DNA methylation mediates the association of prenatal famine exposure with higher adult BMI and serum triglyceride levels.
Abstract
Although it is assumed that epigenetic mechanisms, such as changes in DNA methylation (DNAm), underlie the relationship between adverse intrauterine conditions and adult metabolic health, evidence from human studies remains scarce. Therefore, we evaluated whether DNAm in whole blood mediated the association between prenatal famine exposure and metabolic health in 422 individuals exposed to famine in utero and 463 (sibling) controls. We implemented a two-step analysis, namely, a genome-wide exploration across 342,596 cytosine-phosphate-guanine dinucleotides (CpGs) for potential mediators of the association between prenatal famine exposure and adult body mass index (BMI), serum triglycerides (TG), or glucose concentrations, which was followed by formal mediation analysis. DNAm mediated the association of prenatal famine exposure with adult BMI and TG but not with glucose. DNAm at PIM3 (cg09349128), a gene involved in energy metabolism, mediated 13.4% [95% confidence interval (CI), 5 to 28%] of the association between famine exposure and BMI. DNAm at six CpGs, including TXNIP (cg19693031), influencing β cell function, and ABCG1 (cg07397296), affecting lipid metabolism, together mediated 80% (95% CI, 38.5 to 100%) of the association between famine exposure and TG. Analyses restricted to those exposed to famine during early gestation identified additional CpGs mediating the relationship with TG near PFKFB3 (glycolysis) and METTL8 (adipogenesis). DNAm at the CpGs involved was associated with gene expression in an external data set and correlated with DNAm levels in fat depots in additional postmortem data. Our data are consistent with the hypothesis that epigenetic mechanisms mediate the influence of transient adverse environmental factors in early life on long-term metabolic health. The specific mechanism awaits elucidation.
INTRODUCTION
Since the early 1970s, epidemiological studies have reported associations between an adverse prenatal environment and an increased disease risk in later life (1). Animal studies provided potential mechanisms for these observations (2), including environmentally induced changes to epigenetic marks during development (3). Epigenetic marks such as DNA methylation (DNAm) influence the transcription potential of genomic regions and, once changed, can result in long-term effects (4). Animal experiments show that epigenetic changes that are established during early development contribute to phenotypes later in life (5, 6). In parallel, human studies show changes in DNAm after exposure to a range of adverse prenatal conditions (7–12). These DNAm differences may mediate part of the association between adverse prenatal conditions and childhood phenotypes (13–15). Systematic epigenome-wide studies investigating the associations among these adverse conditions, DNAm changes, and specific phenotypes later in life are still largely lacking (16).
To fill this gap, we examined these associations in the quasi-experimental setting of the Dutch Hunger Winter of 1944–1945 (17), a 6-month famine at the end of World War II. Exposure to famine during gestation is associated with an increased risk of obesity, dyslipidemia, type 2 diabetes, and schizophrenia (18). We focus on our previous studies of the Dutch Hunger Winter, where we documented that prenatal adversity is associated with increases in adverse metabolic phenotypes in adulthood such as body mass index (BMI) and fasting glucose, serum triglycerides (TG), and low-density lipoprotein (LDL) cholesterol concentrations (LDL-C) (19–23). Here, we adopt a genome-wide approach to systematically identify the potential of DNAm to act as a mediator of the relation between prenatal famine exposure and adult outcomes in our study population of prenatally exposed individuals and time and sibling controls (24). We use mediation analysis as a helpful statistical tool to further explore the nature of the relationship between an independent (famine exposure) and dependent variable (metabolic health) through a hypothesized mediator (DNAm).
To formally establish mediation (25), we first re-examined the relation between famine exposure any time during gestation (“famine exposure”) and adult BMI, glucose, TG, and LDL-C outcomes in individuals with genome-wide DNAm data. Next, we examined genome-wide whether DNAm at specific cytosine-phosphate-guanine (CpG) dinucleotides was associated with both famine exposure and the outcome of interest and subjected the identified candidate CpGs to a formal mediation test (26) to determine the extent to which DNAm mediated the association between prenatal famine and adult outcomes. These analyses were repeated for exposure during early gestation, which is an especially sensitive period of gestation (27, 28). To address the potential functional impact of mediating CpGs, we tested for their association with gene expression (29, 30) and correlation in methylation levels across postmortem studies (31, 32).
Using this approach, we show that the increase in BMI and TG among individuals exposed in utero is mediated by DNAm, specifically near genes involved in development and metabolic processes including energy metabolism (PIM3), β cell function (TXNIP), glycolysis (PFKFB3), and adipogenesis (METTL8).
RESULTS
Genome-wide mediation analysis
We first confirmed the previously reported association between famine exposure and BMI, fasted glucose, TG, and LDL-C in the Dutch Hunger Winter Families Study for individuals with complete methylation data (19–23). Individuals with prenatal famine exposure (n = 348) had a 5.6% (+0.36 SD) higher BMI (P = 5.7 × 10−7), a 13.5% (+0.23 SD) higher serum TG (P = 3.8 × 10−3), and a 3.8% (+0.22 SD) higher fasted glucose (P = 0.023) but no difference in LDL-C (P > 0.05), as compared with non-exposed controls (n = 463; Table 1). Therefore, LDL-C was excluded from further analyses. Next, we applied a genome-wide screen (342,596 CpGs) to identify potentially mediating CpGs. We simultaneously tested the association of DNAm with famine exposure and with any of the metabolic outcomes (BMI, fasted glucose, or TG) in a single model. For BMI and TG, this resulted in 8 and 16 associated CpG dinucleotides, respectively (PFDR < 0.05; Fig. 1). These were candidates for mediation and were explored further. No mediation candidates were identified for fasting glucose concentrations. These candidates from the epigenome-wide association study (EWAS) can arise from a strong association with either exposure, outcome, or both (in which case the strength of the two associations individually may be moderate). Only the latter constitute potentially mediating CpGs.
Table 1. Phenotypic differences and famine exposure.
| Controls | Famine exposure | |
| N | 463 | 348 |
| Age (years) [SD]+ | 58.0 [5.4] | 58.9 [0.5]*** |
| Male (%) | 43.0 | 46.0 |
| BMI1 [SD] | 27.0 [4.2] | 28.5 [5.0]*** |
| LDL-C2 [SD] | 3.42 [0.96] | 3.45 [0.97] |
| Triglycerides2 [SD] | 1.48 [0.86] | 1.68 [1.30]** |
| Glucose baseline3 [SD] | 5.32 [0.93] | 5.52 [1.19]* |
Nominal P value either *P < 0.05, **P < 0.01 or ***P < 0.001 from a linear mixed-effects model with the denoted variable as the dependent variable and family identifier as random effect.
+Model included an additional random effect for exposure status to control for the difference in variance in age between groups.
1Model-applied correction for age and gender.
2Model-applied correction for age, gender, and statin used, and individuals who were nonfasting at examination were excluded (excluding two controls and five famine-exposed individuals).
3Model-applied correction for age and gender. Individuals who were nonfasting and had prediagnosed diabetes (thus receiving treatment) before the clinical examination were excluded (excluding 19 controls and 32 famine-exposed individuals).
Fig. 1. Manhattan plots: Outcome genome-wide screens for potential mediators.
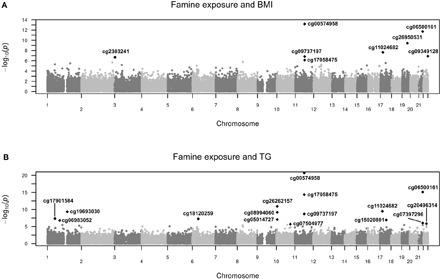
The −log(P value) is shown (y axis) for each CpG relative to its genomic locations (x axis) on the 22 autosomal chromosomes tested for (A) an EWAS on both famine exposure and BMI and (B) an EWAS on both famine exposure and serum TG.
Mediation: Famine exposure and BMI
Of the eight mediation candidates for the EWAS of famine exposure and BMI, only cg09349128 was associated with both exposure and outcome (Pexposure+bmi = 1.3 × 10−7, Pexposure = 1.5 × 10−3, Poutcome = 6.5 × 10−8; Table 2). Of interest, DNAm at this CpG has been consistently reported to be associated with BMI (33–36). A formal mediation analysis showed that DNAm at cg09349128 mediates the association between famine exposure and BMI [βmediation = 0.7%/log(BMI); 95% confidence interval (CI), 0.3 to 1.2%; P = 0.001]. The mediation path explained 13.4% (95% CI, 5 to 28%; P = 0.001) of the association between famine exposure and BMI (R2LMM = 0.27). This finding persisted after statistical adjustment for smoking, socioeconomic status (SES), and reported dietary intake at the time of examination (calories from fat, protein, or carbohydrates or from any source).
