Abstract
Aims
The use of circulatory miRNAs as biomarkers and therapeutic targets for T2DM is an explosive area of study. However, no study has investigated circulatory miRNA expression exclusively in African-American adults. The aim of this study was to identify the expression of nine selected miRNAs in erythrocytes of pre-diabetic and type 2 diabetic African-American adults.
Main Methods
Patients were recruited from the Howard University Hospital Diabetes Treatment Center following an 8 to 10 hour overnight fast. Expression of the nine selected miRNAs (miRNA-499, miRNA-146, miRNA-126, miRNA-223, miRNA-15a, miRNA-15b, miRNA-224, miRNA-326, and miRNA-375) was evaluated using quantitative real time PCR.
Key Findings
miRNA-15a, miRNA-15b, and miRNA-499 were significantly reduced in erythrocytes of pre-diabetic African-American adults. In the T2DM group, we found significant correlations between miRNA-15a and BMI (r=0.59, p=0.04), miRNA-15a and weight (r=0.52, p=0.01), and miRNA-15b and diastolic blood pressure (r=−0.52, p=0.02). In the pre-diabetic group, we found significant correlations between miRNA-15b and weight (r=0.90, p=0.02) and miRNA-499 and HbA1c (r=−0.89, p=0.01).
Significance
To our knowledge, this is the first study investigating miRNA expression in erythrocytes of non-diabetic high-risk obese--pre-diabetic and type 2 diabetic African-American adults. The findings of this study are consistent with previous reports of reduced expression of miRNA-15a, miRNA-15b, and miRNA-499 in human plasma or serum and in animal models. The current findings support the use of circulating miRNA-15a, miRNA-15b, and miRNA-499 as potential biomarkers for T2DM in African-American adults.
Keywords: T2DM, miRNA-15a, miRNA-15b, miRNA-499, African-Americans, Genetic Biomarkers
Introduction
The genetics of T2DM is complex and not clearly understood. The search for diabetes genes and risk markers is complicated by the heterogeneity of the metabolic disease. Genomewide association studies (GWAS) have identified several genetic variants in association with T2DM [1]. However, since the first T2DM GWAS study, identified genetic variants have been modestly associated with T2DM and account for only about 10% of genetic risk [2]. Further complicating the identification of genetic biomarkers for T2DM is ethnicity, as genetic variants may be ethnic specific. Additional studies to identify risk markers in the African-American population are needed.
MiRNAs are a class of small (18–25 nucleotides) non-coding RNAs that are involved in a number of biologically important functions, from cellular development and proliferation, to apoptosis and metabolism [2–4]. Several studies report the functional involvement of intracellular miRNAs in maintaining and regulating β-cell mass, insulin-signaling pathways, and glucose stimulated insulin secretion, making them ideal candidates in unearthing the molecular complexities of T2DM [5–11]. In addition to their intracellular expression and function, miRNAs have also been identified in blood, urine, tears and saliva [2, 3]. These circulating miRNAs are packaged in microvesicles that protect them from endogenous RNase activity and are thought to be involved in cell-to-cell communication [12].
Circulatory miRNAs are stable in circulation, consistently expressed among individuals of the same species, and are differentially expressed between healthy and diseased samples, serving as ideal candidates [13–15]. Additionally, circulating miRNAs can be obtained relatively easy, compared to collecting clinical tissue samples. Several studies have explored the use of circulating miRNAs as potential regulators and biomarkers of metabolic dysfunction in T2DM [2, 10, 17–19]. However, the follow up studies were not consistent with this initial report [19]. These inconsistencies are most likely the result of sample size, experimental protocol, and ethnic differences between study populations.
Erythrocytes make up more than 90% of the cell population in peripheral blood [20]. They are the terminal products of highly regulated cellular differentiation, undergoing enucleation and lacking nucleic acids [4, 20, 21, 25]. However, several reports indicate that RBCs have rather sophisticated extracellular and intracellular environments, containing insulin receptors and abundant miRNAs, respectively [4, 20–22]. Identifying miRNA expression specifically in erythrocytes of African-American adults is a unique approach in identifying genetic risk markers for T2DM. Previous work from our laboratory has explored the role of erythrocytes in the development and progression of T2DM [22]. To date, there are no studies that report circulating miRNA expression exclusively in African-Americans, a high-risk ethnic group. Moreover, few studies report erythrocyte miRNA expression in T2DM. The aim of this pilot and feasibility study was to identify differences in erythrocyte miRNA expression in pre-diabetic and T2DM African-American adults.
