Abstract
There is a critical need for compounds that target cell surface integrin receptors for applications in cancer therapy and diagnosis. We used directed evolution to engineer the Ecballium elaterium trypsin inhibitor (EETI-II), a knottin peptide from the squash family of protease inhibitors, as a new class of integrin-binding agents. We generated yeast-displayed libraries of EETI-II by substituting its 6-amino acid trypsin binding loop with 11-amino acid loops containing the Arg-Gly-Asp (RGD) integrin binding motif and randomized flanking residues. These libraries were screened in a high-throughput manner by fluorescence-activated cell sorting to identify mutants that bound to αvβ3 integrin. Select peptides were synthesized and were shown to compete for natural ligand binding to integrin receptors expressed on the surface of U87MG glioblastoma cells with half-maximal inhibitory concentration (IC50) values of 10-30 nM. Receptor specificity assays demonstrated that engineered knottin peptides bind to both αvβ3 and αvβ5 integrins with high affinity. Interestingly, we also discovered a peptide that binds with high affinity to αvβ3, αvβ5, and α5β1 integrins. This finding has important clinical implications since all three of these receptors can be co-expressed on tumors. In addition, we showed that engineered knottin peptides inhibit tumor cell adhesion to the extracellular matrix protein vitronectin, and in some cases fibronectin, depending on their integrin binding specificity. Collectively, these data validate EETI-II as a scaffold for protein engineering, and highlight the development of unique integrin-binding peptides with potential for translational applications in cancer.
Keywords: protein engineering, directed evolution, tumor targeting agents, integrin binding peptides, yeast surface display, RGD, alternative scaffolds
INTRODUCTION
In cancer, the growth and survival of solid tumors is dependent on their ability to trigger new blood vessel formation to supply nutrients to the tumor cells.1 Several integrin receptors, including αvβ3, αvβ5, and α5β1, have generated clinical interest due to their expression on the surface of cancer cells and the tumor neovasculature, and their proposed role in mediating angiogenesis, tumor growth, and metastasis.2-6 Integrins are a family of adhesion receptors, consisting of α and β subunits that non-covalently associate into heterodimers with distinct ligand binding specificities and cell signaling properties.7 Extracellular matrix (ECM) proteins including fibronectin, vitronectin, osteopontin, laminin, and many others, bind to particular integrin receptors through an Arg-Gly-Asp (RGD) peptide motif.8 The RGD sequence is constrained within a loop structure that presents a particular conformational and stereochemical arrangement of residues critical for optimal integrin binding affinity and specificity.9 In natural ECM ligands, the orientation of the Arg and Asp residues, as well as the residues surrounding the RGD sequence, are critical for mediating binding to certain integrin subtypes, including αvβ3, αvβ5, α5β1, and αiibβ3.10
There are numerous examples of linear peptides,11,12 cyclic peptides,13,14 peptidomimetics,9,15 and antibodies16,17 that have been developed to bind to integrin receptors; however, research exploring the use of small, structured peptide scaffolds for potential drug discovery and development applications has been limited. Here, we developed high affinity integrin binding peptides by engineering optimal RGD-containing epitopes within the solvent-exposed loop of a cystine knot peptide (~3 kDa). Cystine knots, also known as knottins, share a common disulfide-bonded framework and a triple-stranded β-sheet fold, and possess one or more loops that bind diverse targets.18 Knottin family members, which includes protease inhibitors, toxins, and antimicrobials, share little sequence homology apart from their core cysteine residues.19,20As a result, their disulfide-constrained loops tolerate much sequence diversity, which is a critical requirement for protein engineering applications where mutations need to be introduced into a protein without abolishing its three-dimensional fold.
Yeast surface display is a powerful tool for the directed evolution of proteins with altered binding affinity, stability, or expression levels.21,22 We used this technology to engineer mutants of the Ecballium elaterium trypsin inhibitor II (EETI-II)23 [Fig. 1(A)], a 28-amino acid knottin peptide from the squash family of protease inhibitors, that had high affinity binding to integrin receptors. We substituted the 6-amino acid trypsin binding loop of EETI-II with an 11-amino acid loop containing an RGD motif located in different positions along the loop. We generated combinatorial libraries of EETI-II mutants, where the residues flanking the RGD sequence were randomized to be any possible amino acid. These libraries were displayed on the yeast cell surface and screened in a high-throughput manner by flow cytometry to identify mutants that bound tightly to αvβ3 integrin. We discovered several peptides that bound to αvβ3 and αvβ5 integrins with low nanomolar affinity and mediated tumor cell adhesion to the extracellular matrix protein vitronectin. During this process, we also identified a unique peptide that bound to αvβ3, αvβ5, and α5β1 integrins with high affinity and mediated binding to the extracellular matrix proteins vitronectin and fibronectin. Since all three of these integrins can be co-expressed in human malignancies and can contribute to angiogenesis and metastasis, this engineered knottin peptide has potential as a broad spectrum cancer therapeutic or diagnostic agent.
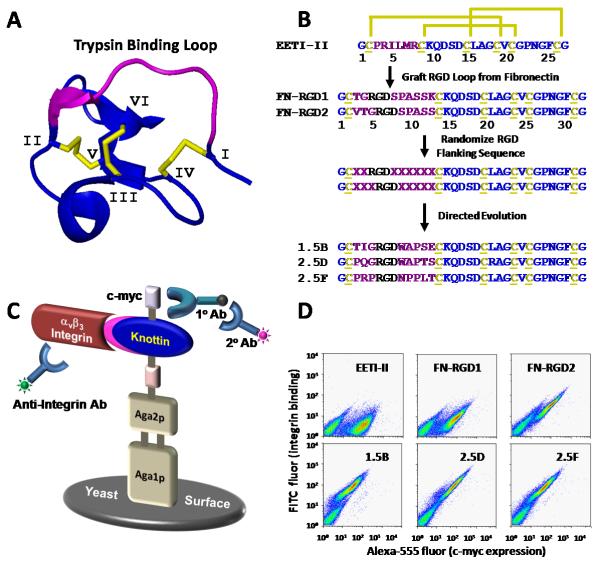
Fig. 1.
EETI-II engineering by yeast surface display. (A) Three dimensional structure of EETI-II (pdb: 2it7). The three disulfide bonds that stabilize the core of the molecule are shown in yellow. Individual cysteine residues are numbered I-VI. (B) Strategy for EETI-II engineering. The primary structure of EETI-II shows the characteristic disulfide bonds of cysteines 1 – 6 (yellow). The sequences of three representative mutants isolated from the first (1.5B) and second (2.5D, 2.5F) rounds of directed evolution are shown. (C) Schematic of the yeast display system. Expression of EETI-II mutants as Aga2p fusions is measured by flow cytometry through detection of a C-terminal c-myc epitope tag using a chicken anti-c-myc antibody and an Alexa 555-labeled secondary antibody. Integrin binding is measured with a FITC-labeled anti-integrin antibody. (D) Flow cytometry dot plots showing c-myc expression (x-axis) and αvβ3 integrin binding (y-axis) of peptides displayed on the surface of yeast.
