ABSTRACT
The fluctuation of tomato's WRKY defense regulators during infection by the root knot nematode Meloidogyne javanica was analyzed: and the spatial and temporal expression of SlWRKY45 was studied in depth with regard to its response to nematode infection, phytohormones, and wounding. Expression of WRKY45 increased substantially within 5 d upon infection and continued through feeding-site development and gall maturation. Histological analysis of nematode feeding sites indicated that WRKY45 was highly expressed within the feeding cells and associated vascular parenchyma cells. Responses of SlWRKY45 promoters to several phytohormones showed that WRKY45 was highly induced by specific phytohormones, including cytokinin, auxin, and the defense-signaling molecule salicylic acid (SA), but not by the jasmonates. Overexpressing tomato lines were generated, and infection tests showed that, significantly, roots over-expressing SlWRKY45 contained substantially increased number of females, indicating that WRKY45 overexpression supported faster nematode development. qRT-PCR tests have shown roots overexpressing WRKY45 suppressed the jasmonic acid and salicylic acid marker genes, proteinase inhibitor (PI), and pathogenesis related protein (PR1), respectively, and also the cytokinin response factors CRF1 and CRF6. Overall, this study indicated SlWRKY45 to be a potential transcription factor whose manipulation by the invading nematode might be critical for coordination of hormone signals supporting favorable condition for nematode development in root tissue.
KEYWORDS: Basal resistance, innate immunity, Meloidogyne javanica, plant defense signaling, phytohormones, root susceptibility, WRKY transcription factor
Introduction
Plant-parasitic nematodes, which fall into the group of sedentary endoparasitic nematodes, require establishment and maintenance of nematode feeding sites (NFS), referred to as giant cells (root knot nematodes, RKN; Meloidogyne) or syncytium (cyst nematodes, CN; Heterodera and Globodera spp.).1
The process of infection by Meloidogyne spp (RKN) starts with nematode migration into a vascular cylinder, once arrival at a suitable site, RKN transforms 4 to 8 root cells into giant cells, from which the nematode feeds.1,2 These potential feeding sites are characterized by very dense cytoplasm, multiple enlarged nuclei, small vacuoles, and proliferation of smooth endoplasmic reticulum; features regarded as the outcome of major developmental reprogramming to ensure the sites' future functioning throughout the period of RKN accommodation in the roots.3,4
Despite extensive studies of how nematodes induce such elaborate cell differentiation in the plant, there is still lack of understanding regards the the entire mechanisms underlying these structural changes.5 Increased availability of emerging transcriptomic technologies led to the use of these technologies in studying plant/nematode interactions, resulting in prolific accumulation of data correlate feeding site establishment and maintenance, with, differential activity of genes related to metabolism, stress responses, protein synthesis, cell division, transport and signal transduction.6-12 Moreover, it was shown that part of the observed changes resulted from injection into plant cells of glandular secretions produced by the nematode that are suspected of interacting, directly or indirectly, with the plant nuclear genome, thereby initiating a cascade of altered gene expression that creates the feeding-site complex.13
Nematode-responsive plant genes can be grouped into categories related to plant developmental pathways and their roles in forming a feeding site.14 Among these groups, the occurrence of transcription factors that act in the transcription-regulating network, and activate and/or repress transcription of their target genes15,16 are considered to be a favored target for the nematode. In the Arabidopsis,17,18 and rice19 genomes, 5.9–8 and 4%, respectively, of the estimated total genes encode to transcription factors. The tomato genome contains 2,505 transcription factors, categorized into 89 families that are distributed among all 12 chromosomes (Tomato Genomic Resources Database; http://59.163.192.91/tomato2/tfs.html). The abundance and importance of plant transcription factors led to their high priority among study targets related to plant responses to biotic stress.20
Recently we elaborated a comprehensive RNAseq analysis of susceptible tomato roots at 2, 5, and 15 d after inoculation (dai) with the RKN M. javanica. This study has revealed, a total of 7,518 differentially expressed genes (DEGs), between uninoculated and inoculated root samples. Among the DEGs, 4.8% were found to be associated with transcription-regulation activity; these genes included the WRKY transcription factors gene family, coding for DNA binding.21 WRKY group is considered to be one of the largest families of transcription factor genes; they modulate many plant processes, including defense mechanisms.22–24 During the last decades a large body of evidence has implicated WRKY proteins in the transcriptional reprogramming that occurs during plant defense responses, and has shown them to be a complex transcriptional network, referred as the WRKY web. However, because of the occurrence of a functional redundancy among the WRKYs, the contributions of individual members of this family to plant immunity is only subtle.25 WRKYs transcription factors are characterized by a highly conserved signature domain, WRKYGQK.26 According to the number of WRKY domains and the features of zinc-finger motifs, the WRKY protein family is categorized into three distinct groups (I, II and III), of which Group II is further divided into five subgroups (IIa to IIe) based on the presence of additional conserved short structural motifs outside the WRKY domain.27
Involvement of WRKY in plant/nematode interactions has been studied mainly in the Cyst nematode Heterodera schachtii and Arabidopsis thaliana system.26,28 Grunewald et al.29 (2008) pointed out that WRKY23 was expressed during the early stages of feeding-site establishment, and that knocking down its expression reduced infection by the cyst nematode Heterodera schachtii. More recently, transcriptome analysis of syncytia induced by the beet cyst nematode Heterodera schachtii in Arabidopsis roots has shown that among thousands of differentially regulated genes in syncytia are many that code for WRKY transcription factors.28 In tomato, a genome-wide identification of WRKYs revealed a total of 81 members of WRKY transcription factors.30 Nevertheless, despite this detailed genomic information, their relation to plant defense signaling against nematode infection received much less attention. In tomato, studies of WRKYs and nematodes mainly addressed the role of the WRKY transcription factor in mediating the Mi-1 resistance gene against the RKN, in which both paralogues SlWRKY72a and b were upregulated during disease resistance mediated by the R gene Mi-1. Whereas virus-induced gene silencing of these two genes in tomato resulted in a clear reduction of Mi-1-mediated resistance against RKN.29 Subsequently, a study by Atamian30 has shown that SlWRKY70 gene-silencing attenuated Mi-1-mediated resistance against RKN, and that SlWRKY70 transcript levels were upregulated by salicylic acid and suppressed by methyl jasmonate.
In light of all available evidence, apart from the information cited above our knowledge about the functions of WRKY proteins in tomato defense against RKN is rather limited. In the present study, 19 WRKY genes from the tomato genome were found to be differentially expressed in response to nematode infection; one of them –WRKY45 – was chosen for detailed studies including phylogeny, conserved motifs, expression profiles elicited by nematode infection, wounding, and phytohormone treatment. Furthermore, SlWRKY45 function analyzing subject WRKY45 as important regulator of hormone signals that facilitate nematode establishment in tomato roots.
Results
Differential regulation of tomato WRKY transcription factor in response to infection with RKNs
To gain a further insight into RKN parasitism on tomato, we used a recent RNAseq analysis of tomato roots following RKN M. javanica infection.21 Among the differentially regulated genes associated with the temporal dynamics of nematode infection, high representation of WRKY transcription factors was noticeable (Table 1). As shown in Table 1, using classification with MapMan 2.0.0 software tool,31 the majority of WRKY genes were recognized as differentially expressed compared with those in uninoculated roots at 15 dai when extensive cellular changes were already impressed as described by Iberkleid et al.21 Among the differentially regulated WRKY genes, WRKY45, which was upregulated upon infection, was chosen for further studies (Table 1). In silico analysis of SlWRKY45 indicated that this protein consists of 301 amino acids from 4 exons encoding 27, 100, 39, and 135 amino acids, respectively, and containing only one WRKY domain and having a zinc-finger-like motif ligand, C-X5-C-X23-H-X1-H (Fig. 1A). The presence of a nuclear localization signal sequence – LSRKRKAEEE at position 83 probably indicates its function as a nuclear transcription factor; in light of these domains and features, this protein was assigned by Huang et al.30 to group II-a of transcription factors. For investigating the relationships among the SlWRKYs genes received from the RNA-seq and other WRKY proteins that already have been reported to be involved in regulating plant responses to nematodes, a total of 43 amino acid sequences from tomato, Arabidopsis, and rice were aligned by using CLUSTALW ClustalX 1.8,32 and hierarchy analysis was applied with the TREEVIEW program (www.sdsc.edu/MEME) (Fig. 1B). As Fig. 1B shows, the analyzed WRKY proteins fell into 5 major clusters, and the following WRKY groups and subgroups were recognized: WRKY N-terminal and C-terminal domains of Group I; Group II, which was further divided into five clades, II-a+b, II-c, II-d+e. Group II-c and Group I-C were clustered together and Group III separately (Fig. 1B). The results showed that SlWRKY45 was clustered to the same branch as – AtWRKY18, AtWRKY40, and AtWRKY60 – from Arabidopsis; all belong to Group II-a, possessing a single WRKY domain (Fig. 1B). Several other SlWRKY transcription factors that are listed in Table 1 as differentially expressed WRKY transcripts were grouped with other WRKY factors that already had been shown to be associated with nematode infection, for example SlWRKY25 and SlWRKY26 have been shown to be closely related to Arabidopsis AtWRKY11 and AtWRKY17, which were shown to be downregulated following Heterodera schachtii infection.28 Similarly, SlWRKY6 clustered to the same branch as tomato SlWRKY72a and SlWRKY72b, which were shown to be transcriptionally upregulated during disease resistance mediated by Mi resistance gene.29
Table 1.
