each of the triplicate gels for each treatment x time group featured a protein extracted from a nubbin generated from a different source coral colony (n = 3 biological replicates per sampling time). As the aforementioned book chapter [35] is not freely available except on ABM’s personal website (coralreefdiagnostics.com), the details have been reiterated in the S1 File. Briefly, isoelectric point (pI) and molecular weight (kDa) were the first and second dimensions, respectively.
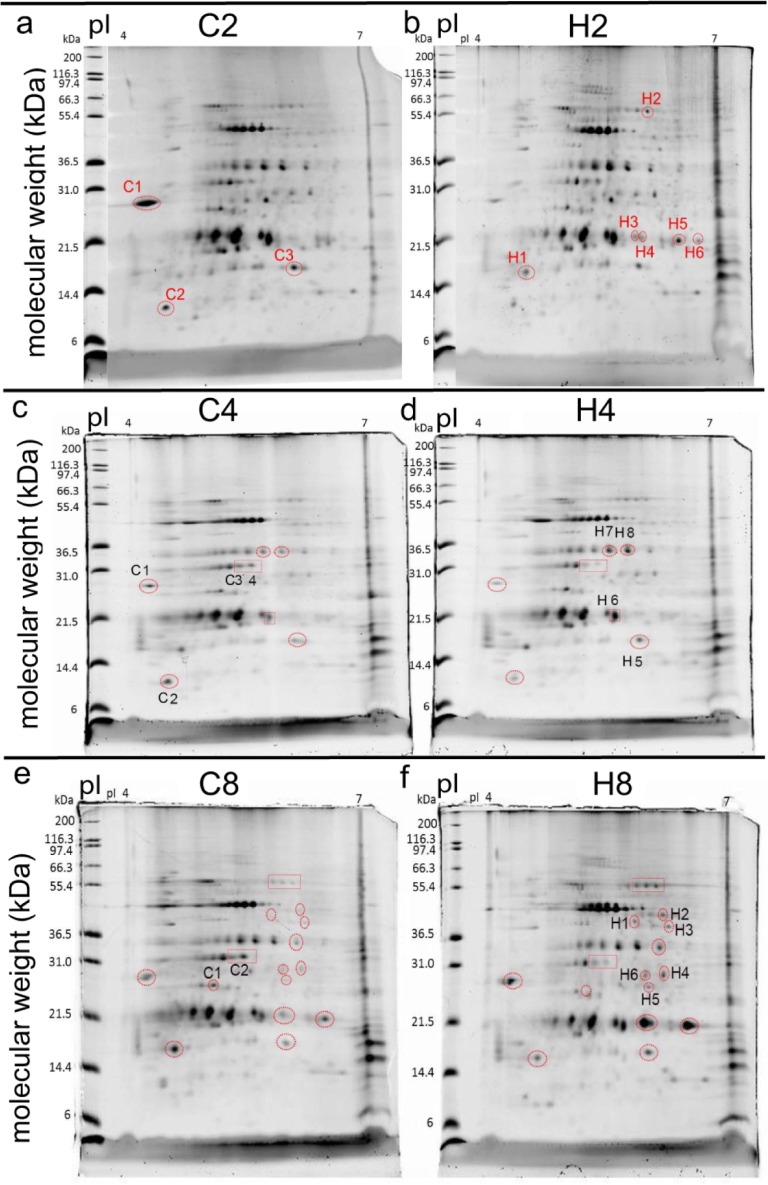
Unique and differentially concentrated protein spots were identified with ImageQuant™ TL software provided with the Typhoon Trio™ scanner (GE Healthcare) used to image the gels (described in detail in the S1 File), and only protein spots that were found by ImageQuant TL to differ in concentration in all three pairs of control vs. high temperature gels for each sampling time were targeted for removal from the representative gel. Proteins that were unique to the proteomes of samples of the control and high-temperature treatments at a particular sampling time were removed from a representative control and high temperature gel, respectively. When no USPs were uncovered, DCPs were instead targeted; in these cases, the protein spot was always removed from the gel in which its concentration was higher. Nine, eight, and eight spots were removed from each representative pair of gels at the two-, four-, and eight-week sampling times, respectively. In certain cases, not every DCP spot was removed from the representative gel due to certain spots being over-saturated; these were likely comprised of a complex mix of a plethora of proteins, and the spot intensity could consequently not be traced to the concentration of a unique protein. Some such protein spots have been encircled in Fig 1 to highlight their differential abundance, though only those given identification codes were removed from the gels.
Fig 1. Two-dimensional gel electrophoresis of proteins from representative control and high- temperature coral samples after two, four, and eight weeks of treatment exposure.
Encircled and labeled protein spots were 1) determined to be differentially concentrated between temperature treatments within a sampling time with image analysis software, 2) removed from the gels, 3) purified, 4) digested with trypsin, and 5) submitted for sequencing by nano-liquid chromatography+mass spectrometry as described in the main text and the S1 File. The isoelectric point (pI) and molecular weight (in kilodaltons [kDa]) were the 1st and 2nd dimensions, respectively. Only labeled spots were removed; in certain cases at the four- and eight-week sampling times, matched (i.e., same pI and molecular weight) spots have been encircled in gels of both treatments to emphasize their differential concentrations, yet they were not sequenced for reasons discussed in the main text. “C2” and "H2" = control and high-temperature samples at the two-week sampling time, respectively. “C4” and "H4" = control and high-temperature samples at the four-week sampling time, respectively. “C8” and "H8" = control and high-temperature samples at the eight-week sampling time, respectively.
USP (n = 8) and DCP (n = 17) spots (Table 1) were removed from the representative gels, digested with trypsin, and prepared for sequencing with MS as described previously [35]. Given the potential inaccessibility of this book chapter, the details are presented again in the S1 File. Digested peptides (2 μl) were sequenced on a nano-liquid chromatography system and detected by an LTQ Orbitrap Discovery Hybrid Fourier Transform Mass Spectrometer (Thermo-Fisher Scientific) at a resolution of 30,000 coupled with a nanospray source that was executed in positive ion mode. The nano-UPLC system (“nanoACQUITY”) was purchased from Waters, as were the desalting (Symmetry C18, 5 μm x 180 μm x 20 mm) and analytical (BEH C18, 1.7 μm x 75 μm x 150 mm) columns. The peptide eluate from the column was directed to the nanospray source, and the MS was operated in data-dependent mode.
Table 1. Summary of two-dimensional gel electrophoresis results.
| Comparison (Fig) |
#C>H spots removed | #C>H USP spots |
#C>H DCP spots | #C>H DCPs identified | # C>H host DCPs | # C>H Sym DCPs | #H>C spots re-moved | #H>C USP spots | #H>C DCP spots | #H>C DCPs identi-fied | #H>C host DCPs | #H>C Sym DCPs | Total host/ Sym DCPs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C2 vs. H2 (Fig 2) |
3 | 0 | 3 | 13 | 7 | 5 | 6 | 4 | 2 | 25 | 8 | 10 | 15/15a |
| C4 vs. H4 (Fig 3) |
4 | 0 | 4 | 7 | 6 | 1 | 4 | 0 | 4 | 3 | 2 | 1 | 8/2b,c |
| C8 vs. H8 (Fig 4) |
2 | 1 | 1 | 6 | 2 | 4 | 6 | 3 | 3 | 16 | 12 | 2 | 14/6c |
| Total (Fig 5) |
9 | 1 | 8 | 26 | 15 | 10 | 16 | 7 | 9 | 44 | 22 | 13 | 37/23 |
In certain cases, the total number of differentially concentrated proteins (DCPs) does not equal the sum of the host coral and Symbiodinium (Sym) DCPs; this is due to either 1) certain proteins being of bacterial origin or 2) the inability to assign a compartment of protein origin in some cases. The five proteins that were documented at different concentrations between temperature treatments at multiple sampling times were counted only at the earlier of the two times at which they were sequenced, leading to a total number of unique host+Sym DCPs of 60 (Fig 5D). For a graphical representation of a portion of the data in this table, please see Fig 6A. C = control treatment. H = high temperature treatment. The numbers after these treatment abbreviations in the “Comparison” column reflect the sampling times (e.g., C2 = control samples of the two-week sampling time). USP = uniquely synthesized protein.
ahost/Sym DCP ratio significantly lower (z-test, p<0.05) than the host/Sym mRNA transcript ratio (1.9±0.4 [std. dev.]).
bhost/Sym DCP ratio significantly higher (z-test, p<0.05) than the host/Sym mRNA transcript ratio.
cHost/Sym DCP ratio differs slightly from that of the respective figure since only unique DCPs were considered in this table; in contrast; all DCPs have been depicted in the compartmental breakdown pie graphs of Fig 4 (10/2 ratio) and 5 (15/7 ratio).
Data analysis
Data files from the spectrometer (.MGF) were used directly as queries of the P. acuta-Symbiodinium transcriptome with “MS-SCAN.” This feature of the transcriptome server was described previously [29], and the program can be accessed on the following website: http://symbiont.iis.sinica.edu.tw/coral_pdltte/static/html/index.html#mscan. Details of the MS-SCAN program can be found in the S1 File, as well as in Kim and Pevzner [36]. The.MGF files produced herein (n = 25) can be downloaded from the following page: http://symbiont.iis.sinica.edu.tw/coral_pdltte/static/html/index.html#stat.MGF files were directly uploaded onto the MS-SCAN program hosted on the transcriptome server, and the digestion enzyme was set to trypsin. In addition to the default parameters for protein identification described in the S1 File and in Kim and Pevzner [36], only those proteins that featured either 1) 15 or more amino acids (AA) sequenced or 2) two peptides mapping to the same reference protein (whose collective length summed to 15 or more AA) were considered. When a protein was documented at relatively higher concentrations in both control and high-temperature gels at the same sampling time, it was discarded from the analysis. These peptide length/number criteria are more stringent than what are commonly employed in the vertebrate (e.g., [37]) and reef coral research ([33,38]) fields, where, furthermore, only one biological replicate is typically used per treatment (but see Weston et al. [39]); however, for those interested in using less stringent peptide inclusion criteria, both peptide length (default = 15 AA) and count (default = 2 peptides mapping to the same reference protein) thresholds can be reduced on the MS-SCAN program implemented on our interactive transcriptome website.
