The ability to characterize phenomes—that is, to acquire in-depth phenotypic profiles at the scale of the whole organism—holds promise to enhance our understanding of genetic variation. Yet, phenomic profiling in vertebrates remains limited. Hur et al.1 developed microCT-based methods and a segmentation algorithm, FishCuT, enabling profiling of hundreds of phenotypic measures at a large number of anatomical sites in the axial skeleton of adult zebrafish (Fig. 1). Approximately 30,000 data points and ∼3500 skeletal elements were analyzed. Vertebral patterns conferred heightened sensitivity, with similar specificity, in discriminating mutant populations compared with analyzing individual vertebrae in isolation. Skeletal phenotypes associated with human brittle bone disease and thyroid stimulating hormone receptor hyperactivity were identified. Finally, allometric modeling-based methods were developed to aid in the discrimination of mutant phenotypes masked by altered growth. This study establishes virtues of microCT-based phenomics in zebrafish. FishCuT is available for download as a beta release.2
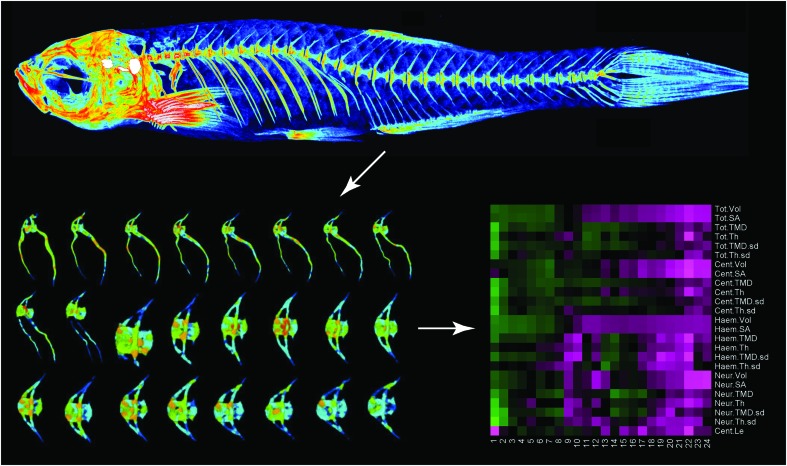
FIG. 1.
Using whole-body microCT scans (top), FishCuT isolates individual vertebrae (bottom left), segments each vertebra into three elements, and computes traits in each element. Standard scores are computed and arranged into “skeletal barcodes” that facilitate data visualization (bottom right). Within the barcode, each column represents a single vertebra and each row represents a phenotypic measure. Color images available online at www.liebertpub.com/zeb
References
- 1.Hur M, et al. MicroCT-based phenomics in the zebrafish skeleton reveals virtues of deep phenotyping in a distributed organ system. Elife 2017;6:e26014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwon RY. FishCuT. Github. 2860a3a. 2017. Available at https://github.com/ronaldkwon/FishCuT