Abstract
HIV-1 Nef is a flexible, multifunctional protein with several cellular targets that is required for pathogenicity of the virus. This protein maintains a high degree of genetic variation among intra- and inter-host isolates. HIV Nef is relevant to HIV-associated neurological diseases (HAND) in patients treated with combined anti-retroviral therapy because of the protein’s role in promoting survival and migration of infected brain macrophages. In this study, we analyzed 2,020 HIV Nef sequences derived from 22 different tissues and 31 subjects using a novel computational approach. This approach combines statistical regression and evolved neural networks (ENNs) to classify brain sequences based on the physical and chemical characteristics of functional Nef domains. Based on training, testing, and validation data, the method successfully classified brain Nef sequences at 84.5% and provided informative features for further examination. These included physicochemical features associated with the Src-homology-3 binding domain, the Nef loop (including the AP-2 Binding region), and a cytokine binding domain. Non-brain sequences from patients with HIV-associated neurological disease were frequently classified as brain, suggesting that the approach could indicate neurological-risk using blood-derived virus or for the development of biomarkers for use in assay systems aimed at drug efficacy studies for the treatment of HIV-associated neurological diseases.
Keywords: HIV, brain, neurological disease, Nef protein, analytical classification tools, macrophages
Introduction
In the HIV-infected population, current combined antiretroviral therapy (cART) can reduce plasma viral loads (pVL) to undetectable levels and restore T-cell counts; however, the spectrum of HIV-associated neurocognitive disorders (HAND) remains a common co-morbidity. Multiple studies have suggested that the HIV Nef protein plays a role in the development of HAND by acting directly and indirectly on brain cells (Khan et al. 2016; Ranki et al. 1995; Acharjee et al. 2014; Saribas, Khalili, and Sariyer 2015; Ghiglione and Turk 2011; Gray et al. 2011b). Nef increases monocyte and macrophage migration in animal models when injected into the brain (Mordelet et al. 2004; Overholser et al. 2003), and modulates cell-signaling pathways (Herbein et al. 2010). Nef contributes to an inflammatory environment by stimulating the production of inflammatory cytokines by macrophages (such as MIP-1α, MIP-1β, TNF-α, IL-1β, and IL-6) (Herbein et al. 2010), and corresponds to activation of the inflammatory factor NF-κB (Overholser et al. 2003; Olivetta et al. 2003). Nef is directly toxic to astrocytes and neurons in vitro, possibly through indirect activity of IP-10 (van Marle et al. 2004), and/or by stimulating the neurotoxin quinolinic acid (Smith et al. 2001). Nef is found in astrocytes of patients with HIV-associated dementia (HAD) and HIV encephalitis (HIVE) (Ranki et al. 1995), and astrocytic expression of Nef impairs spatial and recognition memory and increases neuronal loss (Chompre et al. 2013).
Previously, we compared twelve HIV Nef 3-dimensional (3D) protein structures, generated through sequencing and structural minimization, from AIDS patients with or without severe HIV-associated dementia (HAD) (Lamers, Poon, and McGrath 2011). We found several structural differences within Nef that could alter SH3 binding, change the orientation of the internal core domain, and make available an additional cysteine residue in the variable loop domain, which could alter membrane- or protein-binding potential. We proposed that these changes could play a functional role in the development of HAD. Others have also identified Nef structural-functional relationships (Geyer, Fackler, and Peterlin 2001; Arold and Baur 2001). Interestingly, in another study, we found that host-specific nef DNA and RNA emerged prior to env, and that early (<90 days) convergent evolution in nef, but not env, was evident in brains of simian immunodeficiency virus SIVMac251-infected macaques (Lamers et al. 2015; Strickland et al. 2011). These findings led us to hypothesize that certain HIV nef isolates are better adapted to immune cells in the brain environment (Lewis et al. 2012; Jia et al. 2012; Olivieri et al. 2010; Gray et al. 2011a), which are presumably of myeloid lineage (Williams and Burdo 2012; Williams et al. 2001).
In the current study, we used a newly developed evolved neural network approach to determine if it was possible to classify brain and non-brain Nef sequences, and more specifically, to identify brain-specific Nef (Lamers et al. 2017; Lamers, Fogel, et al. 2016). The approach elucidated structure-function properties of brain Nef that were not discernable by viewing sequence information alone. The study resulted in a comprehensive investigation into the physiochemical properties of brain-adapted Nef.