For all peptides fulfilling our criteria, the respective mRNA sequences from the top hit contigs derived from the MS-SCAN analysis of the P. acuta-Symbiodinium transcriptome were BLASTed against 1) the Stylophora pistillata genome and 2) the Symbiodinium (clade B1) genome (both housed on the NCBI database) to verify protein identity, and proteins were assigned a compartment of origin: unknown, bacteria, host coral, or Symbiodinium. When a contig aligned significantly (e<10−5) to a functionally characterized protein, the respective cellular process (e.g., metabolism) was recorded (if known). The top hit contig sequence, rather than the peptide sequence itself, was used as the BLAST query due to the short nature of the latter (mean length = 20±10 [std. dev. for this and all error terms henceforth] AA) and the consequent inability to generate significant alignments in many cases; that being said, for those peptides >30 AA, the MS-SCAN-derived peptide sequence itself was additionally BLASTed against both the Symbiodinium and S. pistillata genomes to corroborate findings from 1) MS-SCAN and 2) the mRNA-BLAST, and, in the cases when the peptide itself aligned significantly to a presumed homolog in the NCBI database (e<10−5), the first and second NCBI accession numbers in the S1–S3 Tables correspond to the top mRNA-BLAST hit and the top peptide-BLAST hit, respectively.
Expression data (fragments per kilobases mapped) were obtained from the transcriptome server (http://symbiont.iis.sinica.edu.tw/coral_pdltte/static/html/index.html#stat) for each of the mRNAs hypothetically encoding the DCPs uncovered to understand the degree of congruency between mRNA expression and protein concentration for samples of the two-week sampling time only. Details of this analysis can be found in the S1 File. z-tests were used to determine whether the compartmental breakdown of the DCPs deviated significantly from the host/Symbiodinium mRNA ratio of 1.9±0.4 determined previously [29]. Two-sample proportion tests were used to determine whether certain cellular processes (e.g., stress response) in the differentially concentrated proteomes were over-represented relative to their proportional abundance in the transcriptome [29]; this and all other proportion-based analyses were only performed for those cellular processes in which ≥2 DCPs were identified. X2 tests were used to ascertain whether cellular processes were equally represented across sampling times, as well as whether the host/Symbiodinium DCP ratio differed significantly over time. All statistical analyses were performed with JMP® (ver. 12.0.1), and an alpha level of 0.05 was set for all aforementioned tests.
Results
Overview of data
Summaries of the 2D+MS analysis and sequenced peptides for all three sampling times can be found in Tables 1 and 2, respectively, and detailed information on the DCPs uncovered at the two-, four-, and eight-week sampling times (e.g., top hit contigs, accession numbers, peptide lengths, and protein coverage [%]) can be found in the S1 Table, S2 Table and S3 Table, respectively; the respective peptide sequences and associated cellular pathways are located in the S4 Table, S5 Table and S6 Table, respectively.
Table 2. Summary of all unique host coral (Pocillopora acuta) and Symbiodinium differentially concentrated proteins (DCPs) uncovered across temperature treatments.
| time spot |
protein | compartment | cellular process | concentration |
|---|---|---|---|---|
| TWO WEEKS (n = 38) | ||||
| C1-2 | pentraxina | host | immunity | control>high-temp. |
| C1 | transcription factor death-induced obliterator-1 | host | transcription factor | control>high-temp. |
| C1 | protein kinase UbiB | Symbiodinium | protein kinase | control>high-temp. |
| C1 | STI1-like protein | Symbiodinium | stress response | control>high-temp. |
| C1 | myosin-6b | Symbiodinium | actin-based motility | control>high-temp. |
| C2 | WSC domain | host | unknown | control>high-temp. |
| C2 | cadherin EGF LAG seven-pass G-type receptor 2a | host | receptor/signaling | control>high-temp. |
| C2 | pre-mRNA splicing factor SLU7-Ab | Symbiodinium | gene expression/splicing | control>high-temp. |
| C3 | hypothetical protein | host | unknown | control>high-temp. |
| C3 | Pao retrotransposon peptidase | host | unknown/various | control>high-temp. |
| C3 | spectrin alpha chaina | host | cytoskeleton | control>high-temp. |
| C3 | adenylate kinase | Symbiodinium | protein kinase | control>high-temp. |
| C3 | hypothetical protein | unknown | unknown | control>high-temp. |
| H1 | hypothetical protein | host | unknown | high>control temp. |
| H1, H3 | low-density lipoprotein receptor-related protein 4a | host | receptor/transport | high>control temp. |
| H1 | serine/arginine repetitive matrix protein 1a | host | gene expression/splicing | high>control temp. |
| H1 | Rec10 / Red1 | Symbiodinium | meiosis | high>control temp. |
| H1 | VWA domain-containing protein | bacteria | unknown | high>control temp. |
| H2 | hypothetical protein | host | unknown | high>control temp. |
| H2 | histone-lysine N-methyltransferase SETD1B-likea | host | gene expression/splicing | high>control temp. |
| H2 | ribulose-1,5-bisphosphate carboxylase/oxygenaseb | Symbiodinium | photosynthesis | high>control temp. |
| H2-3 | protein with DNAJ and WW domains* | Symbiodinium | stress response | high>control temp. |
| H2 | hypothetical protein | unknown | unknown | high>control temp. |
| H3 | golgin subfamily B member 1a | host | unknown | high temp. only |
| H3 | peptidylprolyl isomerase D* | Symbiodinium | stress response, various | high temp. only |
| H3 | hypothetical protein | unknown | unknown | high temp. only |
| H3 | hypothetical protein | unknown | unknown | high temp. only |
| H4 | E3 ubiquitin protein ligase | Symbiodinium | stress response | high temp. only |
| H4 | hypothetical protein | unknown | unknown | high temp. only |
| H4 | hypothetical protein | unknown | unknown | high temp. only |
| H5-6 | beta-gamma crystallin | host | stress response | high temp. only |
| H5 | sacsina | host | stress response | high temp. only |
| H5 | hypothetical protein | unknown | unknown | high temp. only |
| H6 | ribosome biogenesis protein NSA2-likeb | Symbiodinium | ribosome | high temp. only |
| H6 | pentatricopeptide repeat-containing protein* | Symbiodinium | unknown | high temp. only |
| H6 | voltage-dependent T-type calcium channel subunit α-1H | Symbiodinium | transport | high temp. only |
| H6 | nucleolar protein of 40 kDa | Symbiodinium | unknown | high temp. only |
| H6 | peptidylprolyl isomerase D* (differs from paralog in spot H3) | Symbiodinium | stress response, various | high temp. only |
| FOUR WEEKS (n = 12) | ||||
| C1 | pentraxina,c | host | immunity | control>high-temp. |
| C1 | trichohyalind | host | structural | control>high-temp. |
| C1 | plexina | host | signal transduction/reception | control>high-temp. |
| C2 | avidin | host | reproduction | control>high-temp. |
| C2 | hypothetical protein | host | unknown | control>high-temp. |
| C3 | chloroplast oxygen-evolving enhancerb | Symbiodinium | photosynthesis | control>high-temp. |
| C4 | actin | host | cytoskeleton | control>high-temp. |
| C4 | actin (different from previous paralog) | host | cytoskeleton | control>high-temp. |
| H7 | RNA-directed DNA polymerase from mobile element jockey-likea | host | gene expression/splicing | high>control temp. |
| H7-8 | peridinin-chlorophyll A binding proteinb | Symbiodinium | photosynthesis | high>control temp. |
| H8 | NF-kappa-B inhibitor-like protein 1 isoform X2a | host | immunity | high>control temp. |
| H8 | hypothetical proteinc | host | unknown | high>control temp. |
| EIGHT WEEKS (n = 25) | ||||
| C1 | rho GDP-dissociation inhibitor 1-likea | host | protein homeostasis | control temp. only |
| C1 | hypothetical protein | Symbiodinium | unknown | control temp. only |
| C1 | fucoxanthin-chlorophyll a-c binding protein Fb | Symbiodinium | photosynthesis | control temp. only |
| C1 | hypothetical proteinb | Symbiodinium | unknown | control temp. only |
| C1 | metal transporter Nramp3 | Symbiodinium | transport | control temp. only |
| C2 | centromere-associated protein E-like isoform X1 | host | cell cycle | control>high temp. |
| H1 | trichohyaline | host | structural | high temp. only |
| H1 | hypothetical protein | Symbiodinium | unknown | high temp. only |
| H1 | hypothetical protein | unknown | unknown | high temp. only |
| H1 | hypothetical bacterial proteinc | bacteria | unknown | high temp. only |
| H2 | abhydrolasea | host | metabolism | high temp. only |
| H2 | chromodomain-helicase-DNA-binding protein 1-likea,f | host | DNA repair/transcription | high temp. only |
| H2 | hypothetical proteinb | Symbiodinium | unknown | high temp. only |
| H3 | retrovirus-related Pol polyprotein from transposon 17.6a | host | unknown | high temp. only |
| H3 | guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1a | host | signaling | high temp. only |
| H3 | stabilizer of axonemal microtubules 2-likea | host | cytoskeleton | high temp. only |
| H3 | glycerol-3-phosphate dehydrogenasea | host | metabolism | high temp. only |
| H3 | concanavalin A-like lectin/glucanase superfamily | host | cell membrane binding | high temp. only |
| H4 | glycosyltransferase-like domain-containing protein 1 | host | metabolism | high>control temp. |
| H4 | trichohyalin-like | host | cell structure | high>control temp. |
| H4 | protein w/ DNAJ and WW domainsc | Symbiodinium | stress response | high>control temp. |
| H4 | hypothetical protein | unknown | unknown | high>control temp. |
| H5 | peroxiredoxin-6-like | host | stress response | high>control temp. |
| H5 | hypothetical protein | host | unknown | high>control temp. |
| H6 | endonuclease | host | DNA repair | high>control temp. |
For additional details of the two- (n = 38), four- (n = 12), and eight-week (n = 25) DCPs, please see the S1 Table, S2 Table and S3 Table, respectively. For the respective amino acid sequences, please see the S4 Table, S5 Table and S6 Table, respectively.