Transcriptome-Wide identification of differential expressed WRKY gene members of tomato during M.javanica infection.
| Solyc ID | Solyc Name of WRKY | Name of Similar WRKY | Accession No. | Similarity (%) | 5DAI (Fold Change) | 15DAI (Fold Change) | 28DAI (Fold Change) |
|---|---|---|---|---|---|---|---|
| Solyc08g067360.2.1 | SlWRKY45 | AtWRKY40 | NM_106732 | 39.6% | 0.472721885 | 0.3851854212 | 0.249386 |
| Solyc09g015770.2.1 | SlWRKY81 | AtWRKY70 | NM_115498 | 46.5% | — | 2.223920167 | — |
| Solyc07g056280.2.1 | SlWRKY16 | AtWRKY28 | NM_117927 | 49.7% | — | 8.551544443 | — |
| Solyc06g068460 | SlWRKY40 | OsWRKY76 | DQ298185 | 39.1% | — | 2.495712243 | — |
| Solyc02g080890.2.1 | SlWRKY6 | AtWRKY6 | HM173628 | 50% | — | 3.336627885 | — |
| Solyc04g051540.2.1 | SlWRKY13 | AtWRKY13 | NM_120101 | 36% | — | 3.876912837 | — |
| Solyc12g011200.1.1 | SlWRKY29 | AtWRKY28 | NM_117927 | 45.6% | — | 3.906062138 | — |
| Solyc10g007970.1.1 | SlWRKY5 | AtWRKY65 | NM_102668 | 29.6% | — | 0.150681625 | — |
| Solyc05g012500.2.1 | SlWRKY29 | AtWRKY57 | NM_105598 | 35.5% | — | 2.677541248 | — |
| Solyc09g066010.2.1 | SlWRKY25 | OsWRKY21 | EU230754 | 31.9% | — | 2.543557485 | — |
| Solyc02g088340.2.1 | SlWRKY3 | OsWRKY3 | BK005006 | 29.4% | — | 0.428408556 | — |
| Solyc02g093050.2.1 | SlWRKY26 | OsWRKY15 | BK005018 | 40.3% | — | 2.62928057 | — |
| Solyc05g053380.2.1 | SlWRKY31 | AtWRKY48 | NM_124329 | 36.2% | — | 5.441281248 | — |
| Solyc02g071130.2.1 | SlWRKY29 | AtWRKY71 | NM_102726 | 44.8% | — | 2.593005381 | — |
| Solyc02g067430.2.1 | SlWRKY72 | AtWRKY72 | NM_001343389 | 44.1% | — | 0.459252029 | — |
| Solyc01g079260.2.1 | SlWRKY4 | AtWRKY23 | NM_130294 | 39.5% | — | 5.60113422 | — |
| Solyc03g095770.2.1 | SlWRKY80 | AtWRKY70 | NM_115498 | 41.1% | — | 2.015220496 | — |
| Solyc02g021680.2.1 | SlWRKY35 | AtWRKY35 | NM_001336530 | 39.8% | — | 0.490582202 | — |
| Solyc10g007970.1.1 | SlWRKY77 | AtWRKY65 | NM_102668 | 92.2% | — | 0.150681625 | — |
Figure 1.
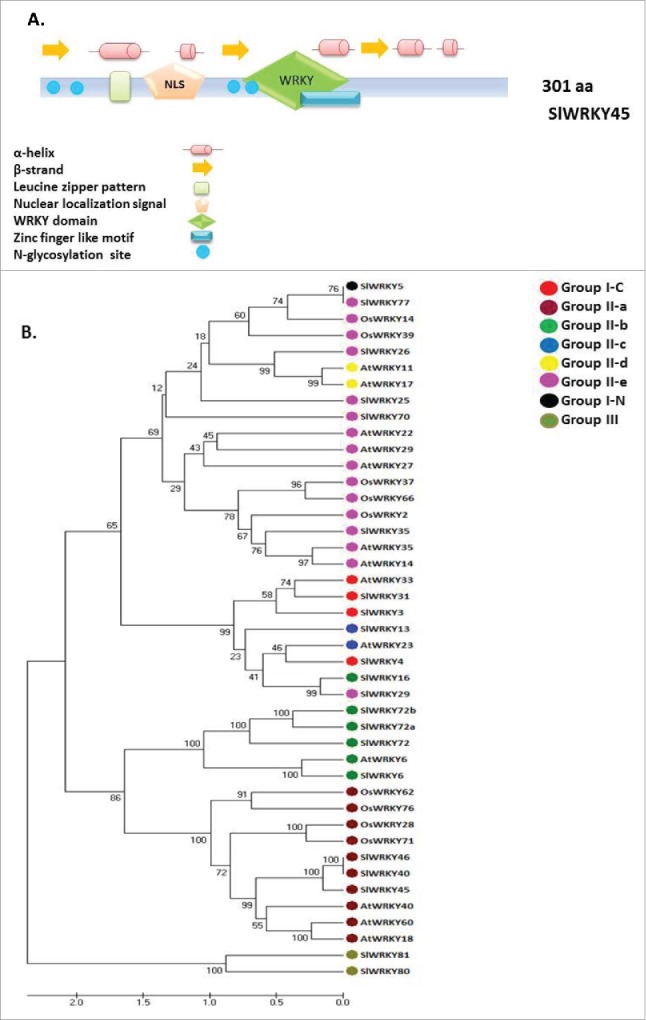
Tomato SlWRKY45 protein structure and phylogenetics. A. Distribution of conserved motifs of the WRKY proteins along WRKY45 protein sequences. The colored boxes placed along the protein sequences are explained in the legend within the bottom of the Figure. A putative nuclear localization signal, as predicted by the SignalP program is shown at positions 83 of WRKY45. B. Unrooted phylogenetic tree representing relationships among WRKYs genes from tomato, Arabidopsis and rice. Members of WRKYs genes from tomato (SlWRKYs), Arabidopsis (AtWRKYs) and rice (OsWRKYs) were subjected to phylogenetic analysis. The amino acid sequences of WRKYs proteins were aligned with aid of ClustalW, and the phylogenetic tree was constructed by means of MEGA 7.0 software by the UPGMA method, with 1,000 bootstrap replicates. The percentage bootstrap scores higher than 50% are indicated on the nodes.
Spatio-temporal expression pattern of SlWRKY45 during M. javanica root infection
The full-length SlWRKY45 promoter fragments, 2001 bp, was isolated from tomato DNA and then used for subsequent cloning steps that resulted in the final binary vectors carrying a promoter: GUS fusion product used to generate SlWRKY45::GUS tomato hairy roots reporter gene line by using Agrobacterium rhizogenes-mediated transformation as detailed in Materials and Methods. To study the expression profiles of SlWRKY45 following inoculation, root tissues were checked for GUS signal at 2, 5, 15, and 28 dai, with second-stage juveniles and compared with GUS expression in respective to uninoculated root lines (Fig. 2A) SlWRKY45 expression pattern: in uninoculated roots a low basal expression level was observed in the root elongation zone, associated with the end of the xylem bundles (Fig. 2A). Two days after the root line SlWRKY45: GUS was challenged with M. javanica J2s no induction was indicated, as observed by a low GUS signal in the root elongation zone, similar to that in the uninoculated root (Fig. 2B). However, a significant elevation of GUS signal was observed at 5 dai (Fig. 2C, D), and through gall formation and maturation (Fig. 2E, F), and in mature galls at 28 dai (Fig. 2G, H), which indicated a highly increased transcript level of SlWRKY45 as the disease advanced. Thus, for SlWRKY45, GUS line confirmed the upregulation observed in inoculated root tissue, compared with uninoculated root tissue, that was found by the RNAseq analysis (Table 1).
Figure 2.
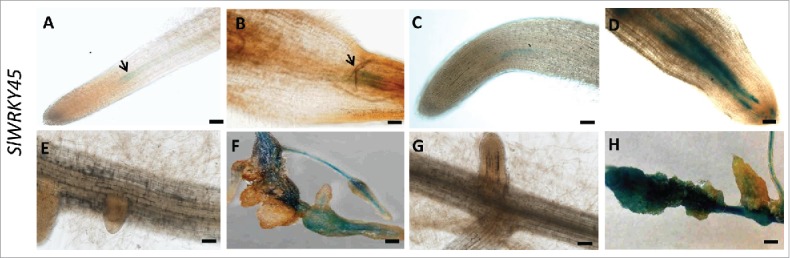
Microscopic analysis of β-glucuronidase (GUS) expression patterns of root-knot nematode (RKN)-infected tomato roots harboring the SlWRKY45 promoter-GUS fusion construct. (A) Uninoculated root harboring the WRKY45:GUS fusion construct exhibited a basal mild GUS signal in the xylem bundles at the root elongation zone; (B) Roots at 2 dai (C) Uninfected roots at 5 d (D) Infected roots at 5 dai. (E) Uninoculated roots at 15 d (F) Developing galls at 15 dai (G) Uninoculated roots at 28 d (H) Mature galls at 28 dai. Arrows indicate nematode. (A-E, G): micrographs as viewed under light microscope. (F, H): bright-field image of galls photographed through a stereomicroscope. Bars: (A-EG) 50 μm; (FH) 500 μm.