Materials and Methods
Ethical Statement
This study was carried out in accordance to the guidelines of the Howard University Institutional Review Board (HUIRB). The protocol was approved by the HUIRB (IRB-13-MED-73). All participants of this study gave written informed consent.
Study Population
Patients were recruited from the Howard University Diabetes Treatment Center (DTC). Whole blood samples were collected from African-American adults between the ages of 18 and 80 years old with or without a family history of T2DM in a parent or grandparent. Patients included both pre-diabetic and T2DM African-American adults. T2DM were previously diagnosed. Non-diabetic high-risk obese—pre-diabetics were identified through the W.E.I.G.H.T study (Working to Engage Insulin-Resistant Group Health Using Technology Study) at the DTC. Control subjects were recruited from the Howard University community. All clinical characteristics for T2DM and pre-diabetic patients were retrieved from electronic files at DTC.
Erythrocyte Isolation
All samples were collected following an overnight fast of 8–10 hours. Venous blood samples were collected in heparinized vacutainer tubes. Erythrocytes were isolated and purified within an hour of the blood collection by Hypaque-Ficoll (HF) gradient centrifugation as previously described [22]. Heparinized blood was centrifuged for 20 minutes at 2,000Xg. The plasma was removed and stored (−80°C) for use in future studies. 2–3 parts isotonic (0.15 mM) choline chloride was added to the cell pellet and mixed gently by inversion. The suspended cells were layered on 3ml Hypaque (33.9%): Ficoll (9%) mixture (1:2.4 ratio) (HF) in a glass tube. The tubes were centrifuge for 20 minutes at 2,000xg. HF and choline chloride layers were aspirated, leaving the erythrocyte pellet. The procedure was repeated. After final centrifugation and aspiration of HF and choline chloride, the erythrocytes were labeled as purified red blood cells [23].
RNA Extraction
Nine T2DM-related miRNAs as reported in previous literature: miRNA-499, miRNA-146, miRNA-126, miRNA-223, miRNA-15a, miRNA-15b, miRNA-224, miRNA-326, and miRNA-375 were selected for this study. These miRNAs were selected as they are reported to be involved in insulin biosynthesis, insulin secretion, and glucose homeostasis. The miRVana miRNA isolation kit obtained from Ambion (Austin, Texas) was used to isolate total RNA from erythrocytes following the procedures outlined by the manufacture. Northern blots were used to provide qualitative and quantitative data of total RNA.
cDNA Synthesis and miRNA Quantification
Reverse transcription and quantitative real time PCR (qRT-PCR) were used for miRNA quantification in erythrocytes. Reverse transcription and PCR reactions were performed using the miSCRIPT II RT Kit (Qiagen) and miSCRIPT SYBR Green PCR Kit (Qiagen), respectively. miSCRIPT II RT system uses total RNA that contains miRNA as the starting material for cDNA synthesis and allows for the detection of multiple miRNAs from a single cDNA preparation. The final reaction volume of reverse transcription was 20µL. Samples were incubated for 60 minutes at 37°C followed by a 5-minute incubation at 95°C. To quantify miRNA expression, a master mix was created for each miSCRIPT primer following the manufactures procedure. For each 10µL reaction: 5µL of SYBR green master mix (iTaq, BIO RAD), 1µL of 10X Universal Primer, 1µL of gene/miRNA specific primer, and 3µL of cDNA template. Amplification was carried out in 96-well plate using the LightCycler 480 (Roche) using the cycling conditions outlined by the manufacture. Ct values >36 were considered to identify miRNA levels below detection limit or absent. qRT-PCR was performed in duplicate for each miRNA. RNU6b was used as a control.
Statistical Analysis
Differences in erythrocyte miRNA levels were analyzed using nonparametric one-way ANOVA. Receiver-operating characteristics (ROC) curves were established to determine the area under the curve (AUC) and to evaluate the ability of circulating miRNAs to discriminate between groups. Statistical analysis was completed using Prism software (GraphPad, La Jolla, CA).
