Abstract
Gaps between molecular ages and fossils undermine the validity of time-calibrated molecular phylogenies. An example of the time gap surrounds the age of angiosperm’s origin. We calculate molecular ages of the earliest flowering plant lineages using 22 fossil calibrations (101 genera, 40 families). Our results reveal the origin of angiosperms at the late Permian, ~275 million years ago. Different prior probability curves of molecular age calculations on dense calibration point distributions had little effect on overall age estimates compared to the effects of altered calibration points. The same is true for reasonable root age constraints. We conclude that our age estimates based on multiple datasets, priors, and calibration points are robust and the true ages are likely between our extremes. Our results, when integrated with the ecophysiological evolution of early angiosperms, imply that the ecology of the earliest angiosperms is critical to understand the pre-Cretaceous evolution of flowering plants.
Keywords: Calibration density, ecophysiology, molecular dating, origin of angiosperms, Permian, rate heterogeneity
Among the major evolutionary events where phylogenetic trees and fossils clash is the age of the origin of the angiosperms (Bell 2015; Herendeen et al. 2017). This remains a mystery because no unambiguous fossil evidence representing an angiosperm stem lineage or link to their sister group exists (Taylor and Taylor 2008; Herendeen et al. 2017). Due to the lack of fossils representing the root as well as high likelihood of numerous extinct stem lineages, an exceptionally long branch between extant angiosperms and their extant sister groups persists. Although Amborella trichopoda Baill. represents the sister group to all other extant angiosperms (Amborella Genome Project 2013), no fossils can be placed near this key divergence. Most of the estimates for this date using molecular data fall in the 250–145 mya range (Bell et al. 2010; Magallón 2010; Moore et al. 2007; Smith et al. 2010; Foster et al. 2017). Hence, calculated ages are considerably older than the oldest accepted angiosperm fossils of about 130 mya (Friis et al. 2011). The age of the earliest angiosperm lineages remains hotly debated (Buerki et al. 2014; Doyle and Endress 2014; Magallón 2014; Beaulieu et al. 2015; Magallón et al. 2015; Foster et al. 2017; Herendeen et al. 2017).
In evolution, timing can be critically important for teasing apart evolutionary processes bearing on major transitions. Framing key evolutionary events in time allows for a clearer perspective on geological, environmental, and ecological contexts bearing on major evolutionary events. Dated fossils that can be placed by phylogenetic reasoning, i. e. that they share unique synapomorphies or combinations of apomorphies with modern taxa, provide the most direct means of determining the age of an evolutionary event (Crepet et al. 2004). Although molecular dating has opened new insights in macroevolution, this approach is not as straightforward as originally envisioned. For instance, numerous quantitative approaches had to be developed to account for heterogeneous rate variation (Rutschmann 2006).
Widely used molecular dating methods are the nonparametric rate smoothing (NPRS; Sanderson 1997), penalized likelihood (PL; Sanderson 2002) and Bayesian relaxed clock (BRC) phylogenetic analyses (Drummond and Rambaut 2007); see Ho and Duchêne (2014), Pirie and Doyle (2012), and Bell (2015) for a review of these approaches. Most critical to all strategies, however, are the pitfalls related to the reliability of the fossil age, and the placement of the fossil on the tree. To estimate molecular ages, in principle a single calibration point is needed. However, ongoing discussions address the number, and distribution of calibration points required to optimize the accuracy of age estimates (Hedges and Kumar 2004; Bell and Donoghue 2005; Meredith et al. 2011; Parham et al. 2011; Sauquet et al. 2012; Bell 2015; Beaulieu et al. 2015). A general consensus is that more calibration points that are widely distributed across the tree will yield a more accurate molecular dating (Hug and Roger 2007). Others, however, argue that a single fossil record is not independent of the complete fossil history and evolution of a clade, but is part of a complex framework including all available fossils (Near et al. 2005; Pyron 2009). If this latter hypothesis is correct, each single fossil calibration will impact the integrity of other calibration points, especially since a descending lineage can never be older than the preceding one. Therefore, a second general consensus is that fewer, but more reliable calibration points, are a better means of obtaining the most reliable dates (Hedges and Kumar 2004; Parham et al. 2011).
Only morphological data can be used to place fossils accurately on the tree. The best placed fossils are ones that have synapomorphies or a combination of apomorphies with extant taxa such that they can be readily resolved as sister to the extant clades in the tree, using phylogenetic analyses (Doyle and Endress 2010; Gandolfo et al. 2004). Unfortunately many fossils are incomplete and often lack the necessary characters to determine their placement, or the interpretation of homology permits multiple placements on the tree such as stem versus crown calibration point (Doyle and Endress 2010; Friis et al. 2011).
Here we provide a reevaluation of the timing of the origin of the first angiosperm lineages using a relatively complete fossil record for calibrations sampled across most extant lineages. We broadly sampled the diversity of extant taxa (in total 104 genera and 43 families, 101 genera and 40 families of angiosperms alone). Our phylogenetic estimates are based on mostly newly generated nuclear and chloroplast derived sequence data. Extant taxon sampling was designed to include taxa that matched known fossils rather than search for fossil calibrations after the phylogenetic estimates were made. Twenty-two fossil calibrations are applied as well as an evaluation of alternative fossil placements such as stem versus crown, as well as the effects of individual calibration points on the entire age estimation. We thus provide an assessment of the impact of the number of fossil calibration points on age estimates and how rate heterogeneity and systematic sampling density among clades impacts the calculated molecular ages. Finally, we discuss what these considerably older age estimates potentially mean in relation to recent discoveries on early angiosperm ecophysiology.
Materials and Methods
Fossils and Sampling
We based our sampling on a screening of paleobotanical literature for fossils of earliest diverging lineages of extant angiosperms, the earliest monocots, and earliest eudicots. Criteria for selecting a fossil were that it could be placed unambiguously among extant taxa according to synapomorphies or a combination of apomorphies, or that confirmation by phylogenetic analysis is available (on morphological data or combined morphological-molecular analyses) (Gandolfo et al. 2008) and an accurate age determination was available at the time of running the analyses. Extant taxa were chosen to ensure optimal fossil calibration especially avoiding long branches (for details see Tables 1, S1 [Salomo et al. 2017]), sampling 163 species corresponding to 104 genera and 43 families (Appendix 1). The dense taxon sampling includes all recognized extant earliest diverging lineages of angiosperms reducing the probability of artificially inflated rate heterogeneity. We did not sample as densely among younger clades within eudicots or monocots because we wanted to clarify dates closest to the origins of angiosperms and we had calibration points for the ages of both eudicots and monocots. Further sampling within these clades would unlikely have impacted ages below these calibrations. For Chloranthistemon P.R. Crane, E.M. Friis & K.R. Pedersen, paleobotanical literature is discordant regarding its fossil placement. To evaluate the effect of its placement, we included the fossil at the stem of extant Chloranthus Sw. (ChlorA), or at the split between Ascarina J.R. Forst. & G. Forst. and Chloranthus-Sarcandra Gardner (ChlorB) (node F and f in Fig. 1).
Table 1.
Fossils used for calibration. All fossils, their corresponding age, and placement as applied to the calibration for phylogenetic hypotheses (nodes refer to Fig. 1) are presented. Details about the references, placement, and phylogenetic relationship of fossils can be found in the Table S1 (Salomo et al. 2017). mrca = most recent common ancestor; min = minimum calibration age, mean = mean in million years applied to the calibration point distribution.
| Node | Fossil and Reference | Period | Placement | Calibration | |
|---|---|---|---|---|---|
| min | mean | ||||
| A | Pluricarpellatia peltata B. Mohr, Bernardes-de-Oliveira and David W. Taylor (Mohr et al. 2008) | Aptian | crown Nymphaeales | 113.0 | 6.0 |
| B | Monetianthus mirus Friis, Pedersen, von Balthazar, Grimm and Crane (Friis et al. 2009) | late Aptian- early Albian | mrca Cabombaceae, Nymphaeaceae | 106.0 | 7.0 |
| C | Microvictoria svitkoana Nixon, Gandolfo and Crepet (Gandolfo et al. 2004) | Turonian | sister to Victoria Lindl. and Euryale Salisb. | 89.8 | 2.0 |
| D | Anacostia Friis, Crane and Pedersen (Friis et al. 1997) and fossil Trimenia Seem. seed (Yamada et al. 2008) | Albian | mrca Trimenia, Schisandraceae | 100.5 | 6.0 |
| E | Asteropollis Hedlund and Norris (Walker and Walker 1984) | Barremian-Aptian | crown Chloranthaceae | 113.0 | 9.0 |
| F f |
Chloranthistemon crossmanensis Crane, Friis and Pedersen (Crane et al. 1989) | Turonian | A. mrca Ascarina and Chloranthus B. stem of extant Chloranthus |
89.8 | 2.0 |
| G | Walkeripollis gabonensis Doyle, Hotton and Ward (Doyle et al. 1990) | late Barremian | stem Winteraceae | 125.0 | 2.5 |
| H | Lactoripollenites africanus Zavada and Benson (Zavada and Benson 1987) | early Turonian-mid Campanian | stem Lactoridaceae | 92.0 | 1.5 |
| I | Saururus tuckerae S.Y. Smith and Stockey (Smith and Stockey 2007) | middle-Eocene | sister to Saururus L. | 46.0 | 2.0 |
| J | Endressinia brasiliana Mohr and Bernardes de Oliveira (Mohr and Bernardes-de-Oliveira 2004) | late Aptian | crown Magnoliales | 113.0 | 3.0 |
| K | Archaeanthus linnenbergeri Dilcher and Crane (Dilcher and Crane 1984) | latest Albian | mrca Annonaceae, Magnoliaceae, Eupomatiaceae, Himantandraceae | 100.5 | 1.5 |
| L | Liriodendroidea E. Knobloch and D.H. Mai, seed (Frumin 1996) | Cenomanian/Turonian | crown Magnoliaceae | 89.8 | 4.1 |
| M | Futabanthus asamigawaensis Takahashi, Friis, Uesugi, Suzuki and Crane (Takahashi et al. 2008) | lower Coniacean | crown Annonaceae | 86.3 | 3.7 |
| N | Virginianthus Friis, Eklund, Pedersen and Crane (Friis et al. 1994) | early – middle Albian | stem Calycanthaceae | 105.0 | 1.5 |
| O | Jerseyanthus calycanthoides Crepet, Nixon and Gandolfo (Crepet et al. 2005) | Turonian | mrca Calycanthus L., Chimonanthus Lindl. | 86.3 | 3.5 |
| P | Potomacanthus lobatus von Balthazar, Pedersen, Crane, Stampanoni and Friis (von Balthazar et al. 2007) | early – middle Albian | mrca Atherospermataceae, Lauraceae, Monimiaceae, Hernandiaceae | 105.0 | 1.5 |
| Q | Mayoa portugallica Friis, Pedersen and Crane (Friis et al. 2004) | Barremian-Aptian | crown Araceae | 113.0 | 6.0 |
| R | tricolpate pollen (Hughes and McDougall 1990) | late Barremian | stem eudicots | 125.0 | 2.5 |
| S | Teixeiraea lusitanica von Balthazar, Pedersen and Friis (von Balthazar et al. 2005) | early Albian | crown Ranunculales | 106.0 | 7.0 |
| T | Nelumbites extenuinervis Upchurch, Crane, and Drinnan, Nelumbites minimus Vakhrameev (Upchurch et al. 1994; Vakhrameev 1952) | early – middle Albian | stem Nelumbonaceae | 105.0 | 1.5 |
| U | Platanocarpus brookensis Crane, Pedersen, Friis and Drinnan (Crane et al. 1993) | early – middle Albian | stem Platanaceae | 105.0 | 1.5 |
Fig. 1.