Materials and Methods
Sequences and alignment
Sequences were downloaded from either GenBank or the HIV Database at Los Alamos (www.lanl.HIV.gov), and included 2,020 unique Nef sequences derived from the anatomical tissues of 31 subjects (Fig I; Supplemental Table I). All 31 subjects in the study had multiple sequences and 26 subjects had sequences from multiple tissues. For 23 subjects, the primary pathology of the subjects at death was known (other than HIV-1 infection) (Fig IA). No cause of death was associated with sequence data for 8 subjects. Sequences from 4 subjects were derived from a longitudinally studied cohort of donors who were cART adherent and who had an undetectable plasma viral load at death (C02*, C04*, C05* and C09*) (Lamers, Rose, et al. 2016). Most brain sequences available for the study were derived from HAD or HAND patients; however, there were 25 sequences generated from brain tissue of patient (AZ) who died from a brain hemorrhage due to severe atherosclerosis, and two sequences were derived from brain tissue of a patient with lung cancer (C05*). Only two subjects included in the study (CB1 and CB3) had sequences from anatomical tissues paired with peripheral blood mononuclear cells (PBMC)-derived sequences at death. Overall, 512 brain sequences and 1,359 non-brain sequences derived from other lymphoid tissues were available for study (Fig IB). We did not categorize a subset of sequences as “brain” or “non-brain” that were obtained from CSF, spinal cord, and meninges because they can share similarity with either brain or lymphoid tissues (Lamers et al. 2011; Salemi et al. 2009; Gonzalez-Perez et al. 2012). These sequences were not used for neural network training or testing but were used as a validation set (as further described below) (Fig IB). Nine molecular clone sequences were included in the study (ADA, CAM1, HCB2, JRCSF, JRFL, MANC and 3 sequences derived from BAL). All amino acid sequences were aligned using Geneious software (ver. 10) (www.geneious.com) followed by manual editing to correct for any obvious alignment errors.
Fig I. Number of nef sequences in study derived from anatomical tissues from subjects used in study.
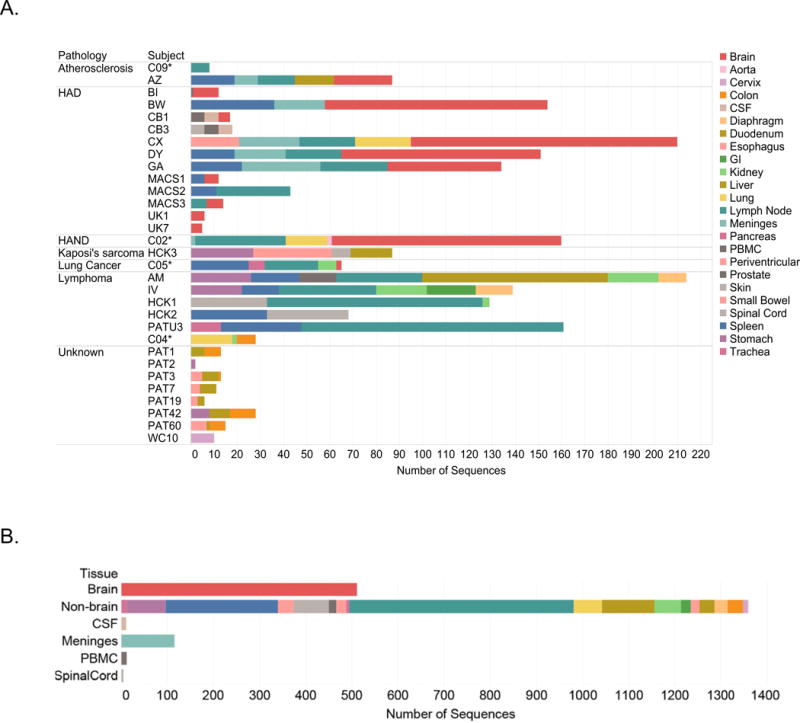
Panel A Subject IDs are on the left and are sorted by primary disease pathology (if known) at death. The number of sequences for each subject are on the y-axis. Asterisks indicate subjects who died while on cART with no measurable plasma viral load. Bars are colored by specific tissue of origin for each sequence. Panel B. Sequences in Panel A are sorted by major categories used for training and testing (brain and non-brain) and validation (CSF, meninges, PBMC and spinal cord) of the ENN.
Feature generation and correlations
72 different features describing various physicochemical characteristics of amino acids were identified from the available literature and resources such as ProtScale and ProtParam (www.expasy.org) (Wilkins et al. 1999) (Table I). These features were grouped into six major classes: amino acid size, shape or structure (n=24), polarity (n=6), composition (n=5), hydrophobicity (n=26), and miscellaneous other features such as those associated with HPLC and pKa (n=8). For each translated sequence, all features were calculated for the complete Nef protein and 10 specific functional domains in separate analyses. The chosen domains were already known as important for proper or altered Nef function (Fig II). For example the myristoylation domain, known for its high degree of variation, is required for the association of Nef with cellular lipid membranes (Bentham, Mazaleyrat, and Harris 2006). The conserved polyproline motif within the MHC1-DM domain is responsible for a number of functions, including MHC-1 down-modulation and positions associated with SH3 binding (Kuo et al. 2012; Jung et al. 2011). Other structural motifs were also considered in the study, including two alpha helix motifs in the center of the structure and another region near the 3′ end; a flexible loop domain, flanked by two beta-sheets that is associated with AP2 binding; and a region associated with altered cytokine immune-system signaling responses (Percario et al. 2015). This effort generated a total set of 72 features calculated for 11 total regions, or 792 total features.