Results
Clinical Characteristics
The clinical characteristics of the study population are presented in Table 1. Body weight was significantly higher in the T2DM (96 ± 4.721 kg, n=31) when compared to the control group (83.13 ± 4.563 kg, n=14), p=0.04. Body weight was not significantly different in the pre-diabetic group when compared to both T2DM and the control groups. BMI was significantly higher in the T2DM (34.45 ± 1.81 kg/m2) and pre-diabetic (35.10 ± 2.66 kg/m2) when compared to the control group (28.22 ± 1.59 kg/m2), p=0.01 and p=0.04, respectively. The control group was overweight (BMI= 25–29.9). Both the T2DM and pre-diabetic group were obese (BMI>30). HbA1c was significantly higher in the T2DM (8.6 ± 0.384) when compared to pre-diabetic (5.14 ± 0.1420) African-American adults.
Table 1.
Study population clinical characteristics.
| T2DM | Pre-diabetics | Control | |
|---|---|---|---|
|
| |||
| Weight (kg) | 96 ± 4.7 | 94.7 ± 8.5 | 83.1 ± 4.6 |
| n=31 | n=6 | n=14 | |
|
| |||
| BMI (kg/m2) | 34.5 ± 1.8 | 35.1 ± 2.6 | 28.2 ± 1.6 |
| n=30 | n=7 | n=13 | |
|
| |||
| HbA1c | 8.6 ± 0.4 | 5.14 ± 0.1 | N/D |
| n=24 | n=8 | ||
|
| |||
| Systolic Blood Pressure (mmHg) | 137.0±3.4 | N/D | N/D |
| n=26 | |||
|
| |||
| Diastolic Blood Pressure (mmHg) | 72.3±1.9 | N/D | N/D |
| n=26 | |||
|
| |||
| Age (Years) | 56.3±2.00 | 21.3±0.92 | 31.5±3.01 |
| n=31 | n=6 | n=14 | |
|
| |||
| Gender | F n=22 | F n=8 | F n=8 |
| M n=9 | M n=1 | M n=9 | |
Patient information was retrieved from electronic files in the Diabetes Treatment Center at Howard University Hospital for both T2DM and pre-diabetic participants. For pre-diabetic participants, only age, BMI, and HbA1C were available, as participants were part of the W.E.I.G.H.T Study and did not require extensive clinical examination, as noted by N/D. Data presented as mean ± SE. p ≤ 0.05 considered significant. F=Female; M=Male.
MiRNA Expression
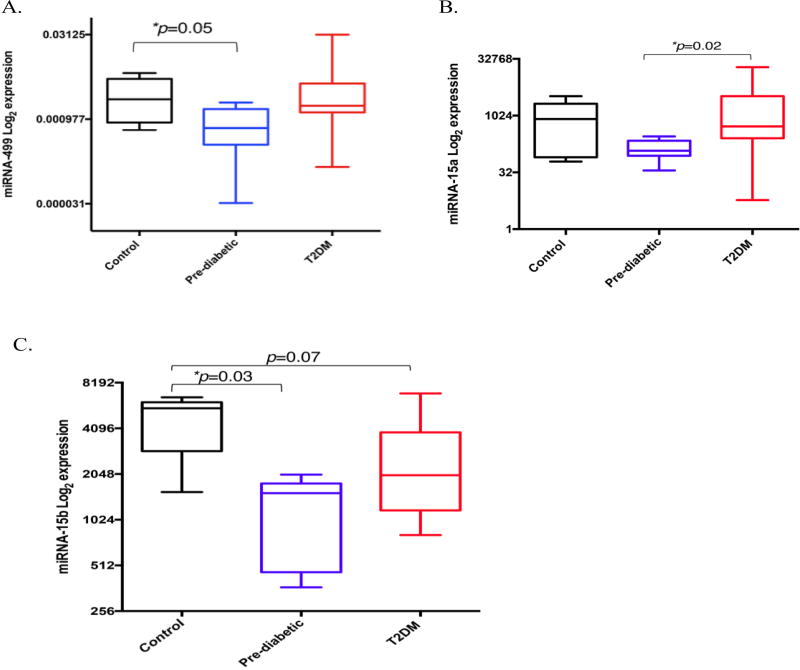
We selected nine miRNAs to be measured in erythrocytes obtained from control, pre-diabetic and T2DM African-American adults. These miRNAs were reported to be associated with T2DM development and progression. To identify if erythrocyte miRNA expression differed between healthy, pre-diabetic and T2DM African-American adults, we isolated total RNA from purified erythrocytes and quantified miRNA expression using qRT-PCR. Of the nine selected miRNAs, only miRNA-499, miRNA-15a, and miRNA-15b were differentially expressed. miRNA-499 was significantly reduced in the pre-diabetic group when compared to the control group (p=0.05), but not the T2DM group (Fig 1A). miRNA-15a was significantly reduced in the pre-diabetic group, when compared to the T2DM group (p=0.02) (Fig 1B). MiRNA-15b was significantly reduced in the pre-diabetic group when compared to controls. Reduced miRNA-15b expression in the T2DM group was reaching statistical significance (p=0.07) when compared to the control group (Fig 1C).