Table 2. Genome-wide screen for potential mediators: famine exposure and BMI.
| CpG characteristics | Mediation EWAS | Associations with either famine exposure or BMI* | |||||||||
| CpG | Location (hg19) |
Nearest gene (expression)† |
Methylation (SD)‡ | Rank | EWAS P§ | EWAS PFDR | βfamine | Pfamine | βBMI | PBMI | Previous EWAS |
| cg00574958 | chr11:68607622 | CPT1A | 13.6 (2.6) | 1 | 6.7 × 10−14 | 2.3 × 10−8 | −0.2 | 0.19 | −4.0 | 1.7 × 10−16 | (33, 56, 72, 73) |
| cg06500161 | chr21:43656587 | ABCG1 | 65.9 (3.0) | 2 | 2.0 × 10−12 | 3.3 × 10−7 | −0.1 | 0.75 | 4.3 | 2.5 × 10−13 | (33, 43, 56, 73) |
| cg26950531 | chr19:38704515 | DPF1 | 32.5 (6.1) | 3 | 2.2 × 10−10 | 4.3 × 10−5 | 0.3 | 0.50 | −8.9 | 1.3 × 10−11 | (56) |
| cg11024682 | chr17:17730094 | SREBF1 | 53.2 (3.2) | 4 | 2.2 × 10−8 | 1.9 × 10−3 | −0.1 | 0.70 | 3.3 | 1.2 × 10−9 | (33, 43, 56) |
| cg09349128 | chr22:50327986 | CRELD2 | 39.0 (3.8) | 5 | 1.3 × 10−7 | 8.3 × 10−3 | −0.7 | 1.5 × 10−3 | −3.5 | 6.5 × 10−8 | (33–36, 56) |
| cg09737197 | chr11:68607675 | CPT1A | 20.4 (4.6) | 6 | 1.5 × 10−7 | 8.3 × 10−3 | −0.4 | 0.093 | −4.4 | 3.2 × 10−9 | (33, 73) |
| cg23032421 | chr3:3152038 | IL5RA | 73.9 (4.1) | 7 | 2.1 × 10−7 | 0.01 | −0.3 | 0.18 | −3.4 | 5.8 × 10−9 | (33) |
| cg17058475 | chr11:68607737 | CPT1A | 16.1 (4.1) | 8 | 7.4 × 10−7 | 0.032 | −0.1 | 0.66 | −4.2 | 2.3 × 10−8 | (30, 56, 72–74) |
| cg15659713¶ | chr8:38586183 | TACC1 | 24.6 (3.9) | 9 | 1.7 × 10−7 | 0.064 | 1.3 | 1.7 × 10−7 | −0.4 | 0.63 | (28) |
| cg26199857¶ | chr12:54764265 | ZNF385A | 68.9 (5.7) | 14 | 5.2 × 10−7 | 0.13 | 2.0 | 3.1 × 10−7 | 1.3 | 0.28 | (28) |
*The estimate and (nominal) P value belonging to the EWAS for famine exposure (β = exposed − unexposed) or BMI (β/log(BMI)).
†Nearest gene within 100 kb.
‡The Illumina 450k array β value (ranging from 0 to 1) multiplied by 100 for easy interpretation. This is done throughout the presented work.
§The P value belonging to an analysis of variance (ANOVA) test (χ2, df = 2) between a generalized estimating equations (GEE) model with and without both famine exposure and BMI.
¶The two CpGs identified in a previous EWAS on famine exposure (28).
Genomic annotation of cg09349128 revealed that it mapped to an enhancer region in multiple cell types (37). The enhancer is linked to PIM3 in epigenome reference data, a gene implicated in cell growth and energy metabolism (38), stem cell renewal (39), and glucose-stimulated insulin secretion in β cells (40). Analysis of 2044 whole blood samples with both genome-wide DNAm (Illumina 450k) and expression [RNA sequencing (RNA-seq)] data (29, 30) showed that methylation of cg09349128 was associated with the expression of PIM3 and also with two additional genes in cis, namely, ZBED4 and CRELD2 (table S1).
Mediation: Famine exposure and TG
Of the 16 mediation candidates for the association between famine exposure and TG, 6 CpGs were associated with both exposure and outcome (table S2). With the exception of cg06983052, methylation at these CpGs was previously reported to be associated with TG or other metabolic traits in EWASs (30, 41–46). All six CpGs mediated the relation between famine exposure and TG (P < 0.007; Table 3). The mediation path of each CpG explained between 19.6 and 28.0% of the association between famine exposure and TG (R2LMM = 0.30). Although not mapping close together and often located on different chromosomes, DNAm levels of the six CpGs were correlated (ρ = 0.17 to 0.73, P < 0.001). We therefore tested mediation for the mean standardized DNAm level across all six CpGs. This aggregate measure likewise mediated the association between famine exposure and TG [βmediation = 0.075 SD/log(TG); 95% CI, 0.047 to 0.104; P < 0.001]. The mediation path explained 80.0% (95% CI, 38.5 to 100%; P = 0.02) of the association. The role as mediator persisted, for both individual CpGs and their aggregate, after additional adjustment for smoking, SES, and current diet. Adjustment for BMI did somewhat attenuate this association [βmediation = 0.053 SD/log(TG); 95% CI, 0.03 to 0.08; P < 0.001], and as a result, the proportion mediated was affected (P = 0.27). Because an additional analysis showed that the CpGs involved did not mediate the association between famine exposure and BMI (P > 0.076), it is unlikely that an extended mediation path including BMI is involved in terms of DNAm.
Table 3. Mediation analysis: DNAm and the association between famine exposure and triglycerides.
| CpG |
Location (hg19) |
Nearest gene* |
Methylation (SD)† |
Rank |
EWAS PFDR |
Pfamine | PTG |
Previous studies |
βmediation‡ |
P mediation |
Proportion mediated (%) [95% CI]§ |
Pproportion |
| cg19693031 | chr1:145441552 | TXNIP | 77.5 (4.3) | 6 | 2.6 × 10−5 | 4.8 × 10−3 | 2.3 × 10−11 | (41–45) | 2.6 [0.7–4.8] | 0.005 | 28.0 [5.7–100] | 0.026 |
| cg18120259 | chr6:43894639 | LOC100132354 | 60.4 (4.7) | 10 | 1.8 × 10−3 | 6.6 × 10−4 | 6.4 × 10−8 | (44) | 2.3 [0.8–4.1] | 0.001 | 24.9 [7.5–100] | 0.021 |
| cg15020801 | chr17:46022809 | PNPO | 36.1 (3.4) | 12 | 3.5 × 10−3 | 7.1 × 10−4 | 6.0 × 10−8 | (30) | 2.3 [0.9–4.2] | 0.001 | 25 [7.0–100] | 0.022 |
| cg06983052 | chr1:90288099 | LRRC8D | 64.8 (3.8) | 13 | 4.2 × 10−3 | 1.0 × 10−5 | 5.3 × 10−6 | 2.6 [1.1–4.5] | <0.001 | 28.0 [8.8–100] | 0.024 | |
| cg07397296 | chr21:43655316 | ABCG1 | 26.9 (3.8) | 14 | 0.021 | 5.1 × 10−3 | 1.9 × 10−7 | (49) | 1.9 [0.6–3.6] | 0.005 | 20.5 [4.6–97.4] | 0.027 |
| cg20496314 | chr22:39759864 | SYNGR1 | 40.2 (4.3) | 15 | 0.032 | 3.9 × 10−3 | 1.5 × 10−7 | (45, 46) | 1.8 [0.5–3.5] | 0.007 | 19.6 [3.6–88.3] | 0.026 |
*Nearest gene within 100 kb.
†The Illumina 450k array β value (ranging from 0 to 1) multiplied by 100 for easy interpretation.
‡This is the estimate and CI based on 10K Monte Carlo simulations of the indirect effect or mediation effect, which is often referred to as the “a × b” effect.
§The percentage of the total exposure-phenotype relationship explained by the indirect (mediated) effect as based on 10K Monte Carlo simulations.