MATERIALS AND METHODS
Materials, cell lines, and reagents
The U87MG human glioblastoma cell line was obtained from American Type Culture Collection. Detergent-solubilized αvβ3 and αvβ5 integrin receptors (octyl-beta-D-glucopyranoside formulations) and α5β1 integrin (Triton X-100 formulation) were purchased from Millipore, and αiibβ3 (Triton-X100 formulation) was purchased from Enzyme Research Laboratories. 125I-labeled echistatin and c(RGDyK) were purchased from Amersham Biosciences, and Peptides International, respectively. Phosphate buffered saline (PBS) was from Invitrogen. All other chemicals were purchased from Fisher Scientific unless otherwise specified. Integrin binding buffer (IBB) was composed of 25 mM Tris pH 7.4, 150 mM NaCl, 2mM CaCl2, 1 mM MgCl2, 1 mM MnCl2, and 0.1% bovine serum albumin (BSA).
Yeast surface display
The open reading frames of wild-type EETI-II and FN-RGD mutants were cloned by overlapping extension PCR using yeast optimized codons. To create FN-RGD1 and FN-RGD2, the trypsin binding sequence PRILMR of wild-type EETI-II was substituted with TGRGDSPASSK or VTGRGDSPASS from the 10th domain of fibronectin. DNA was subcloned into display plasmid pCT30221 using NheI and BamHI restriction sites, and electroporated into yeast strain EBY100.24 A C-terminal c-myc tag was included for detection of cell surface proteins by flow cytometry. For the first round of directed evolution, PCR-amplified libraries based on FN-RGD1 and FN-RGD2 were constructed with NNS degenerate codons (N = A, T, C, or G and S = C or G), which encoded for all 20 amino acids and the TAG stop codon at each position flanking the RGD motif in the grafted loop. For each library, 40 μg of DNA insert and 4 μg of linearized pCT vector were electroporated into EBY100 electrocompetent yeast for homologous recombination as described.24 The two libraries (~ 5 × 106 transformants each) were combined for a total diversity of ~ 107 clones as estimated by serial dilution plating and colony counting. This library size is much smaller than the theoretical diversity of 208 clones that would be needed to fully sample a loop with eight randomized amino acid residues. For the second round of directed evolution, a biased library (~107 transformants) was generated using degenerate oligonucleotides that mostly encoded for amino acid mutations identified in the first round of directed evolution (theoretical diversity of ~1.3 × 106; Supplementary Tables SI and SII). Individual yeast clones and libraries were grown and induced for protein expression as described.24
Flow cytometry and library screening
Protein expression levels and αvβ3 integrin binding were determined by flow cytometry. Detergent-solubilized αvβ3 integrin was added to yeast cells in binding buffer (PBS with 0.1 mM each of MgCl2, CaCl2, and MnCl2, and 1 mg/mL BSA) for 1.5 h at room temperature. Next a 1:250 dilution of chicken anti-c-myc IgY antibody (Invitrogen) was added for 1 h at 4 °C. The cells were washed with ice-cold binding buffer and incubated with a 1:25 dilution of fluorescein (FITC)-conjugated anti-αv integrin antibody (mAb 13C2, Millipore) and a 1:100 dilution of Alexa 555-conjugated goat anti-chicken IgG secondary antibody (Invitrogen) or phycoerythrinconjugated goat anti-chicken IgY PE (Santa Cruz Biotechnology) for 30 min at 4 °C. Cells were washed and analyzed by dual-color flow cytometry using a BD FACSCalibur and CellQuest software (Becton Dickinson).
Fluorescence-activated cell sorting (FACS) was used to screen yeast libraries for mutants with increased αvβ3 integrin binding affinity. A Vantage SE/DiVa Vantoo instrument (Stanford FACS Facility) was used to select yeast cells with enhanced integrin binding (FITC fluorescence) for a given protein expression level (Alexa 555 fluorescence). For the first directed evolution library, approximately 2 × 107 yeast clones were screened with 500 nM αvβ3 integrin. Collected yeast cells were cultured, induced for expression, and sorted by subsequent rounds of FACS. To increase the sort stringency, integrin concentrations were decreased in later rounds of selection: round 3= 100 nM; rounds 4-6= 50 nM. For the second directed evolution library, yeast clones were screened in a similar manner using lower integrin concentrations: rounds 1-2= 100 nM; rounds 3-4= 30 nM; rounds 5-6= 10 nM. Plasmid DNA was recovered using a Zymoprep kit (Zymo Research), amplified in XL-1 blue supercompetent E. coli cells (Stratagene) and sequenced (Elim Biopharmaceuticals).
Cell surface integrin receptor competition binding assay
A competition binding assay was performed as previously described25 to measure the relative binding affinities of engineered knottin peptides, echistatin, and c(RGDyK). Briefly, 2 × 105 U87MG cells were incubated with 0.06 nM 125I-labeled echistatin and varying concentrations of peptides in IBB at room temperature for 3 h. The cell-bound radioactivity remaining after washing was determined by gamma-counting. IC50 values were determined by non-linear regression analysis using Kaleidagraph (Synergy Software), and are presented as the average of experiments performed on three separate days.
Solid phase integrin receptor competition binding assay
To measure the integrin binding specificity of engineered knottin peptides, a competition binding assay was performed as previously described.26 Briefly, detergent-solubilized αvβ3, αvβ5, α5β1, and αiibβ3 integrin receptors were diluted to a final concentration of 1 μg/mL in IBB. 100 μL aliquots were used to coat wells of Maxisorb plates (NalgeNunc), overnight at 4 °C. The wells were washed and blocked with IBB containing 1% BSA for 2 h at room temperature. 125I-labeled echistatin (0.06 nM) and varying concentrations of unlabeled peptides were incubated in the wells for 3 h at room temperature with gentle rocking, and washed 3 times in IBB. Plate-bound radioactivity was solubilized with 200 μL of boiling 2N NaOH followed by gamma-counting. Each data point represents the average value of triplicate wells, and error bars represent the standard deviation.
Integrin-dependent cell adhesion assay
Cell adhesion assays were performed using Cytomax fibronectin- and vitronectin-coated strips (Millipore) as described27 and according to the manufacturer's protocol. Briefly, coated strips were rehydrated with PBS. Varying concentrations of peptides were added to 105 U87MG cells in 100 μL of IBB, incubated for 2 h at 37 °C, 5% CO2, and washed with Dulbecco's PBS (DPBS, Invitrogen). Remaining adherent cells were incubated with 100 μL of 0.2% crystal violet and 10% ethanol for 5 min at room temperature, washed in DPBS and solubilized with 100 μL/well of a 50:50 mixture of 100 mM sodium phosphate, pH 4.5 and ethanol for 5 min. The absorbance 600 nm was measured using a SPECTRAmax PLUS (Molecular Devices) microtiter plate reader. IC50 values were determined by non-linear regression analysis using Kaleidagraph, and are the average of experiments performed on three separate days. Data was normalized using samples containing no competing peptide.