Table I.
Physicochemical and structural features used for the study.
| Class | Features | ||
|---|---|---|---|
| A. Size, Shape, Structure | Molecular Weight (Gasteiger et al. 2005) | Bulkiness (Zimmerman, Eliezer, and Simha 1968) | Antiparallel Beta Strand (Lifson and Sander 1979) |
| Beta Turn (Deleage and Roux 1987) | Beta-sheet Levitt (Levitt 1978) | Alpha Chou and Fasman (Chou and Fasman 1978) | |
| Coil (Deleage and Roux 1987) | Beta Strand (Lifson and Sander 1979) | Parallel Beta (Lifson and Sander 1979) | |
| Surface Area (Darby and Creighton 1993) | Volume (Darby and Creighton 1993) | Surface Exposure (Grantham 1974) | |
| Average Flexibility(Grantham 1974) | Mol. Fraction of Buried Res.(Janin 1979) | Beta Chou and Fasman (Chou and Fasman 1978) | |
| % Accessible Residues (Janin 1979) | 2D Propensity (Lamers et al. 2008) | Transmembrane (Zhao and London 2006) | |
| Avg. Area Buried (Rose et al. 1985) | Membership Class (Grantham 1974) | Mass Membership Class (Lamers et al. 2008) | |
| Recognition Factors (Hofmann and Hadge 1987) | Alpha Helix Levitt (Levitt 1978) | Alpha Helix (Deleage and Roux 1987) | |
|
| |||
| B. Polarity | Polarity (Grantham 1974) | Charge1 | Charge Polarity (Zimmerman, Eliezer, and Simha 1968) |
| Charge Scale (Grantham 1974) | Grantham (Grantham 1974) | ||
|
| |||
| C. Composition | Amino Acid Composition (McCaldon and Argos 1988) | Length of V3 | Amino Acid Composition Swiss Prot (Gasteiger et al. 2005) |
| Relative Mutability (Dayhoff, Schwartz, and Orcutt 1978) | Total Sequence Glycosylation (Van Baelen et al. 2007) | ||
|
| |||
| D. Hydrophobicity | Sweet et al. (Sweet and Eisenberg 1983) | Kyte and Doolittle (Kyte and Doolittle 1982) | Hydrophobicity (Grantham 1974) |
| Abraham and Leo (Abraham and Leo 1987) | Bull and Breese (Bull and Breese 1974) | Guy (Guy 1985) | |
| Roseman (Roseman 1988) | Wolfen et al.(Wolfenden et al. 1981) | Wilson et al. (Wilson et al. 1981) | |
| Eisenberg et al. (Eisenberg et al. 1984) | Hopp and Woods (Hopp and Woods 1981) | Manvalan et al. (Manavalan and Ponnuswamy 1978) | |
| Fauchere et al. (Fauchere and Pliska 1983) | Janin (Janin 1979) | Rao and Argos (Mohana Rao and Argos 1986) | |
| Tanford (Tanford 1962) | Welling et al. (Welling et al. 1985) | Chothia (Chothia 1976) | |
| Cowan and Whittaker (Cowan and Whittaker 1990) | Parker (Parker, Guo, and Hodges 1986) | Browne (Browne, Bennett, and Solomon 1982) | |
| Meek (Meek 1980) | Aboderin(Aboderin 1971) | Rose et al. (Rose et al. 1985) | |
| Black and Mould (Black and Mould 1991) | Miyazawa et al. (Miyazawa and Jernigan 1996) | ||
|
| |||
| E. Local Features | Glycosylation at Positions 6–8 (Van Baelen et al. 2007) Charge at Position 30 (Milich, Margolin, and Swanstrom 1993) |
Glycosylation at Positions 5,7,9 (Van Baelen et al. 2007) | Charge at Position 12 (Milich, Margolin, and Swanstrom 1993) |
|
| |||
| F. HPLC and Other | Retention at pH 2.1 (Meek 1980) | HPLC/TFA (Browne, Bennett, and Solomon 1982) | Refractivity (Jones 1975) |
| HP Scale (Grantham 1974) | pKa alpha carboxylate (Anaspec 2013) | Exchange (Lamers et al. 2008) | |
| pKa Amine (Anaspec 2013) | pI at 25°C (Anaspec 2013) | ||
“Charge” feature = [+1] for K and R; [−1] for D and E.