*congruency between mRNA expression and protein concentration. temp. = temperature.
aProtein identity corroborated by directly BLASTing peptide sequence against the Stylophora pistillata genome.
bProtein identity corroborated by directly BLASTing peptide sequence against the Symbiodinium (clade B1) genome.
cAlso identified at the two-week sampling time.
dAlso identified at the eight-week sampling time.
eAlso identified at the four-week sampling time.
fProtein concentration affected by temperature treatment in a study undertaken with the con-familial coral Seriatopora hystrix [32].
Two-week sampling time
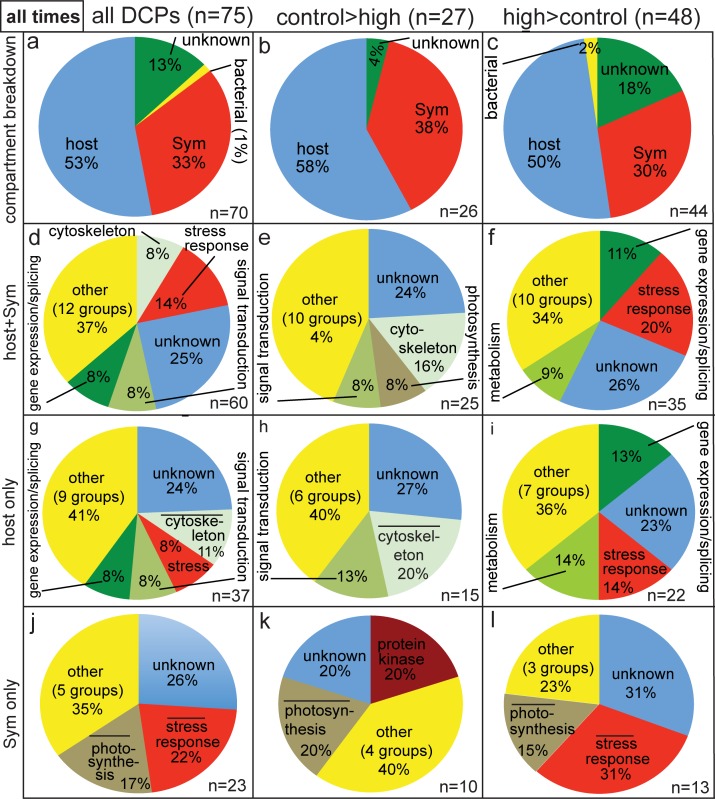
Upon electrophoresing proteins across 2D, there were four protein spots consistently present only in the high-temperature gels of the two-week sampling time (Fig 1B). All four were removed from the representative gel and sequenced, as were three and two additional protein spots documented at higher concentrations in the C2 (Fig 1A) and H2 (Fig 1B) gels, respectively (n = 9 sequenced spots). Upon querying the P. acuta-Symbiodinium transcriptome with MS-SCAN, 38 unique proteins were sequenced across these nine spots, 13 and 25 of which were at higher concentrations in the control and high-temperature gels, respectively (Table 2, S1 Table and S4 Table). Eight of these DCPs (Fig 2A) were either of bacterial origin or could not be assigned a compartment of origin or function; these proteins are not discussed further herein. There were equal numbers of coral and Symbiodinium DCPs (n = 15); given that the host comprises ~60–65% of the biomass of P. acuta [29], this represents a statistically significant over-representation of Symbiodinium proteins (host/Symbiodinium protein ratio of 1 < host/Symbiodinium mRNA ratio of 1.9±0.4; z-test, p<0.05). A relative over-representation of Symbiodinium DCPs was also documented for the proteins whose concentrations were higher in high-temperature samples (Fig 2C; z-test, p<0.05) but not for those more highly concentrated by the control samples (Fig 2B; z-test, p>0.05).
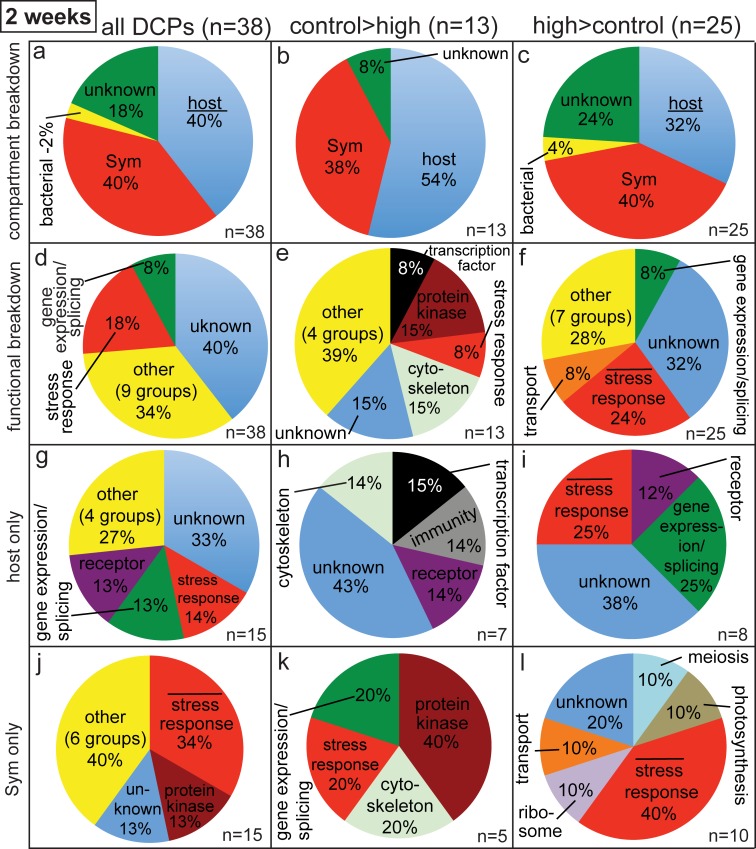
Fig 2. Pie graphs showing breakdown of differentially concentrated proteins (DCPs) between control (C2) and high (H2) temperature samples of the two-week sampling time.
Three and six protein spots were found to be more highly concentrated in the C2 (i.e., C>H) and H2 (H>C) proteomes, respectively, and 13 C>H and 25 H>C DCPs were identified from these nine spots. When the ratio of host/Symbiodinium (Sym) DCPs in panels a and c was significantly lower (z-test, p<0.05) than the Pocillopora acuta/Sym mRNA ratio of 1.9 (i.e., a 65/35% host coral/Sym ratio), the word “host” has been underlined. When the cellular process “stress response” was over-represented in the proteomes of the host coral (g-i) and Sym (j-l) relative to the host coral (6% of the P. acuta host transcriptome) and Sym (4% of the Sym transcriptome) transcriptomes [29], respectively (2-sample proportion test, p<0.05), a bar has been inserted over the term “stress response”.
In terms of the functional breakdown of the two-week sampling time DCPs (Fig 2D–2L), the two most represented categories were the stress response and gene expression/splicing; 18% of all two-week DCPs (n = 7/38) were involved in the former process (Fig 2D). Although “stress response” was not over-represented relative to its proportional abundance in the composite P. acuta-Symbiodinium transcriptome (5%) for proteins more highly concentrated by the control coral samples (8%; Fig 2E), it was for those proteins more highly concentrated by the high- temperature samples (Fig 2F; 2-sample proportion test, p<0.05); nearly 25% of the H>C DCPs (6/25) were involved in the stress response. This relative over-representation of proteins involved in the stress response was mainly driven by the proteomic response of the Symbiodinium compartment; only 2 of the 15 coral host DCPs (Fig 2G) were involved in the stress response (not significantly higher than the 6% value of the P. acuta host transcriptome; 2-sample proportion test, p>0.05), and both were documented at higher concentrations in high-temperature gels than in control ones (Fig 2I). In contrast, one-third of the 15 Symbiodinium DCPs (Fig 2J) were involved in the stress response, and this was significantly over-represented relative to the Symbiodinium transcriptome (4%; 2-sample proportion test, p<0.05). Specifically, one and four Symbiodinium proteins involved in the stress response were documented at higher concentrations in the control (Fig 2K) and high-temperature (Fig 2L) samples, respectively.
Of the 38 DCPs uncovered at the two-week sampling time, only 4 were associated with an mRNA in which a congruent temperature treatment difference was documented; all were of Symbiodinium origin (S1 Table). In all cases, mRNA expression and protein concentration were higher in samples of the high-temperature treatment, and the overall congruency of 10.5% (4/38) was significantly higher than that of a temperature experiment with S. hystrix [32]: 2% (2-sample proportion test, p<0.01). Congruency also differed significantly between compartments (2-sample proportion test, p<0.05): 0% for the coral host (0/15 molecules showed a congruent response between mRNA and protein levels.) vs. 27% (4/15) for Symbiodinium.
Four-week sampling time
At the four-week sampling time, four and four proteins were more highly concentrated by samples of the control (Fig 1C) and high (Fig 1D) temperature treatments, respectively, though none of these were uniquely translated by samples of either (Table 1). From these eight spots, eight C4>H4 and four H4>C4 DCPs were uncovered (Fig 3, Table 2, S2 Table and S5 Table). One DCP identified at the two-week sampling time, host coral pentraxin, was also documented at higher levels in control temperature samples at the four-week sampling time. A hypothetical protein (contig5078) of presumed host origin, in contrast, was more highly concentrated by coral hosts exposed to high temperatures relative to controls at both the two- and four-week sampling times. As such, only 10 of the 12 DCPs identified at the four-week sampling time were unique.