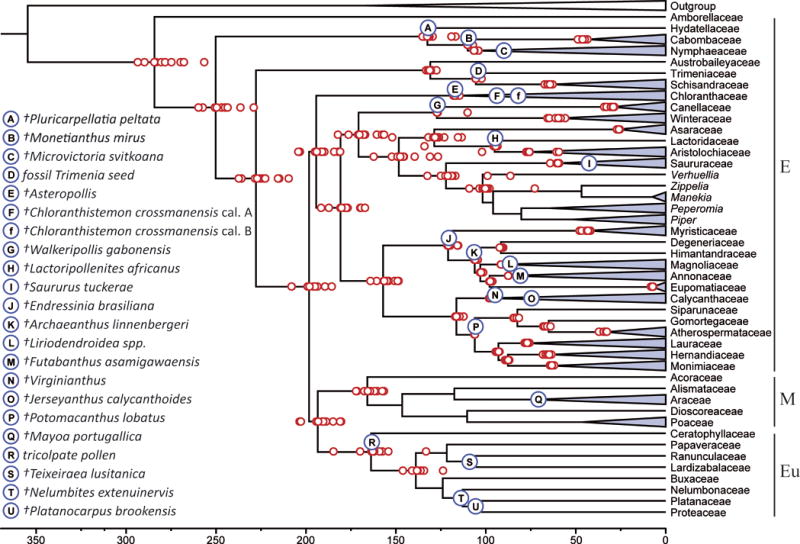
Bayesian relaxed clock suggests earlier origin of flowering plants than that determined by macrofossil evidence. Chronogram of early angiosperms, based on the full sampling of chloroplast and nuclear data using an uncorrelated log-normal clock, GTR + Γ substitution model and a birth-death model for incomplete sampling as implemented in BEAST using 20 fossil calibration points (A-U, without F = Chloranthistemon). Calibration points and corresponding fossils are provided as blue circles (capital letters A-U). The lowercase letter (f) refers to an alternative calibration for Chloranthistemon. Nodes correspond to Bayesian relaxed clock mean age estimates based on the reference analysis (Ref) applying a flowering plant root age constraint (uniform, 400–323 mya). The mean age estimates for all additional performed analyses are provided as red circles (see Table 3 for details). Further details about fossil calibration points are provided in Table 1. Clades have been collapsed at the family level with the exception of Piperaceae where clades are collapsed at the genus level. Letters to the right indicate major clades; E = Earliest diverging lineages, M = Monocots, Eu = Eudicots. Full taxon sampling can be found in Fig. S1.
Phylogenetics
Phylogenetic reconstructions are based on cpDNA (trnK-intron, matK, trnK-psbA spacer) and low copy nuclear DNA (phyA). We kept the plastid data as one region although it contains protein coding portions and non-coding portions. We decided to simplify the number of partitions because increasing the number of partitions exponentially increased computational time. Most sequences were newly generated. DNA isolation, amplification, sequencing, and alignment generation followed Wanke et al. (2007) and Smith et al. (2004). Regions excluded from the analyses and the justification for their removal is provided in Table S2. Primer sequences are listed in Table S3. Potential for paralagous copies of phyA were assessed by a comparison of maximum parsimony analyses that included only phyA sequences to those that included only cpDNA data. Paralogs would likely result in placement of taxa as sister to the duplicated region and be in conflict with the cpDNA. The combined dataset resulted in 7,778 aligned positions excluding regions of uncertain homology outlined in Table S2. Length mutations were coded following Wanke et al. (2007) using SeqState (Müller 2005). Congruence between the DNA regions was assessed using the incongruence length difference test (ILD; Farris et al. 1994). The best fitting model was chosen using jModeltest v0.1.1 (Posada 2008) partitioning the data into cpDNA and phyA. RAxML version 7.7.0 (Stamatakis 2014) was used to calculate 5,000 rapid bootstrap inferences and thereafter a thorough ML search was performed. All free model parameters were estimated by RAxML and the GTR + Γ model of rate heterogeneity was applied. Preliminary analyses indicated that some clades in our dataset that are unequivocally supported in APG III and APG IV (Angiosperm Phylogeny Group 2009, 2016) were not recovered in all analyses especially within BEAST (see methods below). This is true for the position of Chloranthaceae which sometimes resolved as sister to monocots as well as the position of Asaraceae which was sometimes sister to Saururaceae and Piperaceae. Therefore, five individual monophyly constraints were set to enforce outgroup position, APG III and APG IV conformity, and comparability between individual analyses comprising: 1) angiosperms, 2) Nymphaeaceae, 3) monocots, Ceratophyllales, eudicots, 4) Magnoliids, and 5) Aristolochiaceae, Lactoridaceae, Asaraceae.
Molecular Dating
All dating analyses were performed on NVidia Fermi GPGPU’s using BEAST v1.7.5 (Drummond and Rambaut 2007) applying the BEAGLE high-performance library v.1.0 (Suchard and Rambaut 2009). Individual BEAST runs were based on partitioned data with unlinked substitution models for chloroplast and nuclear data. The uncorrelated log-normal (UCLN) model, the GTR model and a birth-death process for incomplete sampling as implemented in BEAST were applied to all partitions and analyses. Sampling frequency was fixed to 5,000 for all analyses. The burn-in was removed after convergence of each Markov chain and was assessed using Tracer v1.5 (Table S4; Rambaut and Drummond 2003). The effective sample size (ESS) for all parameters and analyses was over 100. Consensus trees with mean branch length for each analysis were generated with TreeAnnotator v1.7.5. To test the effect of the applied priors, an additional analysis with empty alignment was performed. The influence of individual prior distributions was tested through analyses comprising both log-normal and exponential priors as well as different mean age estimates (for details see Table 2). Additionally, the influence of individual calibrations was analyzed as single constraints that were independently excluded from the general analysis (Table 3). To test the effect of sampling density within taxa with highly diverse and increased molecular rates we used two additional subsamplings with just 13 and nine Piperales taxa (Table 3). We also ran the analysis with an empty alignment with identical settings to test for the effect of priors on the results.
Table 2.
Calibrations applied to BEAST analyses with reduced fossil density. Independent calibration sets, with replicate sampling (A and B) of five and 10 randomly chosen calibration points, were used to test the effects of fossil calibration point quantity in molecular dating. For each analysis 5A and 5B, as well as 10A and 10B all applied fossil constraints and the corresponding nodes, refer to the letters in Fig. 1 and Table 1, are given. All analyses with reduced fossil density are based on the full taxon sampling and applied the uncorrelated log-normal clock, a GTR + Γ site model, and a birth-death speciation process for incomplete sampling as incorporated in BEAST, as well as a log-normal prior distribution with individual means as shown in Table 1 and a soft, uniform seed plant root constraint of 400–323 MYA.
| 5A | 5B | 10A | 10B | ||||
|---|---|---|---|---|---|---|---|
| fossil | node | fossil | node | fossil | node | fossil | node |
| Asteropollis | E | Archaeanthus linnenbergeri | K | Futabanthus asamigawaensis | M | Archaeanthus linnenbergeri | K |
| Jerseyanthus calycanthoides | O | Mayoa portugallica | Q | Lactoripollenites africanus | H | Asteropollis | E |
| Microvictoria svitkoana | C | Potomacanthus lobatus | P | Mayoa portugallica | Q | Futabanthus asamigawaensis | M |
| Teixeiraea lusitanica | S | Saururus tuckerae | I | Microvictoria svitkoana | C | Liriodendroidea spp. | L |
| fossil Trimenia seed | D | Teixeiraea lusitanica | S | Monetianthus mirus | B | Nelumbites | T |
| Nelumbites extenuinervis, Nelumbites minimus | T | Platanocarpus brookensis | U | ||||
| Pluricarpellatia peltata | A | Pluricarpellatia peltata | A | ||||
| Teixeiraea lusitanica | S | Potomacanthus lobatus | P | ||||
| fossil Trimenia seed | D | Saururus tuckerae | I | ||||
| Virginianthus | N | fossil Trimenia seed | D | ||||
Table 3.
BEAST settings of 16 individual molecular dating analyses. All individual molecular dating analyses applied the uncorrelated log-normal clock, a GTR + Γ site model and a birth-death speciation process for incomplete sampling as incorporated in BEAST. Analysis names corresponding to Fig. 3, letters provided in column “Constraints used” refer to Table S1 (Salomo et al. 2017) and Fig. 1. Individual prior distribution means are provided in detail in Table S1 (Salomo et al. 2017). Further details are provided in Table S3 (Salomo et al. 2017).
| Analysis | Constraints used | Prior distribution | Prior distribution Mean | Seed Plant age constraint | Taxa |
|---|---|---|---|---|---|
| Ref | A, B, C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| Root500 | A, B, C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | Uniform 500–323 MYA | Full sampling |
| NoRoot | A, B, C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | No constraint | Full sampling |
| Mean2 | A, B, C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | All 2 | Uniform 400–323 MYA | Full sampling |
| Exp | A, B, C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Exponential | All 2 | Uniform 400–323 MYA | Full sampling |
| ChlorA | A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| ChlorB | A, B, C, D, E, f, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| NoTricolp | A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P, Q, S, T, U | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| NoWalk | A, B, C, D, E, F, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| NoLac | A, B, C, D, E, G, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| Pip13 | A, B, C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | Uniform 400–323 MYA | All but 13 Piperales excluded: Asarum L., Saruma Oliv., Lactoris Phil., Aristolochia serpentaria L., Thottea Rottb., Houttuynia Thunb., Gymnotheca Decne., Saururus, Verhuellia Miq., Zippelia Blume, Manekia Trel., Piper hostmannianum (Miq.) C. DC., Peperomia hispidula (Sw.) A. Dietr. |
| Pip9 | A, B, C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U | Lognormal | Individual | Uniform 400–323 MYA | All, but 9 Piperales excluded: Asarum, Saruma, Lactoris, Aristolochia serpentaria, Thottea, Houttuynia, Gymnotheca, Saururus, Verhuellia |
| 5A | C, D, E, O, S | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| 5B | I, K, P, Q, S, | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| 10A | A, B, C, D, H, M, N, Q, S, T | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
| 10B | A, D, E, I, K, L, M, P, T, U | Lognormal | Individual | Uniform 400–323 MYA | Full sampling |
Results
Bayesian Phylogenetic Hypotheses
Reconstructed phylogenetic relationships of the cpDNA (trnK-matK-trnK-psbA region) and nDNA (phyA) datasets (missing data < 2%) (for details see Appendix 1; Salomo et al. 2017) did not indicate any incongruence (p < 0.05), each with 158 taxa in the ILD. The congruence implies that orthologs of phyA were included in the analyses. This conclusion is also supported by the fact that only single bands were recovered for phyA amplifications, and sequence reads were clean without multiple peaks. In general, we reconstructed highly supported relationships for all main nodes of early angiosperms (posterior probabilities, PP > 0.95), except for the sister groups Monimiaceae-Hernandiaceae and Degeneriaceae-Magnoliaceae (results not shown).
Temporal Origin of Early Angiosperms
Here we propose molecular age estimates based on our reference (Ref) analysis (Fig. 2, Table 3) with individual means for 20 fossil calibration points (Fig. 1) as specified in Table 1 and a soft, uniform seed plant root age constraint of 400–323 mya (Magallón et al. 2013). The origin of the angiosperm crown group is estimated at (341–)284(–226) mya. Our age estimates for Nymphaeales are (297–)250(–207) mya and (160–)132(–113) mya for the stem and crown age, respectively. Austrobaileyales stem and crown group ages are estimated at (267–)228(–192) and (166–)131(–106) mya, respectively. Crown group magnoliids diverged (208–)181(–156) mya ago. Both crown Magnoliales ([136–]121[–113] mya) and crown Laurales ([129–]116[–107] mya) diversification fall within the Aptian. The age of the most recent common ancestor of Canellales and Piperales is dated in the middle Jurassic (196–)171(–146) mya, with Piperales diversification at the end of the Jurassic (174–)148(–124) mya, followed by Canellales in the Barremian (131–)127(–125) mya. The age of the diversification of Chloranthaceae is estimated to be (125–)117(–113) mya. In summary, Piperales are recovered as the earliest diversifying order of the early angiosperms based on crown age, predating the diversification of eudicots (158–)139(–122) (excl. Ceratophyllum L.) but being second to the diversification of the monocots (192–)166(–141). Additional ages at the family level can be extracted from Figs. 1, 3, S1.