Fig II. Variation in Nef sequence alignment and location of functional domains studied.
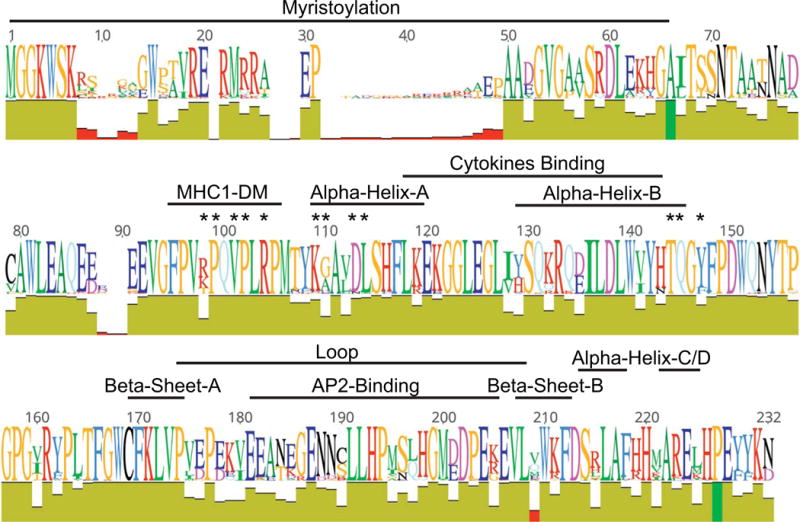
Amino acid sequence variation is presented as a “SeqLogo,” which shows the major amino acids along the alignments; the size of the letter coincides with its relative abundance at each position. Below the SeqLogo is an “identity” panel, where the height is equal to coverage at each position in the alignment and color represent conservation of amino acids with dark green = 100%, light green = 30–99%, red = less than 30%. Functional domains used for ZAPP development are indicated. Asterisks represent individual amino acids associated with SH3 binding.
A standard regression analysis was used to determine which of the 792 features individually maximized class separation between Nef sequences derived from brain and non-brain tissues. 62 of the 792 features had slight class separation (R2>0.8) and are described in Supplemental Table I for each region.
These 62 features were then used as input in the development of evolved neural networks using ZAPP as described below (Fogel 2008; Porto, Fogel, and Fogel 1995; Fogel, Fogel, and Porto 1990). The set of all sequences with known brain or non-brain origin was divided randomly into training (n=1251 sequences) and testing (n=626 sequences). As noted previously, a validation set was generated using sequences derived from CSF, spinal cord, and meninges. The process of generating training and testing sets was repeated three times with random assortments of the sequence data to avoid sampling bias.
ZAPP utilizes an evolutionary algorithm to optimize a population of neural networks using a set of input features on training and testing data (Lamers, Fogel, et al. 2016; Lamers et al. 2017; Fogel et al. 2015; Fogel et al. 2014; Porto, Fogel, and Fogel 1995). A population of neural networks was generated and scored with respect to minimization of mean squared error (MSE) on the output (in this case, the assignment of a sequence to either the brain or non-brain class). At each generation of the evolutionary optimization, models with lowest MSE were saved as “parent” neural networks for the generation of “offspring” neural networks with some variance in the weights associated with connections, or aspects of the neural network architecture itself. For neural network development, MSE is minimized on the training data, while performance on the testing data is measured every 50 generations to avoid over-fitting. The best neural network is then pulled from the population and assayed for performance on the training, testing, and validation sets. For the experiments conducted here, a population of fully-connected, feed-forward neural networks was used, with sigmoid functions for each node, and a fixed architecture. However, given the feature set had a maximum of 62 features, we forced the neural networks to use reduced sets of inputs, where the number of inputs was fixed at either 19 or 20 at a time, with the evolution allowed to randomly choose which of the 62 features to use at input. Evolutionary optimization was used to simultaneously optimize both the weights of the neural network and the features used as input. This approach allowed for the exploration of small feature subsets and their interdependency. In addition, the use of three different divisions of the training and testing data allowed us to determine if the same sub-selected features were obtained for all three replicates or if features were largely specific to each division of the data, which is important because a useful biomarker for brain vs. non-brain Nef would rely on features that were found to be common over as many divisions of the data as possible.
All neural networks were optimized using 50 parents and 50 offspring models in the evolving population, with tournament selection using 4 opponents. The number of generations of evolutionary optimization varied for each neural network architecture and dataset and was chosen as the maximum amount of training generations without increase in testing MSE. Once a best neural network was achieved, a binary discriminating threshold was determined to maximize classification accuracy of the two classes brain and non-brain. This threshold was fixed prior to use with any testing or validation samples. In this way, the neural network output (ZAPP score) could be compared to the threshold for the purpose of binning sequences into one or the other class. The thresholds used for each dataset are provided in Table II.