RESULTS
Loop grafting of fibronectin-derived integrin-binding sequences into EETI-II
Figure 1B outlines our strategy for engineering integrin-binding EETI-II knottin peptides using loop grafting and direction evolution. Amino acids 2-7 of EETI-II form a loop structure that binds to and inhibits trypsin activity28 [Fig. 1(A)]. Previously, several groups demonstrated that other epitopes could be substituted in place of this trypsin binding loop without disrupting the protein fold.29-32 The 10th domain of fibronectin possesses an extended loop containing the RGD recognition motif, which is constrained in a particular conformation for high affinity integrin binding. Using molecular cloning, we grafted sequences comprising this fibronectin loop (TGRGDSPASSK or VTGRGDSPASS) in place of the 6-amino acid EETI-II trypsin binding loop (PRILMR) to recapitulate this integrin binding epitope. These constructs are termed FNRGD1 and FN-RGD2, respectively. Since fibronectin does not contain a disulfide-constrained integrin binding loop for direct transfer into EETI-II, these sequences were selected by measuring the distances between the cysteine residues that constrain the trypsin binding loop of EETI-II (pdb: 2eti) and identifying a relatively comparable distance between residues on the extended fibronectin loop (pdb: 1fna), which was the 11-amino acid sequence TGRGDSPASSK. Concerned about steric constraints, we also shifted the register to move the RGD sequence closer to the center of the loop, resulting in the sequence VTGRGDSPASS.
FN-RGD1, FN-RGD2, and wild-type EETI-II were cloned into the yeast display vector pCT and expressed on the cell wall of S. cerevesiae as protein fusions to the Aga2p agglutinin subunit.21 A schematic of the yeast display system is shown in Figure 1C. These peptides were induced for expression on the yeast cell surface, and display levels were measured through detection of a C-terminal c-myc epitope tag by chicken-anti-c-myc and Alexa 555-labeled anti-chicken antibodies using flow cytometry [Fig. 1(D)]. In addition, we showed that the FN-RGD2 knottin, but not wild-type EETI-II, was able to bind to 50 nM detergent-solubilized αvβ3 integrin, using flow cytometry to detect a FITC-labeled antibody (mAb 13C2) specific for the αv integrin subunit [Fig. 1(D)]. FN-RGD1 exhibited minimal levels of integrin binding compared to FNRGD2, demonstrating that the position of the RGD motif within the loop influences integrin binding. Alexa 488-labeled trypsin was used to verify the expression of folded, functional EETIII wild-type on the surface of yeast (data not shown).
Engineering EETI-II to bind with high affinity to integrin receptors using yeast surface display
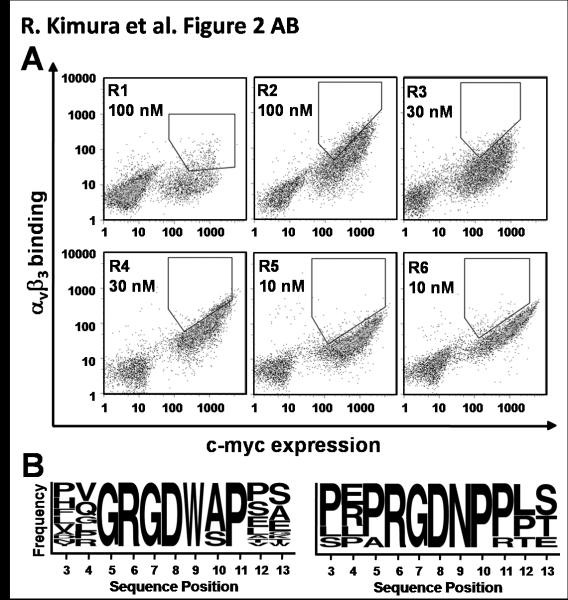
Based on the fibronectin loop grafting experiments described above, combinatorial libraries of EETI-II peptides were created using degenerate oligonucleotides, such that the EETI-II trypsin binding loop contained the randomized sequences XXRGDXXXXXX or XXXRGDXXXXX, where X represents any amino acid. This mutant DNA was transformed into yeast to create libraries of 5 × 106 clones each, which were combined and screened with several rounds of FACS to identify mutants that bound to αvβ3 integrin. Dual-color FACS allowed for quantitative discrimination of clones that bound the highest levels of αvβ3 integrin for a given amount of expression on yeast using a diagonal sort gate (Supplementary Fig. S1). Seven unique clones were isolated from this library (Supplementary Table SI). All of these peptides contained the XXXRGDXXXXX sequence, suggesting that this RGD position was favored for integrin binding over XXRGDXXXX. Pro, Gln, and Arg mutations of Leu 21 were identified; however, these probably arose from primer error or contamination, since mutagenesis was not performed at this position.
A clear integrin binding consensus sequence was not observed with the clones isolated from the first round of directed evolution. Therefore, we created a second library based on these initial hits, using degenerate oligonucleotides to bias the library with amino acid mutations identified from the first round of directed evolution (Supplementary Table SII). Several rounds of FACS were used to isolate peptides with the strongest αvβ3 integrin binding affinities, normalized for c-myc expression levels [Fig. 2(A)]. Twenty two unique clones were identified from this library (Supplementary Table SI). Analysis of the sequences of these isolated clones highlighted amino acid preferences or consensus at particular positions [Fig. 2(B)]. Notably, two distinct families of peptides emerged, one in which the engineered loops favored a XXGRGDW(A/S)PXX sequence, and the other which favored a XX(P/A)RGDNP(P/R)XX sequence.
Fig. 2.
Library sort progressions and sequences from the second round of directed evolution. (A) Density dot plots indicating FITC (y-axis) and Alexa-555 (x-axis) fluorescence on a single-cell level. Yeast-displayed knottin libraries were screened by dual-color FACS for mutants that bound the highest levels of αvβ3 integrin for a given amount of expression. Six rounds of FACS were used to obtain an enriched population of yeast that displayed clones with high αvβ3 integrin binding affinity. Integrin concentrations were reduced in successive rounds of sorting from 100 nM (rounds 1-2), to 30 nM (rounds 3-4), and 10 nM (rounds 5-6). (B) Sequence logos showing the relative frequencies of amino acids present in the engineered EETI-II loop from 22 individual clones. Two distinct families of binders emerged from library sorting and are represented by 17 clones (left) and 5 clones (right). The first amino acid shown is the third amino acid of the parent scaffold EETI-II. This figure was generated using online weblogo software (weblogo.berkeley.edu).