Figure 1.
Erythrocyte miRNA expression in healthy, pre-diabetic, and T2DM African-American adults. (A) miRNA-499 expression control (n=8), pre-diabetic (n=7), and T2DM (n=25) African-American adults, (B) miRNA-15a expression control (n=8), pre-diabetic (n=7), and T2DM (n=24) African-American adults and (C) miRNA-15b expression healthy (n=5), pre-diabetic (n=7), and T2DM (n=23) African-American adults. The Y-axis refers to the miRNA expression ratio (miRNA versus RNU6B) in log2 scale. Expression was measured in duplicate. A non-parametric ANOVA (Kruskal-Wallis test) was used to determine statistical significance between three groups. The Box-Whisker-Plots illustrate the median expression of miRNA-499, miRNA-15a, and miRNA-15b in erythrocytes from healthy, pre-diabetic, and T2DM healthy African-American adults. p ≤ 0.05 considered significant.
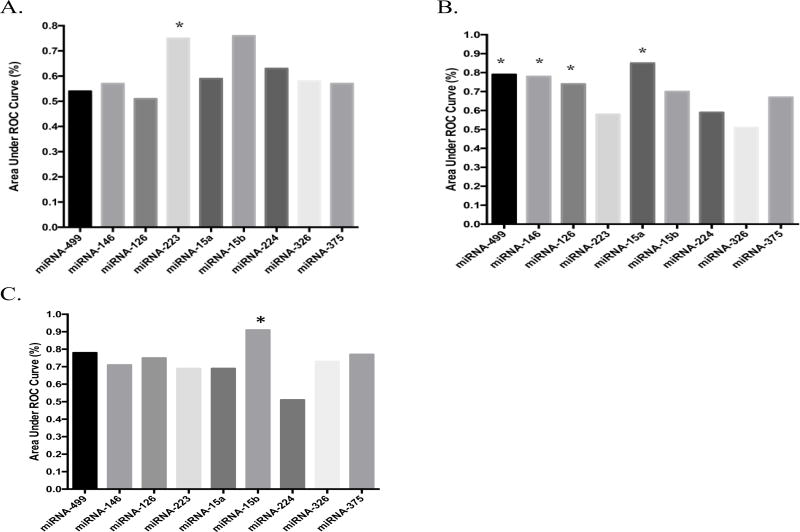
To determine the ability of the selected miRNAs to discriminate between the three groups, we established ROC curves and calculated area under the curve (AUC). AUCs between 0.70 and 0.90 are considered good discriminators. MiRNA-223 was able to discriminate between T2DM and control subjects (AUC=0.7500, p=0.03) (Figure 2A). Both miRNA-499 and miRNA-15a were able to discriminate between T2DM and pre-diabetic African-American adults (AUC=0.7866, p=0.02) and (AUC=0.8512, p=0.05), respectively. Additionally, miRNA-146 (AUC=0.7829, p=0.02) and miRNA-126 (AUC=0.7429, p=0.05) were able to discriminate between these two groups (Figure 2B). Only miRNA-15b was able to discriminate between pre-diabetic and healthy African-American adults (AUC=0.9143, p=0.01) (Figure 2C).
Figure 2.
AUC Calculations for healthy, pre-diabetic, and T2DM African-American Adults. (A) AUC calculations for T2DM versus healthy African-American adults, (B) AUC calculations for T2DM versus pre-diabetic African-American adults, and (C) AUC calculations for pre-diabetic versus healthy African-American adults. p ≤ 0.05 considered significant.