Table II.
Thresholds generated on the output of the best neural network on the training samples only, used for classification of all subsequent sequences in the testing and validation sets.
| Neural Network Architecture (number of input-hidden-output nodes) | DS1 | DS2 | DS3 |
|---|---|---|---|
| 62-10-1 | 0.82 | 0.87 | 0.77 |
| 20-10-1 | 0.72 | 0.76 | 0.79 |
| 10-10-1 | 0.75 | 0.77 | 0.78 |
| 10-5-1 | 0.73 | 0.72 | 0.76 |
| 5-3-1 | 0.78 | 0.78 | 0.74 |
VESPA, a web-based tool used to identify signature patterns in aligned sequence data (www.lanl.gov), was applied to two aligned sequence populations: those classified as brain and those classified as non-brain by the best neural network. VESPA calls a signature position when the most common character in the query data (in our case brain sequences) differs from the most common character in the background data (in our case non-brain sequences). and provides a frequency score.
Sequence Accession Numbers
AB221005, AB253432, AY314054-AY314063, AY713409, DQ357219-DQ357221, DQ358012-DQ358047, EF656973-EF656985, EF656994, EF656995, EF657017-EF657029, EF657035-EF657045, EF657063-EF657121, U63632, GQ868779-GQ869380, HM002302-HM002466, HQ174334-HQ174415, JQ990945, KU645011- KU645017, KU645030- KU645194, KU709356- KU709831, MF511264- MF511677, MF579824-MF579855, U23487, U66543-U66556.
Results
Amino acid variation along the Nef sequence population used in the study is shown in “SeqLogo” format in Fig II. Only two positions in the alignment (shown as dark green in the identity plot) were conserved at 100% (66A, 227P). The performance of each of five separate neural network architectures was evaluated on both training and testing data for dataset 1 (DS1), dataset 2 (DS2), dataset 3 (DS3)(Supplemental Tables II & III). These results indicate that neural networks with as few as 10 inputs have reasonable accuracy in separating brain from non-brain Nef sequences. The 10-10-1 neural networks retained reasonable mean performance on training and testing while using reduced sets of features and using 10 hidden nodes rather than 5 for improved generality. Among the three best 10-10-1 neural networks from the three datasets, only one feature, AA Comp SwissProt 2 in the SH3 binding domain, was common to all three, nine features were found in two of the three, and another nine features were unique to a single dataset (Table III). The most shared features were largely from the SH3 binding domain, Nef loop, cytokines binding, Alpha CD, and AP2 binding domain, suggesting that these domains have importance in the classification of brain vs. non-brain associated Nef. The 10-10-1 neural network from DS2 had the highest predictive accuracy on training (85.4%) and testing (84.3%) and was then used to process all of the sequences from the validation set. The resulting output scores were used, along with the same discriminating threshold used for the analysis of training and testing data, to assign validation sequences to the class brain or non-brain (Figs III and IV). Note that the distribution of the training and testing sequences was roughly similar across patients and over the final scores.
Table III.
Features identified through three independent optimizations of 10-10-1 neural networks. Model 2 had the highest predictive accuracy on training (85.4%) and testing (84.3%) and was used to process all remaining sequences with unknown etiology as either brain or non-brain.
| Model 1 | Model 2 | Model 3 | |||
|---|---|---|---|---|---|
| Location | Feature | Location | Feature | Location | Feature |
| SH3 Binding | AA Comp SwissProt 2 | SH3 Binding | AA Comp Swiss Prot 2 | SH3 Binding | AA Comp Swiss Prot 2 |
| AP2 Binding | Grantham | AP2Binding | Grantham | ||
| Nef Loop | ChargeConversion | NefLoop | Charge Conversion | ||
| Nef Loop | Polarity 2 | Nef Loop | Polarity 2 | ||
| SH3 Binding | Parker | SH3 Binding | Parker | ||
| Cytokines Binding | Cowan | Cytokines Binding | Wilson | ||
| Cytokines Binding | Welling | Cytokines Binding | Welling | ||
| Alpha CD | Beta Sheet Levett | Alpha CD | Beta Sheet Levett | ||
| AP2 Binding | HPScale | AP2 Binding | HP Scale | ||
| Alpha CD | Transmembrane | Alpha CD | Transmembrane | ||
| AP2 Binding | Tanford | ||||
| Cytokines Binding | Aboderin | ||||
| AP2 Binding | Kyle Doolittle | ||||
| SH3 Binding | Antiparallel Beta | ||||
| AlphaCD | Janin | ||||
| Alpha Helix A | HP Scale | ||||
| AP2 Binding | Browne | ||||
| Alpha Helix A | Kyte Doolittle | ||||
| Nef Loop | Wolfenden | ||||
Fig. III. ZAPP scores for all subjects.
