To the Editor:
The filaggrin gene (FLG) is essential for skin differentiation and epidermal barrier formation. FLG loss-of-function (LoF) variants are associated with ichthyosis vulgaris and the major genetic risk factor for developing atopic dermatitis (AD).1, 2, 3 Genetic stratification of patients with AD according to FLG LoF risk is a common practice for both research and clinical studies; however, few studies comprehensively sequence the entire FLG coding region. Most studies that include FLG genotyping have screened for common predominant LoF variants to report allele frequencies after full Sanger sequencing of a smaller batch of test patient samples or previously published data. This strategy potentially results in underreporting of the genetic contribution especially in ethnicities where FLG LoF variants are highly diverse.4 Distinct LoF variants have been reported for most ethnicities studied to date. For example, 2 predominant sequence variants (p.R501X and c.2282del4) make up approximately 80% of the mutation burden in northern Europeans,5 whereas in East Asian ethnicities, a larger FLG LoF mutation spectrum is found with fewer predominating variants.6, 7 However, routinely Sanger sequencing the entire FLG coding region for large cohorts is not always feasible, although desirable as it is essential to correctly stratify patients. To address this, we developed a robust and cost-effective high-throughput PCR-based method for analyzing the entire coding region of FLG using Fluidigm microfluidics technology and next-generation sequencing (NGS). We have applied this method to fully resequence cohorts of Chinese, Malay, and Indian patients with AD from the Singaporean population.
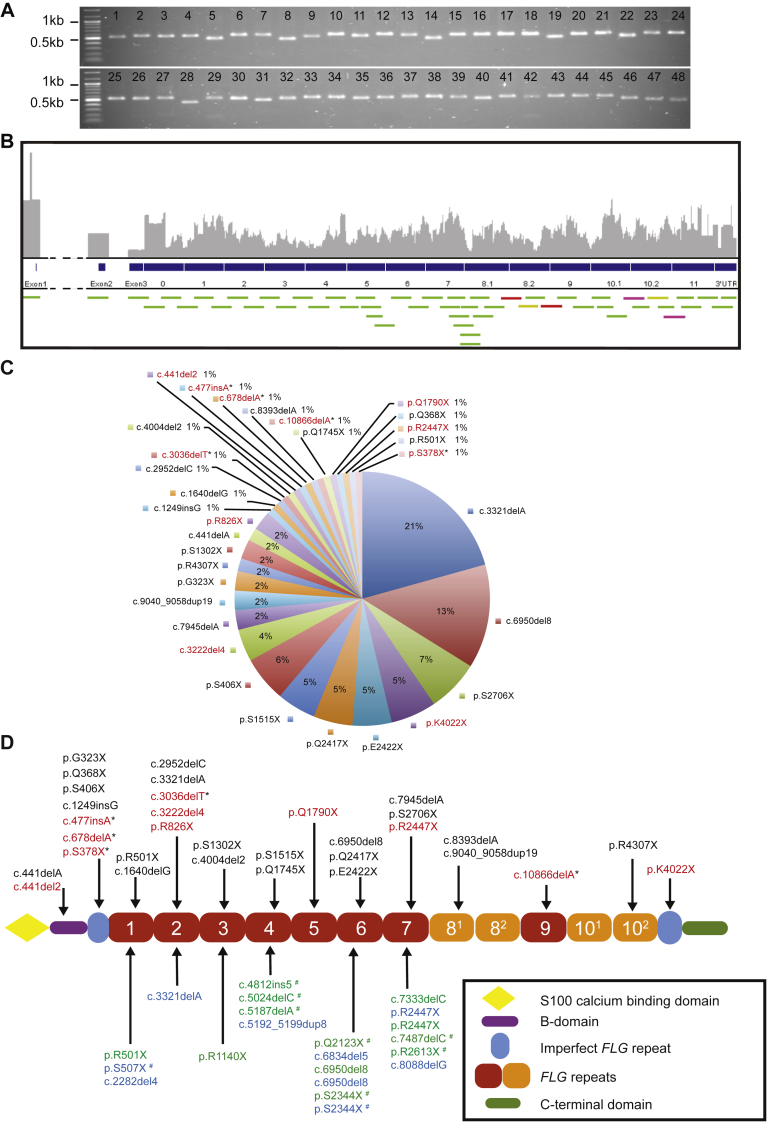
We designed and optimized overlapping FLG-specific primer assays (containing NGS adapters) to span the entire FLG coding region including known intragenic copy number variation (CNV) (see Fig 1, A; see Table E1 in this article's Online Repository at www.jacionline.org). A total of 48 overlapping primer assays, with amplicons of maximum 500 base pairs, provided redundancy for sequencing reads across primer-binding sites and 100% coverage of FLG exon bases (see Fig 1, B). The Fluidigm Access Array 48.48 integrated fluidic circuit (IFC) chip (Fluidigm, San Francisco, Calif) generates 48 amplicons for 48 different DNA samples in parallel, simultaneously thermocycling 2304 PCR reactions at nanoliter volumes (see Fig E1 and this article's Methods section in the Online Repository at www.jacionline.org). Initially 96 DNA samples were assayed in IFC chips before Illumina MiSeq 2x250 bp read mode sequencing (4 samples failed); 14 samples from this batch of 96 were previously Sanger sequenced for the entire FLG coding region.5, 7 The known FLG LoF variant profile was then used to validate LoF variant detection with the IFC and NGS method. We identified all FLG LoF variants originally identified by Sanger in the 14 samples as well as additional variants in 2 samples (see Table I) and documented LoF variants in the remaining 78 samples that passed quality control testing (see Table E2 in this article's Online Repository at www.jacionline.org). LoF variants were all confirmed by visual inspection using Integrated Genome Viewer before Sanger sequencing. In addition, we determined the FLG CNV of repeats 8 and 10 (an important risk factor for AD8) in the 92 samples using relative coverage-based metrics (see Fig E2 in this article's Online Repository at www.jacionline.org).