The six mediating CpGs mapped to (intronic) enhancers, open chromatin regions, and exons. Analysis of blood data (29, 30) showed that all but cg18120259 were associated with the expression of their nearest genes (table S1). This included cg19693031 with TXNIP and cg07397296 with ABCG1. DNAm at both CpGs has been previously associated with TG (30).
Mediation: Early famine exposure
Early gestation appears to be the most vulnerable period in terms of the later-life phenotype (27) as epigenetic (28) consequences of famine exposure. The association between famine exposure and BMI or TG also extended to early gestational exposure but not to preconceptional exposure (table S3). Therefore, we investigated whether mediation by DNAm is also present for the elevated BMI and TG in individuals with “early” exposure. The subsequent genome-wide screens for possible mediators of these associations did not yield candidates for BMI that were associated with both DNAm and outcome (Fig. 2 and table S4).
Fig. 2. Manhattan plot: Outcome genome-wide screen for potential mediators.
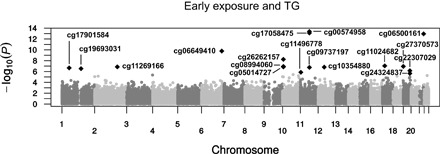
The −log(P value) (y axis) for each CpG relative to its genomic locations (x axis) on the 22 autosomal chromosomes for the EWAS for both early famine exposure and serum TG.
Seventeen CpGs were potential mediators for the association between early exposure and TG. Two of which were associated with both early exposure and TG (table S4). Both cg08994060 [βmediation = 4.1% (95% CI, 0.3 to 8.3%), P = 0.033] and cg11269166 [βmediation = 3.5% (95% CI, 0.4 to 7.2%), P = 0.027] mediated the association between early exposure and TG (R2LMM = 0.32). The proportion mediated was 24.5% for cg08994060 (P = 0.054), whereas cg11269166 mediated 19.4% of the association between exposure and TG (95% CI, 1.5 to 81.3%; P = 0.039). The aggregate measure mediated the association between early exposure and TG [βmediation = 0.069 SD/log(TG) (95% CI, 0.026 to 0.116), P = 0.02], explaining 36.3% (95% CI, 15.7 to 100%; P = 0.02) of the association. Correction for smoking, SES, and current diet had no influence on these outcomes. Additional correction for BMI attenuated the association of cg08994060 [βmediation = 3.2% (95% CI, 0.0 to 7.0%), P = 0.052] but not of cg11269166 or their aggregate.
CpG cg08994060 is located in a deoxyribonuclease I (DNaseI) hypersensitivity cluster in an intron of PFKFB3, a rate-limiting enzyme in glycolysis, and was associated with expression of this gene in whole blood (table S1). Similarly, cg11269166 is located in an enhancer located in an intron of METTL8, a gene involved in adipogenesis (47), and was associated with METTL8 expression in whole blood (table S1).
Cross-tissue comparison
We measured DNAm in whole blood and do not have specimens from other tissues available in our cohort (24). Mean differences in DNAm levels vary between tissues (32), but DNAm level variation in blood may still reflect variation in other tissues (48). We investigated DNAm data across 12 postmortem tissue types from 16 cadavers (30, 31) and found that the DNAm patterns in blood showed broad correspondence with those in internal tissues [Spearman’s ρ = 0.42, P < 10−6; blood, liver, kidney, skin, omentum (visceral fat), subcutaneous fat, and fat from around the kidney], although the correlation between tissues was more modest when CpGs were analyzed individually in this small data set (fig. S3). DNAm in blood was correlated with DNAm in omentum and subcutaneous fat (both ρ = 0.51, P < 10−11) and fat taken from around the kidney (ρ = 0.42, P = 2.7 × 10−7) but not with DNAm in the liver (ρ = 0.14, P = 0.14).
DISCUSSION
A stepwise genome-wide mediation analysis is consistent with the hypothesis that epigenetic mechanisms play a role in mediating the association between prenatal famine exposure and later-life metabolic health. Specifically, our results suggest that DNAm at an enhancer linked to PIM3 expression mediated the association between famine exposure and BMI. PIM3 influences cell growth and energy metabolism, including mitochondrial function (38). DNAm at six CpGs, including at previously TG-associated CpGs at TXNIP and ABCG1 (41–45, 49), mediated the association between famine exposure and TG. DNAm at these CpGs was likewise associated with the expression of genes implicated in cell growth and energy metabolism. Two different CpGs, at PFKFB3 (which plays a key role in glycolysis) and METTL8 (linked to adipogenesis), mediated the association between early exposure and TG. DNAm at the nine identified CpGs correlated with DNAm in various other tissues, including multiple fat deposits. DNAm at the nine CpGs associated with the expression of genes with general roles in growth and metabolism, and although mediators, they are not known to play direct roles in fat and TG metabolism. Therefore, the nine CpGs are unlikely to be directly involved in a mechanistic sense in BMI or TG but may have contributed to adverse morphological or cellular metabolic profiles with an adverse effect on metabolic health in prenatally exposed individuals.
We used a novel approach to identify possible mediators by performing a series of genome-wide analyses that tested for an association with DNAm of both the exposure and a metabolic outcome simultaneously. This resulted in a set of CpGs associated with either famine exposure or a later-life metabolic outcome, or both. The latter were formally tested for mediation. CpGs that associated with (early) famine exposure in our previous EWAS of famine exposure only in the same data set (28) were not identified as potential mediators (that is, were associated with exposure but not with outcome). In concordance with this (28) and earlier work (50–52), all the mediating CpGs overlapped regulatory elements; were linked to the expression of genes involved in growth, differentiation, and metabolism; and were associated with either famine exposure or early famine exposure. It should be noted that the latter analysis was based on a small sample size and had limited power, but the results may point to additional processes unique for those exposed during early gestation.
Several strengths and limitations of this study and mediation analyses in general are important to discuss. We observed that the association between prenatal famine and BMI or TG was mediated by single CpGs (instead of regions). This observation may be related to technical (that is, the sparsity of the Illumina 450k array) and biological factors [differential methylation may be related to very local differences in transcription factor binding, not extending across regions (29)]. We recognize that statistical evidence for mediation does not clarify the nature of the mechanism. However, it does point toward possible pathways that warrant further exploration. It is also relevant to note that DNAm was measured in whole blood, which may not be the most relevant tissue to study for BMI or TG. However, DNAm data from multiple tissues from the same donor (31, 32) show a broad agreement in DNAm patterns of the CpGs in whole blood and various fat deposits. Moreover, two previous studies showed that the associations between DNAm at PIM3 (cg09349128) and BMI (33) and DNAm at ABCG1 (cg07397296) and TG (49), on which we likewise report here, were not only found in blood but could also be validated in DNAm data from adipose tissue. Our findings may therefore point toward more general processes in other tissues, including adipose tissue, and may reflect other epigenetic marks correlated to DNAm.
Mediation analysis is sensitive to unmeasured confounding (53). We address this concern, within the limits of an observational study six decades after the exposure, on several levels with our study design and study execution. First, we analyzed the impact of prenatal famine exposure among same-sex sibling pairs concordant and discordant on famine exposure, thereby mitigating confounding by familial factors. Next, we selected individuals born before and after the famine in the same institutions as controls for those with prenatal famine exposure and rely on the quasi-experimental circumstances of the Dutch Famine, precluding confounding by social characteristics related to wealth and nutrition in pregnancy. The inclusion of individuals either conceived and born before the famine or conceived and born after the famine also allows us to investigate and adjust for the influence of age, which is crucial (54). Last, we could effectively exclude confounding effects of several measured lifestyle factors including diet and smoking.
We recognize that the interpretation of mediation analyses requires prudence (53) because mediation analysis is sensitive to differences in measurement accuracy between observed exposure and an empirically measured mediator. However, this risk is reduced by the quasi-experimental setting of our study. Also, mediation analysis is equally sensitive for spurious associations as regular association analysis (53). Of the nine mediating CpGs identified here, all but cg06983052 (LRRC8D) have been related to obesity, TG, or other metabolic phenotypes in previous studies (Tables 2 and 3). In addition, mediation can also be affected by reverse causation (53). Prenatal famine exposure may be considered a causal anchor that, while not as strong as genetic variants in Mendelian randomization (53), reduces the risk of reverse causation for several of the tests performed within the mediation framework with the exception of the association between mediator and outcome (55). Longitudinal data could be used to refute reverse causation (preferably from conception onward), but by design, we did not sample at birth or any other time at a younger age before the potential onset of the metabolic complications.