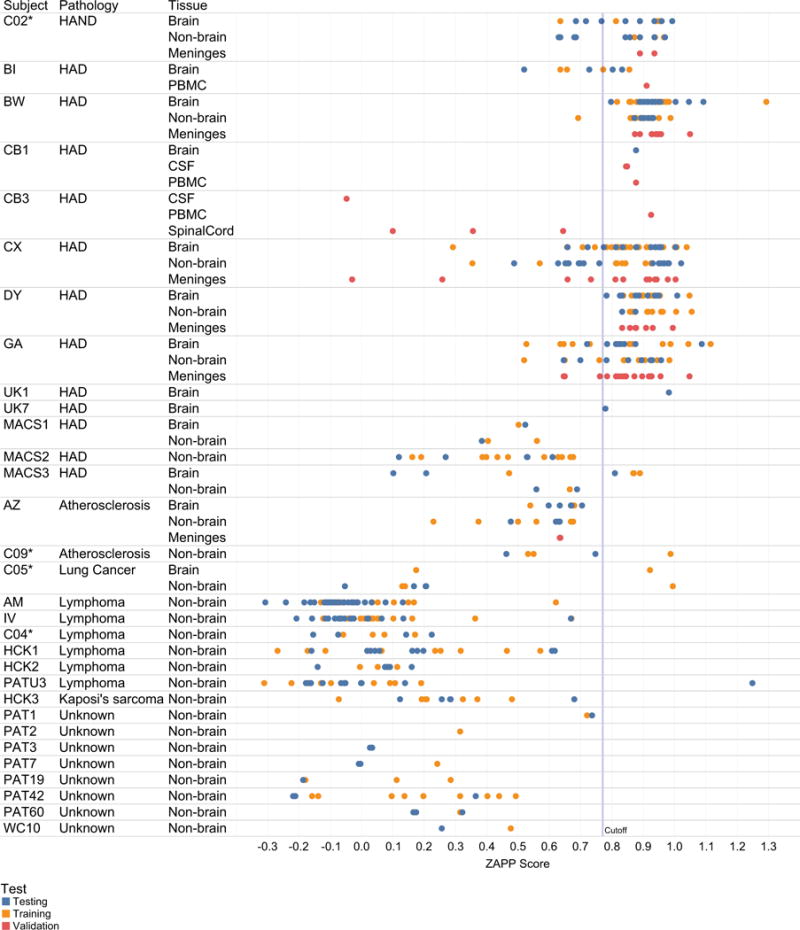
Subject ID, subject pathology and compartments (brain, non-brain, and validation sequence populations from meninges, spinal cord, PBMC, CSF) are listed on left-hand columns. An asterisk indicates subjects who died with no detectable plasma viral load. ZAPP score is on the x-axis, with a calculated discriminatory threshold for Brain and Non-Brain designated as a vertical line at 0.77. Scores for sequence used for testing, training, validation are shown and colored according to legend.
Fig. IV. ZAPP scores for sequences for selected cloned HIV-1 Nef sequences.
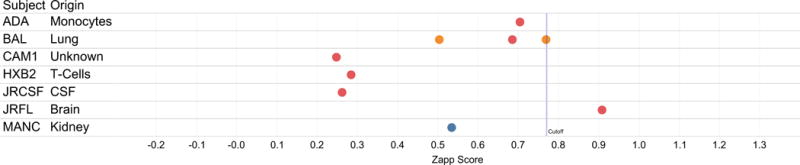
Sequence names are listed on left-hand columns along with the tissue of origin (if known). ZAPP score is shown on x-axis, with a calculated discriminatory threshold for Brain and Non-Brain designated as a vertical line at 0.77. Scores for sequence used for testing, training, validation are colored according to the legend in Fig. 3.
These results indicated that there were underlying physicochemical differences between Nef derived from patients with HAD as compared to other populations, particularly cancer patients (Fig III). 26 out of 27 of brain sequences from the two patients without neurological disease (C05, and AZ) were classified as non-brain. On the other hand, and 309 out of 451 non-brain sequences from patients with HAD/HAND were classified as brain, suggesting that brain-associated features persist in Nef derived from non-brain body tissues of patients with neurological disease. The sequences from one HAND patient in the study who was on cART until death (C02*) scored similarly to patients with HAD. The 55 sequences from HAD subjects MACS1 and MACS2 had an intermediary neural network score and were somewhat differentiated from cancer subjects, whose neural network scores were routinely low (non-brain); however, just 6 of these sequences were derived from actual brain tissues. Only one lymphoma subject (PATU3) had a single sequence that scored as brain; however, all additional sequences from this patient had low scores.