Fig. 2.
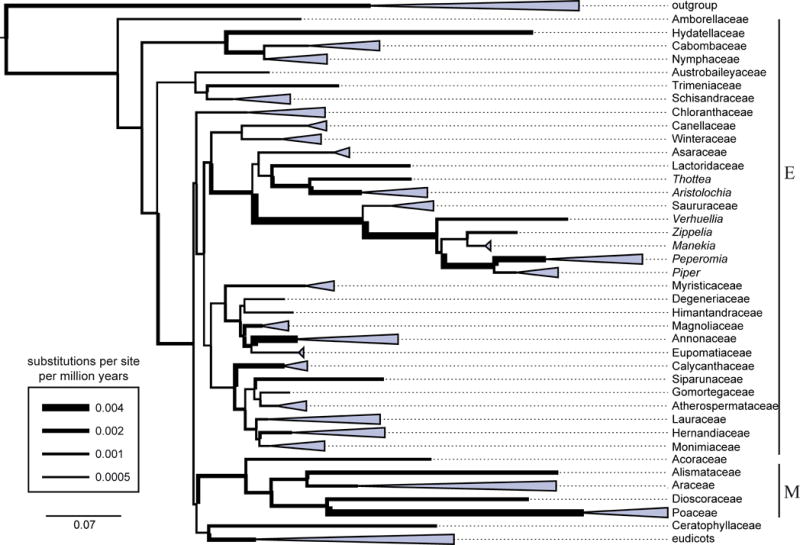
Heatmap showing the molecular age extremes of individual analyses. Mean divergence time estimates (∅) including minimum (min) and maximum (max) 95% confidence interval of main clades for 16 independent molecular dating analyses all applying the uncorrelated log-normal clock, a GTR + Γ site model, and a birth-death speciation process for incomplete sampling as incorporated in BEAST. Node numbers (Node #) correspond to the respective numbers of the phylogram of flowering plants (lower right). Colors are used to highlight classified divergences compared to the reference analysis (Ref). Light blue color indicates 1–2.5% lower values, blue indicates 2.5–5% lower values and dark blue indicates at least 5% lower values, red colors indicate 1–2.5%, 2.5–5% and greater than 5 % higher values, respectively. NA: node not available. Individual settings: Ref: based on full taxon sampling, applying all fossil constraints with the exception of Chloranthistemon (Crane et al. 1989), log-normal prior distribution with individual means (see Table 1) and uniform seed plant root age constraint of 400–323 mya; Root500: same as Ref, but with uniform seed plant root age constraint of 500–323 mya; NoRoot: same as Ref, but without any seed plant root age constraint; Mean2: same as Ref, but with log-normal mean of all individual constraints fixed to 2; Exp: same as Ref, but with exponential prior and exponential mean fixed to 2; ChlorA: same as Ref, but with additional age constraint based on Chloranthistemon as mrca of Ascarina and Chloranthus; ChlorB: same as Ref, but with additional, alternative age constraint based on Chloranthistemon on stem of extant Chloranthus; NoTricolp: same as Ref, but without tricolpate pollen (Hughes and McDougall 1990) constraint; NoWalk: same as Ref, but without Walkeripollis gabonensis (Doyle et al. 1990) constraint; NoLac: same as Ref, but without Lactoripollenites africanus (Zavada and Benson 1987) constraint; Pip13: same as Ref, but with only 13 instead of 65 Piperales taxa; Pip9: same as Ref, but without Piperaceae and only nine Piperales taxa. The datasets 5A and 5B, as well as 10A and 10B use the same settings as Ref, but include only five or 10 randomly chosen fossil constraints respectively. Individual fossil constraints are provided in Table 2. Letters to the right indicate major clades; E = Earliest diverging lineages, M = Monocots.
Fig. 3.
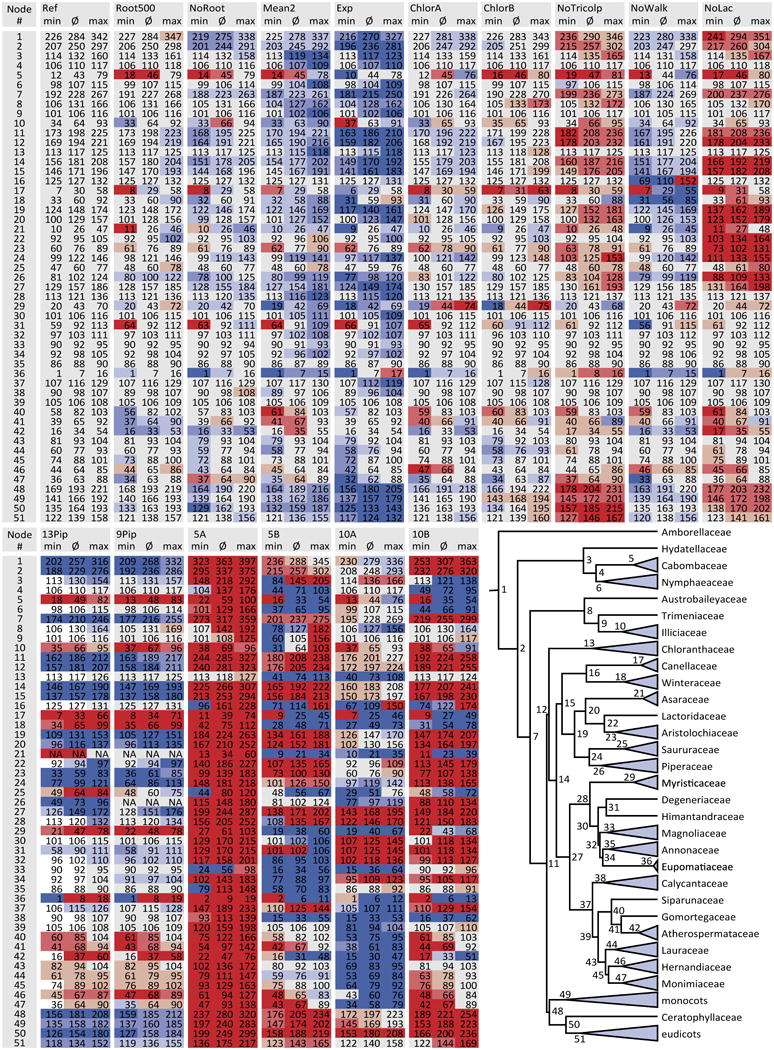
Challenging rate heterogeneity between clades. Phylogram of flowering plants focusing on the earliest angiosperms based on a Bayesian relaxed clock analysis of the full dataset with uncorrelated log-normal clock, GTR + Γ substitution model, applied a birth-death process for incomplete sampling. The mean absolute rates given as substitutions per site per million years corresponding to the thickness of the individual branches are coded in the legend. Clades have been collapsed at the family level with the exception of Piperaceae where clades are collapsed at the genus level.
Calibration Priors
To test the influence of different calibration priors, two additional analyses were performed using either a log-normal prior or an exponential prior both with a fixed mean of two for all constraints (Mean2 and Exp; Fig. 2, Table 3) in contrast to the reference analyses (Ref; Fig. 2, Table 3), used above, with individual log-normal means for all constrains as described in Table 1. The Mean2 analysis predominantly resulted in slightly younger age estimates with the exception of the order Laurales where the opposite trend was observed. The exponential prior (Exp) generally yielded younger age estimates for most nodes and narrower 95% confidence intervals compared to the log-normal priors. However, the different log-normal priors (Ref and Mean2), as well as Exp do not unreasonably differ and therefore bias the analyses. The results of the empty alignment clearly showed that the results are not driven by prior settings (results not shown).
Impact of Individual Fossils
To examine the effect of a particular node calibration within the analysis, single fossils (Walkeripollis gabonensis Doyle, Hotton, and Ward (Doyle et al. 1990), Lactoripollenites africanus Zavada and Benson (Zavada and Benson 1987), and tricolpate pollen (Hughes and McDougall 1990) were individually excluded from the full calibration set (Fig. 2) The exclusion of the tricolpate pollen calibration (NoTricolp; Fig. 2, Table 3) results in a eudicot age estimate of (167–)146(–127) mya and (215–)184(–157) mya for crown and stem respectively, also affecting estimates of the backbone as well as ages within Piperales, with slightly to moderate older estimates. Exclusion of the Walkeripollis constraint (NoWalk; Fig. 2, Table 3) led to an age estimate of (152–)110(–69) mya for crown and (194–)167(–141) mya for stem Canellales, with overall slightly younger backbone estimates. Exclusion of Lactoripollenites from the analysis (NoLac; Fig. 2, Table 3) resulted in a drastically older age within Piperales and angiosperm backbone nodes, but also led to increased age estimates for nearly all nodes of the tree.
Fossil Density
Independent calibration sets with replicate sampling of five and 10 randomly chosen calibration points (5A, 5B, and 10A, 10B; Fig. 2, Table 2) were used to test the effects of fossil calibration point quantity in molecular dating. Our results show no congruent trend of increasing or decreasing ages with respect to fewer versus more fossil calibration points. However fewer calibration points do markedly increase the variance of age estimates. Furthermore, a general trend is that the range of the confidence interval is narrower if more fossil calibration points are used (Fig. 2).
Stem vs. Crown Fossil Placement
The age of the split between Ascarina and Chloranthus-Sarcandra is (97–)92(–90) in analysis ChlorA and (120–)109(–97) mya in analysis ChlorB compared to (119–)94(–53) mya in our reference analysis (Ref) without any Choranthistemon constraint. The split between Chloranthus and Sarcandra was congruently estimated as (79–)49(–21) mya in analysis ChlorA and (84–)48(–15) mya without a Choranthistemon constraint, compared to (94–)91(–90) mya in ChlorB. The age estimate of the diversification of Chloranthaceae without Chloranthistemon is estimated as (125–)117(–113) mya. This age is not affected through the additional Chloranthistemon constraints which result in (123–)117(–113) mya and (128–)118(–113) mya in analysis ChlorA and ChlorB, respectively.
Seed Plant Root Age Constraints
The effect of soft root age constraints for seed plants using either 400–323 mya as in our reference analysis (Ref) or 500–323 mya (Root500; Fig. 2, Table 3) shows a moderate increase in estimated ages for the angiosperm backbone (Figs. 1, 2), with the greatest impact on the flowering plants crown age: (342–)284(–226) mya and (347–)284(–227) mya, for Ref and Root500, respectively, compared to (338–)275(–219) mya without any seed plant root age constraint (NoRoot; Fig. 2, Table 3). In addition, when no seed plant root constraint is used, the seed plant age is calculated to be (441–)338(–238) mya. Hence, root age constraints had surprisingly little effect when a dense calibration point distribution over the tree is used.