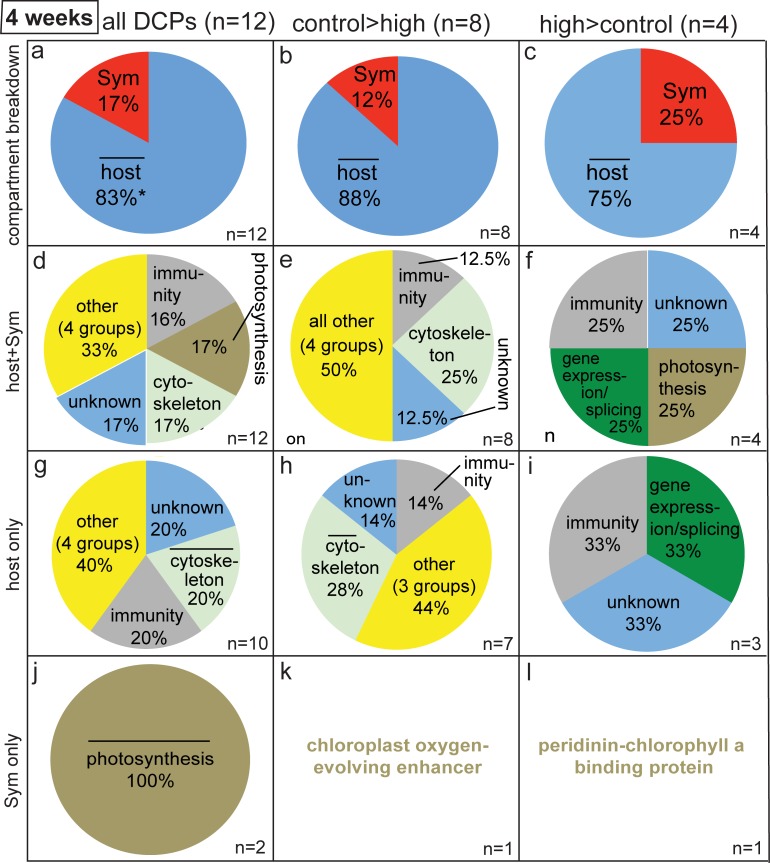
Fig 3. Pie graphs showing breakdown of differentially concentrated proteins (DCPs) between control (C4) and high (H4) temperature samples of the four-week sampling time.
Four and four protein spots were found to be more highly concentrated in the C4 (i.e., C>H) and H4 (H>C) proteomes, respectively, and 8 C>H and 4 H>C DCPs were identified from these eight spots. When the host:Symbiodinium (Sym) ratio was significantly higher (z-test, p<0.05) than the P. acuta:Sym mRNA ratio of 1.9±0.4 (std. dev.), a bar has been inserted over the word “host” in (a-c). Likewise, when a cellular process was over-represented in the proteomes of the host (g-i) and Sym (j-l) relative to the host coral and Sym transcriptomes [29], respectively (2-sample proportion test, p<0.05), a bar has been placed over the category name.
The overall 83/17% host/Symbiodinium DCP ratio (i.e., ~5:1) for the 12 DCPs uncovered at the four-week sampling time (Fig 3A) was significantly higher than the coral/Symbiodinium ratio of 1 documented across the 38 DCPs of the two-week sampling time (Fig 2A; X2 test, p<0.001), and it was also significantly higher than the 1.9:1 host:Symbiodinium mRNA transcript ratio (z-test, p<0.05). When looking only at the eight proteins that were more highly concentrated by samples of the control temperature treatment at the four-week sampling time (Fig 3B), seven and one were from the coral host and dinoflagellate compartments, respectively, a similar proportional breakdown to that observed for the H>C DCPs (Fig 3C). For a detailed treatise on the functional breakdown of all 12 DCPs of the four-week sampling time (Fig 3D), please see the S1 File, as well as Fig 3E–3L, S2 Table and S5 Table.
Eight-week sampling time
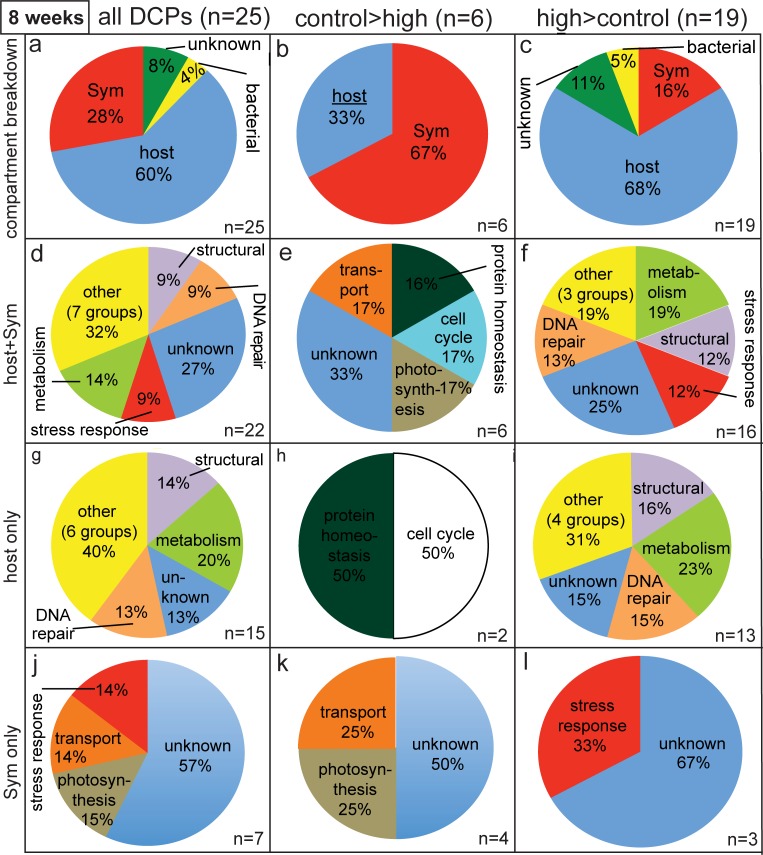
At the eight-week sampling time, two and six protein spots were more highly concentrated by samples of the control (Fig 1E) and high (Fig 1F) temperature treatments, respectively, and one and three of these were uniquely synthesized by samples of the respective treatment (Table 1). From these eight spots, 6 C8>H8 and 19 H8>C8 DCPs were uncovered (Fig 4, Table 2, S3 Table and S6 Table). Two DCPs identified at the two-week sampling time, a Symbiodinium protein with both DNAJ and WW domains and a bacterial protein of unknown function, were also documented at higher abundance in high-temperature gels of the eight-week sampling time. A host coral trichohyalin, in contrast, was more highly concentrated by control samples at the four-week sampling time, though instead by high-temperature samples at the eight-week sampling time. As such, of the 25 DCPs identified at the eight-week sampling time, only 22 were unique; 2 were of unknown origin, resulting in 14 unique host and 6 unique Symbiodinium DCPs at this sampling time (Table 1). The overall host/Sym DCP ratio was, instead, 15:7 (Fig 4A). For a detailed treatise on the functional and compartmental breakdown of the 25 DCPs uncovered at the eight-week sampling time, please see the S1 File, as well as Fig 4B–4L, S3 Table and S6 Table.
Fig 4. Pie graphs depicting breakdown of differentially concentrated proteins (DCPs) between control (C8) and high (H8) temperature samples of the eight-week sampling time.
Two and six spots were more highly concentrated in the C8 (i.e., C>H) and H8 (H>C) proteomes, respectively, and 6 C>H and 19 H>C DCPs were identified from these eight spots. When the host:Symbiodinium (Sym) DCP ratio was significantly lower (z-test, p<0.05) than the P. acuta: Sym mRNA ratio of 1.9±0.4 (std. dev.), a bar has been inserted under the word “host” in (a-c).
All sampling times
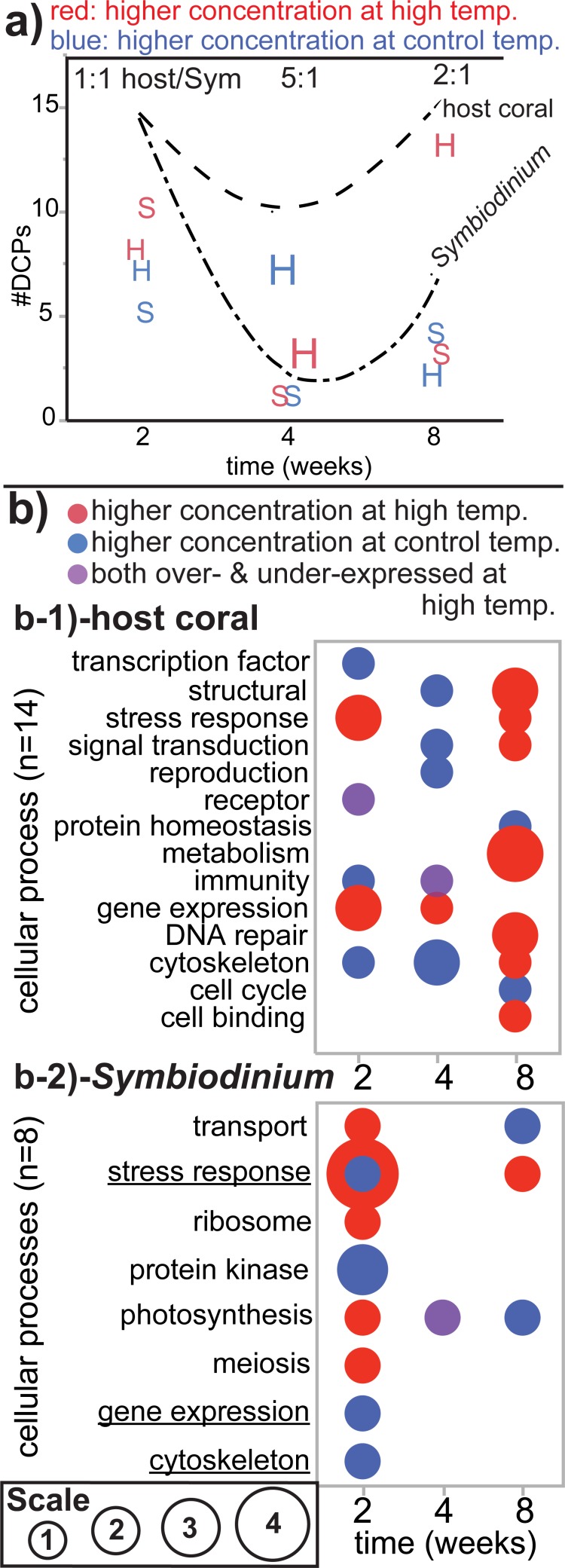
When pooling the data over time (Fig 5), 75 DCPs were sequenced. Since 1) two host coral DCPs were shared between the two- and four-week proteomes, 2) one Symbiodinium and one bacterial DCP were sequenced at both the two- and eight-week sampling times, and 3) one host DCP was sequenced at both the four- and eight-week sampling times, 70 unique DCPs were identified (Fig 5A). Of these, 26 and 44 were more highly concentrated by samples of the control (Fig 5B) and high-temperature (Fig 5C) treatments, respectively. One and nine DCPs were of bacterial and unknown origin, respectively, leading to a total of 37 unique host and 23 unique Symbiodinium DCPs. This 1.6:1 host/Symbiodinium ratio did not differ significantly from the host/Symbiodinium transcript of 1.9 (z-test, p>0.05). The host/Symbiodinium DCP ratio did, however, vary over time (X2 test, p<0.05); this was due to the fact that equal numbers of host coral and Symbiodinium DCPs were uncovered at the two-week sampling time (1:1; Fig 6A), whereas the four- and eight-week DCP pools featured higher host/Symbiodinium DCP ratios (5:1 and 2.1:1, respectively; Fig 6A). Symbiodinium were more likely to increase levels of protein abundance in response to experimentally elevated temperatures (n = 10 H>C DCPs) than the host corals in which they resided (n = 8 H>C DCPs) at the earliest sampling time only (X2 test, p<0.05).