The nine CpGs on which we report include the well-described CpGs mapping to PIM3 (cg09349128) (33–36, 56) and TXNIP (cg19693031) (30, 41–44) that were also reported in large Mendelian randomization studies (30, 33, 56). No evidence was found for an effect of BMI on DNAm at cg09349128 (33, 56), whereas weak evidence was found for an effect of TG on DNAm at cg19693031 (30). However, DNAm at this latter CpG is also associated with prenatal smoke exposure (45) [and not adult smoke exposure (11)], an exposure likewise linked with elevated serum TG (57). DNAm at this CpG is also associated with future type 2 diabetes risk that is independent of the traditional risk factors, which included dyslipidemia (43), analogs to the association between prenatal famine exposure, and type 2 diabetes (20). The overlap with previous EWAS findings highlights not only the fact that we are looking at epigenetic variation with a modest individual impact on variation in metabolic health but also the idea that the relevance of our results may extend beyond prenatal famine exposure.
In summary, using a systematic genome-wide approach, we show that DNAm at specific CpGs mediates a considerable proportion of the associations between prenatal famine exposure and later-life adiposity and serum TG levels. Our data are consistent with the hypothesis that the associations between exposure to an adverse environment during early development and health outcomes in adulthood are mediated by epigenetic factors. The specific causal mechanism awaits elucidation.
MATERIALS AND METHODS
Study setting
The Dutch Hunger Winter was a 6-month famine at the end of World War II that resulted from a combination of punitive measures imposed by occupying German forces after a national railway strike, winter conditions, and fuel shortages related to war operations. Food rations were distributed centrally and rapidly dropped to below 900 kcal/day between 26 November 1944 and 15 May 1945. After 15 May, rations rapidly returned to pre-famine levels. The percentage of calories from proteins, fat, and carbohydrates in the diet was relatively constant as the food rations diminished (17, 58).
Study subjects
The Dutch Hunger Winter Families study is described in detail elsewhere (24). We identified 2417 singleton births between 1 February 1945 and 31 March 1946 at three institutions in famine-exposed cities in the western Netherlands whose mothers were exposed to the famine during or immediately preceding that pregnancy. We selected time controls born in 1943 or in 1947 from the same clinics. We also asked whether a same-sex sibling not exposed to the famine would be willing to participate as a sibling control. Both the famine-exposed and the time controls can therefore have a same-sex sibling as family control. Overall, 1075 interviews and 971 clinical examinations were performed six decades after the exposure. Following the Helsinki guidelines, we obtained ethical approval, both from the Institutional Review Board of Columbia University Medical Center and from the Medical Ethical Committee of the Leiden University Medical Center. The study participants provided verbal consent in a telephone interview, and in case of clinical examinations, a written informed consent was obtained.
Famine exposure definitions
We defined famine exposure by the number of weeks during which the mother was exposed to <900 kcal/day after the last menstrual period (LMP) recorded on the birth record. Missing or implausible LMP records (12%) were (re-)estimated from the birth weights and the date of birth (24). We considered somebody exposed to famine in gestational weeks 1 to 10, 11 to 20, 21 to 30, or 31 to delivery if such a gestational time window was entirely contained within this period and had an average exposure of <900 kcal/day during this 10-week period. As a result of the 6-month duration, some individuals were exposed to two adjacent 10-week periods. In short, pregnancies with an LMP between 26 November 1944 and 4 March 1945 were exposed in weeks 1 to 10, those with an LMP between 18 September 1944 and 24 December 1944 were exposed in weeks 11 to 20, those with an LMP between 10 July 1944 and 15 October 1944 were exposed in weeks 21 to 30, and those with an LMP between 2 May 1944 and 24 August 1944 were exposed in weeks 31 to delivery. We defined individuals exposed in any of these periods as “famine-exposed,” and those exposed in gestational weeks 1 to 10 were defined as early exposed. In addition, individuals with an LMP between 1 February and 12 May 1945 were exposed to an average of <900 kcal/day for 10 weeks before conception and were defined as the preconception exposure group.
Characteristics
Medical examinations were scheduled early in the morning under fasting conditions. Measurement of height was carried out to the nearest millimeter using a portable stadiometer (Seca), and body weight was measured to the nearest 100 g by a portable scale (Seca). BMI was calculated from these measures. A blood draw was performed at the start of a 75-g oral glucose test, and glucose was quickly assessed in serum by hexokinase reaction on a Modular P800 (Roche). Cholesterol measures were reported earlier (19) and were assessed using standard enzymatic assays. LDL-C was calculated for individuals with a TG concentration lower than 400 mg/dl using the Friedewald formula. Dietary intake in the last 12 months was ascertained from a 140-item food frequency questionnaire developed to assess dietary habits in an elderly Dutch population (59). This questionnaire provided estimates of macronutrient intake (total energy and the percentage of fat, protein, and carbohydrate thereof). As a measure of SES, we classified study participants on a five-level education scale, identifying individuals with primary, lower- or middle-level vocational, secondary, higher vocational, and university education.
DNAm data
DNAm was measured using the Illumina Infinium Human Methylation 450k BeadChip, and preprocessing was previously described by us in detail (28). Briefly, samples were randomly distributed, ensuring similar distributions of exposure periods, sex ratios, and mean ages per 96-well plate and 450k array, keeping sibling pairs together, but were randomly assigned to either the left or right column of the 450k array. We assessed data quality using both sample-dependent and sample-independent quality metrics using the R package MethylAid (60). Bisulfite conversion efficiency was assessed using the dedicated 450k probes and sequencing the IGF2 DMR0 of a random set of samples. We remeasured a subset of the genotypes measured on the 450k array with MassARRAY and checked the gender of samples using all X-chromosomal CpGs to exclude sample swaps. We used noob and Functional Normalization as implemented in the minfi package (61) using six principal components to normalize for batch effects, dye color intensity differences, and background signal. Individual measurements with a detection P value of >0.01 or zero-intensity value in one of the used color channels were set as missing. The measurement success rate per sample was >99%. Next, we removed a-specific/polymorphic and non-autosomal probes, probes with <95% success rate, and those probes that were completely methylated or unmethylated in all major cell types in whole blood. Methylation percentages in text, figures, and tables reflect microarray β-value estimates (which range from close to zero to close to one or 0 to 100%, as denoted throughout). For the individuals for whom we have genome-wide DNAm data (Illumina 450k array), 348 individuals met our definition of famine exposure, of whom 73 also meet the definition of exposure during weeks 1 through 10 of gestation (early exposure). In total, 94 individuals met our preconception exposure definition. In addition, we have 463 individuals with no famine exposure directly before conception and during gestation. DNAm data are available upon request.
In addition, we used DNAm data from 16 obductions (31, 32) from which samples were collected within 12 hours of death (mean age, 62.8 years). In concordance with the ethical guidelines in the Code for Proper Secondary Use of Human Tissue in the Netherlands (Dutch Federation of Medical Scientific Societies), these samples were anonymized, and raw data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GSE78743). To explore the relation with gene expression and validate associations with BMI or TG, we referred to the DNAm data of 3296 individuals (30) measured within the Genome of the Netherlands reference project [GoNL; deposited at the European Genome-phenome Archive (EGA) under accession number EGAC00001000277]. Preprocessing and normalization were done as described above for both data sets. R Code for the quality control and normalization is provided at https://git.lumc.nl/molepi/DNAmArray.
Gene expression data
We used RNA-seq data created from total RNA extracted from whole blood using the TruSeq v2 library protocol and 2 × 50–base pair paired-end sequencing on an Illumina HiSeq2000 for 2044 of the 3296 individuals for which we have DNAm data from the GoNL reference project (likewise under EGA accession number EGAC00001000277). Data processing is detailed elsewhere (29) and consisted in removal of barcoded adapters, low-quality reads, alignment against genome build NCBI37, and gene-annotation Ensembl v.71, normalization, and GC-bias correction.
Statistics
All analyses were performed in the R programming environment (R.3.2.2). Reanalysis of the associations between phenotypes and famine exposure was performed with linear mixed-effects models (LMMs) with one of the phenotypes (BMI, TG, glucose, and LDL-c) as the dependent variable using the R lme4 package (62). The sibling-pair identifier was included as a random effect, and where appropriate, we corrected for gender and age at examination. We used the restricted maximum likelihood method and variance components for the covariance matrix structure. The degree of freedom (df) for a variable of interest was calculated with Satterthwaite’s approximation, as implemented in the R package lmerTest (63). The R2 for the LMMs was assessed using the R MuMIn package (64). In line with previous reports (19, 20, 23, 65, 66), we included statin usage when assessing cholesterol variables, excluded nonfasting individuals for the cholesterol analyses, and excluded individuals with diagnosed diabetes (and therefore on medication) before the clinical examinations for the analyses of fasting serum glucose levels.