Sequences from seven molecular clones available from the NCBI AIDS reagent resource were included in the study (Fig IV). Through random allocation, two were used in training, one was used in testing, and six of these isolates were used in the validation set. Notably. the only molecular clone sequence (JR-FL) validated as brain by ZAPP was derived from brain tissue. JR-FL is derived from the same patient as the sequence JR-CSF, which was obtained from cerebral spinal fluid, and classified by ZAPP as non-brain.
The most important features for classification were the composition of SH3 binding positions, the Welling hydrophobicity scale applied to the cytokines binding domain, and the polarity of the Nef Loop and AP-1 binding domains (Table III). Each of these features have important biological functions associated to the function of Nef in the brain, specifically to astrocyte and macrophage infection. These biological implications are considered in greater detail in the discussion.
VESPA (www.lanl.gov) was applied to two aligned sequence populations: brain and non-brain as classified by the best 10-10-1 neural network from DS2. The program identified 13 positions in the brain sequence alignment that differed from the most common amino acids in each position in the non-brain sequence population (Fig V). Of the 13 total positions, six occurred in the highly variable myristoylation domain; however, most of these were had relatively low frequency scores (<22%). Note that the neural networks also did not identify the myristoylation domain important for brain vs non-brain separation in any of the best evolved networks. This result is similar to reports by others (Johnson et al. 2016). Five other positions had a difference in frequency score between 35–53% and occurred in regions of Nef that were identified by the neural networks as important for brain classification (corresponding to Fig 2 alignment positions 110, 128, 129, 160, 190) (Fig V). We sampled 7 brain and non-brain sequences from our alignment to view these 13 positions directly in sequences with known tissue origin; overall, it appeared unlikely that this signature alone could be used to correctly classify brain sequences (Fig V).
Fig. V. Signature analysis of predicted brain and predicted non-brain sequence populations.
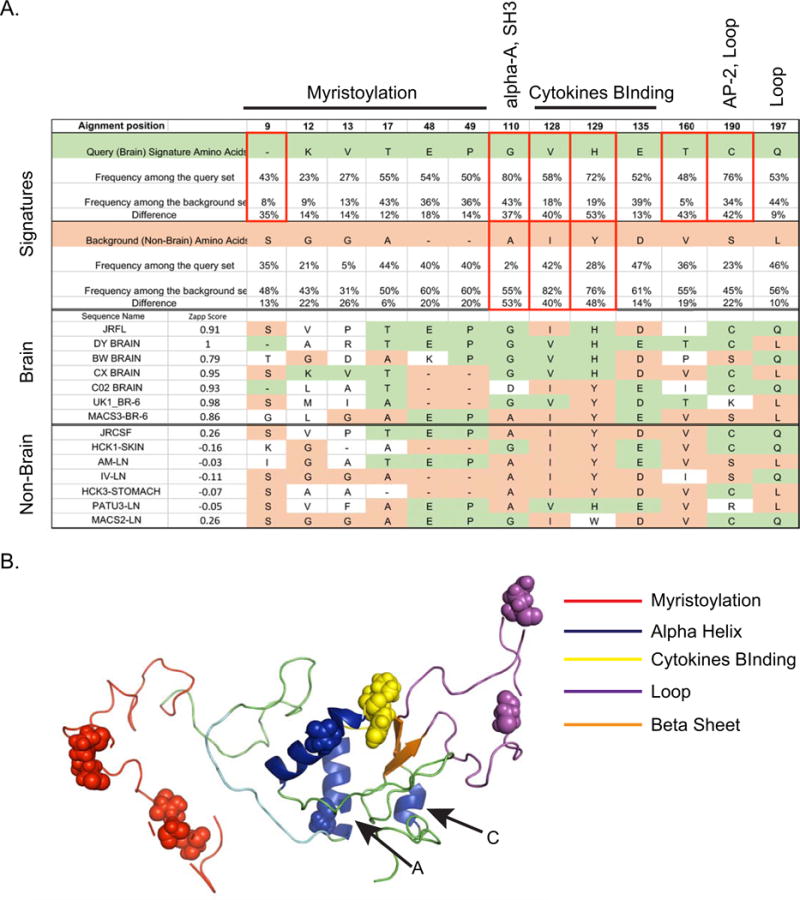
Panel A. A sequence alignment containing all DS2-predicted brain sequences was used to query against the background alignment containing all DS2-predicted non-brain sequences. A signature position (top row) is called when the most common character in the query data differs from the most common character in the background data. Signature amino acids for the brain alignment are colored green and for the non-brain alignment are colored light orange. Positions where the query differed from the background more than 30% are indicated with red boxes. In the middle and lower panels, a selection of brain and non-brain signature positions derived from Nef sequences with known tissue origin are shown. The ZAPP scores for each particular sequences are shown in the second column. The signature positions are colored according to a brain signature (green) or a non-brain signature (salmon). Positions where the amino acid did not match either signature are not colored. Panel B. 3D Nef JRCSF in “cartoon” format showing selected structural and functional regions. Identified signature positions are shown as “spheres”. Alpha Helices A and C, which were identified as useful for brain classification, are indicated.