SlWRKY45 expression is induced upon feeding site development and maintenance
To gain further insights concerning SlWRKY45 expression during nematode infection thin sections of galls expressing WRKY45 promoter-GUS constructs at 15 and 28 dai were analyzed. Thin sections of galls of SlWRKY45:GUS lines clearly show high expression of WRKY45 at 15 and 28 dai, as indicated by a remarkable GUS signal associated with the layer of pericycle cells adjacent to the protoxylem poles and surrounding the developing giant cells, as observed by light- and dark-field microscopy. Feeding sites associated with the developing nematodes at 15 and 28 dai also were observed to have strong GUS signal in giant cells (Fig. 3A, B), which might indicate that SlWRKY45 continued to function through this period of nematode development.
Figure 3.
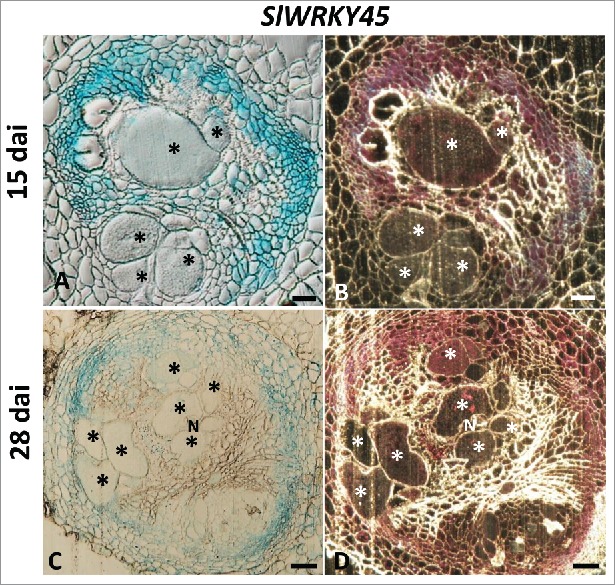
Activity of WRKY45 promoter within thin sections of galls analyzed by GUS staining in representative transformed tomato lines infected with Meloidogyne javanica. Microscopic analyses of β-glucuronidase (GUS) activity in cross-sections of tomato root gall expressing WRKY45 promoter-GUS constructs at 15 and 28 dai. At 15 and 28 dai all observed giant cells were already mature. ((A, B) Galls sections induced on WRKY45:GUS line at 15 dai. (C, D) Galls sections induced on WRKY45:GUS line at 28 dai. (N) The female body of the nematode can be seen at the edge of the giant cells (*). Bars, 200 μm. GUS staining is observed as blue color in whole mounts, and as a red precipitate in the dark field micrographs of the sections.
WRKY45:GUS fusion transgenic root lines showed an early response to wounding
We investigated the effects of wounding on the respective expressions of SlWRKY45 genes in tomato roots. Fig. 4 shows that line harboring the SlWRK45:GUS construct showed a remarkable increase of the GUS signal associated with the vascular bundles as early as 9 h after wounding, and this signal remained at 24 h after wounding (Fig. 4A-C). These results suggest that WRKY45 is a transcriptional factor that, among other functions, regulates early gene expression in response to wounding. Thus, transcription of the GUS fusion was found not only at the local wound site but, moreover, also was induced systemically in the vasculature of distant unwounded tissue.
Figure 4.
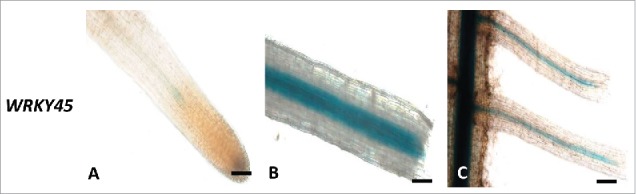
Wounding-induced expression of WRKY45 promoter activity. (A) Intact WRKY45:GUS root line. (B) WRKY45:GUS root line at 9 after wounding (C) at 24 h after wounding (Bars, 100 μm).
Effects of plant hormones on SlWRKY45 gene expression in tomato roots
To investigate the subtle impact of plant hormones salicylic acid (SA), jasmonic acid (JA), methyl jasmonate (MeJa), N6-benzyladenine (BA), indole-3-acetic acid (IAA), and indole-3-butyric acid (IBA) on WRKY45 gene expression in tomato roots, the histochemical GUS bioassay was applied to an intact root reporter line. For that purpose plant hormones were applied to 2-week-old dark-grown tomato roots, and GUS expression was monitored in these treated roots. Fig. 5 shows the results of this study: GUS signal in WRKY45:GUS line was induced by SA only at 5 µM concentration (Fig. 5a-c). The promoters of WRKY45 responded to IBA treatment, as shown by a very strong GUS signal that was observed in roots of WRKY45 at 10 µM (Fig. 5d-f). Application of exogenous IAA increased the promoter activities of WRKY45 (Fig. 5g-i). Upon application of the cytokinin BA, promoter activities were clearly indicated in the vascular tissue of the root tip in root line WRKY45 at 0.1 and 0.5 µM, (Fig. 5j-l). These observation strongly suggest that BA signaling pathways are in part mediated by SlWRKY45 TF. Among the tested jasmonates neither MeJA nor JA induced expression of GUS reporter gene at the root tips of WRKY45:GUS line; in fact, repression of WRKY45 by JA and MeJa is indicated by disappearance of the basal GUS signal at the root tip (Fig. 5m-r). Analysis of mature root tissue of WRKY45 GUS lines showed a similar tendency in WRKY45 GUS responses (Fig. S1) except for line SlWRKY45:GUS, in which expression of WRKY45 was observed in the vasculature upon exposure to MeJA treatment (Fig. S1 p-r).
Figure 5.
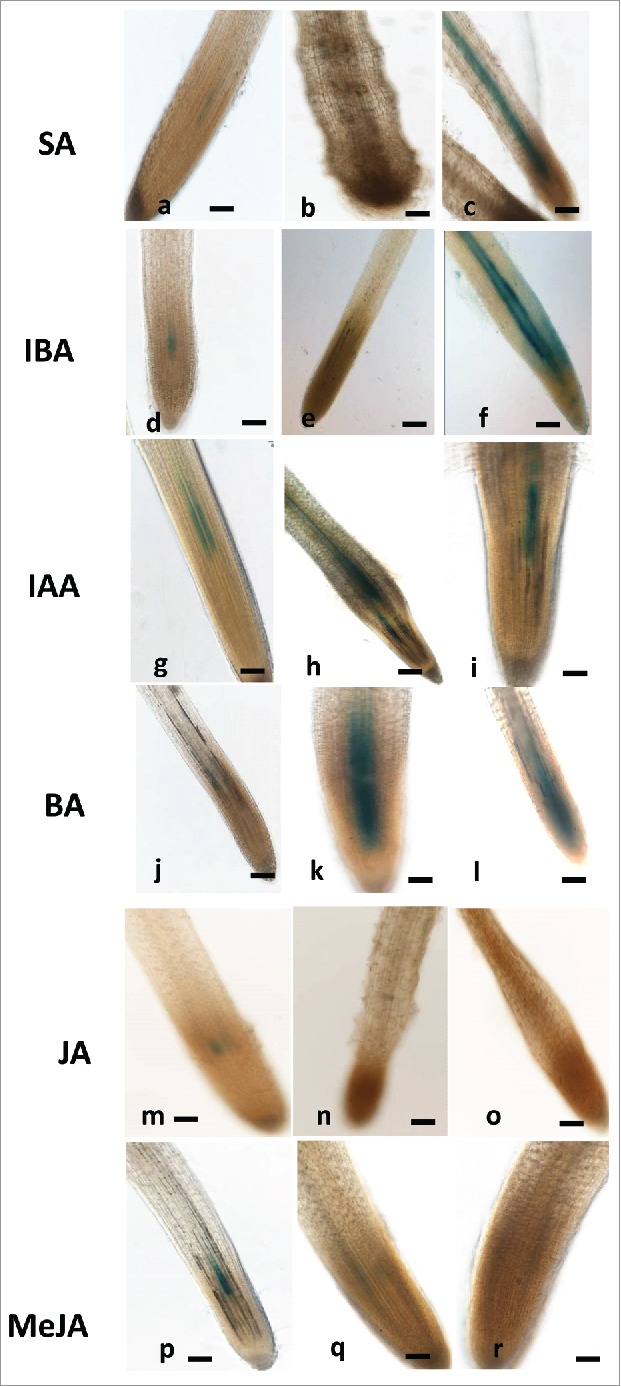
Effect of exogenous phytohormones application on GUS expression in root line WRKY45:GUS. Seven-days-old subcultured roots were transferred to GB as control (a,d,g,j,m,p) or to GB containing SA (1, 5mM) (a-c), IBA (1, 10 µM) (d-f), IAA (1, 5, µM) (g-i), and BA (0.1, 0.5, µM) (j-l) JA (10, 20 µM (m-o), MeJA (0.01, 0.1mM (p-r) for 16 hr before root staining. GUS was detected histochemically and roots were monitored. The figures are representative of at least 5 independent experiments. Scale bar = 0.5 mm.