Fig 5. Compartmental and functional breakdown of all differentially concentrated proteins (DCPs).
Across the nine, eight, and eight differentially concentrated/uniquely synthesized protein spots removed from representative gels of the two-, four-, and eight-week sampling times, respectively, 75 DCPs were uncovered; upon counting five proteins sequenced at multiple time points only once, 37 and 23 of the 70 unique DCPs were found to be of host coral (Pocillopora acuta) and Symbiodinium (Sym) origin, respectively. When a cellular process was over-represented in the differentially concentrated proteomes of the host coral (g-i) and Sym (j-l) relative to the host coral and Sym transcriptomes [29], respectively (2-sample proportion test, p<0.05), a bar has been inserted above the category name.
Fig 6. Summary of the dataset.
The number of differentially concentrated proteins (DCPs) was plotted over time for both the host coral (“H”) and Symbiodinium (“S;” Sym) for proteins documented at higher concentrations at high temperature (red icons) and those over-expressed at the control temperature (blue icons). The sizes of the icons are proportional to the host/Sym DCP ratio at each sampling time (listed at the top of the plot). The upper and lower hatched lines represent the total number of DCPs at each sampling time for the host coral and Sym, respectively. In (b) the sizes of the bubbles are proportional to the number of DCPs involved in each cellular process for both the host coral (b-1) and Sym (b-2). Purple bubbles represent those cellular processes for which one protein was over-expressed at high temperature, whereas another was documented at higher concentrations at the control temperature. This annotation is not used for the Sym “stress response” category at the two-week sampling time (b-2), in which 1 and 4 DCPs were more highly concentrated by the control and high-temperature samples, respectively. Underlined functional categories in b-2 reflect processes that were also temperature-sensitive in the host coral compartment (b-1).
The most represented functional categories amongst the 60 host+Symbiodinium DCPs were the stress response, cytoskeleton, signal transduction, and gene expression/splicing (Fig 5D and Fig 6B). A more detailed discussion of the compartmental and functional breakdown of these 60 DCPs can be found in the S1 File, Figs 5 and 6, and Table 2. In general, few cellular processes were represented in the proteomes of both compartments. Of the 19 cellular processes featuring at least one DCP, only three were identified in the differentially concentrated proteomes of both host coral and Symbiodinium: the stress response, gene expression/splicing, and the cytoskeleton (Fig 6B). Similarly, few cellular processes were represented at more than one sampling time. In the host coral compartment (Fig 6B-1), structural proteins, stress response proteins, signal transduction proteins, and proteins involved in immunity and the cytoskeleton were identified in the differentially concentrated proteomes of multiple sampling times, while for Symbiodinium (Fig 6B-2) only three cellular processes were identified at multiple sampling times: transport, the stress response, and photosynthesis. Only proteins involved in the stress response were sequenced (i.e., differentially concentrated) in both cellular compartments of the coral-dinoflagellate endosymbiosis at multiple sampling times (Fig 6B).
Discussion
Differential proteomic responses of host corals and their in hospite Symbiodinium populations to experimentally elevated temperatures
When attempting to track the responses of multi-compartmental organisms, such as reef-building corals [40–41] or symbiotic forams [42–43], to changes in their environments, it is important to focus not only on the hosts, but also their symbionts [44]. Although Symbiodinium comprise ~30–40% of the live biomass of P. acuta, they contributed an equal number of DCPs as their coral hosts at the two-week sampling time. This suggests that Symbiodinium were more strongly affected at the protein level by a two-week elevated temperature exposure than their anthozoan hosts, as was also documented in these same samples at the mRNA level [29]. This does not mean that Symbiodinium were more stressed than their cnidarian hosts, since they were ultimately found to have acclimated to 30°C [9]. Furthermore, when pooling data across all three sampling times, the host/Symbiodinium DCP ratio of 1.6 did not differ significantly from this holobiont’s biomass ratio of ~2:1, meaning that, in contrast to our hypothesis, Symbiodinium were only more responsive at the protein-level to elevated temperature exposure than their hosts over a relatively short-term timescale.
Despite the fact that the host coral and Symbiodinium exhibited a similar degree of differential protein regulation upon having pooled data across all three sampling times, the cellular processes affected by elevated temperatures tended to differ between the two compartments, and only proteins involved in the stress response were identified at multiple sampling times in both the host coral and Symbiodinium. The latter compartment up-regulated stress protein levels to a greater extent than did their coral hosts. Such Symbiodinium stress proteins are discussed in more detail below. In contrast, the anthozoan compartment was more likely to differentially concentrate proteins involved in the cytoskeleton, metabolism, and, more generally, processes likely to be involved in osmoadaptation (discussed in more detail below with respect to mechanisms of high-temperature acclimation). Although these data highlight the fact that, in general, different cellular processes were affected by high temperature exposure in each compartment, protein localization and activity data are necessary to more conclusively model the cellular response of the model reef coral P. acuta and its in hospite Symbiodinium populations to elevated temperatures over multi-week timescales.
Molecular mechanisms of coral acclimation to elevated temperatures
In general, the proteomes of the three sampling times appeared similar on the 2D gels, though the DCPs differed markedly; only 5 of the 75 DCPs (7%) were sequenced at multiple sampling times. Of these five, only host coral pentraxin was down-regulated at high temperature at multiple sampling times. Based on homology searches and studies of vertebrates, pentraxin is likely involved in immunity [45]. A relative decrease in abundance of a single protein putatively involved in immunity at two sampling times does not, in and of itself, suggest that host coral immunity was compromised by high temperature exposure (especially since the corals ultimately acclimated to such elevated temperatures); however, others have instead documented induction of immuno-targeted genes in temperature-stressed corals [46], meaning that future works should take a more detailed look at proteins involved in the coral immune system (particularly in corals that are ultimately found to bleach). The decrease in available energy stores coincident with coral bleaching could indeed be predicted to lead to an immuno-compromised state driven by coral malnourishment [47].
In addition to immunity, a number of other cellular processes were affected by elevated temperature exposure. We now use our proteome-scale data to propose two, possibly related cellular processes that may have been involved in the high-temperature acclimation of the experimental corals. We first discuss the role of proteins involved in the stress response and protein homeostasis/turnover. We then turn to the potential importance of osmoregulation in coral acclimation to high temperatures. Regarding the former process, 25 and 40% of the host coral and Symbiodinium H>C DCP pools, respectively, comprised proteins involved in the stress response at the two-week sampling time; given that exposure to 30°C was hypothesized to elicit stress, this result is unsurprising. The two host coral H>C stress proteins identified at the two-week sampling time included beta-gamma crystallin and sacsin. Although the former has only ever been hypothesized to be involved in the stress response (as well as calcium binding; [48]), it was found to be down-regulated in S. hystrix specimens exposed to a variable temperature regime for one week [32]; given its proven temperature sensitivity in multiple experiments featuring different coral models, we recommend that this protein, which is water soluble and frequently documented in the lenses of vertebrate eyes, be functionally characterized in the near future.
With respect to the second of the two host coral proteins up-regulated at high temperatures at the two-week sampling time, sacsin is a co-chaperone of heat shock protein 70 (HSP70), the most well characterized molecular chaperone. Curiously, the expression of neither the host nor Symbiodinium hsp70 mRNA is affected by short- or long-term high-temperature exposure in Taiwanese corals, presumably because both are synthesized at high levels at all times in preparation for the frequent upwelling events that characterize the reefs of Southern Taiwan [3]. That being said, there is little congruency between gene and protein expression in reef corals or Symbiodinium [32]. Therefore, it is possible that the HSP70 proteins with which sacsin associates were synthesized at higher levels in the high-temperature samples analyzed herein; they may not have been sequenced due to, for instance, low cellular concentrations (few 70-kDa spots were evident in any of the gels [Fig 1].). Regardless of this discrepancy, a more thorough elucidation of the role of co-chaperones like sacsin and STl1, which was actually down-regulated at high temperatures in Symbiodinium, in acclimation to high temperatures represents a worthy endeavor for future study.
This STl1-like protein was the only Symbiodinium stress protein down-regulated at high temperatures; in contrast, four Symbiodinium stress proteins were up-regulated after two weeks of high-temperature exposure: an uncharacterized protein with DNAJ and WW domains, two peptidylprolyl isomerase D paralogs, and an E3 ubiquitin protein ligase. The former protein has not been characterized and was only hypothesized to be involved in the stress response given its possession of a DNAJ domain, while peptidylprolyl isomerase D proteins are involved in a diverse array of cellular processes aside from the stress response; therefore, it is too speculative at the current time to predict the specific cellular roles of these proteins as they relate to the stress response. It is worth noting that these three proteins were amongst only four (the final being a pentatricopeptide repeat-containing protein) in the entire dataset encoded by mRNAs that also underwent increases in expression in response to exposure to elevated temperatures.