For their robustness, we used generalized estimating equations as implemented in the R geepack package (67) for the series of EWAS for potential mediators. We used a Gaussian link function to model DNAm (reflected by the 450k array β value). In each EWAS, we added the following as covariates: gender, age at examination (scaled and centered), row on the 450k array, one variable denoting the batch (unique combination of bisulfite conversion plate and scan batch), the first three principal variance components as a proxy measure for cell heterogeneity, and, where appropriate, additional corrections specific for the phenotype of interest (BMI, TG, and glucose). We controlled for correlation within sibships in the GEE, thereby addressing (unmeasured) confounding on the familial level. We compared this model with an extension additionally containing both the phenotype of interest and a zero-one variable denoting the famine exposure status. Model fits were compared using analysis of variance (ANOVA), yielding a χ2 statistic for evaluation (df = 2). Each EWAS was corrected for genome-wide inflation by dividing the χ2 statistic by the inflation factor (λ) (68), and results were adjusted for multiple testing using false discovery rate (FDR) correction (69) where appropriate. The λ ranged between 1.12 and 1.19. Inflation was caused by the fact that sibling pairs were measured on the same microarrays (28). When sibling pairs and unrelated individuals are analyzed separately, the λ is close to one.
To quantify the mediation effect of DNAm, we performed model-based mediation analysis using the method originally developed by Imai et al. (25) in the R mediation package (26), which implements LMM and other advanced statistical models in a causal mediation testing framework. Mediation analysis on multiple CpGs was simultaneously performed on the mean of the standardized β values that were multiplied by the directionality of the association for each CpG (times −1 for negative relationships between DNAm and famine exposure and the later-life phenotype). We report on the βmediation, more often referred to as the estimate of the indirect/mediation path or “a × b” effect, and the proportion mediated, the percentage of the total effect on the outcome explained by the indirect/mediation path. CIs and test statistics for these measures were estimated by 10,000 Monte Carlo simulations.
DNAm was linked to gene expression in cis (<100 kb) by first performing an EWAS using the R package CATE (70) with DNAm of one mediating CpG as the dependent variable and correcting for age and gender and five latent variables (using the “ed” eigenvalue difference method). Possible inflation and bias inherent to the correlation structure of both DNAm and gene expression data were corrected using the R package bacon (71). Corrected estimates, standard errors, and test statistics for only those genes within 100 kb were then assessed and corrected by FDR for multiple testing. All P values reported were two-sided.
Supplementary Material
Acknowledgments
We express our gratitude to the participants of the Dutch Hunger Winter Families study, the staff of TNO Quality of Life, the staff of the Department of Gerontology and Geriatrics Study Center at the Leiden University Medical Center (LUMC), and the LUMC Central Clinical Chemical Laboratory. Funding: This study was supported by a VENI grant by the Netherlands Organization for Scientific Research (91617128 to E.W.T.), the U.S. National Institute of Aging (R01AG042190 to L.H.L. and B.T.H.), the European Union’s Seventh Framework Program IDEAL (259679), and the U.S. NIH (R01-HL067914 to L.H.L.). The funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript. Author contributions: Conceptualization: E.W.T., L.H.L., and B.T.H. Methodology: E.W.T., K.M.X., and E.W.v.Z. Investigation: E.W.T., R.C.S., A.D.S., and L.H.L. Formal analysis: E.W.T., R.L., and K.F.D. Validation: K.F.D. Software: E.W.T., R.L., and E.W.v.Z. Resources: A.D.S., Biobank-based Integrative Omics Studies (BIOS) Consortium, and L.H.L. Data curation: E.W.T., R.C.S., R.L., BIOS Consortium, A.D.S., and L.H.L. Writing (original draft): E.W.T., L.H.L., and B.T.H. Writing (review and editing): R.C.S., R.L., K.F.D., A.D.S., and P.E.S. Visualization: E.W.T., R.L., and K.F.D. Supervision: P.E.S., L.H.L., and B.T.H. Project administration: L.H.L. and B.T.H. Funding acquisition: E.W.T., P.E.S., L.H.L., and B.T.H. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Members of the BIOS Consortium: Peter A. ‘t Hoen, René Pool, Marleen M. van Greevenbroek, Coen D. Stehouwer, Carla J. van der Kallen, Casper G. Schalkwijk, Cisca Wijmenga, Sasha Zhernakova, Ettje F. Tigchelaar, Marian Beekman, Joris Deelen, Diana van Heemst, Jan H. Veldink, Leonard H. van den Berg, Cornelia M. van Duijn, Albert Hofman, André G. Uitterlinden, P. Mila Jhamai, Michael Verbiest, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Hailiang Mei, Jan Bot, Dasha V. Zhernakova, Peter Van ‘t Hof, Patrick Deelen, Irene Nooren, Matthijs Moed, Martijn Vermaat, Marc Jan Bonder, Freerk van Dijk, Michiel van Galen, Wibowo Arindrarto, Szymon M. Kielbasa, Morris A. Swertz, Aaron Isaacs, and Lude Franke.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/1/eaao4364/DC1
fig. S1. Quantile-quantile (QQ) plots for EWAS on famine exposure and phenotypes.
fig. S2. QQ plots for EWAS on early exposure and phenotypes.
fig. S3. Correlation between tissues for the nine mediating CpG dinucleotides.
table S1. Associations with gene expression and phenotypes.
table S2. Genome-wide screen for potential mediators: famine exposure and TG.
table S3. Early exposure.
table S4. Genome-wide screen for potential mediators: early exposure and triglycerides.
Reference (75)
REFERENCES AND NOTES
- 1.D. J. P. Barker, Fetal and Infant Origins of Adult Disease (British Medical Journal, 1992). [Google Scholar]
- 2.Rinaudo P., Wang E., Fetal programming and metabolic syndrome. Annu. Rev. Physiol. 74, 107–130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waterland R. A., Michels K. B., Epigenetic epidemiology of the developmental origins hypothesis. Annu. Rev. Nutr. 27, 363–388 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Jaenisch R., Bird A., Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Bogdarina I., Haase A., Langley-Evans S., Clark A. J., Glucocorticoid effects on the programming of AT1b angiotensin receptor gene methylation and expression in the rat. PLOS ONE 5, e9237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waterland R. A., Jirtle R. L., Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allard C., Desgagné V., Patenaude J., Lacroix M., Guillemette L., Battista M. C., Doyon M., Ménard J., Ardilouze J. L., Perron P., Bouchard L., Hivert M. F., Mendelian randomization supports causality between maternal hyperglycemia and epigenetic regulation of leptin gene in newborns. Epigenetics 10, 342–351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez-Salas P., Moore S. E., Baker M. S., Bergen A. W., Cox S. E., Dyer R. A., Fulford A. J., Guan Y., Laritsky E., Silver M. J., Swan G. E., Zeisel S. H., Innis S. M., Waterland R. A., Prentice A. M., Hennig B. J., Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat. Commun. 5, 3746 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guénard F., Deshaies Y., Cianflone K., Kral J. G., Marceau P., Vohl M.