Individual clones were displayed on the yeast cell surface and tested to identify peptides with the highest αvβ3 integrin binding affinities to be synthesized for further characterization. Complete binding titrations of yeast-displayed knottin peptides could not be performed due to lack of sufficient quantities of detergent-solubilized αvβ3 integrin receptor. Therefore, relative binding was measured at several integrin concentrations by flow cytometry as described above, and was normalized to c-myc expression levels (data not shown). All mutants tested exhibited increased integrin binding affinity over the fibronectin-derived sequences FN-RGD1 and FNRGD2 (Figure 1D and data not shown). We chemically synthesized mutant 1.5B, which was the highest affinity binder from the first round of directed evolution, and mutants 2.5D and 2.5F, which appeared to be the highest affinity binders from each distinct peptide family identified in the second round of directed evolution. Since all mutants contained the XXXRGDXXXXX sequence, we synthesized FN-RGD2 for comparison to the evolved knottin peptides, and did not move forward with FN-RGD1, which did not appear to present the RGD motif in an optimal conformation for high affinity integrin binding [see also Fig. 1(D)].
Knottin peptides were prepared using standard Fmoc-based solid phase synthesis methods (Supplementary Methods). In all synthetic peptides, the lysine at position 15 was mutated to serine to facilitate site-specific chemical coupling of molecular imaging probes or chemotherapeutic agents to the N-terminus in future experiments. This Lys to Ser mutation did not affect αvβ3 integrin binding to yeast-displayed FN-RGD2 (data not shown). Since the Leu21 mutations identified through directed evolution did not appear to influence integrin binding, peptides were synthesized with leucine at this position. Crude peptides were purified by reversed-phase HPLC, and molecular masses were verified using electrospray mass spectrometry (Supplementary Table SIII and Supplementary Fig. S2). Peptides were folded in the presence of dimethyl sulfoxide and glutathione. Folded peptides exhibited distinct chromatographic profiles that allowed them to be purified from unfolded and misfolded isomers (Supplementary Fig. S2), in agreement with previous studies on wild-type EETI-II.33 Mass spectrometry analysis on oxidized peptides demonstrated a loss of 6 Da, indicating the formation of three disulfide bonds (Supplementary Table SIII).
Binding of engineered knottin peptides to integrin-expressing tumor cells
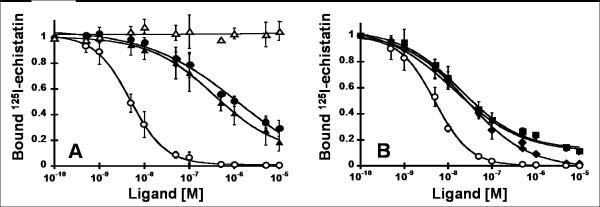
Knottin peptides were tested for their ability to compete for cell surface integrin binding with 125I-labeled echistatin, Echistatin, a protein from snake venom that binds to αvβ3 integrin with a KD of 0.36 nM,34 has an extended loop containing an RGD motif, although its C-terminal strand is also involved in mediating integrin binding affinity and specificity.35,36 U87MG glioblastoma cells, which express ~105 αvβ3 integrin receptors per cell,37 were used for these studies. We compared the relative receptor binding affinities of unlabeled echistatin, FN-RGD2, and knottin mutants 1.5B, 2.5D, or 2.5F, to that of c(RGDyK), a backbone cyclized pentapeptide that has been extensively characterized and used to image integrin expression in vivo (reviewed in 38,39). A version of the FN-RGD2 peptide containing a scrambled RGD amino acid sequence, designated FN-RDG2, was used as a negative control and was not able to compete for 125I-echistatin binding to U87MG cells [Fig. 3(A)]. All of the RGD-containing peptides inhibited the binding of 125I-echistatin to U87MG cells in a dose dependent manner (Fig. 3). Their relative binding affinities are depicted as IC50 values in Table I. Knottin peptides engineered by directed evolution were shown to bind to U87MG cells with a significantly higher affinity than both the loop-grafted FN-RGD2 and c(RGDyK) peptides.
Fig. 3.
Competition binding of peptides to tumor cell surface integrins. Varying concentrations of unlabeled peptides were incubated with 125I-labeled echistatin and allowed to compete for binding to integrin receptors expressed on the surface of U87MG glioblastoma cells. The fraction of 125I-echistatin bound to the cell surface is plotted versus the concentration of unlabeled (A) c(RGDyK) (●), FN-RGD2 (▲), and FN-RDG2 (△), and (B) evolved mutants 1.5B (▼), 2.5D (■), and 2.5F (◆). (A,B) Unlabeled echistatin (○) was used as a positive control to compare binding data from different experiments. Data shown is the average of triplicate values and error bars represent standard deviations. IC50 values are shown in Table I.
Table I.
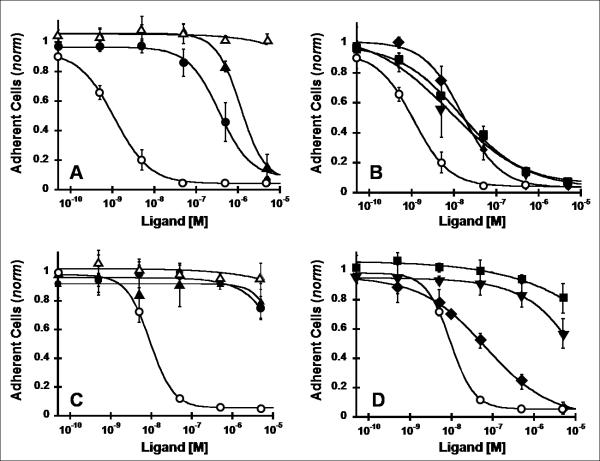
U87MG cell binding and adhesion data. IC50 values from competition binding assays (Fig. 3) and cell adhesion assays (Fig. 5) are summarized.
| Binding Assay | Cell Adhesion Assay | ||
|---|---|---|---|
| Ligand | (fibronectin) | (vitronectin) | |
| Echistatin | 4.9 ± 1.0 nM | 9.2 ± 1.5 nM | 0.90 ± 0.23 nM |
| c(RGDyK) | 860 ± 400 nM | (--- ) | 1100 ± 200 nM |
| FN-RGD2 | 370 ± 150 nM | (--- ) | 350 ± 220 nM |
| 1.5B | 13 ± 3 nM | (--- ) | 9.1 ± 2.9 nM |
| 2.5D | 19 ± 6 nM | (--- ) | 10 ± 2 nM |
| 2.5F | 26 ± 5 nM | 77 ± 28 nM | 17 ± 6 nM |
| FN-RDG2 | (--- ) | (--- ) | (--- ) |
(---) very weak to no competition.