We next sought to identify relationships between the selected miRNAs and clinical characteristics associated with T2DM development and progression, such as BMI and HbA1c. We did not find any significant relationships between the selected miRNAs and clinical characteristics in the control group. However, we did find strong significant correlations between miRNA-15b and weight (r=0.92, p=0.02) and miRNA-499 and HbA1c (r=−0.89, p=0.01) in the pre-diabetic group (Table 2). In the T2DM group, we found significant correlations between miRNA-15a and BMI (r=0.59, p=0.004), miRNA-15a and weight (r=0.52, p=0.01), and miRNA-15b and diastolic blood pressure (r=−0.5247, p=0.02) (Table 3).
Table 2.
Correlations between miRNA and Clinical Characteristics in Pre-Diabetic African-American Adults.
| miRN-15a | miRNA-15b | miRNA-499 | |
|---|---|---|---|
|
| |||
| Body Weight (kg) | r=0.164 | r=0.902 | r=0.509 |
| p=0.791 | p=0.02* | p=0.381 | |
|
| |||
| BMI (kg/m2) | r=0.232 | r=0.359 | r=0.186 |
| p=0.65 | p=0.48 | p=0.72 | |
|
| |||
| HbA1c | r=−0.752 | r=−0.515 | r=−0.891 |
| p=0.08 | p=0.29 | p=0.01* | |
Pearson’s correlation was used to identify relationships.
p ≤ 0.05 considered significant.
Table 3.
Correlations between miRNA and clinical characteristics in T2DM African-American adults.
| miRNA-15a | miRNA-15b | miRNA-499 | |
|---|---|---|---|
|
| |||
| Body Weight (kg) | r=0.52 | r=0.12 | r=0.28 |
| p=0.01* | p=0.59 | p=0.18 | |
|
| |||
| BMI (kg/m2) | r=0.59 | r=0.15 | r=0.23 |
| p=0.004** | p=0.521 | p=0.29 | |
|
| |||
| HbA1c | r=−0.36 | r=−0.46 | r=0.07 |
| p=0.12 | p=0.053 | p=0.74 | |
Pearson’s correlation was used to identify relationships.
p ≤ 0.05 considered significant.
Discussion
The potential use of circulatory miRNAs as biomarkers has been extensively explored in cancer. Current research explores the use of these circulatory regulatory RNAs as biomarkers and indicators of metabolic dysfunction in T2DM. However, no study reports findings exclusively from the African-American population. Furthermore, few studies have explored miRNA expression specifically in erythrocytes. To our knowledge, this is the first study to investigate the expression of miRNAs in erythrocytes of pre-diabetic and T2DM African-American adults. We report that miRNA-499, miRNA-15a, and miRNA-15b are reduced in the erythrocytes of pre-diabetic African-American adults, findings consistent with current literature. We also found that the expression of these miRNAs correlate with several clinical characteristics associated with T2DM, including HbA1c and BMI, making them ideal candidates for genetic risk markers in African-Americans. The pre-diabetic group did not follow typical diagnostic characteristics, as these patients were recruited from an ongoing early intervention study at DTC.
Erythrocyte miRNA expression could be used in addition to glycated protein analysis in plasma to identify persons of African descent at risk of T2DM. Africa has the highest prevalence of undiagnosed hyperglycemia in the world [24]. However, the ability to diagnosis pre-diabetes in this population is complicated because HbA1c cannot be measured in the absence of hemoglobin [24]. The reduced expression of miRNA-15a, miRNA-15b, and miRNA-499 in combination with glycated albumin could be beneficial in identifying persons at risk for T2DM in the African-American population.
Each of the identified miRNAs in our study are reported to be involved in key processes in T2DM development and progression, such as glucose homeostasis, insulin signaling, and insulin biosynthesis. miRNA-499 is a cardio- or skeletal muscle specific miRNA and is elevated in the plasma of patients diagnosed with acute myocardial infarction [25]. Recent reports reveal that miRNA-499 is reduced in the livers of db/db mice and targets PTEN, negatively regulating AKT/GSK activation and impairing glucose and insulin tolerance [6]. In addition to reduced miRNA-499 expression in the pre-diabetic group, we also found a strong significant correlation between miRNA-499 and HbA1c in this group, further supporting that miRNA-499 targets PTEN and is involved in both insulin and glucose tolerance.