Overexpressing of WRKY45 promotes RKN M. javanica infection
To investigate whether SlWRKY45 is involved in regulation of plant responses to nematode infection, a plant binary vector containing a 902-bp SlWRKY45 ORF under the control of the CaMV 35S promoter was introduced into tomato by using A. rhizogene-mediated transformation. A total of 20 kanamycin-resistant putative transformants were obtained and maintained on selective media, the resulting transgenic plants did not show phenotypic change compared with the WT. Presence of the transgenes in the transformed plants was confirmed by PCR analysis using gene-specific primers. From all of the independent kanamycin-resistant transgenic lines harboring the 35S:SlWRKY45 construct, two lines (oe:wk-03, oe:wk-04) with high SlWRKY45 transcript levels according to qRT-PCR analysis were selected for further analysis (Fig. S2). To determine the effect of SlWRKY45 overexpression on disease development in tomato, hairy roots from transgenic and pControl roots were inoculated with J2s, and the nematode developmental stages were monitored at 28 dai (Fig. 6). Disease development, as indicated by galling on oe:wk-03, oe:wk-04 and Control lines is shown in Fig. 6A; it indicates some advantages that could be seen on overexpressing root lines. Regarding nematode development: at 28 dai, compared with control roots, increased numbers of nematodes molted into the female stages in root lines oe:wk-03 and oe:wk-04 (Fig. 6B). Thin sections of overexpressing lines did not indicate any alteration in numbers of giant cells (Fig. 6C); however, it was observed that at 28 dai the area of giant cells in the overexpressing lines oe:wk-03 and oe:wk-04 increased, suggesting that the process of gall formation was accelerated in these lines (Fig. 6C, D). These results indicate that overexpression of SlWRKY45 resulted in accelerated disease development.
Figure 6.
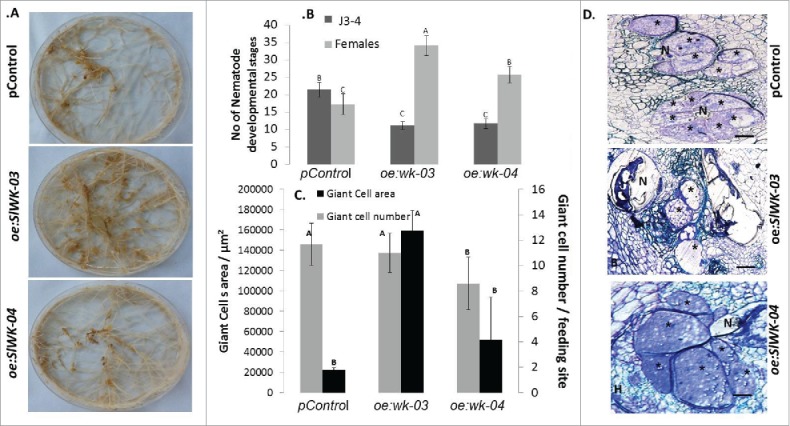
Overexpression of SlWRKY45 in tomato hairy roots promote RKN M. javanica development. A. Increased susceptibility of tomato hairy roots expressing SlWRKY45 (oe:wk-03, oe:wk-04) is accompanied by increased galling occurrence compared with pControl line. B. Meloidogyne susceptibility/resistance of transgenic tomato roots expressing SlWRKY45 compared with pControl line, All roots lines were inoculated with 300 sterile pre-parasitic J2s and the infected roots were assessed for J3 and J4 and mature females development at 28 dai through observation under the dissecting microscope following staining with acid fuchsin dye. Note the significant (P < 0.05) increase in percentage of mature females in oe:wk-03 and oe:wk-04 root lines in comparison with vector control roots. Data are expressed as means of 25 plants from each line; the experiment was repeated 3 times, giving consistent results. The percentage of each developmental stage is represented by a mean standard error. Different letters above the bars denote a significant difference (P ≤ 0.05, analysis of variance) between hairy roots lines analyzed by Tukey-Kramer multiple comparison tests. C. Longitudinal sections of Meloidogyne javanica feeding sites developed in root lines oe:wk-03, oe:wk-04 and pControl. Average GC area was measured on 50 GC systems and average number of GCs of each feeding site system as measured on 60 gall cross sections for each root line and measurements are given as mean ± standard error. Different letters above the bars denote statistically significant differences (P ≤ 0.05, analysis of variance) determined by Tukey-Kramer multiple comparison tests. Giant-cell (GC) area of plants carrying the SlWRK45 gene were more extensive compared with pControl line. D. Thin sections of oe:wk-03, oe:wk-04 and control line were stained with toluidine blue; * = GC, N = nematode, bar = 100 μm.
Increased susceptibility of SlWRKY45-overexpressing roots line induce alteration in hormone-responsive gene expression
To evaluate the contributions of biosynthesis and regulation of hormones, including JA, SA, BA, and IAA, to the observed increased susceptibility of oe:wk-03- and oe:wk-04-overexpressing roots, expression of a set of gene markers involved in these various pathways was investigated by means of qRT-PCR. Transcription of genes whose induction patterns are often used as molecular markers for the activation of the SA signaling pathways were analyzed; as pathogenesis-related (PR-1; accession no. M69247) and phenylalanine ammonia lyase (PAL5; accession number M90692.1). Although no differences between PAL5 expression in pControl and oe:wk-03-infected lines were observed, a significant decrease was observed in PR-1 expression in the oe:wk-03-infected line compared with the pControl line (Fig. 7). The expression of the JA biosynthesis genes OPR (accession number A1486721) did not show any changes in its profile, whereas the JA marker gene proteinase inhibitor II (Pin2; accession numberL21194) clearly exhibited repression in oe:wk-03 compared with pControl, following infection. Next, the cytokinin response factors (CRFs) – part of the cytokinin signal transduction pathway – were analyzed. Both CRF1 (accession number NM001247062) and CRF6 (accession number XM004241080) have been downregulated in oe:wk-03 compared with pControl, following infection, which suggests suppression of the cytokinin signaling pathway in the overexpressing infected line (Fig. 7). For all qRT-PCR analyses, transcripts were normalized against the geometric mean of the expression levels of the 3 most stable endogenous tomato reference genes: 18S, β-actin, and tubulin. Overall, our results suggest that overexpression of WRKY45 decreased transcript accumulation of 3 marker genes: PR1, Pin2 – both associated with defense-related pathways – along with suppression of the cytokinin signaling pathway, as indicated by suppression of CRF1 and CRF6.
Figure 7.
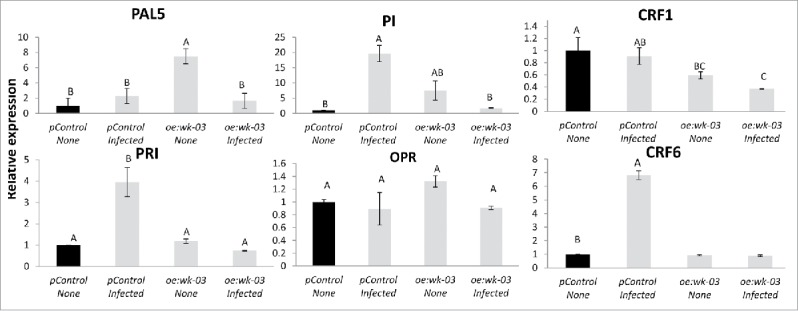
Analyzing the effect of WRKY45 overexpression in manipulating hormonal pathways response. Expression level of defense-related target genes in oe:wk-03 expressing root line compared with vector control prior and 5 dai with Meloidogyne javanica. Total RNA was prepared from pControl transformed control roots and roots expressing SlWRKY45 with/without infection. The graph shows the mean and standard error of the relative amount of transcripts of these genes in SlWRKY45 expressing roots (oe:wk-03) in comparison with vector transformed control roots (pControl) growing under the same conditions (vector control expression level set at zero). All target genes were normalized using the normalization factor calculated as the geometric mean of the expression levels of 3 tomato endogenous reference genes 18S, β-actin and β-tubulin. Each reaction was performed in triplicate and the results represented the mean of 2 independent biologic replicates. Statistical significance of the differences between oe:wk-03 and pControl transformed control roots were determined by Tukey-Kramer multiple comparison test, and significant differential expression (P≤ 0.05) is indicated with asterisks. The experiments were repeated three times with similar results.