Integrin binding specificities of engineered knottin peptides
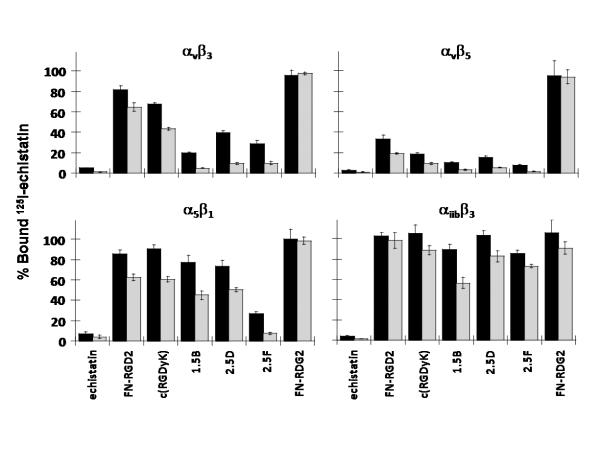
Since U87MG cells have been shown to co-express αvβ3, αvβ5, and α5β1 integrins,40 we measured integrin binding specificity by competition of 125I-echistatin to detergent-solubilized αvβ3, αvβ5, α5β1, and αiibβ3 integrin receptors coated onto microtiter plates (Fig. 4). Echistatin bound strongly to all of the tested integrins, in agreement with previous reports.41 The scrambled FN-RDG2 knottin peptide, our negative control, did not bind to any of the integrins used in this study. All RGD-containing peptides bound to αvβ3 and αvβ5 integrins to some degree, with the affinity-matured knottin peptides 1.5B, 2.5D, and 2.5F showing the strongest levels of binding compared to FN-RGD2 and c(RGDyK) peptides (Fig. 4). Interestingly, the 2.5F mutant bound with strong affinity to α5β1 integrin, while mutants 1.5B and 2.5D exhibited weaker binding to this receptor. With the exception of echistatin, all of the RGD-containing peptides bound weakly (with micromolar affinities) to the αiibβ3 integrin receptor (Fig. 4, and Supplementary Fig. S3). This result has important implications for in vivo tumor-targeting applications, since the αiibβ3 integrin is widely expressed on platelet cells and is involved in mediating the blood clotting process.42
Fig. 4.
Competition binding of peptides to surface-immobilized integrins. To determine integrin binding specificity, 125I-labeled echistatin and 5 nM (black bars) or 50 nM (grey bars) unlabeled peptides were added to microtiter plates coated with detergent-solubilized integrin receptor subtypes αvβ3, αvβ5, α5β1, and αiibβ3. Unbound 125I-echistatin was removed, and the amount of plate-bound 125I-echistatin remaining was measured. Unlabeled echistatin, which binds to all four integrins with high affinity, was used as a positive control. Error bars represent the standard deviation of measurements performed in triplicate.
Engineered knottin peptides mediate tumor cell adhesion to extracellular matrix proteins
We next measured the ability of engineered knottin peptides to block U87MG cell adhesion to vitronectin- and fibronectin-coated microtiter plates. Vitronectin is a natural ligand for several integrins, including αvβ3 and αvβ5.7 The RGD-containing peptides were all able to inhibit U87MG cell adhesion to vitronectin-coated plates in a dose-responsive manner [Fig. 5(A,B)]. Interestingly, the IC50 values of cell adhesion correlated with the 125I-echistatin competition binding data (Table I), indicating that inhibition of cell adhesion is directly related to integrin binding events. Fibronectin also binds to several integrins, (including αvβ3 and α5β1), but the α5β1 integrin receptor is generally selective for fibronectin.7 We found that only echistatin and knottin peptide 2.5F were able to block U87MG cell adhesion to fibronectin-coated plates [Fig. 5(C,D)], consistent with their ability to bind both αv and α5β1 integrins with high affinity. The FN-RDG2 negative control was not able to inhibit U87MG cell adhesion to vitronectin or fibronectin, as expected.
Fig. 5.
Inhibition of integrin-dependent cell adhesion. Vitronectin (A,B) or fibronectin (C,D) coated strips were incubated with U87MG cells for 2 h with the indicated concentrations of peptides. Adherent cells remaining after several wash steps were quantified with crystal violet staining by absorbance at 600 nm. Values were normalized to a negative control containing no competing peptide. Symbols are: (A,C) c(RGDyK) (●), FN-RGD2 (▲), and FN-RDG2 (△), and (B,D) evolved mutants 1.5B (▼), 2.5D (■), and 2.5F (◆). Unlabeled echistatin (○) was used as a positive control to compare cell adhesion data from different experiments. Data shown is the average of triplicate values and error bars represent standard deviations. IC50 values are shown in Table I.
Engineered knottin peptides exhibit high stability in serum
Finally, we tested the stability of the knottin peptide 2.5D upon exposure to human or mouse serum at 37 °C. Reversed-phase HPLC was used to quantify the amount of intact knottin peptide remaining at various times post incubation. We found that approximately 90% of the peptide remained after incubation for 24 h in human and mouse serum, with approximately 70% remaining after 96 h (Supplementary Fig. S4).
DISCUSSION
Previous studies validated the EETI-II knottin, a trypsin-binding protease inhibitor, as a scaffold for protein engineering by demonstrating that biologically-active loops could be substituted in place of the trypsin binding epitope.29-32 Here, we used yeast surface display to engineer EETI-II mutants that bound to αvβ3/αvβ5 or αvβ3/αvβ5/α5β1 integrin receptors with high affinity and modulated tumor cell adhesion to the extracellular matrix proteins fibronectin and/or vitronectin. Yeast surface display, a eukaryotic platform, allowed cell surface integrin binding to be normalized with knottin expression levels during library screens by single-cell flow cytometric analysis, and may have facilitated the folding of disulfide-rich knottin peptides. However, bacterial cell surface display or mRNA display might also have been successful as these technologies have previously been used to present folded EETI-II variants for high-throughput screening.29,33,43
Our results demonstrate that modification of a single loop can lead to EETI-II-based peptides with antibody-like affinities against a target of interest. Although we chose to modify the trypsin binding loop for this protein engineering study, we are currently investigating the tolerance of other EETI-II loops to mutagenesis. The disulfide-bonded core of EETI-II and its analogues impart high thermal stability and resistance to human proteases,44 which was attractive to us for the goal of developing peptides with potential in vivo tumor targeting applications. In our experiments, engineered knottin peptides exhibited high stability upon exposure to serum proteases, indicating that this scaffold is well-suited for in vivo applications such as molecular imaging or targeted delivery of therapeutic payloads to tumors.