In contrast to the three proteins mentioned in the previous paragraph, E3 ubiquitin protein ligase is both well characterized and strongly implicated in the cellular stress response; specifically, this enzyme aids in the process by which an E2 ubiquitin-conjugating enzyme transfers a ubiquitin tag to a functionally compromised protein slated to be degraded by the proteasome [49]. As such, this protein is involved in protein homeostasis and turnover, a process we hypothesized previously [29] (reiterated in the Introduction) to be important in coral high-temperature acclimation. The elevated concentrations of E3 ubiquitin protein ligase and HSP70 co-chaperones in both compartments of P. acuta-Symbiodinium endosymbioses exposed to high temperatures for two weeks suggests that protein turnover in these same samples was relatively high. This may be indicative of incomplete acclimation or even baseline levels of stress (e.g., due to photoinhibition [50], reactive oxygen species production [51], or osmotic pressure decreases [discussed in more detail below]). However, as corals survived for nine months at 30°C, this protein degradation/folding-mediated stress response was clearly able to restore homeostasis to the extent that sufficient energy could be shuttled to growth-related processes (growth was similar between treatments [9].). A look at the additional cellular processes affected by elevated temperature exposure may further elucidate how such high-temperature acclimation occurred.
Maintenance of osmotic pressure is the most energetically expensive task a cell undertakes [52], and coral-Symbiodinium osmoregulation has been hypothesized to be impacted by elevated temperature exposure [53]. This hypothesis stems from the notion that cessation of osmolyte flux from Symbiodinium to the host coral as a result of high temperature-induced photoinhibition [50] could result in a drop in the osmotic pressure of the host gastrodermal cell. Indeed, Symbiodinium photosynthesis may have been influenced by high temperature exposure herein given the number of differentially concentrated photosynthesis-associated proteins uncovered (e.g., ribulose-1,5-bisphosphate carboxylase/oxygenase, fucoxanthin-chlorophyll a-c binding protein F, chloroplast oxygen-evolving enhancer, and peridinin-chlorophyll A binding protein). Such a decrease in osmotic pressure could lead to a consequent collapse of the coral plasma membrane upon the Symbiodinium cell [54]. This would necessitate a regulatory volume increase, in which, amongst a plethora of processes, the cytoskeleton must be rebuilt [55]. This may explain why a number of actins and other cytoskeleton proteins (e.g., spectrin, trichohyalin, and stabilizer of axonemal microtubules 2-like) differed in concentration between temperature treatments in the coral host in particular; such has also been documented at the mRNA-level in thermally challenged corals [56]. Re-establishment of osmotic pressure, then, may have allowed for these corals to have maintained cellular homeostasis at the experimentally elevated temperature of 30°C, though gastrodermal cell-specific proteomic approaches (sensu [10]) are needed to substantiate this hypothetical role of osmoadaptation, as well as the protein turnover-related processes described above, in coral acclimation to elevated temperatures. It is worth noting that, since Symbiodinium possess rigid, durable cell walls, osmotic stress is not hypothesized to affect their cell volume [53].
Low congruency between gene expression and protein concentration
At the two-week sampling time, only 4 of the 38 DCPs were associated with an mRNA whose expression also differed significantly between treatments. All four of these molecules were expressed at higher levels by Symbiodinium within corals exposed to elevated temperatures, and Symbiodinium were significantly more likely to be characterized by congruency between mRNA expression and protein concentration (27%) than the coral hosts in which they resided (0%). This 27% congruency is significantly higher than that of another study; not a single Symbiodinium molecule showed a similar temperature-related difference at the mRNA and protein levels in cells housed within the gastroderms of S. hystrix [32].
The immense sizes of the P. acuta and Symbiodinium transcriptomes (>100,000 contigs each) may account for, in part, the low degree of congruency between mRNA expression and protein abundance; for many of the genes of both P. acuta and Symbiodinium, multiple splice variants exist [29]. Given that the actual translation of these mRNAs was not observed, it cannot be known which splice variants were actually translated into proteins. Like Mascot (Matrix Sciences), MS-SCAN/MS-GF+ makes guesses about protein identity based on querying translated transcriptomes; no coral proteomes have been sequenced at present, and only a partial sea anemone proteome (~3,000 proteins from Aiptasia sp.) currently exists [57]. Simply because MS-SCAN yields a top hit for a particular peptide sequence does not mean that the “correct” splice variant was targeted; the peptides sequenced by MS are typically short (10–15 AA) and so may align to multiple splice variants. It is possible that improvements in protein sequencing will lead to longer AA “reads,” in which case there will be greater confidence in having isolated the splice variant encoding the protein of interest [58]. This will allow us to know whether the congruency between mRNA and protein concentration is indeed only 0–27% and 0–2% for Symbiodinium and pocilloporid corals, respectively. Until such can be verified, it is unadvised to use mRNA-level data alone to make inferences about the cellular behavior of reef corals, especially with respect to their ability to acclimate to environmental change (sensu [59–60]).
Supporting information
(DOCX)
Underlined spots represent uniquely synthesized proteins (see Fig 1 and Table 1.). Congruent and marginally congruent results between gene expression and protein concentration are highlighted in green and purple, respectively. Proteins involved in the stress response are highlighted in red. Proteins highlighted in blue and yellow were documented at different concentrations between treatments at the four- and eight-week sampling times, respectively (Table 2). Please see the S4 Table for peptide sequences. “C” and “H” in the “Spot” column correspond to spots removed from the control and high-temperature treatment gels, respectively. In the “mRNA effect” column, “C2” and “H2” correspond to control and high-temperature samples at the two-week sampling time, respectively, while “C36” and “H36” correspond to control and high-temperature samples at the 36-week sampling time, respectively. AA = amino acid. kDa = kilodalton. MW = molecular weight. NS = not significant (neither temperature nor time effect in the repeated measures ANOVA [p>0.05]). pI = isoelectric point. Sym = Symbiodinium.
(DOCX)
Proteins highlighted in blue and grey also differed in concentration between treatments at the two- and eight-week sampling times, respectively (Table 2). Please see the S5 Table for hypothetical functions and peptide sequences. “C” and “H” in the “Spot” column correspond to spots removed from the control and high-temperature treatment gels, respectively. AA = amino acids. DCP = differentially concentrated protein. kDa = kilodalton. MW = molecular weight. pI = isoelectric point. Sym = Symbiodinium.
(DOCX)
Underlined spots represent uniquely synthesized proteins (see Fig 1 and Table 1.). Proteins highlighted in yellow and grey also differed in concentration between treatments at the two- and four-week sampling times, respectively (Table 2), and those highlighted in red were hypothesized to be involved in the stress response. Please see the S6 Table for hypothetical functions and peptide sequences. “C” and “H” in the “Spot” column correspond to spots removed from the control and high-temperature treatment gels, respectively. AA = amino acids. kDa = kilodalton. MW = molecular weight. pI = isoelectric point. Sym = Symbiodinium.
(DOCX)
The 13 and 25 unique proteins whose concentrations were higher in samples of the control (C) and high temperature (H) treatments, respectively, at the two-week sampling time were included. Additional details of the sequenced proteins can be found in the S1 Table. Although multiple missed cleavages exist in certain peptide sequences (more than the two allowed by the MS-GF+ script [described in the supplemental methods section of the S1 file]), these peptides were nevertheless included provided that they were either 1) 15 or more amino acids (AA) in length or 2) paired with one or more additional peptides that mapped to the same reference protein (whose collective length summed to 15 or more AA). The compartment of origin has been mentioned next to the spot number when it could be determined. Of the 38 proteins identified by BLAST analysis of the top hit contig (mRNA) derived from MS-SCAN analysis of the Pocillopora acuta-Symbiodinium (“Sym”) transcriptome (see the S1 Table for contig accession numbers.), the identities of 12 were further verified by directly BLASTing the MS-SCAN-derived peptide sequences against the Stylophora pistillata (n = 8) and Symbiodinium (clade B1) genomes (n = 4).
(DOCX)
The eight and four proteins whose concentrations were higher in samples of the control (C) and high temperature (H) treatments, respectively, at the four-week sampling time were included. Additional details of the sequenced proteins can be found in the S2 Table. Although multiple missed cleavages exist in certain peptide sequences (more than the two allowed by the MS-GF+ script [desribed in the supplemental methods outlined in the S1 file]), these peptides were nevertheless included provided that they were either 1) 15 or more amino acids (AA) in length or 2) paired with one or more additional peptides that mapped to the same reference protein (whose collective length summed to 15 or more AA). No unique, differentially concentrated proteins were identified from spots H5 and H6. The compartment of origin has been included next to the spot number. Of the 12 proteins identified by BLAST analysis of the top hit contig derived from MS-SCAN analysis of the Pocillopora acuta-Symbiodinium (“Sym”) transcriptome (see the S2 Table for contig accession numbers.), the identies of half were further verified by directly BLASTing the MS-SCAN-derived peptide sequences against the Stylophora pistillata (n = 4) and Symbiodinium (clade B1) genomes (n = 2).
(DOCX)
The 6 and 19 proteins whose concentrations were higher in samples of the control (C) and high (H) temperature treatments, respectively, at the eight-week sampling time were included. Additional details of the sequenced proteins can be found in the S3 Table. Although multiple missed cleavages exist in certain peptide sequences (more than the two allowed by the MS-GF+ script [described in the supplemental methods outlined in the S1 file]), these peptides were nevertheless included provided that they were either 1) 15 or more amino acids (AA) in length or 2) paired with one or more additional peptides that mapped to the same reference protein (whose collective length summed to 15 or more AA). The lysine- and arginine-rich nature of the Pocillopora acuta and Symbiodinium (“Sym”) proteomes of this study, as well as another carried out with the con-familial pocilloporid coral Seriatopora hystrix [32], suggests that alternative protein digest enzymes may serve better for future proteomic analyses of pocilloporid corals. The compartment of origin has been included next to the spot number when it could be determined. Of the 25 proteins identified by BLAST analysis of the top hit contig derived from MS-SCAN analysis of the P. acuta-Symbiodinium transcriptome (see the S3 Table for contig accession numbers.), the identities of 11 were further verified by directly BLASTing the MS-SCAN-derived peptide sequences against the Stylophora pistillata (n = 8) and Symbiodinium (clade B1) genomes (n = 3). bact = bacterial.
(DOC)
Acknowledgments
Thanks are given to Dr. Yu-Bin Wang for implementation of the MS-SCAN function on the P. acuta-Symbiodinium transcriptome server and Dr. Tung-Yung Fan for acquiring the collection permits on our behalf.
Data Availability
The mass spectrometry (.MGF) files produced herein (n = 25) can be downloaded from the following page: http://symbiont.iis.sinica.edu.tw/coral_pdltte/static/html/index.html#stat.