Both miRNA-15a and miRNA-15b belong to the miRNA-15 superfamily [26, 27]. miRNA-15a was initially discovered in the plasma of T2DM subjects by Zampetaki and colleagues [18]. The findings of our study are consistent with the original report of reduced expression of circulating miRNA-15a in persons at risk for developing T2DM. Our findings also suggest that circulatory miRNA-15a may not differ between African-Americans and Italian populations. However, future studies are needed to confirm our findings. These findings are also consistent with several studies reporting that miRNA-15a is reduced in both diet induced obesity and in prolonged hyperglycemia, both hallmarks of T2DM development [28, 29]. We did identify significant relationships between miRNA-15a and BMI and miRNA-15a and body weight in the T2DM group. miRNA-15a is reported to inhibit uncoupling protein-2 (UCP2), regulating β-cell function and insulin biosynthesis [29]. miRNA-15b is predicted to target several components of the insulin signaling pathway, including TNFα and SOCS3. Recently, Ye and Steinle investigated the expression of miRNA-15b and miRNA-16 in human retinal endothelial cells (REC) in a hyperglycemic state. The findings of their study revealed that miRNA-15b and miRNA-16 are reduced in hyperglycemia [30]. As a result, both TNFα and SOCS3 protein expression were increased in human REC, ultimately resulting in insulin resistance.
Conclusions
For the first time we report a reduced expression of miRNA-15a, miRNA-15b and miRNA-499 in erythrocytes of pre-diabetic African-American adults. Reduced expression of these circulatory miRNAs could serve as prognostic and diagnostic markers for T2DM in persons of African descent. This exploratory study is limited by sample size and subjects were not age and sex-matched. Additionally, we did not require an extensive clinical examination for either the pre-diabetic and control groups. However, the findings of this study do support previous studies in other ethnic groups and cell studies. Future research will explore the molecular mechanisms contributing to key relationships with clinical characteristics and hormones of energy metabolism. Additionally, future work will explore how diabetes drug therapies alter circulatory miRNA expression, as drug therapies may result in improved or altered expression.
Acknowledgments
The authors would like to acknowledge all the participants of this study, the Endocrine Fellows and Physicians of the Howard University Hospital Diabetes Treatment Center, and the phlebotomist that contributed to this study. The authors would also like to acknowledge the Nekhai Laboratory.
Funding: This work was supported by the JHU-UMD DRC Grant P30DK079637-08, Grant #4525 and the NIH Research Grant 5G12MD007597. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Prasad RB, Groop L. Genetics of type 2 diabetes—pitfalls and possibilities. Genes. 2015;(6):87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Connell TM, Markunas CA. DNA methylation and microrna-based biomarkers for risk of type 2 diabetes. Curr Diabetes Rev. 2016;1(12):20–29. doi: 10.2174/1573399811666150515125557. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Sundquist J, Zoller B, Memon A, Palmer K, et al. Determination of 14 circulating micrornas in swedes and iraqis with and without diabetes mellitus type 2. PLoS One. 2014;(9):e86792. doi: 10.1371/journal.pone.0086792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leidinger P, Backes C, Dahmke I, Galata V, Huwer H, Stehle I, et al. What makes a blood cell based mirna expression pattern disease specific? – a mirnome analysis of blood cell subsets in lung cancer patients and healthy controls. Oncotarget. 2015;19(5):9484–9497. doi: 10.18632/oncotarget.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poy M, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, et al. miR-375 maintains normal pancreatic α- and β-cell mass. PNAS. 2009;106(14):5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Zhang N, Pan H, Wang Z, Cao Z. Mir-499-5p contributes to hepatic insulin resistance by suppressing pten. Cell Physiol and Biochem. 2015;(36):2357–2365. doi: 10.1159/000430198. [DOI] [PubMed] [Google Scholar]