The high number of αvβ3-binding peptides we identified is most likely due to the promiscuity of this integrin in binding many natural RGD-containing ligands, including fibronectin, fibrinogen, vitronectin, osteopontin, von Willebrand factor, and thrombospondin.7 In contrast, the α5β1 integrin is generally selective for fibronectin, and requires synergistic binding epitopes (i.e. the sequence PHSRN on the 9th domain of fibronectin) in addition to the RGD motif for ligand binding and activity.45 Interestingly, the knottin peptide 2.5F, whose integrin binding loop contains the sequence CPRPRGDNPPLTC, does not appear to require these additional contacts to bind with high affinity to the α5β1 integrin. The proline-rich sequence may constrain the binding epitope into a favorable conformation for these high affinity interactions. We are now pursuing molecular modeling and structural studies to provide insight into the unique binding integrin specificity of this peptide.
Existing antibody, peptide, and small molecule inhibitors under clinical development target αvβ3, αvβ3/αvβ5 or α5β1 integrins (reviewed in2,46,47). An exception is the peptide ATN-161 (Ac-PHSCN-NH2), which appears to bind both αvβ3 and α5β1 integrins, albeit with micromolar affinity, ultimately limiting its therapeutic potential. Although αvβ3, αvβ5, or α5β1 integrins are expressed at low levels on normal tissues, such as growth factor-stimulated endothelial cells involved in angiogenesis or wound healing, their overexpression on tumors and the tumor vasculature has generally allowed integrin-targeting agents to achieve tumor-specific effects.5 However, these agents seem to have stalled in clinical trials in recent years.47 As multiple integrin receptors (e.g. αvβ3, αvβ5, and α5β1) can be co-expressed on tumors and contribute to angiogenesis,5 blocking only one integrin receptor subtype may not be an optimal therapeutic strategy, and agents that target multiple integrin receptors may be more efficacious.47 The engineered knottin peptide 2.5F, which targets all three integrins, could offer significant benefits for cancer diagnostic and therapeutic applications. In addition, to improve clinical outcome there is a critical need for molecular imaging agents that can be used to identify patients who will respond to integrin-targeted treatments and monitor their disease progression.38 In a follow-up study, we showed that the engineered knottin peptides described here can be used to image integrin expression in living subjects using near-infrared fluorescence imaging and positron emission tomography.48 High affinity integrin-binding knottin peptides 2.5D and 2.5F, conjugated to a small molecule fluorochrome or radiolabel, exhibited a significant increase in tumor uptake in murine U87MG tumor xenograft models compared to weaker binding peptide conjugates, including FN-RGD2 and c(RGDyK). Moreover, radiolabeled knottin peptides had favorable biodistribution profiles (high tumor uptake, kidney clearance, and low uptake in other tissues) demonstrating great potential as clinical diagnostics for a variety of cancers.48
RGD motifs have been previously inserted into other proteins to demonstrate that RGD residues presented within a constrained molecular scaffold could bind integrins and mediate their function.32,43,49,50 However, in some cases these studies did not generate proteins with sufficient integrin binding affinities or biological activities for therapeutic applications,32,50 presumably because simple loop grafting did not result in optimal presentation of residues within the RGD-containing loop to integrin receptors. When high affinity was achieved, the ligand was shown to bind promiscuously to αvβ3 and αiibβ3 integrins.49 Directed evolution, using phage display, has been combined with RGD loop grafting to engineer large protein domains that bind to integrins with high affinity.51,52 These studies highlighted the importance of long loop length and proper positioning of the RGD motif within a given loop for high affinity integrin binding. Our results were consistent with these studies in that introduction of a short fibronectin-derived sequence (GRGDSP) into EETI-II did not result in binding to αvβ3 integrin (data not shown). Moreover, we did not isolate any clones from the XXRGDXXXXX library when screening for mutants with increased αvβ3 integrin binding affinity. Although we limited our study to EETI-II libraries with randomized XXRGDXXXXXX and XXXRGDXXXXX loop sequences, it is possible that alternate loop lengths and RGD positions would have also resulted in high affinity integrin binding mutants.
Richards, et al., have used phage display to engineer the naturally-occurring RGD loop within the 10th domain of fibronectin for high affinity and specific binding to the αvβ3 integrin receptor.27 In this study, an RGDWXE consensus sequence was identified after screening a library based on the randomization of the loop residues XRGDXXXX. Alanine scanning suggested that the Trp residue may be important for conferring specificity to αv over αiib, while the Glu residue may confer specificity to β3 over β5.27 Here, EETI-II knottin 2.5D contained a Trp residue immediately following the RGD sequence, which may contribute to its lack of binding to αiibβ3 integrin. However, EETI-II knottin 2.5F possessed an Asn residue at this position, suggesting that residues other than Trp are involved in mediating integrin binding specificity. Since the three mutant knottin peptides we tested all contained a Pro instead of the Glu residue at the third position following the RGD sequence, it would be interesting in future studies to see if this substitution would reduce their binding to αvβ5 integrin.
In comparison to these previous directed evolution studies, which used large protein domains to develop integrin binding agents, our work provides an interesting example where small, constrained peptide scaffolds have been engineered to target tumor-specific integrins with low nanomolar affinities and unique specificities. Although yeast surface display has previously been used to engineer alternative scaffolds based on the 10th domain of fibronectin (10FN3),53-56 our work demonstrates that this technology can be used to engineer new recognition properties into a molecular scaffold with a non-antibody (Ig)-like fold, and complements our recent study where a truncated form of the Agouti-related protein was engineered to bind specifically to αvβ3 integrin with single-digit nanomolar affinities.57 The potential to introduce binding epitopes into EETI-II and evolve them for high affinity binding may prove useful as a general strategy for engineering cystine knot peptides against other clinically-relevant targets.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Frank V. Cochran for help with peptide synthesis and purification and Bertrand Lui and Hen-Tzu Jill Lin for help with statistical analysis. We acknowledge the Stanford FACS Core Facility for access to high-speed flow cytometric sorters, and the Stanford Bio-X program for providing a BD FACSCalibur flow cytometer for analysis. Funded by the NIH/NCI Howard Temin Award 5K01 CA104706 (to J.R.C.), and a Stanford Molecular Imaging Scholars postdoctoral fellowship NIH 5R25 CA118681 (to R.H.K.). A.M.L was partially supported by a Stanford Dean's Fellowship.
Abbreviations
- EETI-II
Ecballium elaterium trypsin inhibitor
- RGD
Arg-Gly-Asp
- IC50
half-maximal inhibitory concentration
- ECM
extracellular matrix
- IBB
integrin binding buffer
- PBS
phosphate buffered saline
- FITC
fluorescein
- FACS
fluorescence-activated cell sorting
- DPBS
Dulbecco's phosphate buffered saline
Footnotes
Potential conflicts of interest. The authors have declared that no competing interests exist.