Discussion
Networks of WRKY transcription factors as part of signaling machinery regulating RKN M. javanica disease progress in tomato
To further understand the role of WRKY TFs during the tomato/RKN M. javanica interaction, data from recent transcriptome-wide analyses21 were examined for WRKY genes induced or repressed following nematode infection. Our present analysis revealed that among the 81 WRKY members that were previously found in tomato,30 19 WRKYs members have been identified, through RNAseq analysis of tomato (Solanum lycopersicum), as differentially expressed transcripts upon RKN infection. To date, involvement of WRKYs TF in RKN disease development in tomato has been demonstrated, mainly in relation to the Mi resistance gene: both paralogous SlWRKY72a and SlWRKY72b as well as SlWRKY70 were shown to contribute to basal immunity in tomato required for implementing gene-for-gene resistance mediated by the tomato Mi-1 gene.29,30 Other WRKYs implicated in plant-host/nematode interactions were mainly concentrated in Arabidopsis and the cyst nematode Heterodera schachtii systems.33 Amjad et al.35 showed that out of the 59 WRKYs present on the ATH1 GeneChip, 28 were significantly downregulated and 6 upregulated, as compared with control root segments. It is intriguing that a similar tendency was observed in the present study: a higher proportion of downregulated than upregulated SlWRKYs transcripts following M. javanica infection –13 downregulated and 6 upregulated genes compared with control uninoculated root segments (Table 1). In light of their structural and phylogenetic features, the differentially regulated WRKYs transcripts were all classified among 8 groups and subgroups: I-C, I-N, II-a, II-b, II-c, II-d, II-e, and III. Among the differentially expressed WRKYs a high representation of WRKYs that belong to subfamily II was observed. Interestingly, in rice OsWRKYs,34 a higher percentage of rice WRKY Group II proteins are involved in plant defense responses to pathogens, among them transcriptional activator or repressor regulators of the immune response. Thus, the finding that the majority of differentially expressed SlWRKYs genes belonged to group II (Table 1) provides further confirmation of their possible function in mediating plant defense responses against plant parasitic nematodes in tomato. In support of this notion, other WRKYs that were implicated in regulating plant responses to nematodes, and that were assigned to Group II include AtWRKY23 (Group II-c),29 AtWRKY11, AtWRKY17 (Group II-d+e), and AtWRKY6 (Group II-b).33 An additional member of Group II-c, AtWRKY48 was shown to act as a repressor of basal defense in Arabidopsis, therefore its upregulation would favor of nematode development.35 Phylogenetic analysis following alignment of all differentially expressed SlWRKYs against other WRKYs revealed that several differential SlWRKYs were clustered along with other studied WRKYs whose involvement in the plant response to nematodes was already deciphered. For instance, SlWRKY72 exhibited 78% identity to SlWRKY72a, which was shown to be transcriptionally upregulated during disease-resistance-mediated Mi-1, which, in turn, boosts basal defense responses.29,36 Another member of WRKY, SlWRKY3 that was shown to be upregulated during infection was clustered with AtWRKY33; both belong to group I-C. Previously, Zheng et al.39 have shown that AtWRKY33 functioned as a positive regulator of JA/ET-mediated defense-response signaling, and a negative regulator of the SA-mediated defense response. In relation to nematode infection, Amjad et al.35 have shown that overexpression of AtWRKY33 with various promoters resulted in enhanced resistance against H. schachtii, whereas the WRKY33 mutant became more susceptible. As already well documented, JA and SA are important signaling molecules that are involved in plant-defense responses along with other phytohormones, all of which might influence the outcome of nematode-caused diseases.37-39 Among the Arabidopsis WRKYs that are the most closely clustered with differentially regulated SlWRKYs are many that participate in various hormone signaling pathways or whose genes are regulated in response to various phytorhormones.40 For instance, Arabidopsis WRKY18, 40 and 60, which clustered to subgroup II-a, participate in JA, SA, and ABA signaling. These results highlight the possible function of differentially regulated SlWRKYs TFs in regulating phytohormone-mediated signal pathways during nematode infection.
In the present study we chose to focus on SlWRKY45WRKY TF from tomato that was shown to be upregulated upon infection, and about which there was no previously known information:. The GUS histochemical bioassay supported RNAseq results and provided further evidence that SlWRKY45 participate in the parasitic interaction. By performing wounding experiments on promoter:GUS fusion lines we have shown that SlWRKY45 TF might be involved in the wound response accompanying the infection process. However, the long duration of GUS signal suggests that SlWRKY45 member has additional function(s). Similar studies have shown that several wound-responsive WRKY genes are also regulated by pathogen infection.41,42
SlWRKY45
For WRKY45-overexpressing mutant, compared with the control line, we found significant increases in apparent galling formation and number of developed females, and also in the overall feeding site area, as measured on thin sections of galls of oe:wk-03 and oe:wk-04 overexpression lines. Although, hitherto, no study has been focused on understanding the role of SlWRKY45, in the present study we found high similarity among the function of SlWRKY45 and other WRKYs members from Arabidopsis and rice, which are all clustered to the subfamily WRKYs II-a. For example, we have shown here that similarly to its relative member OsWRKY71 from rice,43–45 SlWRKY45 might be involved in cytokinin regulation. Present findings clearly show that SlWRKY45 expression was induced by application of the cytokinin N6-benzyladenine (BA) as indicated by the high GUS signal in the WRKY45:GUS line that followed exposure to the cytokinin treatment. Furthermore, expression of cytokinin-responsive factors encoding genes CRF1 and CRF6 were significantly repressed in the nematode-infected line oe:wrky-03 overexpressing SlWRKY45, compared with infected control lines, which suggests that WRKY45 acts as a transcriptional repressor of these cytokinin-responsive factors.
The necessity for cytokinin signaling in regulating RKN disease has led to the speculation that nematodes produce cytokinin(s) that might be injected into plants to establish parasitism.46 These findings were further supported by Kammerhofer et al.40 who provided genetic evidence that nematode-derived cytokinin is involved in activating the host cell cycle during infection.47,48 In a more recent study Shanks et al.55 found that Arabidopsis lines with reduced cytokinin sensitivity showed reduced susceptibility to nematode infection, indicating that cytokinin signaling is required for optimal nematode development. Taken together, it might be suggested that plant-parasitic nematodes carry the ability to synthesize and secrete a functional plant hormone and thereby manipulate WRKYs TF transcription. Similar to our observation of increased susceptibility of oe:wk-03 and oe:wk-04 lines overexpressing WRKY45, Liu et al.56 showed that closely related members from rice OsWRKY62 and OsWRKY76 enhanced plant susceptibility to the blast fungus Magnaporthe oryzae and to the leaf blight bacterium Xanthomonas oryzae pv. oryzae. Moreover, Liu et al.56 demonstrated that knocking out both OsWRKY62 and OsWRKY76 also elicited greatly increased expression of defense-related genes, and the resulting accumulation of phytoalexins led to increased resistance.
Conclusion
We have provided evidence that during RKN Meloidogyne javanica infection of tomato roots expression of several WRKYs TFs was changed; however, the contribution of each WRKY to promotion of nematode development is not yet known. Given that plant-parasitic nematodes depend on prolonged nutrient delivery by the host, it is tempting to speculate that this requires continuous repression of defense pathways, through activation of repressive activity. In general, increased transcript abundance of SA- and JA-responsive genes is essential for inducing the resistance conferred by the two signaling pathways.49–51 Our present results suggest that upregulation of WRKY45 in feeding sites and in associated neighboring dividing cells might, suggest this member is involved in repressing plant-defense responses to nematode infection. Moreover, given that WRKY45 was continuously expressed from 5 dai onward, it might be deduced that WRKY45 could be activated – directly or indirectly – by nematode secretions. Taken together, these findings indicate that the prominent role of SlWRKY45 in signaling during nematode accommodation in plant roots provides a promising target for manipulation. In future studies, it will be intriguing to identify plant components that directly interact with WRKY45.
Materials and methods
WRKYs comparisons, domains and phylogenetic analyses
Conserved secondary structure features along with predicted potential motifs for SlWRKY45 protein were analyzed by using PHD,52 Jpred algorithms,53 and the MEME (Multiple Expectation Maximization for Motif Elicitation) program 4.11.1, available at http://meme-suite.org/tools/meme.