-C., Differential methylation in glucoregulatory genes of offspring born before vs. after maternal gastrointestinal bypass surgery. Proc. Natl. Acad. Sci. U.S.A. 110, 11439–11444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heijmans B. T., Tobi E. W., Stein A. D., Putter H., Blauw G. J., Susser E. S., Slagboom P. E., Lumey L. H., Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. U.S.A. 105, 17046–17049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joehanes R., Just A. C., Marioni R. E., Pilling L. C., Reynolds L. M., Mandaviya P. R., Guan W., Xu T., Elks C. E., Aslibekyan S., Moreno-Macias H., Smith J. A., Brody J. A., Dhingra R., Yousefi P., Pankow J. S., Kunze S., Shah S. H., McRae A. F., Lohman K., Sha J., Absher D. M., Ferrucci L., Zhao W., Demerath E. W., Bressler J., Grove M. L., Huan T., Liu C., Mendelson M. M., Yao C., Kiel D. P., Peters A., Wang-Sattler R., Visscher P. M., Wray N. R., Starr J. M., Ding J., Rodriguez C. J., Wareham N. J., Irvin M. R., Zhi D., Barrdahl M., Vineis P., Ambatipudi S., Uitterlinden A. G., Hofman A., Schwartz J., Colicino E., Hou L., Vokonas P. S., Hernandez D. G., Singleton A. B., Bandinelli S., Turner S. T., Ware E. B., Smith A. K., Klengel T., Binder E. B., Psaty B. M., Taylor K. D., Gharib S. A., Swenson B. R., Liang L., DeMeo D. L., O’Connor G. T., Herceg Z., Ressler K. J., Conneely K. N., Sotoodehnia N., Kardia S. L., Melzer D., Baccarelli A. A., van Meurs J. B., Romieu I., Arnett D. K., Ong K. K., Liu Y., Waldenberger M., Deary I. J., Fornage M., Levy D., London S. J., Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 9, 436–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soubry A., Murphy S. K., Huang Z., Murtha A., Schildkraut J. M., Jirtle R. L., Wang F., Kurtzberg J., Demark-Wahnefried W., Forman M. R., Hoyo C., The effects of depression and use of antidepressive medicines during pregnancy on the methylation status of the IGF2 imprinted control regions in the offspring. Clin. Epigenetics 3, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao-Lei L., Dancause K. N., Elgbeili G., Massart R., Szyf M., Liu A., Laplante D. P., King S., DNA methylation mediates the impact of exposure to prenatal maternal stress on BMI and central adiposity in children at age 13½ years: Project Ice Storm. Epigenetics 10, 749–761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao-Lei L., Veru F., Elgbeili G., Szyf M., Laplante D. P., King S., DNA methylation mediates the effect of exposure to prenatal maternal stress on cytokine production in children at age 13½ years: Project Ice Storm. Clin. Epigenetics 8, 54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Küpers L. K., Xu X., Jankipersadsing S. A., Vaez A., la Bastide-van G. S., Scholtens S., Nolte I. M., Richmond R. C., Relton C. L., Felix J. F., Duijts L., van Meurs J. B., Tiemeier H., Jaddoe V. W., Wang X., Corpeleijn E., Snieder H., DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int. J. Epidemiol. 44, 1224–1237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds R. M., Jacobsen G. H., Drake A. J., What is the evidence in humans that DNA methylation changes link events in utero and later life disease? Clin. Endocrinol. 78, 814–822 (2013). [DOI] [PubMed] [Google Scholar]
- 17.G. C. E. Burger, J. C. Drummond, H. R. Sandstead, in Malnutrition and Starvation in Western Netherlands, September 1944-July 1945, vol. 1 (General State Printing Office, 1948). [Google Scholar]
- 18.Lumey L. H., Stein A. D., Susser E., Prenatal famine and adult health. Annu. Rev. Public Health 32, 237–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lumey L. H., Stein A. D., Kahn H. S., Romijn J. A., Lipid profiles in middle-aged men and women after famine exposure during gestation: The Dutch Hunger Winter Families Study. Am. J. Clin. Nutr. 89, 1737–1743 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lumey L. H., Stein A. D., Kahn H. S.. Food restriction during gestation and impaired fasting glucose or glucose tolerance and type 2 diabetes mellitus in adulthood: Evidence from the Dutch Hunger Winter Families Study. J. Dev. Orig. Health Dis. 1, S164 (2009). [Google Scholar]
- 21.Ravelli A. C., van der Meulen J. H., Michels R. P., Osmond C., Barker D. J., Hales C. N., Bleker O. P., Glucose tolerance in adults after prenatal exposure to famine. Lancet 351, 173–177 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Ravelli A. C., van der Meulen J. H., Osmond C., Barker D. J., Bleker O. P., Obesity at the age of 50 y in men and women exposed to famine prenatally. Am. J. Clin. Nutr. 70, 811–816 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Stein A. D., Kahn H. S., Rundle A., Zybert P. A., van der Pal-de Bruin K., Lumey L. H., Anthropometric measures in middle age after exposure to famine during gestation: Evidence from the Dutch famine. Am. J. Clin. Nutr. 85, 869–876 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Lumey L. H., Stein A. D., Kahn H. S., van der Pal-de Bruin KM, Blauw G. J., Zybert P. A., Susser E. S., Cohort profile: The Dutch Hunger Winter families study. Int. J. Epidemiol. 36, 1196–1204 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Imai K., Keele L., Tingley D., A general approach to causal mediation analysis. Psychol. Methods 15, 309–334 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Tingley D., Yamamoto T., Hirose K., Keele L., Imai K., mediation: R Package for Causal Mediation Analysis. J. Stat. Softw. 59, 1–38 (2014).26917999 [Google Scholar]
- 27.Roseboom T. J., Painter R. C., van Abeelen A. F., Veenendaal M. V., de Rooij S. R., Hungry in the womb: What are the consequences? Lessons from the Dutch famine. Maturitas 70, 141–145 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Tobi E. W., Slieker R. C., Stein A. D., Suchiman H. E., Slagboom P. E., van Zwet E. W., Heijmans B. T., Lumey L. H., Early gestation as the critical time-window for changes in the prenatal environment to affect the adult human blood methylome. Int. J. Epidemiol. 44, 1211–1223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonder M. J., Luijk R., Zhernakova D. V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J., Slieker R. C., Jhamai P. M., Verbiest M., Suchiman H. E., Verkerk M., van der Breggen R., van Rooij J., Lakenberg N., Arindrarto W., Kielbasa S. M., Jonkers I., van ’t Hof P., Nooren I., Beekman M., Deelen J., van Heemst D., Zhernakova A., Tigchelaar E. F., Swertz M. A., Hofman A., Uitterlinden A. G., Pool R., van Dongen J., Hottenga J. J., Stehouwer C. D., van der Kallen C. J., Schalkwijk C. G., van den Berg L. H., van Zwet E. W., Mei H., Li Y., Lemire M., Hudson T. J.; BIOS Consortium, Slagboom P. E., Wijmenga C., Veldink J. H., van Greevenbroek M. M., van Duijn C. M., Boomsma D. I., Isaacs A., Jansen R., van Meurs J. B., ‘t Hoen P. A., Franke L., Heijmans B. T., Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 49, 131–138 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Dekkers K. F., van Iterson M., Slieker R. C., Moed M. H., Bonder M. J., van Galen M., Mei H., Zhernakova D. V., van den Berg L. H., Deelen J., van Dongen J., van Heemst D., Hofman A., Hottenga J. J., van der Kallen C. J., Schalkwijk C. G., Stehouwer C. D., Tigchelaar E. F., Uitterlinden A. G., Willemsen G., Zhernakova A., Franke L., ‘t Hoen P. A., Jansen R., van Meurs J., Boomsma D. I., van Duijn C. M., van Greevenbroek M. M., Veldink J. H., Wijmenga C.; BIOS Consortium, van Zwet E. W., Slagboom P. E., Jukema J. W., Heijmans B. T., Blood lipids influence DNA methylation in circulating cells. Genome Biol. 17, 138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Relton C. L., Gaunt T., McArdle W., Ho K., Duggirala A., Shihab H., Woodward G., Lyttleton O., Evans D. M., Reik W., Paul Y.-L., Ficz G., Ozanne S. E., Wipat A., Flanagan K., Lister A., Heijmans B. T., Ring S. M., Davey Smith G., Data resource profile: Accessible resource for integrated epigenomic studies (ARIES). Int. J. Epidemiol. 44, 1181–1190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slieker R. C., Bos S. D., Goeman J. J., Bovee J. V., Talens R. P., van der Breggen R., Suchiman H. E., Lameijer E.-W., Putter H., van den Akker E. B., Zhang Y., Jukema J. W., Slagboom P. E., Meulenbelt I., Heijmans B. T., Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 6, 26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahl S., Drong A., Lehne B., Loh M., Scott W. R., Kunze S., Tsai P.-C., Ried J. S., Zhang W., Yang Y., Tan S., Fiorito G., Franke L., Guarrera S., Kasela S., Kriebel J., Richmond R. C., Adamo M., Afzal U., Ala-Korpela M., Albetti B., Ammerpohl O., Apperley J. F., Beekman M., Bertazzi P. A., Black S. L., Blancher C., Bonder M.