REFERENCES
- 1.Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol. 1992;3:65–71. [PubMed] [Google Scholar]
- 2.Alghisi GC, Ruegg C. Vascular integrins in tumor angiogenesis: mediators and therapeutic targets. Endothelium. 2006;13:113–135. doi: 10.1080/10623320600698037. [DOI] [PubMed] [Google Scholar]
- 3.Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 4.Kim S, Bell K, Mousa SA, Varner JA. Regulation of angiogenesis in vivo by ligation of integrin alpha5beta1 with the central cell-binding domain of fibronectin. Am J Pathol. 2000;156:1345–1362. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizejewski GJ. Role of integrins in cancer: survey of expression patterns. Proc Soc Exp Biol Med. 1999;222:124–138. doi: 10.1177/153537029922200203. [DOI] [PubMed] [Google Scholar]
- 6.Stupack DG, Cheresh DA. Integrins and angiogenesis. Curr Top Dev Biol. 2004;64:207–238. doi: 10.1016/S0070-2153(04)64009-9. [DOI] [PubMed] [Google Scholar]
- 7.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 8.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 9.Haubner R, Finsinger D, Kessler H. Stereoisomeric peptide libaries and peptidomimetics for designing selective inhibitors of the avb3 integrin for a new cancer therapy. Angew Chem Int Ed. 1997;36:1374–1389. [Google Scholar]
- 10.Haas TA, Plow EF. Integrin-ligand interactions: a year in review. Curr Opin Cell Biol. 1994;6:656–662. doi: 10.1016/0955-0674(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 11.Healy JM, Murayama O, Maeda T, Yoshino K, Sekiguchi K, Kikuchi M. Peptide ligands for integrin alpha v beta 3 selected from random phage display libraries. Biochemistry. 1995;34:3948–3955. doi: 10.1021/bi00012a012. [DOI] [PubMed] [Google Scholar]
- 12.Koivunen E, Gay DA, Ruoslahti E. Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J Biol Chem. 1993;268:20205–20210. [PubMed] [Google Scholar]
- 13.Koivunen E, Wang B, Ruoslahti E. Isolation of a highly specific ligand for the alpha 5 beta 1 integrin from a phage display library. J Cell Biol. 1994;124:373–380. doi: 10.1083/jcb.124.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koivunen E, Wang B, Ruoslahti E. Phage libraries displaying cyclic peptides with different ring sizes: ligand specificities of the RGD-directed integrins. Biotechnology (N Y) 1995;13:265–270. doi: 10.1038/nbt0395-265. [DOI] [PubMed] [Google Scholar]
- 15.Curley GP, Blum H, Humphries MJ. Integrin antagonists. Cell Mol Life Sci. 1999;56:427–441. doi: 10.1007/s000180050443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikecz K. Vitaxin applied molecular evolution. Curr Opin Investig Drugs. 2000;1:199–203. [PubMed] [Google Scholar]
- 17.Trikha M, Zhou Z, Nemeth JA, Chen Q, Sharp C, Emmell E, Giles-Komar J, Nakada MT. CNTO 95, a fully human monoclonal antibody that inhibits alphav integrins, has antitumor and antiangiogenic activity in vivo. Int J Cancer. 2004;110:326–335. doi: 10.1002/ijc.20116. [DOI] [PubMed] [Google Scholar]
- 18.Pallaghy PK, Nielsen KJ, Craik DJ, Norton RS. A common structural motif incorporating a cystine knot and a triple-stranded beta-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiche L, Heitz A, Gelly JC, Gracy J, Chau PT, Ha PT, Hernandez JF, Le-Nguyen D. Squash inhibitors: from structural motifs to macrocyclic knottins. Curr Protein Pept Sci. 2004;5:341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- 20.Craik DJ, Daly NL, Waine C. The cystine knot motif in toxins and implications for drug design. Toxicon. 2001;39:43–60. doi: 10.1016/s0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 21.Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- 22.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favel A, Mattras H, Coletti-Previero MA, Zwilling R, Robinson EA, Castro B. Protease inhibitors from Ecballium elaterium seeds. Int J Pept Protein Res. 1989;33:202–208. doi: 10.1111/j.1399-3011.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 24.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 25.Cheng Z, Wu Y, Xiong Z, Gambhir SS, Chen X. Near-infrared fluorescent RGD peptides for optical imaging of integrin alphavbeta3 expression in living mice. Bioconjug Chem. 2005;16:1433–1441. doi: 10.1021/bc0501698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlando RA, Cheresh DA. Arginine-glycine-aspartic acid binding leading to molecular stabilization between integrin alpha v beta 3 and its ligand. J Biol Chem. 1991;266:19543–19550. [PubMed] [Google Scholar]
- 27.Richards J, Miller M, Abend J, Koide A, Koide S, Dewhurst S. Engineered fibronectin type III domain with a RGDWXE sequence binds with enhanced affinity and specificity to human alphavbeta3 integrin. J Mol Biol. 2003;326:1475–1488. doi: 10.1016/s0022-2836(03)00082-2. [DOI] [PubMed] [Google Scholar]
- 28.Kratzner R, Debreczeni JE, Pape T, Schneider TR, Wentzel A, Kolmar H, Sheldrick GM, Uson I. Structure of Ecballium elaterium trypsin inhibitor II (EETI-II): a rigid molecular scaffold. Acta Crystallogr D Biol Crystallogr. 2005;61:1255–1262. doi: 10.1107/S0907444905021207. [DOI] [PubMed] [Google Scholar]
- 29.Christmann A, Walter K, Wentzel A, Kratzner R, Kolmar H. The cystine knot of a squash-type protease inhibitor as a structural scaffold for Escherichia coli cell surface display of conformationally constrained peptides. Protein Eng. 1999;12:797–806. doi: 10.1093/protein/12.9.797. [DOI] [PubMed] [Google Scholar]
- 30.Hilpert K, Wessner H, Schneider-Mergener J, Welfle K, Misselwitz R, Welfle H, Hocke AC, Hippenstiel S, Hohne W. Design and characterization of a hybrid miniprotein that specifically inhibits porcine pancreatic elastase. J Biol Chem. 2003;278:24986–24993. doi: 10.1074/jbc.M212152200. [DOI] [PubMed] [Google Scholar]
- 31.Krause S, Schmoldt HU, Wentzel A, Ballmaier M, Friedrich K, Kolmar H. Grafting of thrombopoietin-mimetic peptides into cystine knot miniproteins yields high-affinity thrombopoietin antagonists and agonists. Febs J. 2007;274:86–95. doi: 10.1111/j.1742-4658.2006.05567.x. [DOI] [PubMed] [Google Scholar]