The SignalP (http://www.cbs.dtu.dk/services/SignalP) program was used to determine the subcellular localization of both WRKYs genes. WRKYs sequences from Arabidopsis, rice, and tomato were obtained by applying BLAST searches to a range of available online databases, such as Sol Genomics Network (https://solgenomics.net/), the Arabidopsis Information Resource TAIR (https://www.arabidopsis.org/), the Rice Gene annotation Project (http://rice.plantbiology.msu.edu/), and the NCBI (www.ncbi.nlm.nih.gov). Phylogenetic analysis was inferred from the unweighted pair grouping by means of the arithmetic-averaging (UPGMA) method.54 The optimal tree with the sum of branch length = 8.36747947 is shown (Fig. 1); the percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those used for the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by the Poisson correction method and are expressed in terms of the number of amino acid substitutions per site. The analysis involved 43 amino acid sequences. All positions with less than 100% site coverage were eliminated, i.e., we allowed only positions with less than 0% of alignment gaps, missing data, or ambiguous bases, and there was a total of 77 positions in the final data set. Evolutionary analyses were conducted in MEGA7.55 All WRKYs accession numbers and SolyC IDs that were used for the phylogenetic analyses comprise the following: SlWRKY46(Solyc08g067340.2.1), SlWRKY40(Solyc06g068460),SlWRKY45 (Solyc08g067360.2.1), SlWRKY6 (Solyc02g080890.2.1), SlWRKY72 (Solyc02g067430.2.1), SlWRKY72b(GU017422), SlWRKY72a (GU017421), SlWRKY3 (Solyc02g088340.2.1), SlWRKY13 (Solyc04g051540.2.1), SlWRKY35 (Solyc02g021680.2.1), SlWRKY4(Solyc01g079260.2.1), SlWRKY31(Solyc05g053380.2.1), SlWRKY16 (Solyc07g056280.2.1), SlWRKY29(Solyc12g011200.1.1), SlWRKY26 (Solyc02g093050.2.1), SlWRKY25 (solyc09g066010.2.1), SlWRKY5 (Solyc10g007970.1.1), SlWRKY77(Solyc10g007970.1.1),SlWRKY81 (Solyc09g015770.2.1), SlWRKY80 (Solyc03g095770.2.1), SlWRKY70 (Solyc05g014040.1.1); from Arabidopsis thaliana: AtWRKY40 (AT1G80840.1), AtWRKY60 (AT2G25000.1), AtWRKY18 (AT4G31800.1), AtWRKY6 (AT1G62300), AtWRKY33 (AT2G38470.1), AtWRKY23 (AT2G47260.1), AtWRKY11 (AT4G31550), AtWRKY17(AT2G24570), AtWRKY29 (AT4G23550), AtWRKY27 (AT5G52830.1), AtWRKY22 (AT4G01250), AtWRKY35 (AT2G34830), AtWRKY14 (AT1G30650); and from rice: OsWRKY76 (ABC02813), OsWRKY28 (BK005031.1), OsWRKY71 (BAF80893), OsWRKY62 (ABC02810), OsWRKY14 (DAA05079), OsWRKY39 (DAA05104), OsWRKY2 (AHM24028), OsWRKY3 (DAA05068), OsWRKY66(DAA05131).
Nematode infection procedure
Meloidogyne javanica was propagated on greenhouse-grown tomato Solanum lycopersicon cv. Avigail (870). Nematode egg masses were extracted from roots with 0.05% (v/v) sodium hypochlorite (NaClO), followed by sucrose flotation.56 For sterilization, the eggs were placed on a sterile WhatmanH filter holder (Whatman International, Dassel, Germany) with a cellulose acetate filter membrane of pore size 5 µm (Sartorius Stedim Biotech, Gottingen, Germany). Eggs on the filter were exposed for 10 min to 0.01% (w/v) mercuric chloride (HgCl2) (Sigma-Aldrich, St Louis, MO, USA), followed by 0.7% streptomycin solution (Sigma-Aldrich, St Louis, MO, USA), and three washes with 50 mL of sterilized distilled water.57 Sterile eggs were then collected from the membrane and placed on 30-µm-opening sieves in sterile 0.01 M MES (Sigma-Aldrich, St Louis, MO, USA) buffer under dark condition for 3 d. Freshly hatched preparasitic J2s were then collected in a 50-mL Falcon tube. For nematode infection tests, wild-type (WT) tomato roots and transgenic lines, growing on standard Gambourg's B5 salt medium (Duchefa, Haarlem, the Netherlands) were inoculated with 300 sterile freshly hatched M. javanica preparasitic J2s. The plates were left uncovered in a laminar Air flow hood until water had completely soaked into the medium58 The inoculated and uninoculated roots were left to grow horizontally in darkness, and root samples were taken for either RNA extraction or assessment of nematode development at predesignated time points.
Plant materials and growth conditions
Tomato (Lycopersicon esculentum) cv Avigail 870 was used as the background line for the transformation. Tomato seeds were treated with 1.6% NaOCl for 10 min with continuous shaking, washed with sterile water for 5 min, and placed on standard-strength Gambourg's B5 medium supplemented with 2% sucrose and 0.8% Gelrite agar (Duchefa, Haarlem, the Netherlands). The plates were kept in darkness at 26°C for 2 days, followed by 2 weeks under a 16/8-h photoperiod until cotyledons emerged. They then were used immediately for cocultivation.
Plasmid construction and generation of transgenic hairy roots
All PCR amplifications used for plasmids construction were performed with the Recombinant Taq DNA polymerase (Thermo Scientific, Paisley, UK) according to the manufacturer's instructions. For amplifying SlWRKY45 promoter region tomato genomic DNA was used as a template for PCR reactions, with the following primers: WRKY45 Promo F1 (5′- GTTAATCGTCATGGAGCTCCTCGTATAAG -3′) and WRKY45 Promo R1 (5′- CCAGTCAGCTCCCGGGTGAATCTATAAGAAAAATTGTCC -3′) for the 2001-bp WRKY45 amplicon. These were designed to create the SacI and SmaI restriction sites, respectively, at the 5′ and 3′ ends of the promoter. The SmaI restriction site was placed before the ATG start codon of the GUS-encoding gene to guarantee the correct reading frame when the promoter was fused to the GUS gene. WRKY45 promoter amplicons were then cloned into the pUC19_Y vector59 at the SacI and SmaI restriction sites. The whole cassettes containing the specific gene promoters and the GUS reporter gene were then isolated by restriction digestion with SacI and SalI, and cloned into the pCAMBIA2300 binary vector.60 The identity, orientation, and junctions of the resulting pCAM–WRKY45:GUS constructs was confirmed according to its digestion patterns. The pCAMBIA2300 empty-vector control and both constructs were subsequently used for Agrobacterium rhizogenes-mediated transformation, as described below. For overexpression of SlWRKY45 the coding sequence was amplified from cDNA of total RNA isolated from 1-month-old tomato plants by using gene-specific primers with the addition of a KpnI restriction site in the forward primer (OE WRKY45Fp1 5′-GTATGGGTACCATGGATACAAACTTGG) and Hind III in the reverse primer (ExRp1–5′- CCTAGAAGCTTTTATATAATTTTTTGCATTTGA-3′), respectively. A full-length, 902-bp SlWRKY45 PCR product was cloned in pHANNIBAL vector with KpnI and HindIII restriction enzymes. The 3067-bp fragment carrying the CaMV 35S Promoter with SlWRKY45 coding sequence and OCS terminator were digested with Not I restriction enzyme, which then was subcloned into pART27 vector.61 The identity, orientation, and junctions of the resulting pART27+OEWKRY45 construct were confirmed by digestion. Both the empty control pART27 and the constructed pART27 + OEWKRY45 plasmids were subsequently used for Agrobacterium rhizogenes-mediated root transformation as described below.
Agrobacterium rhizogenes-mediated root transformation and production of hairy root cultures
The binary vectors for promoter analysis, d pCAM–WRKY45:GUS, together with pART27+OEWKRY45 and empty vector control pART27 and the pCAMBIA2300 empty-vector control, were transferred into Agrobacterium rhizogenes ATCC 15834 by electro-transformation.62 Individual cotyledons were excised from 15- to 20-day-old tomato seedlings and immersed in a 2-day-old A. rhizogenes suspension for incubation at 28°C for 2 h, with agitation at 100 rpm. The excised cotyledons then were placed on a standard-strength Gambourg's B5 salts medium for 3 d for co-cultivation, and then were transferred to B5 agar media supplemented with the antibiotics kanamycin at 50 mg/mL (Duchefa, Haarlem, the Netherlands) and timentin (15:1) at 300 mg/mL (Duchefa, Haarlem, the Netherlands). After 7–10 d of incubation in darkness at 25°C roots emerged from the wounded surface of the cotyledons. Hairy roots were transferred to Gamborg's B5 medium containing 0.8% gelrite and kanamycin at 50 mg/mL. For nematode infection experiments, transformed roots were subcultured in antibiotic-free media for 2 weeks, 300 freshly hatched sterile M. javanica juveniles then were used to inoculate transgenic root lines, and root samples were taken at designated time points, for GUS assessment or for disease evaluation. The presence and expression of transgenes in the tomato hairy roots overexpressing WRKY45 was confirmed through qRT-PCR analysis. with the primer sets FWRKY45 and RWRKY45, as shown in Table S1.