Discussion
Age Estimates and Additional Fossil Record
The reconstructed topology was nearly identical to previous studies (Soltis et al. 2011). Only the relationships of Myristicaceae as sister to all remaining Magnoliales (Himantandraceae (Degeneriaceae, Magnoliaceae (Annonaceae and Eupomatiaceae)) (Figs. 1, S1) are supported by our data and in conflict with supported relationships published previously. However, our results are in accordance with more recent findings (Massoni et al. 2014). Our age estimates, emerging from multiple calibration sets, converge on an age of extant angiosperm origin in the mid Permian (mean ages of individual analyses 294–257 mya). Such a time estimate is controversial, but is consistent with, or only slightly older than, other recent, fossil-only constraint molecular dating results such as 242 mya for a relaxed analysis (Magallón and Castillo 2009), 275–215 mya (Magallón 2010), 240–175 (Clarke et al. 2011), 240–225 mya (Zeng et al. 2014), or 251–192 (Foster et al. 2017). Interestingly, all these ages are significantly older than the angiosperm fossil record (Friis et al. 2011). Recently, Magallón et al. (2015) recovered an early Cretaceous age for the origin of angiosperms, however it should be noted that they bracketed the age range for the origin of angiosperms to fall within this range (140–136 mya).
Pre-Cretaceous, angiosperm-like fossil pollen from the middle and late Triassic (Cornet 1989; Hochuli and Feist-Burkhardt 2004, 2013) potentially support the hypothesis of an earlier angiosperm origin than currently accepted based on macrofossils (Friis et al. 2011). This fossil pollen has “all the essential features of angiosperm pollen” (p. 1, Hochuli and Feist-Burkhardt 2013). However, there does remain some uncertainty on the assignment of this pollen to angiosperms and it is possible that these fossils have an affinity with Gnetales as well (Herendeen et al. 2017). Furthermore, exceptionally old fossils representing extant, relatively deeply nested lineages in angiosperm phylogeny also support the emerging hypothesis that the phylogenetic root of angiosperms is much older than suspected. These fossils include a Ranunculaceae member (Leefructus, 125.8–122.6 mya) (Sun et al. 2011), and a probable Poaceae-Pooideae fossil from the Early Cretaceous (110–100 mya) (Poinar 2004, 2011). Nevertheless, if angiosperms are as ancient as our age estimates suggest, they still possess a reasonably long stem history considering they are sister to all living gymnosperms (367–306 mya; Clarke et al. 2011). Additionally, our results indicate that a major radiation of extant lineages without doubt occurred in the Cretaceous which is congruent with the accepted macrofossil record for extant angiosperm’s phylogenetic diversification (Fig. 1) (Friis et al. 2011).
The earliest diverging extant lineages of angiosperms have been included in numerous molecular phylogenetic dating studies with most recent studies by Massoni et al. (2015a, b). However, the greater number of calibration points toward the base of the tree included in the present analysis might give more accurate estimates of the crown age and divergence times leading to the eudicot/monocot split. Our age estimates for Nymphaeales are (297–)250(–207) mya and (160–)132(–114) mya for the stem and crown age, respectively. These ages are thus more consistent with additional fossil data for the crown Nymphaeaceae than earlier molecular dating studies suggested (Monetianthus mirus, ~112 mya; Carpestella lacunata, ~108 mya; Doyle and Endress 2014; Friis et al. 2009), as younger ages for Nymphaeaceae are here, and previously, calculated than the aforementioned fossil confirms (Yoo et al. 2005). However, the latter study has been a subject of some discussion (Nixon 2008; Doyle and Endress 2014).
Austrobaileyales stem and crown group ages are estimated at (267–)228(–192) and (166–)131(–106) mya, respectively, being consistent with estimates from Bell et al. (2010) for the crown group age and with Magallón and Castillo (2009) for the stem group age. Crown group magnoliids diverged (208–)181(–156) mya ago and are congruent with the dates proposed by Smith et al. (2010) (198–)163(–138) mya, but older compared to estimates from Bell et al. (2010) (138–)122(–108) mya or the 135–126 mya proposed by Moore et al. (2007).
Molecular Rates and Dates
Molecular rates are the result of substitutions and time constraints and any given branch is predominantly affected by the nearest fossil constraints, potentially resulting in heterogeneous rate distributions across a phylogenetic tree, and potentially hampering or influencing age estimations of other nodes in the tree. We evaluated rate heterogeneity through the coefficient of variation which varied by 24% (95% highest posterior density (HPD) = 71–95%). These values are comparable to previous studies (Smith et al. 2010) and indicate the need for a relaxed clock approach rather than a strict clock. In addition, Fig. 3 shows that rate changes between all nodes fluctuate only within the range of one magnitude, but rate heterogeneity could still represent a potential pitfall as molecular clock models may fail to assess the correct rate of a branches (Rutschmann 2006; Beaulieu et al. 2015). However, it is unclear to what extent an extreme or anomalous rate influences age estimates of more distantly related clades. Furthermore, it is questionable if this effect outbalances fossil calibration point density or distribution (Hug and Roger 2007) as well as the general selection of prior probability curves of fossil calibration points (Heads 2012).
Within our data set the highest substitution rates occur in Poaceae, Calycanthaceae, Annonaceae, and especially Piperales within Piperaceae and Saururaceae (Fig. 3). The covariance parameter in our data was positive (0.139), implying that lineages with fast rates are generally more likely to lead to other lineages with fast rates and vice versa. Although a most comprehensive set of fossils is used to estimate ages, and a single fossil should have little impact on the overall age estimations, single fossil inclusion versus exclusion still results in rate alternation of proximal nodes and thus in discrepant age estimates between independent analyses (Figs. 1, 2).
Here the high rate in Piperales and the Lactoripollinites fossil is chosen to circumscribe the impact of heterogeneous rate distributions. In our dataset, a fossil placed at a branch with high rates plays a more important role as a maximum constraint than as a minimum constraint, as high molecular rates would otherwise tend to be evened out through older ages. Given a high rate, such as in Piperales, Lactoripollinites heavily constrained the age of all Piperales by providing a maximum age constraint. Without Lactoripollinites, age estimates are generally older for Piperales. However, the exclusion of Lactoripollinites (NoLac; Fig. 2, Table 3) affects age estimates in general, especially the backbone. The opposite effect is visible at branches with low rates, therefore minimum age constraints are more important at those clades. An example of this is the Walkeripollis fossil which increased the age of the crown Canellales and had a slight influence on nearby nodes, but no significant effect on other age estimates.
Another critical point with respect to heterogeneous rates is the balance of the taxonomic sampling. Our age estimates for two additional reduced datasets, including nine and 13 taxa for Piperales only (Pip9, Pip13; Fig. 2, Table 3), are generally consistent with the full sampling datasets. Nonetheless, the reduction of the heterogeneity, e.g. through removing a portion of a clade that has an accelerated rate such as Piperaceae, resulted in younger age estimates for backbone nodes and taxa within Piperales. For example, Piperales were estimated as (153–)131(–109) mya and (151–)127(–105) mya when 13 and nine Piperales taxa were sampled, respectively, compared to the (174–)148(–124) mya within the full sampling. The greatest effect occurs within Piperaceae with estimates of (121–)99(–77) mya and (113–)86(–64) mya for 13 and nine Piperales taxa for stem Piperaceae compared to (146–)122(–99) mya in the full reference dataset (for more nodes see Figs. 1, 2). Therefore, the age estimates for Piperales and part of the flowering plant backbone scaled with the sampling density of Piperales taxa in our dataset. This undesirable circumstance is especially critical if no suitable minimum and maximum age constraints are available near the lineage in question as posterior inaccuracy in Bayesian dating is increased. The sampling density of taxa with accelerated rates of divergence within a dataset is therefore a critical concern within BRC molecular dating. This inaccuracy is a limitation of current methods that we tried to assess and balance using different taxon sets and priors. The most accurate ages are thus likely between our extremes.
Beaulieu et al. (2015) found that ages for the origin of angiosperms that were substantially older than indicated by the fossil record were largely the result of rate heterogeneity among early lineages and calibration sampling. In conclusion, sampling strategies for dating studies should not only include taxa with respect to taxonomy and calibration points, but also with respect to rate heterogeneity within a particular clade, testing for the true range of possible ages, and unraveling potential bias of abnormal rates. Because our age estimates are not totally discrepant when multiple sets are used, we can conclude that the age estimates are robust and not unreasonably affected by taxon sampling or fossil choice.
Evolutionary Implications in Relation to Early Angiosperm Ecology
These findings underscore some vexing questions: why was the radiation of the earliest angiosperms delayed for nearly 100 mya after the split from the most recent common ancestor (mrca), and why has the angiosperm lineage not been detected in the known Pre-Cretaceous fossil record? If angiosperm origin occurred in the Jurassic or even earlier, as our data and others suggest, such a temporal pattern raises the possibility that the ecological context in which the earliest angiosperms diversified is poorly sampled by the known geological record. Such a conclusion recalls the conclusions of Axelrod (1952, 1970) in his upland hypothesis, which proposed that the angiosperm line exhibited a long pre-Cretaceous history out of fossil capturing lowland basins. It is difficult to reconcile our findings with the hypothesis that angiosperms originated with an ensemble of highly weedy reproductive and vegetative features that are responsible for their modern dominance in high productivity habitats (Royer et al. 2010; Taylor and Hickey 1996; Wing and Boucher 1998). Such is the case because we would then expect the angiosperm line to be readily detectable in fossil forming sediments through the Mesozoic.
Currently, no fossil data are available that can draw a detailed picture of how the earliest angiosperms functioned in the context of their ancestral environment. Hence, any consideration on the biology of the pre-Cretaceous angiosperms is hypothetical. Nonetheless, others have opined that the persistent time gap between molecular and fossil data may be telling us a fundamental aspect about early angiosperm ecology (Smith et al. 2010). Indeed, functional analyses illuminating the capacities for Early Cretaceous angiosperm fossil leaves to transport water and photosynthesize, comparative ecophysiological studies across extant earliest diverging lineages of angiosperms, as well as the integration of these results with paleoclimatic data on Late Paleozoic to Mesozoic global climate change frame a testable hypothesis to explain the prolonged, ecological marginality of the pre-Cretaceous angiosperm line.
The oldest known angiosperm fossil leaves possess low densities of vein branching, a trait marking low transpirational capacity and therefore the possession of far lower photosynthetic capacity and productivity relative to modern, derived angiosperms (Brodribb and Feild 2010; Feild et al. 2011). However, vein densities of early angiosperms fall within the low and limited range for all other known Paleozoic to Mesozoic non-angiosperms (Boyce et al. 2009; Feild et al. 2011). Hence, low productivity capacity by itself cannot act as the primary ecological limitation that restricted diversification of the putative pre-Cretaceous angiosperm radiation. Instead, we suggest that the critical ecological constraint is that acute drought-intolerance (i.e. ancestral xerophobia of angiosperms [Feild et al. 2009a]) greatly accentuated the ecological constraints of low productivity capacity of early angiosperms. A hypothesis of ancestral angiosperm xerophobia flows from demonstration that core functions, including growth and reproduction, of extant earliest diverging lineages of angiosperm lineages depend upon copious and reliable supplies of water. Outstanding features of angiosperms used to reconstruct xerophobia include a deep phylogenetic persistence of drought-vulnerable xylem vessels with long scalariform perforation plates throughout the extant phylogenetic tree, reliance on root pressure for growth, and flowers that require stable hydration to bear fruits (Carlquist 2009; Feild et al. 2009a, b; Feild and Wilson 2012; Sperry et al. 2007). Extant angiosperm phylogeny also suggests that drought niche evolution began in sites of extremely low evaporative demand and high water availability such as in the forest understory or in aquatic zones with unlimited water. Angiosperms then transitioned into canopy exposed and open disturbed zones in wet forests. True shifts into habitats where high evaporative demand and soil aridity were combined occurred much later, perhaps by the Late Cretaceous and Early Paleocene depending on the clade (Bond and Scott 2010; Feild et al. 2009a; Sauquet et al. 2008).