-J., Brosch M., Carstensen-Kirberg M., de Craen A. J. M., de Lusignan S., Dehghan A., Elkalaawy M., Fischer K., Franco O. H., Gaunt T. R., Hampe J., Hashemi M., Isaacs A., Jenkinson A., Jha S., Kato N., Krogh V., Laffan M., Meisinger C., Meitinger T., Mok Z. Y., Motta V., Ng H. K., Nikolakopoulou Z., Nteliopoulos G., Panico S., Pervjakova N., Prokisch H., Rathmann W., Roden M., Rota F., Rozario M. A., Sandling J. K., Schafmayer C., Schramm K., Siebert R., Slagboom P. E., Soininen P., Stolk L., Strauch K., Tai E.-S., Tarantini L., Thorand B., Tigchelaar E. F., Tumino R., Uitterlinden A. G., van Duijn C., van Meurs J. B. J., Vineis P., Wickremasinghe A. R., Wijmenga C., Yang T.-P., Yuan W., Zhernakova A., Batterham R. L., Davey Smith G., Deloukas P., Heijmans B. T., Herder C., Hofman A., Lindgren C. M., Milani L., van der Harst P., Peters A., Illig T., Relton C. L., Waldenberger M., Järvelin M.-R., Bollati V., Soong R., Spector T. D., Scott J., McCarthy M. I., Elliott P., Bell J. T., Matullo G., Gieger C., Kooner J. S., Grallert H., Chambers J. C., Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 541, 81–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al Muftah W. A., Al-Shafai M., Zaghlool S. B., Visconti A., Tsai P.-C., Kumar P., Spector T., Bell J., Falchi M., Suhre K., Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin. Epigenetics 8, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demerath E. W., Guan W., Grove M. L., Aslibekyan S., Mendelson M., Zhou Y.-H., Hedman Å. K., Sandling J. K., Li L.-A., Irvin M. R., Zhi D., Deloukas P., Liang L., Liu C., Bressler J., Spector T. D., North K., Li Y., Absher D. M., Levy D., Arnett D. K., Fornage M., Pankow J. S., Boerwinkle E., Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum. Mol. Genet. 24, 4464–4479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dick K. J., Nelson C. P., Tsaprouni L., Sandling J. K., Aïssi D., Wahl S., Meduri E., Morange P.-E., Gagnon F., Grallert H., Waldenberger M., Peters A., Erdmann J., Hengstenberg C., Cambien F., Goodall A. H., Ouwehand W. H., Schunkert H., Thompson J. R., Spector T. D., Gieger C., Trégouët D.-A., Deloukas P., Samani N. J., DNA methylation and body-mass index: A genome-wide analysis. Lancet 383, 1990–1998 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Chadwick L. H., The NIH roadmap epigenomics program data resource. Epigenomics 4, 317–324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beharry Z., Mahajan S., Zemskova M., Lin Y.-W., Tholanikunnel B. G., Xia Z., Smith C. D., Kraft A. S., The Pim protein kinases regulate energy metabolism and cell growth. Proc. Natl. Acad. Sci. U.S.A. 108, 528–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aksoy I., Sakabedoyan C., Bourillot P.-Y., Malashicheva A. B., Mancip J., Knoblauch K., Afanassieff M., Savatier P., Self-renewal of murine embryonic stem cells is supported by the serine/threonine kinases Pim-1 and Pim-3. Stem Cells 25, 2996–3004 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Vlacich G., Nawijn M. C., Webb G. C., Steiner D. F., Pim3 negatively regulates glucose-stimulated insulin secretion. Islets 2, 308–317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soriano-Tárraga C., Jiménez-Conde J., Giralt-Steinhauer E., Mola-Caminal M., Vivanco-Hidalgo R. M., Ois A., Rodríguez-Campello A., Cuadrado-Godia E., Sayols-Baixeras S., Elosua R., Roquer J.; GENESTROKE Consortium Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum. Mol. Genet. 25, 609–619 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Florath I., Butterbach K., Heiss J., Bewerunge-Hudler M., Zhang Y., Schöttker B., Brenner H., Type 2 diabetes and leucocyte DNA methylation: An epigenome-wide association study in over 1,500 older adults. Diabetologia 59, 130–138 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Chambers J. C., Loh M., Lehne B., Drong A., Kriebel J., Motta V., Wahl S., Elliott H. R., Rota F., Scott W. R., Zhang W., Tan S.-T., Campanella G., Chadeau-Hyam M., Yengo L., Richmond R. C., Adamowicz-Brice M., Afzal U., Bozaoglu K., Mok Z. Y., Ng H. K., Pattou F., Prokisch H., Rozario M. A., Tarantini L., Abbott J., Ala-Korpela M., Albetti B., Ammerpohl O., Bertazzi P. A., Blancher C., Caiazzo R., Danesh J., Gaunt T. R., de Lusignan S., Gieger C., Illig T., Jha S., Jones S., Jowett J., Kangas A. J., Kasturiratne A., Kato N., Kotea N., Kowlessur S., Pitkaniemi J., Punjabi P., Saleheen D., Schafmayer C., Soininen P., Tai E.-S., Thorand B., Tuomilehto J., Wickremasinghe A. R., Kyrtopoulos S. A., Aitman T. J., Herder C., Hampe J., Cauchi S., Relton C. L., Froguel P., Soong R., Vineis P., Jarvelin M.-R., Scott J., Grallert H., Bollati V., Elliott P., McCarthy M. I., Kooner J. S., Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: A nested case-control study. Lancet Diabetes Endocrinol. 3, 526–534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen A.-K., Zeilinger S., Kastenmüller G., Römisch-Margl W., Brugger M., Peters A., Meisinger C., Strauch K., Hengstenberg C., Pagel P., Huber F., Mohney R. P., Grallert H., Illig T., Adamski J., Waldenberger M., Gieger C., Suhre K., Epigenetics meets metabolomics: An epigenome-wide association study with blood serum metabolic traits. Hum. Mol. Genet. 23, 534–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joubert B. R., Felix J. F., Yousefi P., Bakulski K. M., Just A. C., Breton C., Reese S. E., Markunas C. A., Richmond R. C., Xu C. J., Küpers L. K., Oh S. S., Hoyo C., Gruzieva O., Söderhäll C., Salas L. A., Baiz N., Zhang H., Lepeule J., Ruiz C., Ligthart S., Wang T., Taylor J. A., Duijts L., Sharp G. C., Jankipersadsing S. A., Nilsen R. M., Vaez A., Fallin M. D., Hu D., Litonjua A. A., Fuemmeler B. F., Huen K., Kere J., Kull I., Munthe-Kaas M. C., Gehring U., Bustamante M., Saurel-Coubizolles M. J., Quraishi B. M., Ren J., Tost J., Gonzalez J. R., Peters M. J., Håberg S. E., Xu Z., van Meurs J. B., Gaunt T. R., Kerkhof M., Corpeleijn E., Feinberg A. P., Eng C., Baccarelli A. A., Benjamin Neelon S. E., Bradman A., Merid S. K., Bergström A., Herceg Z., Hernandez-Vargas H., Brunekreef B., Pinart M., Heude B., Ewart S., Yao J., Lemonnier N., Franco O. H., Wu M. C., Hofman A., McArdle W., Van der Vlies P., Falahi F., Gillman M. W., Barcellos L. F., Kumar A., Wickman M., Guerra S., Charles M.-A., Holloway J., Auffray C., Tiemeier H. W., Smith G. D., Postma D., Hivert M. F., Eskenazi B., Vrijheid M., Arshad H., Antó J. M., Dehghan A., Karmaus W., Annesi-Maesano I., Sunyer J., Ghantous A., Pershagen G., Holland N., Murphy S. K., DeMeo D. L., Burchard E. G., Ladd-Acosta C., Snieder H., Nystad W., Koppelman G. H., Relton C. L., Jaddoe V. W., Wilcox A., Melén E., London S. J., DNA methylation in newborns and maternal smoking in pregnancy: Genome-wide consortium meta-analysis. Am. J. Hum. Genet. 98, 680–696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah S., Bonder M. J., Marioni R. E., Zhu Z., McRae A. F., Zhernakova A., Harris S. E., Liewald D., Henders A. K., Mendelson M. M., Liu C., Joehanes R., Liang L.; BIOS Consortium, Levy D., Martin N. G., Starr J. M., Wijmenga C., Wray N. R., Yang J., Montgomery G. W., Franke L., Deary I. J., Visscher P. M., Improving phenotypic prediction by combining genetic and epigenetic associations. Am. J. Hum. Genet. 97, 75–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jakkaraju S., Zhe X., Pan D., Choudhury R., Schuger L., TIPs are tension-responsive proteins involved in myogenic versus adipogenic differentiation. Dev. Cell 9, 39–49 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Talens R. P., Boomsma D. I., Tobi E. W., Kremer D., Jukema J. W., Willemsen G., Putter H., Slagboom P. E., Heijmans B. T., Variation, patterns, and temporal stability of DNA methylation: Considerations for epigenetic epidemiology. FASEB J. 24, 3135-3144 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Pfeiffer L., Wahl S., Pilling L. C., Reischl E., Sandling J. K., Kunze S., Holdt L. M., Kretschmer A., Schramm K., Adamski J., Klopp N., Illig T., Hedman Å. K., Roden M., Hernandez D. G., Singleton A. B., Thasler W. E., Grallert H., Gieger C., Herder C., Teupser D., Meisinger C., Spector T. D., Kronenberg F., Prokisch H., Melzer D., Peters A., Deloukas P., Ferrucci L., Waldenberger M., DNA methylation of lipid-related genes affects blood lipid levels. Circ. Cardiovasc. Genet. 