Histochemical localization and microscopic analysis of GUS activity
Two-week-old promoter GUS hairy root lines were inoculated as described above, and assayed histochemically for GUS activity at the designated times after infection. A set of uninfested plants served as controls. For GUS assays, infected and uninfected transgenic root tissues were removed from the Petri dishes at specific time points after inoculation, and were infiltrated with GUS staining buffer containing 50 mM sodium phosphate pH 7.0, 10 mM EDTA, 5 mM K4[Fe2(CN)6], 5 mM K3[Fe2(CN)6], 0.2% (v/v) Triton X-100, and 2 mM 5-bromo-4-chloro-3-indolyl β-D-glucuronide (X-Gluc). GUS staining was performed for 12 h at 37°C, followed by 2 washes in water. At each designated time point at least 15 infected or uninfected transgenic roots were assayed for GUS expression. To determine the cellular localization of GUS expression in developing GCs, semi-thin gall sections were investigated microscopically according to Mitchum et al.63 For sectioning, a GUS-stained roots were fixed in 0.25% (w/v) glutaraldehyde, 4% (v/v) paraformaldehyde in 50 mM PBS at pH 7.2, and then dehydrated and embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) according to the manufacturer's instructions. Embedded tissues were sectioned at 3 μm and posteriorly mounted in Depex (Sigma-Aldrich, St Louis, MO, USA). For observation and documentation, GUS-stained roots mounted on microscope slides, and semi-thin sections were photographed with either a Leica DMLB light microscope or a Nikon D5 Ri2 microscope equipped with accessories for dark-field illumination (Leica Microsystems, Wetzlar, Germany; Nikon Corporation, Tokyo, Japan). In addition, a Leica MZFLIII stereomicroscope (Leica Microsystems) equipped with a Nikon DS-Fi1 camera was used for GUS observation and documentation.
Evaluation of tomato roots response to nematode infection
To monitor nematode development in control and SlWRKY45-overexpressing root lines, root systems grown in monoxenic culture were harvested at 28 dai. Infected roots were stained with acid fuchsin solution (Sigma-Aldrich, St Louis, MO, USA) (17.5 mg acid fuchsin, 500 mL ethanol and 500 mL acetic acid) for overnight. Stained roots were washed three times in distilled water and stored according to Bybd et al.64 The destained roots then were mounted with tap water and the galls were dissected under an SZX12 stereo-microscope (Olympus, Shinjuku, Tokyo, Japan). For evaluation of nematode development, the numbers of J3, J4, and mature females, with or without egg masses, were counted at 28 dai. Mean numbers of sedentary J3 and J4 juveniles, and females were obtained by screening 10 replicates in each line. Infection tests were performed twice and similar results were obtained, those of only one experiment are presented. Statistical differences were determined independently for each experiment by all-mean comparison by means of the Tukey-Kramer test at an α level of 0.05, on JMP Pro 10 software (SAS Inc., Cary, NC, USA).
Genomic DNA, RNA extraction and cDNA isolation
Genomic DNA was isolated from 1-month-old soil-grown tomato seedlings, cv Avigail 870by the cetyltrimethylammonium bromide (CTAB) method described by Goetz et al.65 To obtain the full-length SlWRKY45 cDNA, tomato roots and leaves were homogenized in liquid nitrogen, following total RNAs extraction with the GeneJET Plant RNA Purification Mini Kit (ThermoScientific, Vilnius, Lithuania). To remove contaminating genomic DNA, the RNA samples were incubated in the presence of 10 units of TURBO DNA-freeTM DNASE (Applied Biosystems, Foster City, CA, USA), and the DNA-free RNA was converted into first-strand cDNA with the Verso cDNA Kit (Thermo Scientific).
qPCR analysis
Gene transcripts were quantified by real-time RT-PCR on total RNA extracted from uninoculated and inoculated root lines at 5 dai. Five 200-mg root systems from each corresponding time point were pooled, uninfected roots were used as controls, and RNA was extracted as described above. The qRTPCR reactions used the SYBR-Green ROX Mix (ABgene). All the primers used for gene quantification are shown in Table S1; they were designed with the aid of the PrimerExpress software (Applied Biosystems). The subsequent real-time PCR reaction contained 3.4 µL of the cDNA in a total volume of 10 µL that contained SYBR-Green ROX Mix (ABgene), 150 nM of forward primer, and 150 nM of reverse primer. The reaction was performed in real-time PCR plastic ware (Axygen, Union City, CA, USA). All PCR cycles comprised 2 min at 50°C and10 min at 95°C, followed by 40 2-step cycles of 10 s at 95°C and 1 min at 60°C.
After the PCR reaction, a melting curve was generated by gradually increasing the temperature to 95°C, to test for amplicon specificity. For qPCR a mixture of all the cDNAs was used for all treatments, as a template for calibration curves designed for each pair of primers. Three constitutively expressed genes – actin (ACT – GenBank accession number U60482.1); b-tubulin (TUB – GenBank accession number NM_001247878.1); and 18S gene (GenBank accession number BH012957.1) – were the endogenous controls for tomato gene expression analysis (Table S1). Transcript levels were normalized for each sample by comparison with the geometric mean of the corresponding selected housekeeping genes. It previously had been confirmed that all of the housekeeping genes displayed minimal variation across treatments and that they were the most stable housekeeping genes among a set of tested genes in a given cDNA sample.66 Values were expressed as the increase or decrease in levels, relative to a calibration sample. The following control reactions were included: PCR negative control without a cDNA template, to confirm that there were no nonspecific PCR products (NTC); and an NRT control reaction which contained cDNA that had not been subjected to a reverse transcriptase reaction. Statistical differences between treatments and/or root lines were calculated by LSD, according to the Tukey-Kramer multiple comparison test at P ≤ 0.05. For confirmation of all qRT-PCR results, expression of the subset of genes was analyzed in another two independent experiments, which yielded the same results.
Supplementary Material
References
- 1.Kyndt T, Vieira P, Gheysen G, de Almeida-Engler J. Nematode feeding sites: unique organs in plant roots. Planta. 2013;238:807–818. doi: 10.1007/s00425-013-1923-z. [DOI] [PubMed] [Google Scholar]
- 2.de Almeida Engler J, Engler G, Gheysen G. Unravelling the plant cell cycle in nematode induced feeding sites In: Jones J, Gheysen G, Fenoll C, eds. Genomics and Molecular Genetics of Plant-Nematode Interactions: Dordrecht: Springer Science+Business Media, 2011:349–368. [Google Scholar]
- 3.Bartlem DG, Jones MGK, Hammes UZ. Vascularization and nutrient delivery at root-knot nematode feeding sites in host roots. J Exp Botany. 2013;65:1789–1798. doi: 10.1093/jxb/ert415. [DOI] [PubMed] [Google Scholar]
- 4.Gheysen G, Fenoll C. Gene expression in nematode feeding sites. Annu Rev Phytopathol. 2002;40:191–219. doi: 10.1146/annurev.phyto.40.121201.093719. [DOI] [PubMed] [Google Scholar]
- 5.Portillo M, Cabrera J, Lindsey K, Topping J, Andres MF, Emiliozzi M, Oliveros JC, García-Casado G, Solano R, Koltai H, et al.. Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 2013;197:1276–1290. doi: 10.1111/nph.12121. [DOI] [PubMed] [Google Scholar]
- 6.Barcala M, Garcia A, Cabrera J, Casson S, Lindsey K, Favery B, García-Casado G, Solano R, Fenoll C, Escobar C, et al.. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 2010;61:698–712. doi: 10.1111/j.1365-313X.2009.04098.x. [DOI] [PubMed] [Google Scholar]
- 7.Bar-Or C, Kapulnik Y, Koltai H. A broad characterization of the transcriptional profile of the compatible tomato response to the plant parasitic root knot nematode Meloidogyne javanica. Eur J Plant Pathol. 2005;111:181–192. doi: 10.1007/s10658-004-2134-z. [DOI] [Google Scholar]
- 8.Grundler FMW, Sobczak M, Golinowski W. Formation of wall openings in root cells of Arabidopsis thaliana following infection by the plant-parasitic nematode Heterodera schachtii. Eur J Plant Pathol. 1998;104:545–551. doi: 10.1023/A:1008692022279. [DOI] [Google Scholar]
- 9.Hammes UZ, Schachtman DP, Berg RH, Nielsen E, Koch W, McIntyre LM, Taylor CG. Nematode-induced changes of transporter gene expression in Arabidopsis roots. Mol Plant Microbe In. 2005;18:1247–1257. doi: 10.1094/MPMI-18-1247. [DOI] [PubMed] [Google Scholar]
- 10.Jammes F, Lecomte P, Almeida-Engler J, Bitton F, Martin-Magniette ML, Renou JP, Abad P, Favery B. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 2005;44:447–458. doi: 10.1111/j.1365-313X.2005.02532.x. [DOI] [PubMed] [Google Scholar]
- 11.Puthoff DP, Nettleton D, Rodermel SR, Baum TJ. Arabidopsis gene expression changes during cyst nematode parasitism revealed by statistical analyses of microarray expression profiles. Plant J. 2003;33:911–921. doi: 10.1046/j.1365-313X.2003.01677.x. [DOI] [PubMed] [Google Scholar]
- 12.Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Bohlmann H. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 2009;57:771–784. doi: 10.1111/j.1365-313X.2008.03727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanholme B, De Meutter J, Tytgat T, Van Montagu M, Coomans A, Gheysen G. Secretions of plant-parasitic nematodes: a molecular update. Gene. 2004;332:13–27. doi: 10.1016/j.gene.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 14.Gheysen G, Fenoll C. Gene expression in nematode feeding sites. Annu Rev Phytopathol. 2002;40:191–219. doi: 10.1146/annurev.phyto.40.121201.093719. [DOI] [PubMed] [Google Scholar]