Critically, the geological timeframe bounded by the close of the Permian to the Early Cretaceous, which encapsulates our reconstructed long phylogenetic fuse of early angiosperm evolution, marks a global pattern of aridity across equatorial to mid-latitude wet zones which consisted of a large geographic area (Chaboureau et al. 2014; Ziegler et al. 2003). The best evidence for stable, wet, non-freezing regions comes from coals that are geographically restricted to high latitude and near boreal zones (Boyce and Lee 2010; Ziegler et al. 2003). It is possible that moist mountain tops under the influence of oceanic orographic clouds also supported localized wet forest pockets at mid to low paleolatitudes (Feild et al. 2009a). Therefore, a hypothesis that the early angiosperm may have been a rare, geographically localized line that was highly dependent upon water for millions of years can explain why the angiosperm line has so far escaped fossil detection. With the widespread aridity, at levels that would be lethal to all extant earliest diverging lineages of angiosperm clades, during much of the Mesozoic, the angiosperm line may have scraped by in localized wet patches. However, later spread of global moisture in the mid- to Late Cretaceous, perhaps bolstered, in part, by angiosperms themselves through the evolution of densely-veined leaves that fuelled increased transpiration fed raincycles, opened up hydraulically permissive environments on a larger geographical scale where angiosperms could diversify and dominate (Boyce and Lee 2010; Boyce et al. 2009). Later emerging global aridity (mid to Late Cenozoic) catalyzed diverse angiosperm and non-angiosperm radiations in dry environments, clearly indicating that angiosperms had fully broken out of drought constraints by at least these times (Boyce and Lee 2010; Feild et al. 2009a; Pittermann et al. 2012). Future testing of this hypothesis will have to look to the fossil record, but there are now tools at hand to read the hydrological preferences of early angiosperms based on fossilized leaves and pollen (Feild et al. 2009a).
Supplementary Material
Acknowledgments
We thank U. Markwardt and G. Juckeland for access to computational resources and support through the Center for Information Services and High Performance Computing (ZIH) and CUDA Research Center, TU-Dresden. We are grateful to the botanical gardens of Ghent University, Dresden University, Bonn University, Duke University, Brisbane, Sydney, and Singapore and the following colleagues for providing material and research support: P. Asselman, B. Oelschlägel, S. Wagner, A. Barniske, T. Heidcamp, P. Manos, D. Crayn, E. Rabakonandriania, M. Quijano, Á. Idárraga, and P. Knopf. We appreciate detailed comments by James A. Doyle and a anonymous reviewers that largely improved our manuscript. This work was co-supported by the DFG (NE681/5-2, NE681/10-1) and the DAAD (PPP Colombia and China). Research was further supported by the INBRE Program, NIH Grant Nos. P20 RR016454 (National Center for Research Resources) and P20 GM103408 (National Institute of General Medical Sciences).
Appendix 1
List of taxa and GenBank accessions. Data are in the order of family, species sample, GenBank accessions for the chloroplast trnK-matK-trnK-psbA (CP) and nuclear phyA region (phyA), voucher and herbarium, and species name for that gene, NA is used if the sequence was not generated or available, na indicates a different species or individual was used for the two DNA regions. In some cases, different species were either used, or were already available in GenBank for a genus. In those instances, “sp.” is used in the sample, and the species is cited for each DNA region when known. Where both sequences were obtained from GenBank a “G” is included for the voucher. Some collections were unvouchered, but are noted for their Botanical Garden collection numbers (BG).
Cycadaceae: Cycas sp., NC 009618.1, na, G, Cycas taitungensis C.F. Shen, K.D. Hill, C.H. Tsou & C.J. Chen, Cycas sp., na, Y07571, Cycas revoluta Thunb.; Ginkgoaceae: Gingko biloba, MF287374, AJ286638, BG Dresden (voucher DR s.n.), Gingko biloba L.; Pinaceae: Pinus sp., NC 011155.3, na, G, Pinus krempfii Lecomte, Pinus sp., na, EU203182, Pinus sylvestris L.; Amborellaceae: Amborella trichopoda, NC 005086.1, AF190062, G, Amborella trichopoda Baill.; Hydatellaceae: Trithuria filamentosa, MF287375, NA, DP12 (Mt. Field, Lake Dobson, 1,000 m), Trithuria filamentosa Rodway; Nymphaeaceae: Euryale ferox, DQ185537.1, MF287517, BG, Euryale ferox Salisb. ex K.D. Koenig & Sims; Nuphar advena, DQ185531.1, MF287518, BG Dresden DE-0-DR-001787, Nuphar advena R. Br.; Nymphaea odorata, MF287376, AF190098, BG Dresden DE-0-DR-002378, Nymphaea odorata Aiton; Brasenia schreberi, MF287377, MF287519, BA112, Brasenia schreberi J. F. Gmel.; Cabomba caroliniana, MF287378, MF287520, commercial source, (voucher DR s.n.), Cabomba caroliniana A. Gray; Victoria crusiana, MF287379, MF287521, BG Dresden xx-0-DR-005032, Victoria crusiana Orb.; Illiciaceae: Kadsura japonica, DQ185525.1, MF287522, BG Dresden xx-0-DR-008506, Kadsura japonica (L.) Dunal; Schisandra rubriflora, DQ185526.1, MF287523, Paul Goetghebeur 12984 (GENT), Schisandra rubriflora (Turcz.) Baill.; Illicium henryi, MF287380, MF287524, BG DD 003357-18 (DR), Illicium henryi Diels; Trimeniaceae: Trimenia neocaledonica, MF287381, NA, Feild s.n. NC 2011 (Mt. Aoupinier, 900m), Trimenia neocaledonica Baker f.; Austrobaileyaceae: Austrobaileya scandens, MF287382, MF287525, BG Bonn AU-0-BONN-881, Austrobaileya scandens C.T. White; Chloranthaceae: Chloranthus spicatus, NC 009598.1, AF190076, G, Chloranthus spicatus (Thunb.) Makino; Ascarina solmsiana, MF287383, NA, Feild s. n. (Mt. Dzumac stopa, 10.15.01), Ascarina solmsiana Schltr.; Hedyosmum sp., MF287384, na, Feild s. n., Hedyosmum goudotianum Solms, Hedyosmum sp., na, MF287526, Feild s. n. (Costa Rica), Hedyosmum mexicanum C. Cordem.; Sarcandra glabra, MF287385, AF276741, BG, Sarcandra glabra (Thunb.) Nakai; Acoraceae: Acorus sp., NC 007407.1, na, G, Acorus calamus L., Acorus sp., na, AF190060, G, Acorus gramineus Sol. ex Aiton; Alismataceae: Sagittaria sp., MF287386, na, BA104, Sagittaria montevidensis Cham. & Schltdl., Sagittaria sp., na, AF190102, G, Sagittaria sp.; Araceae: Lemna sp., NC 010109.1, na, G, Lemna minor L., Lemna sp., na, U08168.1, G, Lemna gibba L.; Spathiphyllum sp., AM920559.1, na, G, Spathiphyllum wallisii Regel, Spathiphyllum sp., na, AF276745.1, G, Spathiphyllum sp.; Dioscoreaceae: Dioscorea elephantipes, NC 009601.1, AF276720.1, G, Dioscorea elephantipes (L’Hér.) Engl.; Poaceae: Hordeum vulgare, NC 008590, DQ201140.1, G, Hordeum vulgare L.; Sorghum bicolor, NC 008602, AY466073.1, G, Sorghum bicolor (L.) Moench; Ceratophyllaceae: Ceratophyllum demersum, NC 009962.1, AF276716, G, Ceratophyllum demersum L.; Fumariaceae: Dicentra sp., MF287387, na, N201, Dicentra eximia (Ker Gawl.) Torr., Dicentra sp., na, MF287527, Smith s. n. (SRP), Dicentra spectabilis (L.) Lem.; Lardizabalaceae: Lardizabala biternata, MF287388, AF276730, R28, Lardizabala biternata Ruiz & Pav.; Ranunculaceae: Aquilegia sp., MF287389, na, C. Neinhuis s. n. (DR), Aquilegia ecalcarata Maxim., Aquilegia sp., na, AF190066, G, Aquilegia sp.; Buxaceae: Buxus sp., NC 009599, na, G, Buxus microphylla Siebold & Zucc., Buxus sp, na, MF287528, BG Dresden (voucher DR s.n.), Buxus balearica Lam.; Nelumbonaceae: Nelumbo sp., MF287390, na, NEL–N179, Nelumbo lutea Willd., Nelumbo sp., na, AF190096, G, Nelumbo nucifera Gaertn.; Platanaceae: Platanus occidentalis, NC 008335, EU642773, G, Platanus occidentalis L.; Proteaceae: Grevillea sp., MF287391, na, BG, Grevillea banksii R. Br., Grevillea sp, na, EU642771, G, Grevillea caleyi R. Br.; Magnoliaceae: Liriodendron tulipifera, NC 008326, MF287529, G, Liriodendron tulipifera L.; Liriodendron chinense, MF287392, EU849909, Paul Goetghebeur 12976 (GENT), Liriodendron chinense (Hemsl.) Sarg.; Magnolia wilsonii, MF287393, MF287530, BG Dresden xx-0-DR-006957, Magnolia wilsonii (Finet & Gagnep.) Rehder; Magnolia denudata, MF287394, MF287531, BG Dresden xx-0-DR-008250, Magnolia denudata Desr.; Myristicaceae: Myristica castaneifolia, MF287395, MF287532, BG Dresden (voucher DR s.n.), Myristica castaneifolia A. Gray; Myristica cylindrocarpa, MF287396, MF287533, BA92, Myristica cylindrocarpa J. Sinclair; Virola flexuosa, MF287397, MF287534, DP78, Virola flexuosa A. C. Sm.; Iryanthera ulei, MF287398, MF287535, DP74, Iryanthera ulei Warb.; Compsoneura mutisii, MF287399, MF287536, DP79, Compsoneura mutisii A. C. Sm.; Degeneriaceae: Degeneria vitiense, MF287400, AF190078, Pak 149 (BOCH), Degeneria vitiense I.W. Bailey & A.C. Sm.; Himantandraceae: Galbulimima baccata, MF287401, MF287537, Weston 929 (NSW), Galbulimima baccata F.M. Bailey; Eupomatiaceae: Eupomatia benettii, MF287402, MF287538, Endress 5197 (Z), Eupomatia benettii F. Muell.; Eupomatia laurina, MF287403, MF287539, BG Dresden AU-0-DR-010625, Eupomatia laurina R. Br.; Annonaceae: Uvaria chamae, MF287404, MF287540, BG Dresden TG-0-DR-014576, Uvaria chamae P. Beauv.; Anaxagorea javanica, MF287405, NA, BG Singapore, 24.08.2010, Anaxagorea javanica Blume; Polyalthia suberosa, MF287406, MF287541, BG, Polyalthia suberosa (Roxb.) Thwaites; Annona sp., MF287407, na, BJBN 26291, Annona muricata L., Annona sp., na, AF190064, G, Annona sp.; Monodora crispata, MF287408, MF287542, BG, Monodora crispata Engl.; Asimina triloba, MF287409, MF287543, BG, Asimina triloba (L.) Dunal; Isolona cooperi, MF287410, MF287544, Frida Billet S3778 (BR), Isolona cooperi Hutch. & Dalziel ex G.P. Cooper & Record; Popowia whytei, MF287411, MF287545, Frida Billet S2460 (BR), Popowia whytei Stapf; Artabotrys hexapelatus, MF287412, MF287546, BG, Artabotrys hexapelatus (L. f.) Bhandari; Rollinea sieberi, MF287413, MF287547, Frank Van Caekenberghe S0049 (BR), Rollinea sieberi A. DC.; Desmos dasymaschalus, MF287414, MF287548, BG, Desmos dasymaschalus (Blume) Saff.; Calycanthaceae: Chimonanthus praecox, MF287415, MF287549, BG, Chimonanthus praecox (L.) Link; Calycanthus floridus, NC 004993, AF190072, G, Calycanthus floridus L.; Sinocalycanthus chinensis, MF287416, MF287550, Stefan Wanke priv. coll. (voucher DR s.n.), Sinocalycanthus chinensis W.C. Cheng & S.Y. Chang; Idiospermum australiense, MF287417, MF287551, BG