Funding Statement
This work was funded by the United States National Science Foundation (postdoctoral research fellowship to ABM; OISE-0852960) and the Khaled bin Sultan Living Oceans Foundation (postdoctoral research fellowship to ABM). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoegh-Guldberg O, Bruno JF. The impact of climate change on the world’s marine ecosystems. Science. 2010;328: 1523–1528. doi: 10.1126/science.1189930 [DOI] [PubMed] [Google Scholar]
- 2.Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, et al. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318: 1737–1742. doi: 10.1126/science.1152509 [DOI] [PubMed] [Google Scholar]
- 3.Mayfield AB, Wang LH, Tang PC, Hsiao YY, Fan TY, Tsai CL, et al. Assessing the impacts of experimentally elevated temperature on the biological composition and molecular chaperone gene expression of a reef coral. PLoS ONE. 2011;e26529 doi: 10.1371/journal.pone.0026529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown BE. Coral bleaching: causes and consequences. Coral Reefs. 1997;16: s129–s138. [Google Scholar]
- 5.Putnam HM, Edmunds PJ. The physiological response of reef corals to diel fluctuations in seawater temperature. J Exp Mar Biol Ecol. 2011;396: 216–223. [Google Scholar]
- 6.Barshis DJ, Stillman JH, Gates RD, Toonen RJ, Smith LW, Birkeland C. Protein expression and genetic structure of the coral Porites lobata in an environmentally extreme Samoan back reef: does host genotype limit phenotypic plasticity? Mol Ecol. 2010;19: 140297–140297. [DOI] [PubMed] [Google Scholar]
- 7.Krueger T, Horwitz N, Bodin J, Giovani ME, Escrig S, Meibom A, et al. Common reef-building coral in the Northern Red Sea resistant to elevated temperature and acidification. R Soc Open Sci. 2017;4: 170038 doi: 10.1098/rsos.170038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver TA, Palumbi SR. Do fluctuating temperature environments elevate coral thermal tolerance? Coral Reefs. 2011;30: 429–440. [Google Scholar]
- 9.Mayfield AB, Fan TY, Chen CS. Physiological acclimation to elevated temperature in a reef-building coral from an upwelling environment. Coral Reefs. 2013;32: 909–921. [Google Scholar]
- 10.Peng SE, Chen WNU, Chen HK, Lu CY, Mayfield AB, Fang LS, et al. Lipid bodies in coral-dinoflagellate endosymbiosis: ultrastructural and proteomic analyses. Proteomics. 2011;17: 3540–3455. [DOI] [PubMed] [Google Scholar]
- 11.Chen WNU, Kang HJ, Weis VM, Mayfield AB, Fang LS, Chen CS. Diel rhythmicity of lipid body formation in a coral-Symbiodinium endosymbiosis. Coral Reefs. 2012;31: 521–534. [Google Scholar]
- 12.Mayfield AB, Hsiao YY, Fan TY, Chen CS. Temporal variation in RNA/DNA and protein/DNA ratios in four anthozoan-dinoflagellate endosymbioses of the Indo-Pacific: implications for molecular diagnostics. Platax. 2012;9: 1–24. [Google Scholar]
- 13.Wang LH, Lee HH, Fang LS, Mayfield AB, Chen CS. Normal fatty acid and phospholipid synthesis are prerequisites for the cell cycle of Symbiodinium and their endosymbiosis with sea anemones. PLoS ONE. 2013;e72486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayfield AB, Hsiao YY, Chen HK, Chen CS. Rubisco expression in the dinoflagellate Symbiodinium sp. is influenced by both photoperiod and endosymbiotic lifestyle. Mar Biotech. 2014;16: 371–384. [DOI] [PubMed] [Google Scholar]
- 15.Chen HK, Song SN, Wang LH, Mayfield AB, Chen YJ, Chen WNU, et al. A compartmental comparison of major lipid species in a coral-Symbiodinium endosymbiosis: evidence that the coral host regulates lipogenesis of its cytosolic lipid bodies. PLoS ONE. 2015;e0132519 doi: 10.1371/journal.pone.0132519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin S, Cheng S, Song B, Zhong Z, Lin X, Li W, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350: 691–694. doi: 10.1126/science.aad0408 [DOI] [PubMed] [Google Scholar]
- 17.Chen WNU, Hsiao YJ, Mayfield AB, Young R, Hsu L, Chen CS, et al. Vertical transmission of heterologous clade C Symbiodinium in a model anemone infection system. Peer J. 2016;4: e2358 doi: 10.7717/peerj.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen HK, Mayfield AB, Wang LH, Chen CS. Coral lipid bodies as the relay center interconnecting diel-dependent lipidomic changes in different cellular compartments. Sci Reports. 2017;7: 3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putnam HM, Stat M, Pochon X, Gates RD. Endosymbiotic flexibility associates with environmental sensitivity in scleractinian corals. Proc Royal Soc B. 2012;279: 4352–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putnam HM, Mayfield AB, Fan TY, Chen CS, Gates RD. The physiological and molecular responses of larvae from the reef-building coral Pocillopora damicornis exposed to near-future increases in temperature & pCO2. Mar Biol. 2013;160: 2157–2173. [Google Scholar]
- 21.Mayfield AB, Chen M, Meng PJ, Lin HJ, Chen CS, Liu PJ. The physiological response of the reef coral Pocillopora damicornis to elevated temperature: results from coral reef mesocosm experiments in Southern Taiwan. Mar Environ Res. 2013;86: 1–11. doi: 10.1016/j.marenvres.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 22.Mayfield AB, Fan TY, Chen CS. Real-time PCR-based gene expression analysis in the model reef-building coral Pocillopora damicornis: insight from a salinity stress study. Platax. 2013;10: 1–29. [Google Scholar]
- 23.Mayfield AB, Chen CS, Liu PJ. Decreased green fluorescent protein-like chromoprotein gene expression in specimens of the reef-building coral Pocillopora damicornis undergoing high temperature-induced bleaching. Platax. 2014;11: 1–23. [Google Scholar]
- 24.Mayfield AB, Chen YH, Dai CF, Chen CS. The effects of temperature on gene expression in the reef-building coral Seriatopora hystrix: insight from aquarium studies in Southern Taiwan. Int J Mar Sci. 2014;4(50): 1–23. [Google Scholar]
- 25.Mayfield AB. Uncovering spatio-temporal and treatment-derived differences in the molecular physiology of a model coral-dinoflagellate mutualism with multivariate statistical approaches. J Mar Sci Eng. 2016;4: 63. [Google Scholar]
- 26.Mayfield AB, Chen CS, Dempsey AC, Bruckner AW. The molecular ecophysiology of closely related pocilloporids from the South Pacific: a case study from the Austral and Cook Islands. Platax. 2016;13: 1–25. [Google Scholar]
- 27.Mayfield AB, Chen CS, Dempsey AC. Identifying corals displaying aberrant behavior in Fiji’s Lau Archipelago. PLoS ONE. 2017;e0177267 doi: 10.1371/journal.pone.0177267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayfield AB, Chen CS, Dempsey AC. Biomarker profiling in reef corals of Tonga’s Ha’apai and Vava’u archipelagos. PLoS ONE. 2017;e0185857 doi: 10.1371/journal.pone.0185857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayfield AB, Wang YB, Chen CS, Chen SH, Lin CY. Compartment-specific transcriptomics in a reef-building coral exposed to elevated temperatures. Mol Ecol. 2014;23: 5816–5830. doi: 10.1111/mec.12982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gates RD, Edmunds PJ. The physiological mechanisms of acclimatization in tropical reef corals. Integr Comp Biol. 1999;39: 30–43. [Google Scholar]
- 31.Barshis DJ, Ladner JT, Oliver TA, Seneca FO, Traylor-Knowles N, Palumbi SR. Genomic basis for coral resilience to climate change. Proc Natl Acad Sci USA. 2013;110: 1387–1392. doi: 10.1073/pnas.1210224110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayfield AB, Wang YB, Chen CS, Chen SH, Lin CY. Dual-compartmental transcriptomic+proteomic analysis of a marine endosymbiosis exposed to environmental change. Mol Ecol. 2016;25: 5944–5958. doi: 10.1111/mec.13896 [DOI] [PubMed] [Google Scholar]
- 33.Weston AJ, Dunlap WC, Beltran VH, Starcevic A, Hranueli D, Ward M, et al. Proteomics links the redox state to calcium signaling during bleaching of the scleractinian coral Acropora microphthalma on exposure to high solar irradiance and thermal stress. Mol Cell Proteomics. 2015;14: 585–595. doi: 10.1074/mcp.M114.043125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayfield AB, Chan PH, Putnam HM, Chen CS, Fan TY. The effects of a variable temperature regime on the physiology of the reef-building coral Seriatopora hystrix: results from a laboratory-based reciprocal transplant. J Exp Biol. 2012;215: 4183–4195. doi: 10.1242/jeb.071688 [DOI] [PubMed] [Google Scholar]
- 35.Mayfield AB, Chen YJ, Lu CY, Chen CS. Proteins responsive to variable temperature exposure in the reef-building coral Seriatopora hystrix In: Ortiz S, editor. Coral reefs: ecosystems, environmental impact and current threats. New York: NOVA Publishing; 2016. pp. 1–72. [Google Scholar]
- 36.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nature Commun. 2014;5: 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mente E, Pierce GJ, Antonopoulou E, Stead D, Martin SAM. Postprandial hepatic protein expression in trout Oncorhynchus mykiss: a proteomics examination. Biochem Biophys Res. 2017;9: 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuichi T, Yamada L, Shinzato C, Sawada H, Satoh N. Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PLoS ONE. 2017;e0156424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weston AJ, Dunlap WC, Shick JM, Klueter A, Iglic K, Vukelic A, et al. A profile of an endosymbiont-enriched fraction of the coral Stylophora pistillata reveals proteins relevant to microbial-host interactions. Mol Cell Proteomics. 2012;11: M111.015487 doi: 10.1074/mcp.M111.015487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayfield AB, Hirst MB, Gates RD. Gene expression normalization in a dual-compartment system: a real-time PCR protocol for symbiotic anthozoans. Mol Ecol Res. 2009;9: 462–470. [DOI] [PubMed] [Google Scholar]
- 41.Mayfield AB, Fan TY, Chen CS. The physiological impact of ex situ transplantation on the Taiwanese reef-building coral Seriatopora hystrix. J Mar Biol. 2013;Article ID 569361. [Google Scholar]
- 42.Doo SS, Mayfield AB, Nguyen HD, Chen HK. 2013. Protein analysis in large benthic foraminifera In Kitazato H and Bernhard J, editors. Approaches to study living foraminifera. New York: Springer; 2016. pp. 71–89. [Google Scholar]