Fig 1.
FLG primer validation, amplicon coverage, and FLG LoF variant analysis in Singaporean cohorts with AD. A, A total of 48 FLG-specific primer assays were validated by PCR and gel electrophoresis to confirm expected amplicon size and absence of nonspecific products. B, Schematic visualization of overlapping amplicon design across 12-repeat FLG coding region (green, yellow, red, purple bars). Primer assay 34 (yellow bars), 35 (red bars), and 41 (purple bars) produced multiple distinct amplicons. C, Spectrum of disease-associated FLG LoF variants identified in the Singaporean Chinese IV and/or AD population; 11 additional variants (highlighted in red) were identified in addition to those from our previous survey, of which 5 variants have not been previously reported (*). D, Profilaggrin schematic showing LoF FLG variants domain positions—Singaporean Chinese samples are present above the schematic, variants not previously reported in Singapore Chinese samples are highlighted (red) including FLG LoF variants not previously published (*). Positions of FLG LoF variants identified from Singapore Malays are highlighted in blue and Singapore Indians in green below the schematic. FLG LoF variants not previously identified in AD patient-based studies are marked with #. IV, Ichthyosis vulgaris.
Table I.
Concordance of FLG LoF variant detection for 14 previously fully Sanger-sequenced AD samples using our MiSeq 2x250 bp protocol
| Sample ID | Sanger sequencing | Illumina MiSeq assay |
|---|---|---|
| IA-P003 | p.S1515X | p.S1515X |
| IA-P009 | No LoF detected | No LoF detected |
| IA-P014 | No LoF detected | No LoF detected |
| IA-P017 | p.E2422X | p.E2422X |
| IA-P021 | p.S406X; c.6950_6957del8 | p.S406X; c.6950_6957del8 |
| IA-P024 | c.1640delG | c.1640delG |
| IA-P025 | p.Q368X; c.3321delA | p.Q368X; c.3321delA |
| IA-P028 | c.7945delA | c.7945delA |
| IA-P062 | p.Q2417X | p.Q2417X |
| IA-P063 | c.2952delC | c.2952delC |
| IA-P083∗ | c.9040_9058dup19 | c.9040_9058dup19; p.Q1790X |
| IA-P084∗ | p.S1302X | p.S1302X; p.S1515X |
| IA-P090 | c.4004del2 | c.4004del2 |
| IA-P094 | c.2282del4; p.R2447X | c.2282del4; p.R2447X |
Our NGS protocol detected additional LoF variants in 2 samples.
The IFC and NGS sequencing method was then used to analyze a further 334 Singaporean ichthyosis vulgaris and/or AD patient samples to obtain estimates of disease-associated LoF allele frequency in the 3 major ethnicities of Singapore—Chinese, Malay, and Indian. In 279 Chinese Singaporean samples (see Table E3 in this article's Online Repository at www.jacionline.org), we identified a further 11 additional LoF variants, raising the total number identified in this population to 33 (an increase from 22 variants identified in our previous study7) with 5 not previously reported in the literature (see Fig 1, C and D); 85 of these samples had also been previously Sanger sequenced7 and the concordance profile was near identical (see Table E4 in this article's Online Repository at www.jacionline.org). A total of 14 LoF variants reached significance using Fisher exact test (P < .05) compared with population control data derived from ExAC (version 0.3.1) exome database (see Table E5 and the Methods section in this article's Online Repository at www.jacionline.org). The combined FLG LoF mutation allele frequency for Chinese Singaporean patients with AD is now 32.3%, an increase from 20.2% in our previous survey of 425 patient samples further supporting the biological importance of FLG mutations in AD.7 Smaller cohorts of 19 Indian and 36 Malay patients were analyzed and we identified FLG LoF variants in 9 Indian samples and 9 Malay samples (see Fig 1, D; see Tables E6 and E7 in this article's Online Repository at www.jacionline.org). The identification of unreported FLG LoF variants in Indian and Malay ethnicities from Singapore confirms the diversity of FLG variants in AD between different ethnic groups. In total we identified 18 variants with limited overlap with the Chinese samples (4 out of 18) and a number of these are not present in the ExAC database, highlighting the contribution of rare, family-specific mutations in AD (11 of 18). This small but well-characterized study of Indian and Malay Singaporeans highlights the variation in combined allele frequencies between ethnicities, with 47.4% of Indian patients with AD and 25% of Malay patients with AD harboring FLG LoF variants. The presence of FLG LoF variants was strongly associated with AD; however, it was not significantly associated with increasing severity in the 334 patients analyzed in this study, possibly due to the small number of mild cases analzyed (mild cases in this cohort 5.3%; see Table E8 in this article's Online Repository at www.jacionline.org).