Dresden (voucher DR s.n.), Idiospermum australiense (Diels) S.T. Blake; Siparunaceae: Siparuna guianensis, MF287418, MF287552, BG Dresden (voucher DR s.n.), Siparuna guianensis Aubl.; Gomortegaceae: Gomortega keule, MF287419, MF287553, BG Dresden, (voucher DR 25.09.11), Gomortega keule (Molina) Baill.; Atherospermataceae: Doryphora sassafras, MF287420, MF287554, BG, Doryphora sassafras Endl.; Daphnandra micrantha, MF287421, MF287555, Goodwin s. n., Daphnandra micrantha (Tul.) Benth.; Laurelia sempervirens, MF287422, MF287556, BG Dresden (voucher DR 25.09.12), Laurelia sempervirens (Ruiz & Pav.) Tul.; Laureliopsis phillipiana, MF287423, MF287557, BG Dresden (voucher DR 25.09.09), Laureliopsis phillipiana (Looser) Schodde; Monimiaceae: Peumus boldus, MF287424, MF287558, Paul Goetghebeur 12976 (GENT), Peumus boldus Molina; Kibara sp., MF287425, MF287560, BG, Kibara sp.; Hedycarya arborea, AJ627927.1, MF287559, Quandt s. n., Hedycarya arborea J.R. Forst. & G. Forst.; Mollinedia tomentosa, MF287426, MF287561, BG, Mollinedia tomentosa (Benth.) Tul.; Hernandiaceae: Hernandria bivalvis, MF287427, MF287562, BG, Hernandia bivalvis Benth.; Hernandia peltata, MF287428, MF287563, Paul Goetghebeur 12977 (GENT), Hernandia peltata Meisn.; Gyrocarpus americanus, MF287429, MF287564, BG, Gyrocarpus americanus Jacq.; Sparattanthelium tarapotanum, MF287430, NA, Frank Van Caekenberghe S0050 (BR), Sparattanthelium tarapotanum Meisn.; Lauraceae: Cinnamomum aromaticum, MF287431, MF287565, BG, Cinnamomum aromaticum Zoll.; Cryptocarya laevigata, MF287432, MF287566, Paul Goetghebeur 8880 (GENT), Cryptocarya laevigata Blume; Cassytha filiformis, MF287439, MF287573, A. Chanderbali et al. 205 (MO), Cassytha filiformis L.; Lindera benzoin, MF287433, MF287567, BG, Lindera benzoin (L.) Blume; Lindera obtusiloba, MF287434, MF287568, BG, Lindera obtusiloba Blume; Chlorocardium venenosum, MF287435, MF287569, R. Vásquez et al. 25236 (MO), Chlorocardium venenosum (Kosterm. & Pinkley) Rohwer, H.G. Richt. & van der Werff; Hypodaphnis zenkeri, MF287436, MF287570, G. McPherson 16184 (MO), Hypodaphnis zenkeri (Engl.) Stapf; Neocinnamomum mekongense, MF287437, MF287571, L. Heng 8547 (MO), Neocinnamomum mekongense (Hand.-Mazz.) Kosterm.; Anaueria brasiliensis, MF287438, MF287572, R. Vásquez et al. 25235 (MO), Anaueria brasiliensis Kosterm.; Neolitsea sericea, MF287440, MF287574, BG, Neolitsea sericea (Blume) Koidz.; Aniba affinis, MF287441, MF287575, BG, Aniba affinis (Meisn.) Mez; Persea americana, MF287442, MF287576, Frida Billet S638 (BR), Persea americana Mill.; Sassafras albidum, MF287443, MF287577, BG, Sassafras albidum (Nutt.) Nees; Winteraceae: Takhtajania perrieri, MF287444, MF287578, BG, Takhtajania perrieri (Capuron) Baranova & J.-F. Leroy; Bubbia queenslandiana, MF287445, MF287579, BG, Bubbia queenslandiana Vink; Pseudowintera sp., MF287446, MF287580, BG, Pseudowintera sp.; Tasmannia insipida, MF287447, MF287581, no voucher, Tasmannia insipida R. Br. ex DC.; Tasmannia lanceolata, MF287448, MF287582, Stefan Wanke 16999, Tasmannia lanceolata (Poir.) A.C. Sm.; Drimys winterana, MF287449, AF190080, BG, Drimys winterana Thell.; Cannellaceae: Canella winterana, MF287450, AF190074, BG Dresden (voucher DR 25.09.10), Canella winterana (L.) Gaertn.; Cinnamosma fragrans, MF287451, MF287583, DP03 (PJH034, Madagascar, Diego Suarez), Cinnamosma fragrans Baill.; Lactoridaceae: Lactoris fernandeziana, MF287452, AF190091, BG, Lactoris fernandeziana Phil.; Aristolochiaceae: Aristolochia arborea, MF287453, AB206923, Samain et al. 2009-292 (MEXU, DR), Aristolochia arborea Linden; Aristolochia clematitis, MF287454, AB206925, W. Starmühler s. n. (DR), Aristolochia clematitis L.; Aristolochia fimbriata, MF287455, MF287584, BG Dresden (voucher DR s.n.), Aristolochia fimbriata Cham. & Schltdl.; Aristolochia gigantea, MF287456, MF287585, S.T. Wagner 18 (DR), Aristolochia gigantea Mart.; Aristolochia grandiflora, MF287457, MF287586, Isnard et al. 01 (MEXU), Aristolochia grandiflora Sw.; Aristolochia kaempferi, MF287458, AB353504, Neinhuis sn (DR), Aristolochia kaempferi Willd.; Aristolochia maxima, MF287459, AB206934, Gonzalez 4018 (COL) Aristolochia maxima Jacq.; Aristolochia pentandra, MF287460, AB206942, Cola de Caballo, Mexico, privat coll. B. Westlund, Aristolochia pentandra Jacq.; Aristolochia praevenosa, MF287461, AB206946, (commercial source, voucher DR s.n.), Aristolochia praevenosa F. Muell.; Aristolochia promissa, MF287462, AB206947, Neinhuis 118 (DR), Aristolochia promissa Mast.; Aristolochia serpentaria, MF287463, AB206940, Westlund sn (DR), Aristolochia serpentaria L.; Thottea tomentosa, MF287468, AB206951, Pradeep s. n. (TBGT), Thottea tomentosa (Blume) Ding Hou; Asaraceae: Asarum canadense, MF287464, MF287587, Gonzalez 3577 (COL), Asarum canadense L.; Asarum chingchengense, MF287465, MF287588, BG, Asarum chingchengense C.Y. Cheng & C.S. Yang; Asarum europaeum, MF287466, MF287589, BG, Asarum europaeum L.; Saruma henryi, MF287467, AF190104, Neinhuis 120 (DR), Saruma henryi Oliv.; Piperaceae: Manekia incurva, MF287469, MF287590, BG, Manekia incurva (Sieber ex Schult.) T. Arias, Callejas & Bornst.; Manekia sydowii, MF287470, NA, Samain & Wanke 2010-011 (HUA), Manekia sydowii (Trel.) T. Arias, Callejas & Bornst.; Verhuellia lunaria, MF287471, MF287591, Jimenez & Garcia 3560 (GENT), Verhuellia lunaria (Desv. ex Ham.) C. DC.; Zippelia begoniifolia, MF287472, na, Shao Wu Meng s.n. (KUN), Zippelia begoniifolia Blume, Zippelia begoniifolia, na, MF287592, Regalado 1675 (MO), Zippelia begoniifolia Blume; Piper aduncum, MF287473, MF287593, Bornstein 765 (SEMO), Piper aduncum L.; Piper aequale, MF287474, MF287594, Bornstein 716 (SEMO), Piper aequale Vahl; Piper amalago, MF287475, MF287595, Bornstein 712 (SEMO), Piper amalago L.; Piper arboreum, MF287476, MF287596, Bornstein 699 (SEMO), Piper arboreum Aubl.; Piper auritum, MF287478, MF287598, Rincon et al. 2409 (XAL), Piper auritum Kunth; Piper austrocaledonicum, MF287477, MF287597, McPherson 1919 (MO), Piper austrocaledonicum C. DC.; Piper borbonense, MF287479, MF287599, 87.3.616 for Conservatoire et Jardins Botaniques de Nancy, Piper borbonense (Miq.) C. DC.; Piper betle, MF287480, MF287600, Smith 5808 (SRP), Piper betle L.; Piper caninum, MF287482, na, Jaramillo 218 (DUKE), Piper caninum Blume, Piper caninum, na, MF287602, Flynn 6750 (PTBG), Piper caninum Blume; Piper capense, MF287481, MF287601, Smith 4925 (SCA), Piper capense L. f.; Piper comptonii, MF287483, MF287603, Munzinger 4788 (NOU), Piper comptonii S. Moore; Piper cinereum, MF287484, MF287604, Jaramillo 653 (DUKE), Piper cinereum C. DC.; Piper darienense, MF287485, MF287605, Aguilar 111197 (NY), Piper darienense C. DC.; Piper excelsum, MF287498, MF287617, Wanke 071 (DR), Piper excelsum G. Forst.; Piper flaviflorum, MF287486, MF287606, Li 06171 (PE), Piper flaviflorum C. DC.; Piper guineense, MF287487, MF287607, Smith 4923 (SCA), Piper guineense Schumach. & Thonn.; Piper guahamense, MF287488, MF287608, Flynn 6748 (PTBG), Piper guahamense C. DC.; Piper hostmannianum, MF287489, MF287609, Tepe 599 (MU), Piper hostmannianum (Miq.) C. DC.; Piper humistratum, MF287490, MF287618, Tepe 542 (MU), Piper humistratum Görts & K.U. Kramer; Piper methysticum, MF287491, MF287610, Morden 2975 (HAW), Piper methysticum L. f.; Piper rothianum, MF287492, MF287611, Smith 6545 (SRP), Piper rothianum F. M. Bailey; Piper sanctum, MF287493, MF287612, Domingues 251 (HEM), Piper sanctum (Miq.) Schltdl. ex C. DC.; Piper semiimersum, MF287494, MF287613, Li 06161 (PE), Piper semiimersum C. DC.; Piper subpenninerve, MF287495, MF287614, Wong 1 (SRP), Piper subpenninerve Ridl.; Piper urophyllum, MF287496, MF287615, Davidson 10902 (SRP), Piper urophyllum C. DC.; Piper wallichii, MF287497, MF287616, Li 06212 (PE), Piper wallichii (Miq.) Hand.-Mazz.; Peperomia argyreia, MF287499, MF287619, Symmank & Mathieu 2008-052 (GENT, LPB), Peperomia argyreia (Miq.) E. Morren; Peperomia blanda, MF287500, na, BG, Peperomia blanda, (Jacq.) Kunth, Peperomia blanda, na, MF287620, Burrows 8521 (herbarium at Buffelskloof Private Nature Reserve, Mpumalanga, South Africa), Peperomia blanda, (Jacq.) Kunth; Peperomia dolabriformis, MF287501, MF287621, Samain 022 (GENT), Peperomia dolabriformis Kunth; Peperomia emarginella, MF287502, na, Samain & Vanderschaeve 2005-010 (GENT), Peperomia emarginella (Sw. ex Wikstr.) C. DC., Peperomia emarginella, na, MF287622, Davidson 10867 (CR), Peperomia emarginella (Sw. ex Wikstr.) C. DC.; Peperomia galioides, MF287503, na, Samain et al. 2008-128 (GENT, LPB), Peperomia galioides Kunth, Peperomia galioides, na, MF287623, 1978-1274 (BG Gent, not vouchered), Peperomia galioides Kunth; Peperomia hartwegiana, MF287504, MF287624, 2007-0833 (BG Gent, not vouchered), Peperomia hartwegiana Miq.; Peperomia hispidula, MF287505, na, Samain et al. 2007-074 (GENT, MEXU), Peperomia hispidula (Sw.) A. Dietr., Peperomia hispidula, na, MF287625, Samain 052 (GENT), Peperomia hispidula (Sw.) A. Dietr.; Peperomia lancifolia, MF287506, MF287626, Samain et al. 2007-094 (GENT, LPB), Peperomia lancifolia Hook.; Peperomia magnifoliiflora, MF287507, na, Symmank & Mathieu 2008-062 (GENT, LPB), Peperomia magnifoliiflora A. Dietr., Peperomia magnifoliiflora, na, MF287627, Samain 136 (GENT), Peperomia magnifoliiflora A. Dietr.; Peperomia maypurensis, MF287508, MF287628, Wanke 006 (DD), Peperomia maypurensis Kunth; Peperomia parvifolia, MF287509, na, Samain et al. 2009-027 (GENT, USM), Peperomia parvifolia C. DC., Peperomia parvifolia, na, MF287629, Samain 011 (GENT), Peperomia parvifolia C. DC.; Peperomia pellucida, MF287510, na, Symmank & Mathieu 2008-082 (GENT, LPB), Peperomia pellucida (L.) Kunth, Peperomia pellucida, na, MF287630, Smith 4929 (SCA), Peperomia pellucida (L.) Kunth; Peperomia serpens, MF287511, na, BG, Peperomia serpens (Sw.) Loudon, Peperomia serpens, na, MF287631, Tepe 548 (MU), Peperomia serpens (Sw.) Loudon; Saururaceae: Houttuynia cordata, MF287512, AF276726, BG, Houttuynia cordata Thunb.; Anemopsis californica, MF287513, MF287632, BG, Anemopsis californica Hook. & Arn.; Gymnotheca chinensis, MF287514, MF287633, BG, Gymnotheca chinensis Decne.; Saururus cernuus, MF287515, AF190106, BG, Saururus cernuus L.; Saururus chinensis, MF287516, MF287634, Paul Goetghebeur 12898 (GENT), Saururus chinensis (Lour.) Baill.