- 7.Locke JM, Xavier G, Dawe HR, Rutter GA, Harries LW. Increased expression of mir-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia. 2014;122(57):128. doi: 10.1007/s00125-013-3089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karolina D, Armugan A, Tavintharan S, Wong M, Lim S, Sum C, et al. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS ONE. 2011;6:e22839. doi: 10.1371/journal.pone.0022839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Latreille M, Herrmanns K, Renwick N, Tuschl T, Malecki MT, McCarthy MI, et al. mir-375 gene dosage in pancreatic β-cells: implications for regulation of β-cell mass and biomarker development. J Mol Med. 2015 doi: 10.1007/s00109-015-1296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bork-Jensen J, Scheel C, Christophersen DV, Nilsson E, Friedrichsen M, Fernandez-Twinn DS, et al. Glucose tolerance is associated with differential expression of micrornas in skeletal muscle: results from studies of twins with and without type 2 diabetes. Diabetologia. 2015;(58):363–373. doi: 10.1007/s00125-014-3434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;(474):649–654. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 12.Zen C, Zhang C. Circulating MicroRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;(32):326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Ba Y, Cai X, et al. Characterication of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;(18):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell P, Parkin R, Kroh E, Fritz B, Wyman S, Pogosova E, et al. Circulating microRNAs as stable blood-base markers for cancer detection. PNAS. 2009;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilad S, Meiri E, Yogev Y, Benjamin S, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rong Y, Bao W, Shan Z, Liu J, Yu X, Xia S, et al. Increased microrna-146a levels in plasma of patients with newly diagnosed type 2 diabetes mellitus. PLOS. 2013;9(8):e73272. doi: 10.1371/journal.pone.0073272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong L, Zhu J, Han W, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48(1):61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 18.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma mircoRNA profiling reveals loss of endothelial mir-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 19.Zhang T, Lv C, Li L, et al. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. BioMed Res Int. 2013;2013:761617. doi: 10.1155/2013/761617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Wang Y, Telen M, Chi J. The genomic analysis of erythrocyte microrna expression in sickle cell diseases. PLoS One. 2008;4(6):e2360. doi: 10.1371/journal.pone.0002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kannan M, Aterya C. Differntial profiling of human red blood cells during storage for 52 selected micrornas. Transfusion. 2010;50(7):1581–1588. doi: 10.1111/j.1537-2995.2010.02585.x. [DOI] [PubMed] [Google Scholar]
- 22.Gambhir KK, Archer JA, Bradley CJ. Characteristics of human erythrocyte insulin receptors. Diabetes. 1978;27(7):701–708. doi: 10.2337/diab.27.7.701. [DOI] [PubMed] [Google Scholar]
- 23.Gambhir KK, Ornasir J, Headings V, Bonar A. Decreased total carbonic anhydrase esterase activity and decreased levels of carbonic anhydrase1 isozyme in erythrocytes of type II diabetic patients. Biochem Genetics. 2007;45(5–6):431–439. doi: 10.1007/s10528-007-9086-x. [DOI] [PubMed] [Google Scholar]
- 24.Sumner AE, Duong MT, Aldana PC, Ricks M, Tulloch-Reid MK, Lozier JN, et al. A1c combined with glycated albumin improves detection of prediabetes in Africans: the afriacns in america study. Diabetes Care. 2016;39(2):271–277. doi: 10.2337/dc15-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Chen X, Su T, Li H, Huang Q, Wu D, et al. Circulating mir-499 are novel and sensitive biomarker of acute myocardial infarction. J Thorac Dis. 2015;3(7):303–308. doi: 10.3978/j.issn.2072-1439.2015.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Cheng X, Lu Z, Wang J, Chen H, Fan W, et al. Upregualtion of mir-15b in nafld models and in the serum of patients with fatty liver disease. Diabetes Res and Clinic Pract. 2013;(99):327–334. doi: 10.1016/j.diabres.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 27.Finnerty JR, Wang W, Hebert SS, Wilfred BR, Mao G, Nelson PT. The mir-15/107 group of microrna genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;(3):491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee IS, Park KC, Yang K, Choi H, Jang YS, Lee JM, Kim H. Exentaide reverse dysrefulated microRNAs in high-fat diet-induced obese mice. Obes Res Clin Pract. 2015 doi: 10.1016/j.orcp.2015.07.011. http://dx.doi.org/10.1016/j.arcp.2015.07.011. [DOI] [PubMed]
- 29.Sun L, Jiang B, Li W, Zou J, Shi Y, Liu Z. Microrna-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res and Clin Pract. 2011;91(1):94–100. doi: 10.1016/j.diabres.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Ye E, Steinle J. mir-15b/16 protects primary human retinal microvascular endothelial cells against hyperglycemia-induced increases in tumor necrosis factor alpha and suppressor of cytokine signaling 3. J of Neuroinflammation. 2015;44(12) doi: 10.1186/s12974-015-0265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]