- 32.Reiss S, Sieber M, Oberle V, Wentzel A, Spangenberg P, Claus R, Kolmar H, Losche W. Inhibition of platelet aggregation by grafting RGD and KGD sequences on the structural scaffold of small disulfide-rich proteins. Platelets. 2006;17:153–157. doi: 10.1080/09537100500436663. [DOI] [PubMed] [Google Scholar]
- 33.Wentzel A, Christmann A, Kratzner R, Kolmar H. Sequence requirements of the GPNG beta-turn of the Ecballium elaterium trypsin inhibitor II explored by combinatorial library screening. J Biol Chem. 1999;274:21037–21043. doi: 10.1074/jbc.274.30.21037. [DOI] [PubMed] [Google Scholar]
- 34.Kumar CC, Nie H, Rogers CP, Malkowski M, Maxwell E, Catino JJ, Armstrong L. Biochemical characterization of the binding of echistatin to integrin alphavbeta3 receptor. J Pharmacol Exp Ther. 1997;283:843–853. [PubMed] [Google Scholar]
- 35.Marcinkiewicz C, Vijay-Kumar S, McLane MA, Niewiarowski S. Significance of RGD loop and C-terminal domain of echistatin for recognition of alphaIIb beta3 and alpha(v) beta3 integrins and expression of ligand-induced binding site. Blood. 1997;90:1565–1575. [PubMed] [Google Scholar]
- 36.Wierzbicka-Patynowski I, Niewiarowski S, Marcinkiewicz C, Calvete JJ, Marcinkiewicz MM, McLane MA. Structural requirements of echistatin for the recognition of alpha(v)beta(3) and alpha(5)beta(1) integrins. J Biol Chem. 1999;274:37809–37814. doi: 10.1074/jbc.274.53.37809. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Xiong Z, Wu Y, Cai W, Tseng JR, Gambhir SS, Chen X. Quantitative PET imaging of tumor integrin alphavbeta3 expression with 18F-FRGD2. J Nucl Med. 2006;47:113–121. [PMC free article] [PubMed] [Google Scholar]
- 38.Cai W, Gambhir SS, Chen X. Multimodality tumor imaging targeting integrin alphavbeta3. Biotechniques. 2005;39:S6–S17. doi: 10.2144/000112091. [DOI] [PubMed] [Google Scholar]
- 39.Liu S. Radiolabeled multimeric cyclic RGD peptides as integrin alphavbeta3 targeted radiotracers for tumor imaging. Mol Pharm. 2006;3:472–487. doi: 10.1021/mp060049x. [DOI] [PubMed] [Google Scholar]
- 40.Bruning A, Runnebaum IB. CAR is a cell-cell adhesion protein in human cancer cells and is expressionally modulated by dexamethasone, TNFalpha, and TGFbeta. Gene Ther. 2003;10:198–205. doi: 10.1038/sj.gt.3301887. [DOI] [PubMed] [Google Scholar]
- 41.Pfaff M, McLane MA, Beviglia L, Niewiarowski S, Timpl R. Comparison of disintegrins with limited variation in the RGD loop in their binding to purified integrins alpha IIb beta 3, alpha V beta 3 and alpha 5 beta 1 and in cell adhesion inhibition. Cell Adhes Commun. 1994;2:491–501. doi: 10.3109/15419069409014213. [DOI] [PubMed] [Google Scholar]
- 42.Pytela R, Pierschbacher MD, Ginsberg MH, Plow EF, Ruoslahti E. Platelet membrane glycoprotein IIb/IIIa: member of a family of Arg-Gly-Asp--specific adhesion receptors. Science. 1986;231:1559–1562. doi: 10.1126/science.2420006. [DOI] [PubMed] [Google Scholar]
- 43.Zhao B, Helms LR, DesJarlais RL, Abdel-Meguid SS, Wetzel R. A paradigm for drug discovery using a conformation from the crystal structure of a presentation scaffold. Nat Struct Biol. 1995;2:1131–1137. doi: 10.1038/nsb1295-1131. [DOI] [PubMed] [Google Scholar]
- 44.Werle M, Schmitz T, Huang HL, Wentzel A, Kolmar H, Bernkop-Schnurch A. The potential of cystine-knot microproteins as novel pharmacophoric scaffolds in oral peptide drug delivery. J Drug Target. 2006;14:137–146. doi: 10.1080/10611860600648254. [DOI] [PubMed] [Google Scholar]
- 45.Aota S, Nomizu M, Yamada KM. The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J Biol Chem. 1994;269:24756–24761. [PubMed] [Google Scholar]
- 46.Meyer A, Auernheimer J, Modlinger A, Kessler H. Targeting RGD recognizing integrins: drug development, biomaterial research, tumor imaging and targeting. Curr Pharm Des. 2006;12:2723–2747. doi: 10.2174/138161206777947740. [DOI] [PubMed] [Google Scholar]
- 47.Tucker GC. Integrins: molecular targets in cancer therapy. Curr Oncol Rep. 2006;8:96–103. doi: 10.1007/s11912-006-0043-3. [DOI] [PubMed] [Google Scholar]
- 48.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Engineered knottin peptides: A new class of agents for imaging integrin expression in living subjects. Cancer Res. doi: 10.1158/0008-5472.CAN-08-2495. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith JW, Tachias K, Madison EL. Protein loop grafting to construct a variant of tissue-type plasminogen activator that binds platelet integrin alpha IIb beta 3. J Biol Chem. 1995;270:30486–30490. doi: 10.1074/jbc.270.51.30486. [DOI] [PubMed] [Google Scholar]
- 50.Yamada T, Matsushima M, Inaka K, Ohkubo T, Uyeda A, Maeda T, Titani K, Sekiguchi K, Kikuchi M. Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme. J Biol Chem. 1993;268:10588–10592. [PubMed] [Google Scholar]
- 51.Hufton SE, van Neer N, van den Beuken T, Desmet J, Sablon E, Hoogenboom HR. Development and application of cytotoxic T lymphocyte-associated antigen 4 as a protein scaffold for the generation of novel binding ligands. FEBS Lett. 2000;475:225–231. doi: 10.1016/s0014-5793(00)01701-4. [DOI] [PubMed] [Google Scholar]
- 52.Li R, Hoess RH, Bennett JS, DeGrado WF. Use of phage display to probe the evolution of binding specificity and affinity in integrins. Protein Eng. 2003;16:65–72. doi: 10.1093/proeng/gzg002. [DOI] [PubMed] [Google Scholar]
- 53.Gilbreth RN, Esaki K, Koide A, Sidhu SS, Koide S. A dominant conformational role for amino acid diversity in minimalist protein-protein interfaces. J Mol Biol. 2008;381:407–418. doi: 10.1016/j.jmb.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci U S A. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lipovsek D, Lippow SM, Hackel BJ, Gregson MW, Cheng P, Kapila A, Wittrup KD. Evolution of an interloop disulfide bond in high-affinity antibody mimics based on fibronectin type III domain and selected by yeast surface display: molecular convergence with single-domain camelid and shark antibodies. J Mol Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 57.Silverman AP, Levin AM, Lahti JL, Cochran JR. Engineered cystine-knot peptides that bind alpha(v)beta(3) integrin with antibody-like affinities. J Mol Biol. 2009;385:1064–1075. doi: 10.1016/j.jmb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.