Literature Cited
- Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society. 2016;181:1–20. [Google Scholar]
- Axelrod DI. A theory of angiosperm evolution. Evolution. 1952;6:29–60. [Google Scholar]
- Axelrod DI. Mesozoic paleogeography and early angiosperm history. The Botanical Review. 1970;36:277–319. [Google Scholar]
- Bell CD. Between a rock and a hard place: Applications of the “molecular clock” in systematic biology. Systematic Botany. 2015;40:6–13. [Google Scholar]
- Bell CD, Donoghue MJ. Dating the Dipsacales: Comparing models, genes, and evolutionary implications. American Journal of Botany. 2005;92:284–296. doi: 10.3732/ajb.92.2.284. [DOI] [PubMed] [Google Scholar]
- Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. American Journal of Botany. 2010;97:1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, O'Meara BC, Crane P, Donoghue MJ. Heterogeneous rates of molecular evolution and diversification could explain the Triassic age estimate for angiosperms. Systematic Biology. 2015;64:869–878. doi: 10.1093/sysbio/syv027. [DOI] [PubMed] [Google Scholar]
- Bond WJ, Scott AC. Fire and the spread of flowering plants in the Cretaceous. New Phytologist. 2010;188:1137–1150. doi: 10.1111/j.1469-8137.2010.03418.x. [DOI] [PubMed] [Google Scholar]
- Boyce CK, Lee JE. An exceptional role for flowering plant physiology in the expansion of tropical rainforests and biodiversity. Proceedings of the Royal Society B: Biological Sciences. 2010;277:3437–3443. doi: 10.1098/rspb.2010.0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce CK, Brodribb TJ, Feild TS, Zwieniecki MA. Angiosperm leaf vein evolution was physiologically and environmentally transformative. Proceedings of the Royal Society B: Biological Sciences. 2009;276:1771–1776. doi: 10.1098/rspb.2008.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, Feild TS. Leaf hydraulic evolution led a surge in leaf photosynthetic capacity during early angiosperm diversification. Ecology Letters. 2010;13:175–183. doi: 10.1111/j.1461-0248.2009.01410.x. [DOI] [PubMed] [Google Scholar]
- Buerki S, Forest F, Alvarez N. Proto-South-East Asia as a trigger of early angiosperm diversification. Botanical Journal of the Linnean Society. 2014;174:326–333. [Google Scholar]
- Carlquist S. Xylem heterochrony: An unappreciated key to angiosperm origin and diversifications. Botanical Journal of the Linnean Society. 2009;161:26–65. [Google Scholar]
- Chaboureau A-C, Sepulchre P, Donnadieu Y, Franc A. Tectonic-driven climate change and the diversification of angiosperms. Proceedings of the National Academy of Sciences USA. 2014;111:14066–14070. doi: 10.1073/pnas.1324002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JT, Warnock RCM, Donoghue PCJ. Establishing a time-scale for plant evolution. New Phytologist. 2011;192:266–301. doi: 10.1111/j.1469-8137.2011.03794.x. [DOI] [PubMed] [Google Scholar]
- Cornet B. Late Triassic angiosperm-like pollen from the Richmond rift basin of Virginia, U.S.A. Palaeontographica Abteilung B. 1989;213:37–87. [Google Scholar]
- Crane PR, Friis EM, Pedersen KR. Reproductive structure and function in Cretaceous Chloranthaceae. Plant Systematics and Evolution. 1989;165:211–226. [Google Scholar]
- Crane PR, Pedersen KR, Friis EM, Drinnan AN. Early Cretaceous (early to middle Albian) platanoid inflorescences associated with Sapindopsis leaves from the Potomac Group of eastern North America. Systematic Botany. 1993;18:328. [Google Scholar]
- Crepet WL, Nixon KC, Gandolfo MA. Fossil evidence and phylogeny: The age of major angiosperm clades based on mesofossil and macrofossil evidence from Cretaceous deposits. American Journal of Botany. 2004;91:1666–1682. doi: 10.3732/ajb.91.10.1666. [DOI] [PubMed] [Google Scholar]
- Crepet WL, Nixon KC, Gandolfo MA. An extinct calycanthoid taxon, Jerseyanthus calycanthoides, from the Late Cretaceous of New Jersey. American Journal of Botany. 2005;92:1475–1485. doi: 10.3732/ajb.92.9.1475. [DOI] [PubMed] [Google Scholar]
- Dilcher DL, Crane PR. Archaenthus: An early angiosperm from the Cenomanian of the western interior of North America. Annals of the Missouri Botanical Garden. 1984;71:351–383. [Google Scholar]
- Doyle JA, Endress PK. Integrating Early Cretaceous fossils into the phylogeny of living angiosperms: Magnoliidae and eudicots. Journal of Systematics and Evolution. 2010;48:1–35. [Google Scholar]
- Doyle JA, Endress PK. Integrating Early Cretaceous fossils into the phylogeny of living angiosperms: ANITA lines and relatives of Chloranthaceae. International Journal of Plant Sciences. 2014;175:555–600. [Google Scholar]
- Doyle JA, Hotton CL, Ward JV. Early Cretaceous tetrads, zonasulculate pollen, and Winteraceae taxonomy, morphology, and ultrastructure. American Journal of Botany. 1990;77:1544–1557. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS, Källersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. [Google Scholar]
- Feild TS, Wilson JP. Evolutionary voyage of angiosperm vessel structure-function and its significance for early angiosperm success. International Journal of Plant Sciences. 2012;173:596–609. [Google Scholar]
- Feild TS, Chatelet DS, Brodribb TJ. Ancestral xerophobia: A hypothesis on the whole plant ecophysiology of early angiosperms. Geobiology. 2009a;7:237–264. doi: 10.1111/j.1472-4669.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- Feild TS, Chatelet DS, Brodribb TJ. Giant flowers of southern Magnolia are hydrated by the xylem. Plant Physiology. 2009b;150:1587–1597. doi: 10.1104/pp.109.136127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feild TS, Upchurch GR, Chatelet DS, Brodribb TJ, Grubbs KC, Samain M-S, Wanke S. Fossil evidence for low gas exchange capacities for Early Cretaceous angiosperm leaves. Paleobiology. 2011;37:195–213. [Google Scholar]
- Foster CSP, Sauquet H, van der Merwe M, McPherson H, Rossetto M, Ho SYW. Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Systematic Botany. 2017;66:338–351. doi: 10.1093/sysbio/syw086. [DOI] [PubMed] [Google Scholar]
- Friis EM, Eklund H, Pedersen KR, Crane PR. Virginianthus calycanthoides gen. et sp. nov. -A calycanthaceous flower from the Potomac Group (Early Cretaceous) of eastern North America. International Journal of Plant Sciences. 1994;155:772–785. [Google Scholar]
- Friis EM, Crane PR, Pedersen KR. Anacostia, a new angiosperm from the Early Cretaceous of North America and Portugal with trichotomocolpate/monocolpate pollen. Grana. 1997;36:225–244. [Google Scholar]
- Friis EM, Pedersen KR, Crane PR. Araceae from the Early Cretaceous of Portugal: Evidence on the emergence of monocotyledons. Proceedings of the National Academy of Sciences USA. 2004;101:16565–16570. doi: 10.1073/pnas.0407174101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis EM, Pedersen KR, von Balthazar M, Grimm GW, Crane PR. Monetianthus mirus gen. et sp. nov., a nymphaealean flower from the Early Cretaceous of Portugal. International Journal of Plant Sciences. 2009;170:1086–1101. [Google Scholar]
- Friis EM, Crane PR, Pedersen KR. Early flowers and angiosperm evolution. Cambridge, U. K, and New York: Cambridge University Press; 2011. [Google Scholar]
- Frumin S. Liriodendroid seeds from the Late Cretaceous of Kazakhstan and North Carolina, USA. Review of Palaeobotany and Palynology. 1996;94:39–55. [Google Scholar]
- Gandolfo MA, Nixon KC, Crepet WL. Cretaceous flowers of Nymphaeaceae and implications for complex insect entrapment pollination mechanisms in early angiosperms. Proceedings of the National Academy of Sciences USA. 2004;101:8056–8060. doi: 10.1073/pnas.0402473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandolfo MA, Nixon KC, Crepet WL. Selection of fossils for calibration of molecular dating models. Annals of the Missouri Botanical Garden. 2008;95:34–42. [Google Scholar]
- Heads M. Bayesian transmogrification of clade divergence dates: A critique. Journal of Biogeography. 2012;39:1749–1756. [Google Scholar]
- Hedges SB, Kumar S. Precision of molecular time estimates. Trends in Genetics. 2004;20:242–247. doi: 10.1016/j.tig.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Herendeen PS, Friis EM, Pedersen KR, Crane PR. Palaeobotanical redux: Revisiting the age of the angiosperms. Nature Plants. 2017;3:17015. doi: 10.1038/nplants.2017.15. [DOI] [PubMed] [Google Scholar]
- Ho SY, Duchêne S. Molecular-clock methods for estimating evolutionary rates and timescales. Molecular Ecology. 2014;23:5947–5965. doi: 10.1111/mec.12953. [DOI] [PubMed] [Google Scholar]
- Hochuli PA, Feist-Burkhardt S. A boreal early cradle of Angiosperms? Angiosperm-like pollen from the Middle Triassic of the Barents Sea (Norway) Journal of Micropalaeontology. 2004;23:97–104. [Google Scholar]
- Hochuli PA, Feist-Burkhardt S. Angiosperm-like pollen and Afropollis from the Middle Triassic (Anisian) of the Germanic Basin (Northern Switzerland) Frontiers in Plant Science. 2013;4:1–14. doi: 10.3389/fpls.2013.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Roger AJ. The impact of fossils and taxon sampling on ancient molecular dating analyses. Molecular Biology and Evolution. 2007;24:1889–1897. doi: 10.1093/molbev/msm115. [DOI] [PubMed] [Google Scholar]
- Hughes N, McDougall A. Barremian-Aptian angiospermid pollen records from southern England. Review of Palaeobotany and Palynology. 1990;65:145–151. [Google Scholar]
- Magallón S. Using fossils to break long branches in molecular dating: A comparison of relaxed clocks applied to the origin of angiosperms. Systematic Biology. 2010;59:384–399. doi: 10.1093/sysbio/syq027. [DOI] [PubMed] [Google Scholar]
- Magallón S. A review of the effect of relaxed clock method, long branches, genes, and calibrations in the estimation of angiosperm age. Botanical Sciences. 2014;92:1–22. [Google Scholar]