- 43.Doo SS, Mayfield AB, Byrne M, Chen HK, Nguyen H, Fan TY. Reduced expression of the rate-limiting carbon fixation enzyme RuBisCO in the benthic foraminifer Baculogypsina sphaerulata holobiont in response to heat shock. J Exp Mar Biol Ecol. 2012;430–431: 63–67. [Google Scholar]
- 44.Putnam HM, Barott K, Ainsworth TD, Gates RD. The vulnerability and resilience of reef-building corals. Current Biol. 2017;27: R528–R540. [DOI] [PubMed] [Google Scholar]
- 45.Du Clos TW. Pentraxins: structure, function, and role in inflammation. ISRN Inflamm. 2013;Article ID 379040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinzón JH, Kamel B, Burge CA, Harvell CD, Medina M, Weil E, et al. Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. Royal Soc Open Sci. 2015;2: 140214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer CV, Traylor-Knowles N. Towards an integrated network of coral immune mechanisms. Proc Royal Soc B. 2012;279: 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundgren L, Vera JC, Peplow L, Manel S, van Oppen MJH. Genotype-environment correlations in corals from the Great Barrier Reef. BMC Genetics. 2013;14: 9 doi: 10.1186/1471-2156-14-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nature Rev Mol Cell Biol. 2005;6: 599–609. [DOI] [PubMed] [Google Scholar]
- 50.Jones RJ, Hoegh-Guldberg O, Larkum AWD, Schreiber U. Temperature-induced bleaching of corals begins with impairment of the CO2 fixation metabolism in zooxanthellae. Plant Cell Environ.1998;21: 1219–1230. [Google Scholar]
- 51.Lesser MP. Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in symbiotic dinoflagellates. Limnol Oceanogr. 1996;41: 271–283. [Google Scholar]
- 52.Hochachka PW, Somero GN. Biochemical adaptation Oxford: Oxford University Press; 2002. [Google Scholar]
- 53.Mayfield AB, Gates RD. Osmoregulation in anthozoan-dinoflagellate symbiosis. Comp Biochem Physiol. 2007;147A: 1–10. [DOI] [PubMed] [Google Scholar]
- 54.Mayfield AB, Hsiao YY, Fan TY, Chen CS, Gates RD. Evaluating the temporal stability of stress-activated protein kinase and cytoskeleton gene expression in the Pacific corals Pocillopora damicornis and Seriatopora hystrix. J Exp Mar Biol Ecol. 2010;395: 215–222. [Google Scholar]
- 55.Thirone ACP, Speight P, Zulys M, Rotstein OD, Szaszi K, Pedersen SF, et al. Hyperosmotic stress induces Rho/Rho kinase/LIM kinase-mediated cofilin phosphorylation in tubular cells: key role in the osmotically triggered F-actin response. Am J Physiol Cell Physiol. 2009;296: CC463–C475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desalvo MK, Voolstra CR, Sunagawa S, Schwarz J, Stillman J, Coffroth MA, et al. Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol Ecol. 2008;17: 3952–3971. doi: 10.1111/j.1365-294X.2008.03879.x [DOI] [PubMed] [Google Scholar]
- 57.Oakley CA, Ameismeier MF, Peng L, Weis VM, Grossman AR, Davy SK. Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia. Cell Microbiol. 2016;18(7): 1009–1023. doi: 10.1111/cmi.12564 [DOI] [PubMed] [Google Scholar]
- 58.Shalit T, Elinger D, Savidor A, Gabasgvili A, Levin Y. MS1-based label-free proteomics using a quadrupole orbitrap mass spectrometer. J Proteome Res. 2016;14(4): 1979–1985. [DOI] [PubMed] [Google Scholar]
- 59.Palumbi SR, Barshis DJ, Traylor-Knowles N, Bay RA. Mechanisms of reef coral resistance to future climate change. Science. 2014;344: 895–898. doi: 10.1126/science.1251336 [DOI] [PubMed] [Google Scholar]
- 60.Ruiz-Jones LJ, Palumbi SR. Tidal heat pulses on a reef trigger a fine-tuned transcriptional response in corals to maintain homeostasis. Science Advances. 2017;e1601298 doi: 10.1126/sciadv.1601298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Underlined spots represent uniquely synthesized proteins (see Fig 1 and Table 1.). Congruent and marginally congruent results between gene expression and protein concentration are highlighted in green and purple, respectively. Proteins involved in the stress response are highlighted in red. Proteins highlighted in blue and yellow were documented at different concentrations between treatments at the four- and eight-week sampling times, respectively (Table 2). Please see the S4 Table for peptide sequences. “C” and “H” in the “Spot” column correspond to spots removed from the control and high-temperature treatment gels, respectively. In the “mRNA effect” column, “C2” and “H2” correspond to control and high-temperature samples at the two-week sampling time, respectively, while “C36” and “H36” correspond to control and high-temperature samples at the 36-week sampling time, respectively. AA = amino acid. kDa = kilodalton. MW = molecular weight. NS = not significant (neither temperature nor time effect in the repeated measures ANOVA [p>0.05]). pI = isoelectric point. Sym = Symbiodinium.
(DOCX)
Proteins highlighted in blue and grey also differed in concentration between treatments at the two- and eight-week sampling times, respectively (Table 2). Please see the S5 Table for hypothetical functions and peptide sequences. “C” and “H” in the “Spot” column correspond to spots removed from the control and high-temperature treatment gels, respectively. AA = amino acids. DCP = differentially concentrated protein. kDa = kilodalton. MW = molecular weight. pI = isoelectric point. Sym = Symbiodinium.
(DOCX)
Underlined spots represent uniquely synthesized proteins (see Fig 1 and Table 1.). Proteins highlighted in yellow and grey also differed in concentration between treatments at the two- and four-week sampling times, respectively (Table 2), and those highlighted in red were hypothesized to be involved in the stress response. Please see the S6 Table for hypothetical functions and peptide sequences. “C” and “H” in the “Spot” column correspond to spots removed from the control and high-temperature treatment gels, respectively. AA = amino acids. kDa = kilodalton. MW = molecular weight. pI = isoelectric point. Sym = Symbiodinium.
(DOCX)
The 13 and 25 unique proteins whose concentrations were higher in samples of the control (C) and high temperature (H) treatments, respectively, at the two-week sampling time were included. Additional details of the sequenced proteins can be found in the S1 Table. Although multiple missed cleavages exist in certain peptide sequences (more than the two allowed by the MS-GF+ script [described in the supplemental methods section of the S1 file]), these peptides were nevertheless included provided that they were either 1) 15 or more amino acids (AA) in length or 2) paired with one or more additional peptides that mapped to the same reference protein (whose collective length summed to 15 or more AA). The compartment of origin has been mentioned next to the spot number when it could be determined. Of the 38 proteins identified by BLAST analysis of the top hit contig (mRNA) derived from MS-SCAN analysis of the Pocillopora acuta-Symbiodinium (“Sym”) transcriptome (see the S1 Table for contig accession numbers.), the identities of 12 were further verified by directly BLASTing the MS-SCAN-derived peptide sequences against the Stylophora pistillata (n = 8) and Symbiodinium (clade B1) genomes (n = 4).
(DOCX)
The eight and four proteins whose concentrations were higher in samples of the control (C) and high temperature (H) treatments, respectively, at the four-week sampling time were included. Additional details of the sequenced proteins can be found in the S2 Table. Although multiple missed cleavages exist in certain peptide sequences (more than the two allowed by the MS-GF+ script [desribed in the supplemental methods outlined in the S1 file]), these peptides were nevertheless included provided that they were either 1) 15 or more amino acids (AA) in length or 2) paired with one or more additional peptides that mapped to the same reference protein (whose collective length summed to 15 or more AA). No unique, differentially concentrated proteins were identified from spots H5 and H6. The compartment of origin has been included next to the spot number. Of the 12 proteins identified by BLAST analysis of the top hit contig derived from MS-SCAN analysis of the Pocillopora acuta-Symbiodinium (“Sym”) transcriptome (see the S2 Table for contig accession numbers.), the identies of half were further verified by directly BLASTing the MS-SCAN-derived peptide sequences against the Stylophora pistillata (n = 4) and Symbiodinium (clade B1) genomes (n = 2).
(DOCX)
The 6 and 19 proteins whose concentrations were higher in samples of the control (C) and high (H) temperature treatments, respectively, at the eight-week sampling time were included. Additional details of the sequenced proteins can be found in the S3 Table. Although multiple missed cleavages exist in certain peptide sequences (more than the two allowed by the MS-GF+ script [described in the supplemental methods outlined in the S1 file]), these peptides were nevertheless included provided that they were either 1) 15 or more amino acids (AA) in length or 2) paired with one or more additional peptides that mapped to the same reference protein (whose collective length summed to 15 or more AA). The lysine- and arginine-rich nature of the Pocillopora acuta and Symbiodinium (“Sym”) proteomes of this study, as well as another carried out with the con-familial pocilloporid coral Seriatopora hystrix [32], suggests that alternative protein digest enzymes may serve better for future proteomic analyses of pocilloporid corals. The compartment of origin has been included next to the spot number when it could be determined. Of the 25 proteins identified by BLAST analysis of the top hit contig derived from MS-SCAN analysis of the P. acuta-Symbiodinium transcriptome (see the S3 Table for contig accession numbers.), the identities of 11 were further verified by directly BLASTing the MS-SCAN-derived peptide sequences against the Stylophora pistillata (n = 8) and Symbiodinium (clade B1) genomes (n = 3). bact = bacterial.
(DOC)
Data Availability Statement
The mass spectrometry (.MGF) files produced herein (n = 25) can be downloaded from the following page: http://symbiont.iis.sinica.edu.tw/coral_pdltte/static/html/index.html#stat.