- 15.Burley SK, Kamada K. Transcription factor complexes. Curr Opin Struct Biol. 2002;12:225–30. doi: 10.1016/S0959-440X(02)00314-7. [DOI] [PubMed] [Google Scholar]
- 16.Pan YP, Tsai CJ, Ma BY, Nussinov R. Mechanisms of transcription factor selectivity. Trends Genet. 2010;26:75–83. doi: 10.1016/j.tig.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Rodriguez P, Riano-Pachon DM, Correa LG, Rensing SA, Kersten B, Mueller-Roeber B. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38:D822–D827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al.. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 19.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al.. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 20.Alves MS, Dadalto SP, Gonçalves AB, de Souza GB, Barros VA, Fietto LG. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes. 2014;2:85–106. doi: 10.3390/proteomes2010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iberkleid I, Sela N, Brown Miyara S. Meloidogyne javanica fatty acid- and retinol-binding protein (Mj-FAR-1) regulates expression of lipid-, cell wall-, stress- and phenylpropanoid-related genes during nematode infection of tomato. BMC Genomics. 2015;16:272. doi: 10.1186/s12864-015-1426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eulgem T, Tsuchiya T, Wang XJ, Beasley B, Cuzick A, Tor M, Zhu T, McDowell JM, Holub E, Dangl JL. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 2007;49:829–839. doi: 10.1111/j.1365-313X.2006.02999.x. [DOI] [PubMed] [Google Scholar]
- 23.Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Eulgem T. Dissecting the WRKY web of plant defense regulators. PLoS Pathog. 2006;2:e126. doi: 10.1371/journal.ppat.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, et al.. Solution structure of an arabidopsis WRKY DNA binding domain. Plant Cell. 2005;17:944–956. doi: 10.1105/tpc.104.026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 28.Ali MA, Wieczorek K, Kreil DP, Bohlmann H. The Beet Cyst Nematode Heterodera schachtii modulates the expression of WRKY transcription factors in syncytia to favour its development in arabidopsis roots. Plos One. 2014;9:e102360. doi: 10.1371/journal.pone.0102360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T. WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J. 2010;63:229–240. doi: 10.1111/j.1365-313X.2010.04232.x. [DOI] [PubMed] [Google Scholar]
- 30.Atamian HS, Eulgem T, Kaloshian I. SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta. 2012;235:299–309. doi: 10.1007/s00425-011-1509-6. [DOI] [PubMed] [Google Scholar]
- 31.Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al.. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. Clustal-W - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amjad Ali M, Wieczorek K, Kreil DP, Bohlmann H. The beet cyst nematode Heterodera schachtii modulates the expression of WRKY transcription factors in syncytia to favour its development in arabidopsis roots. Plos One. 2014;9:e102360. doi: 10.1371/journal.pone.0102360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng X, Wang H, Jang J, Xiao T, He H, Jiang D, Tang X. OsWRKY80-OsWRKY4 module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice (N Y). 2016;9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing DH, Lai ZB, Zheng ZY, Vinod KM, Fan BF, Chen ZX. Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol Plant. 2008;1:459–470. doi: 10.1093/mp/ssn020. [DOI] [PubMed] [Google Scholar]
- 36.Bhattarai KK, Xie QG, Mantelin S, Bishnoi U, Girke T, Navarre DA, Kaloshian I. Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Mol Plant Microbe In. 2008;21:1205–1214. doi: 10.1094/MPMI-21-9-1205. [DOI] [PubMed] [Google Scholar]
- 37.Kammerhofer N, Radakovic Z, Regis JMA, Dobrev P, Vankova R, Grundler FMW, Siddique S, Hofmann J, Wieczorek K. Role of stress-related hormones in plant defence during early infection of the cyst nematode Heterodera schachtii in Arabidopsis. New Phytol. 2015;207:778–789. doi: 10.1111/nph.13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumari C, Dutta TK, Banakar P, Rao U. Comparing the defence-related gene expression changes upon root-knot nematode attack in susceptible versus resistant cultivars of rice. Sci Rep-Uk. 2016;6:2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nahar K, Kyndt T, De Vleesschauwer D, Höfte M, Gheysen G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011;157:305–316. doi: 10.1104/pp.111.177576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakshi M, Oelmüller R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 2014;9:e27700. doi: 10.4161/psb.27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheong YH, Chang H-S, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Bai X, Wang X, Chu C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol. 2007;164:969–979. doi: 10.1016/j.jplph.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Raines T, Blakley IC, Tsai Y-C, Worthen JM, Franco-Zorrilla JM, Solano R, Schaller GE, Loraine AE, Kieber JJ. Characterization of the cytokinin-responsive transcriptome in rice. BMC Plant Biol. 2016;16:260. doi: 10.1186/s12870-016-0932-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z-L, Xie Z, Zou X, Casaretto J, Ho T-hD, Shen QJ. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004;134:1500–1513. doi: 10.1104/pp.103.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Meutter J, Tytgat T, Witters E, Gheysen G, Van Onckelen H, Gheysen G. Identification of cytokinins produced by the plant parasitic nematodes Heterodera schachtii and Meloidogyne incognita. Mol Plant Pathol. 2003;4:271–277. doi: 10.1046/j.1364-3703.2003.00176.x. [DOI] [PubMed] [Google Scholar]
- 47.de Almeida Engler J, De Vleesschauwer V, Burssens S, Celenza JL, Inzé D, Van Montagu M, Engler G, Gheysen G. Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode-induced galls and syncytia. Plant Cell. 1999;11:793–807. doi: 10.1105/tpc.11.5.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goverse A, Overmars H, Engelbertink J, Schots A, Bakker J, Helder J. Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol Plant Microbe In. 2000;13:1121–1129. doi: 10.1094/MPMI.2000.13.10.1121. [DOI] [PubMed] [Google Scholar]
- 49.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 50.Penninckx I, Eggermont K, Terras F, Thomma B, De Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Penninckx IA, Thomma BP, Buchala A, Métraux J-P, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–13. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rost B. Secondary structure prediction of all-helical proteins in two states. Protein Engineering. 1993;6:831–836. doi: 10.1093/protein/6.8.831. [DOI] [PubMed] [Google Scholar]
- 53.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 54.Sokal RR, Sneath PH. The estimation of taxonomic resemblance. Principles of Numerical Taxonomy. 1963;123-168. doi: 10.2307/1217562. [DOI] [Google Scholar]
- 55.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hussey RS, Baker KR. Comparison of methods of collecting inocula for Meloidogyne spp., including a new technique. Plant Dis Rep. 1973;57:1025–8. [Google Scholar]
- 57.Jansen van Vuuren R, Woodward B. The response of cassava cultivars to root-knot nematode infestation: an in vitro method. Euphytica. 2001;120:109–113. doi: 10.1023/A:1017524210671. [DOI] [Google Scholar]
- 58.Sijmons PC, Grundler FM, Mende N, Burrows PR, Wyss U. Arabidopsis thaliana as a new model host for plant-parasitic nematodes. Plant J. 1991;1:245–254. doi: 10.1111/j.1365-313X.1991.00245.x. [DOI] [Google Scholar]
- 59.Vancanneyt G, Schmidt R, O'Connor-Sanchez A, Willmitzer L, Rocha-Sosa M. Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet. 1990;220:245–250. doi: 10.1007/BF00260489. [DOI] [PubMed] [Google Scholar]
- 60.Remy S, Thiry E, Coemans B, Windelinckx S, Swennen R, Sagi L. Improved T-DNA vector for tagging plant promoters via high-throughput luciferase screening. Biotechniques. 2005;38:763–770. doi: 10.2144/05385RR01. [DOI] [PubMed] [Google Scholar]
- 61.Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al.. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313X.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 62.Shen W-J, Forde BG. Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res. 1989;17:8385. doi: 10.1093/nar/17.20.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitchum MG, Sukno S, Wang X, Shani Z, Tsabary G, Shoseyov O, Davis EL. The promoter of the Arabidopsis thaliana Cel1 endo-1,4-beta glucanase gene is differentially expressed in plant feeding cells induced by root-knot and cyst nematodes. Mol Plant Pathol. 2004;5:175–181. doi: 10.1111/j.1364-3703.2004.00216.x. [DOI] [PubMed] [Google Scholar]
- 64.Bybd DW Jr, Kirkpatrick T, Barker KB. An improved technique for clearing and staining plant tissues for detection of nematodes. J Nematol. 1983;51:142–143. [PMC free article] [PubMed] [Google Scholar]
- 65.Goetz M, Godt DE, Guivarc'h A, Kahmann U, Chriqui D, Roitsch T. Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc Natl Acad Sci. 2001;98:6522–6527. doi: 10.1073/pnas.091097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034-1-0034-11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