In conclusion, we describe a multiplexed targeted resequencing method to study the FLG coding region. We highlight that comprehensive sequencing improves accuracy estimates of genetic contribution from FLG deleterious alleles and CNVs in AD. This strategy to study genetic variation does not rely on previous mutation spectrum information from any given population or ethnicity and therefore can identify rare, family-specific, or de novo variants globally. This approach outperforms exome sequencing because of its throughput and ability to analyze known CNVs that are not currently reflected in NCBI RefSeq for FLG. Amplicon resequencing is robust, reliable, and unbiased and has the potential for scalable sample preparation for small and large research or clinical studies, increasing accurate genotyping for improved outcomes. We have developed a cost-efficient FLG genotyping method (∼10 times cheaper than exome sequencing) for researchers and clinicians studying patients from any ethnicity that is vital to advance a precision medicine approach to AD. This method can facilitate research studies immediately and be developed for clinical genetic diagnostics in the future.
Acknowledgments
We thank all the patients who participated in this study, and the research coordinators at the National Skin Centre, especially Nancy Liew and Veron Lu, for diligently collecting samples. MiSeq quality control was checked with the kind assistance of Christopher Wong, Hui Mann Seah, and Zhengzhong Qu, Polaris, Genome Institute of Singapore (GIS), A*STAR. We thank the GIS Sequencing platform, A*STAR, for its assistance. We also thank Dr Andy South for early discussions regarding Fluidigm Access Array technology for mutation detection.
Footnotes
This work was funded by A*STAR Strategic Positioning Fund (SPF) grants for basic and translational skin research (grant nos. IAF SPF 2013/004 and IAF SPF 2013/005 to J.E.A.C., E.B.L., J.N.F., S.L.I.J.D., and J.L.), and A*STAR SPF grant for genetic orphan diseases (grant no. IAF SPF 2012/005 to X.F.C.C.W.). A.S and F.J.D.S. were supported by a Wellcome Trust Strategic Award (grant no. 098439) to W.H.I.M.
All sequencing files were submitted to NCBI Sequence Read Archive (SRA) and are publicly available under BioProject ID “PRJNA360024.” Accession codes for the individual samples are included in Table E9 in this article's Online Repository at www.jacionline.org.
Disclosure of potential conflict of interest: S. L. I. J. Denil received a grant from A*STAR, Institute of Medical Biology. J. E. A. Common received grants from GSK and A*STAR, Institute of Medical Biology; and payments for lectures from Galderma. C. Wong received a grant from A*STAR, Institute of Medical Biology. E. B. Lane received grants from GSK and A*STAR, Institute of Medical Biology. A. S. L. Tay received a grant from A*STAR, Institute of Medical Biology. A. Sandilands was funded by a grant from the Wellcome Trust (to W.H.I. McLean). W. H. I. McLean and F. J. D. Smith (University of Dundee) have registered patents for genetic testing and sequencing of FLG. The rest of the authors declare that they have no relevant conflicts of interest.
Supplementary data
References
- 1.Palmer C.N.A., Irvine A.D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S.P. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 2.Smith F.J.D., Irvine A.D., Terron-Kwiatkowski A., Sandilands A., Campbell L.E., Zhao Y. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 3.van den Oord R.A.H.M., Sheikh A. Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. BMJ. 2009;339:b2433. doi: 10.1136/bmj.b2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark A.G., Hubisz M.J., Bustamante C.D., Williamson S.H., Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandilands A., Terron-Kwiatkowski A., Hull P.R., O'Regan G.M., Clayton T.H., Watson R.M. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 2007;39:650–654. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- 6.Irvine A.D., McLean W.H.I., Leung D.Y.M. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med. 2011;365:1315–1327. doi: 10.1056/NEJMra1011040. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Common J.E.A., Haines R.L., Balakrishnan A., Brown S.J., Goh C.S.M. Wide spectrum of filaggrin-null mutations in atopic dermatitis highlights differences between Singaporean Chinese and European populations. Br J Dermatol. 2011;165:106–114. doi: 10.1111/j.1365-2133.2011.10331.x. [DOI] [PubMed] [Google Scholar]
- 8.Brown S.J., Kroboth K., Sandilands A., Campbell L.E., Pohler E., Kezic S. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. J Invest Dermatol. 2012;132:98–104. doi: 10.1038/jid.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.