- Magallón S, Castillo A. Angiosperm diversification through time. American Journal of Botany. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- Magallón S, Hilu KW, Quandt D. Land plant evolutionary timeline: Gene effects are secondary to fossil constraints in relaxed clock estimation of age and substitution rates. American Journal of Botany. 2013;100:556–573. doi: 10.3732/ajb.1200416. [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist. 2015;207:437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- Massoni J, Forest F, Sauquet H. Increased sampling of both genes and taxa improves resolution of phylogenetic relationships within Magnoliidae, a large and early-diverging clade of angiosperms. Molecular Phylogenetics and Evolution. 2014;70:84–93. doi: 10.1016/j.ympev.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Massoni J, Couvreur TLP, Sauquet H. Five major shifts of diversification through the long evolutionary history of Magnoliidae (angiosperms) BMC Evolutionary Biology. 2015a;15:49. doi: 10.1186/s12862-015-0320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoni J, Doyle J, Sauquet H. Fossil calibration of Magnoliidae, an ancient lineage of angiosperms. Palaeontologia Electronica. 2015b;18:1–25. [Google Scholar]
- Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TLL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, BurkHerrick A, Westerman M, Ayoub NA, Springer MS, Murphy WJ. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammam diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Mohr BAR, Bernardes-de-Oliveira MEC. Endressinia brasiliana, a Magnolialean angiosperm from the Lower Cretaceous Crato Formation (Brazil) International Journal of Plant Sciences. 2004;165:1121–1133. [Google Scholar]
- Mohr BAR, Bernardes-de-Oliveira MEC, Taylor DW. Pluricarpellatia, a nymphaealean angiosperm from the Lower Cretaceous of northern Gondwana (Crato Formation, Brazil) Taxon. 2008;57:1147–1158. [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K. SeqState - primer design and sequence statistics for phylogenetic DNA data sets. Applied Bioinformatics. 2005;4:65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]
- Near TJ, Meylan PA, Shaffer HB. Assessing concordance of fossil calibration points in molecular clock studies: An example using turtles. The American Naturalist. 2005;165:137–146. doi: 10.1086/427734. [DOI] [PubMed] [Google Scholar]
- Nixon KC. Paleobotany, evidence, and molecular dating: An example from the Nymphaeales. Annals of the Missouri Botanical Garden. 2008;95:43–50. [Google Scholar]
- Parham JF, Donoghue PCJ, Bell CJ, Calway TD, Head JJ, Holroyd PA, Inoue JG, Irmis RB, Joyce WG, Ksepka DT, Patané JSL, Smith ND, Tarver JE, van Tuinen M, Yang Z, Angielczyk KD, Greenwood JM, Hipsley CA, Jacobs L, Makovicky PJ, Müller J, Smith KT, Theodor JM, Warnock RCM. Best practices for justifying fossil calibrations. Systematic Biology. 2011;61:346–359. doi: 10.1093/sysbio/syr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie MD, Doyle JA. Dating clades with fossils and molecules: The case of Annonaceae. Botanical Journal of the Linnean Society. 2012;169:84–116. [Google Scholar]
- Pittermann J, Stuart SA, Dawson TE, Moreau A. Cenozoic climate change shaped the evolutionary ecophysiology of the Cupressaceae conifers. Proceedings of the National Academy of Sciences USA. 2012;109:9647–9652. doi: 10.1073/pnas.1114378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinar GO. Programinis burmitis gen. et sp. nov., and P. laminatus sp. nov., Early Cretaceous grass-like monocots in Burmese amber. Australian Systematic Botany. 2004;17:497. [Google Scholar]
- Poinar GO. Vetuformosa buckleyin. gen., n. sp. (Ephemeroptera: Baetidae; Vetuformosinae n. subfam.), a new subfamily of mayflies in Early Cretaceous Burmese amber. Historical Biology. 2011;23:369–374. [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Pyron RA. A likelihood method for assessing molecular divergence time estimates and the placement of fossil calibrations. Systematic Biology. 2009;59:185–194. doi: 10.1093/sysbio/syp090. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer. 2003 Retrieved from http://beast.bio.ed.ac.uk/tracer.
- Royer DL, Miller IM, Peppe DJ, Hickey LJ. Leaf economic traits from fossils support a weedy habit for early angiosperms. American Journal of Botany. 2010;97:438–445. doi: 10.3732/ajb.0900290. [DOI] [PubMed] [Google Scholar]
- Rutschmann F. Molecular dating of phylogenetic trees: A brief review of current methods that estimate divergence times. Diversity and Distributions. 2006;12:35–48. [Google Scholar]
- Salomo K, Smith JF, Feild TS, Samain M-S, Bond L, Davidson C, Zimmers J, Neinhuis C, Wanke S. Data from: The emergence of earliest angiosperms may be earlier than fossil evidence indicates. Dryad Digital Repository. 2017 doi: 10.5061/dryad.1rd0m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M. Nonparametric approach to estimating divergence times in the absence of rate constancy. Molecular Biology and Evolution. 1997;14:1218–1231. [Google Scholar]
- Sanderson M. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Molecular Biology and Evolution. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sauquet H, Weston PH, Anderson CL, Barker NP, Cantrill DJ, Mast AR, Savolainen V. Contrasted patterns of hyperdiversification in Mediterranean hotspots. Proceedings of the National Academy of Sciences USA. 2008;106:221–225. doi: 10.1073/pnas.0805607106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauquet H, Ho SYW, Gandolfo MA, Jordan GJ, Wilf P, Cantrill DJ, Bayly MJ, Bromham L, Brown GK, Carpenter RL, Lee DN, Murphy DJ, Sniderman JMK, Udovicic F. Testing the impact of calibration on molecular divergence times using a fossil-rich group: The case of Nothofagus (Fagales) Systematic Biology. 2012;61:289–313. doi: 10.1093/sysbio/syr116. [DOI] [PubMed] [Google Scholar]
- Smith JF, Hileman LC, Powell MP, Baum DA. Evolution of GCYC, a Gesneriaceae homolog of CYCLOIDEA, within Gesnerioideae (Gesneriaceae) Molecular Phylogenetics and Evolution. 2004;31:765–779. doi: 10.1016/j.ympev.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Smith SA, Beaulieu JM, Donoghue MJ. An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proceedings of the National Academy of Sciences USA. 2010;107:5897–5902. doi: 10.1073/pnas.1001225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SY, Stockey RA. Establishing a fossil record for the perianthless Piperales: Saururus tuckerae sp. nov. (Saururaceae) from the Middle Eocene Princeton Chert. American Journal of Botany. 2007;94:1642–1657. doi: 10.3732/ajb.94.10.1642. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, Brockington SF, Refulio-Rodriguez NF, Walker JB, Moore MJ, Carlsward BS, Bell CD, Latvis M, Crawley S, Black C, Diouf D, Xi Z, Rushworth CA, Gitzendanner MA, Sytsma KJ, Qiu Y-L, Hilu KW, Davis CC, Sanderson MJ, Beaman RS, Olmstead RG, Judd WS, Donoghue MJ, Soltis PS. Angiosperm phylogeny: 17 genes, 640 taxa. American Journal of Botany. 2011;98:704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- Sperry JS, Hacke UG, Feild TS, Sano Y, Sikkema EH. Hydraulic consequences of vessel evolution in angiosperms. International Journal of Plant Sciences. 2007;168:1127–1139. [Google Scholar]
- Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–1376. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Dilcher DL, Wang H, Chen Z. A eudicot from the Early Cretaceous of China. Nature. 2011;471:625–628. doi: 10.1038/nature09811. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Friis EM, Uesugi K, Suzuki Y, Crane PR. Floral evidence of Annonaceae from the Late Cretaceous of Japan. International Journal of Plant Sciences. 2008;169:908–917. [Google Scholar]
- Taylor DW, Hickey LJ. Evidence for and implications of an herbaceous origin for angiosperms. In: Barker D, Taylor DW, Hickey LJ, editors. Flowering plant origin, evolution, and phylogeny. New York: Chapman and Hall Press; 1996. pp. 232–266. [Google Scholar]
- Taylor EL, Taylor TN. Seed ferns from the late Paleozoic and Mesozoic: Any angiosperm ancestors lurking there? American Journal of Botany. 2008;96:237–251. doi: 10.3732/ajb.0800202. [DOI] [PubMed] [Google Scholar]
- Upchurch G, Crane PR, Drinnan AN. The megaflora from the Quantico locality (upper Albian), Lower Cretaceous Potomac Group of Virginia. Martinsville: Virginia Museum of Natural History; 1994. [Google Scholar]
- Vakhrameev VA. Stratigraphy and fossil flora of the Cretaceous deposits in the western Kazakhstan. Regional Stratigraphy of the USSR. 1952;1:340. [Google Scholar]
- von Balthazar M, Pedersen KR, Friis EM. Teixeiria lusitanica, a new fossil flower from the Early Cretaceous of Portugal with affinities to Ranunculales. Plant Systematics and Evolution. 2005;255:55–75. [Google Scholar]
- von Balthazar M, Pedersen KR, Crane PR, Stampanoni M, Friis EM. Potomacanthus lobatus gen. et sp. nov., a new flower of probable Lauraceae from the Early Cretaceous (Early to Middle Albian) of eastern North America. American Journal of Botany. 2007;94:2041–2053. doi: 10.3732/ajb.94.12.2041. [DOI] [PubMed] [Google Scholar]
- Walker J, Walker A. Ultrastructure of Lower Cretaceous angiosperm pollen and the origin and early evolution of flowering plants. Annals of the Missouri Botanical Garden. 1984;71:464–521. [Google Scholar]
- Wanke S, Jaramillo MA, Borsch T, Samain M-S, Quandt D, Neinhuis C. Evolution of Piperales-matK gene and trnK intron sequence data reveal lineage specific resolution contrast. Molecular Phylogenetics and Evolution. 2007;42:477–497. doi: 10.1016/j.ympev.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Wing SL, Boucher LD. Ecological aspects of the cretacceous flowering plant radiation. Annual Review of Earth and Planetary Sciences. 1998;26:379–421. [Google Scholar]
- Yamada T, Nishida H, Umebayashi M, Uemura K, Kato M. Oldest record of Trimeniaceae from the Early Cretaceous of northern Japan. BMC Evolutionary Biology. 2008;8:135. doi: 10.1186/1471-2148-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M-J, Bell CD, Soltis PS, Soltis DE. Divergence times and historical biogeography of Nymphaeales. Systematic Botany. 2005;30:693–704. [Google Scholar]
- Zavada MS, Benson JM. First fossil evidence for the primitive angiosperm family Lactoridaceae. American Journal of Botany. 1987;74:1590–1594. [Google Scholar]
- Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nature Communications. 2014;5:4956. doi: 10.1038/ncomms5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Eshel G, Rees PM, Rothfus T, Rowley D, Sunderlin D. Tracing the tropics across land and sea: Permian to present. Lethaia. 2003;36:227–254. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