8, 334–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tobi E. W., Lumey L. H., Talens R. P., Kremer D., Putter H., Stein A. D., Slagboom P. E., Heijmans B. T., DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum. Mol. Genet. 18, 4046–4053 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tobi E. W., Slagboom P. E., van Dongen J., Kremer D., Stein A. D., Putter H., Heijmans B. T., Lumey L. H., Prenatal famine and genetic variation are independently and additively associated with DNA methylation at regulatory loci within IGF2/H19. PLOS ONE 7, e37933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tobi E. W., Goeman J. J., Monajemi R., Gu H., Putter H., Zhang Y., Slieker R. C., Stok A. P., Thijssen P. E., Muller F., van Zwet E. W., Bock C., Meissner A., Lumey L. H., Eline P. E., Slagboom B. T., DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat. Commun. 5, 5592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richmond R. C., Hemani G., Tilling K., Davey Smith G., Relton C. L., Challenges and novel approaches for investigating molecular mediation. Hum. Mol. Genet. 25, R149–R156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C., Lumey L. H., Exposure to the Chinese famine of 1959–61 in early life and long-term health conditions: A systematic review and meta-analysis. Int. J. Epidemiol. 46, 1157–1170 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Richmond R. C., Timpson N. J., Sørensen T. I., Exploring possible epigenetic mediation of early-life environmental exposures on adiposity and obesity development. Int. J. Epidemiol. 44, 1191–1198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendelson M. M., Marioni R. E., Joehanes R., Liu C., Hedman Å. K., Aslibekyan S., Demerath E. W., Guan W., Zhi D., Yao C., Huan T., Willinger C., Chen B., Courchesne P., Multhaup M., Irvin M. R., Cohain A., Schadt E. E., Grove M. L., Bressler J., North K., Sundström J., Gustafsson S., Shah S., McRae A. F., Harris S. E., Gibson J., Redmond P., Corley J., Murphy L., Starr J. M., Kleinbrink E., Lipovich L., Visscher P. M., Wray N. R., Krauss R. M., Fallin D., Feinberg A., Absher D. M., Fornage M., Pankow J. S., Lind L., Fox C., Ingelsson E., Arnett D. K., Boerwinkle E., Liang L., Levy D., Deary I. J., Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: A Mendelian randomization approach. PLOS Med. 14, e1002215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cupul-Uicab L. A., Skjaerven R., Haug K., Travlos G. S., Wilson R. E., Eggesbo M., Hoppin J. A., Whitworth K. W., Longnecker M. P., Exposure to tobacco smoke in utero and subsequent plasma lipids, ApoB, and CRP among adult women in the MoBa cohort. Environ. Health Perspect. 120, 1532–1537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.G. Trienekens, The food supply in The Netherlands during the Second World War, in Food, Science, Policy and Regulation in the Twentieth Century: International and Comparative Perspectives, D. F. Smith, J. Phillips, Eds. (Routledge, 2000), pp. 117–133. [Google Scholar]
- 59.Grootenhuis P. A., Westenbrink S., Sie C. M., de Neeling J. N., Kok F. J., Bouter L. M., A semiquantitative food frequency questionnaire for use in epidemiologic research among the elderly: Validation by comparison with dietary history. J. Clin. Epidemiol. 48, 859–868 (1995). [DOI] [PubMed] [Google Scholar]
- 60.van Iterson M., Tobi E. W., Slieker R. C., den Hollander W., Luijk R., Slagboom P. E., Heijmans B. T., MethylAid: Visual and interactive quality control of large Illumina 450k datasets. Bioinformatics 30, 3435–3437 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D., Irizarry R. A., Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bates D., Machler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 63.A. Kuznetsova, P. B. Brockhoff, R. H. B. Christensen, lmerTest: Tests in Linear Mixed Effects Models, 2.0-36 (2016); https://cran.r-project.org/web/packages/lmerTest/index.html.
- 64.K. Barton, MuMIn: Multi-Model Inference, 1.15.6 (2016); https://cran.r-project.org/web/packages/MuMIn/index.html.
- 65.Stein A. D., Rundle A., Wada N., Goldbohm R. A., Lumey L. H., Associations of gestational exposure to famine with energy balance and macronutrient density of the diet at age 58 years differ according to the reference population used. J. Nutr. 139, 1555–1561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stein A. D., Pierik F. H., Verrips G. H., Susser E. S., Lumey L. H., Maternal exposure to the Dutch famine before conception and during pregnancy: Quality of life and depressive symptoms in adult offspring. Epidemiology 20, 909–915 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Halekoh U., Højsgaard S., Yan J, The R Package geepack for generalized estimating equations. J. Stat. Softw. 15, 1–11 (2006). [Google Scholar]
- 68.Devlin B., Roeder K., Genomic control for association studies. Biometrics 55, 997–1004 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995). [Google Scholar]
- 70.J. Wang, Q. Zhao, cate: High Dimensional Factor Analysis and Confounder Adjusted Multiple Testing and Estimation, 1.0.4 (2015); https://cran.r-project.org/web/packages/cate/index.html.
- 71.van Iterson M., van Zwet E. W.; BIOS Consortium, Slagboom P. E., Heijmans B. T., Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 18, 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Das M., Sha J., Hidalgo B., Aslibekyan S., Do A. N., Zhi D., Sun D., Zhang T., Li S., Chen W., Srinivasan S. R., Tiwari H. K., Absher D., Ordovas J. M., Berenson G. S., Arnett D. K., Irvin M. R., Association of DNA methylation at CPT1A locus with metabolic syndrome in the genetics of lipid lowering drugs and diet network (GOLDN) study. PLOS ONE 11, e0145789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irvin M. R., Zhi D., Joehanes R., Mendelson M., Aslibekyan S., Claas S. A., Thibeault K. S., Patel N., Day K., Jones L. W., Liang L., Chen B. H., Yao C., Tiwari H. K., Ordovas J. M., Levy D., Absher D., Arnett D. K., Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid-lowering Drugs and Diet Network study. Circulation 130, 565–572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mamtani M., Kulkarni H., Dyer T. D., Goring H. H., Neary J. L., Cole S. A., Kent J. W., Kumar S., Glahn D. C., Mahaney M. C., Comuzzie A. G., Almasy L., Curran J. E., Duggirala R., Blangero J., Carless M. A., Genome- and epigenome-wide association study of hypertriglyceridemic waist in Mexican American families. Clin. Epigenetics 8, 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rönn T., Volkov P., Gillberg L., Kokosar M., Perfilyev A., Jacobsen A. L., Jørgensen S. W., Brøns C., Jansson P.-A., Eriksson K.-F., Pedersen O., Hansen T., Groop L., Stener-Victorin E., Vaag A., Nilsson E., Ling C., Impact of age, BMI and HbA1c levels on the genome-wide methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum. Mol. Genet. 24, 3792–3813 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/1/eaao4364/DC1
fig. S1. Quantile-quantile (QQ) plots for EWAS on famine exposure and phenotypes.
fig. S2. QQ plots for EWAS on early exposure and phenotypes.
fig. S3. Correlation between tissues for the nine mediating CpG dinucleotides.
table S1. Associations with gene expression and phenotypes.
table S2. Genome-wide screen for potential mediators: famine exposure and TG.
table S3. Early exposure.
table S4. Genome-wide screen for potential mediators: early exposure and triglycerides.
Reference (75)