Discussion
HIV-associated neurocognitive changes have been reported since early in the AIDS epidemic (Snider et al. 1983). These CNS symptoms were initially called “HIV Associated Dementia” or HAD (Navia, Jordan, and Price 1986; Antinori et al. 2007). HAD was associated with one or more histopathological abnormalities on autopsy that were unique to HIV patients, such as HIV encephalitis (HIVE), HIV leukoencephalopathy, and diffuse poliodystrophy (Budka 1991; Budka et al. 1991; Budka et al. 1987). HIVE was defined as multiple disseminated foci of microglia, macrophages, and multinucleated giant cells. HIV leukoencephalopathy was defined as diffuse damage to white matter that included myelin loss, reactive astrogliosis, macrophages, and multinucleated giant cells with little or no inflammatory infiltrates. Diffuse poliodystrophy was defined as reactive astrogliosis and microglial activation involving the cerebral gray matter (Budka et al. 1991; Pumarola-Sune et al. 1987). Overall, the postmortem finding most strongly associated with HAD was the number of activated macrophages present within affected areas of the brain (Glass et al. 1995). In addition, dendritic loss (Masliah et al. 1997), neuronal loss (Bell 1998), and brain HIV viral load (Bhaskaran et al. 2008) were associated with HAD.
After the introduction of cART, the incidence of HAD declined dramatically (Bhaskaran et al. 2008); however, milder forms of HIV-associated neurological disorders became prevalent, found in up to 50% of HIV-infected individuals (Heaton et al. 2010) and led to a new nosology called “HIV-Associated Neurocognitive Disorders” (HAND), which includes a diverse spectrum of pathologies, from mild neurocognitive disorders to HAD (Antinori et al. 2007). cART-treated patients with HAND differ from pre-cART patients in many ways. They are less likely to have detectable HIV RNA or have high levels of certain inflammatory biomarkers in their CSF (McArthur et al. 2004). At autopsy, HAND may or may not manifest florid neuropathological changes (Everall et al. 2009; Koedel et al. 1999; Lamers, Rose, et al. 2016). Currently there is a re-evaluation of the pathogenic mechanisms of HAND, including interest in persistent brain viral reservoirs or HIV proteins that can act independently of intact HIV that may drive persistent CNS inflammation and neurotoxicity, all of which are still associated with roles for activated macrophages {Koedel, 1999 #1711;Annunziata, 2003 #1712;Bergonzini, 2009 #1713;Fiala, 1996 #1714;Neumann, 1995 #387;van Marle, 2004 #246;Khan, 2016 #1648;Lenassi, 2010 #1679}.
HIV-1 Nef is a multifunctional protein required for pathogenicity of the virus (Kestler et al. 1991). The protein maintains a high degree of genetic variation among intra- and inter-host isolates. Increased attention has focused on the role of Nef in HAND pathogenesis, as it is expressed abundantly from astrocytes (Sami Saribas et al. 2017) and has been identified in blood and brain exosomes during cART (Raymond et al. 2016; Khan et al. 2016). This could constitute a potential mode of intercellular communication (van Niel et al. 2006) as Nef could prime permissive cells or activate immune responses in myeloid cells (Sami Saribas et al. 2017). Nef also dysregulates autophagy, an adaptive response to stress, in several cell types (Saribas, Khalili, and Sariyer 2015; Gupta et al. 2017). As the actions of Nef continue to grow, we were interested if brain-derived Nef shared characteristics that could allow for its identification and would provide further support for the protein’s role in HAND pathogenesis.
In this study we used statistical analysis combined with ENNs (Lamers et al. 2017; Lamers, Fogel, et al. 2016) to identify physicochemical alterations in Nef sequence domains that could be used to classify Nef sequences into either brain or non-brain classes. ENNs are nonlinear functions (neural networks) that map input variables/features to an output, through a hidden layer of nodes and processing functions. Evolutionary computation is used to search the space of nonlinear functions that has highest accuracy on training examples, while retaining useful generalization on unseen testing data. This approach allows for model optimization and simultaneous sub-selection of the features used as input to the model. In that sense, the approach provides a unique understanding of the features that are best suited for classification. This is in direct contrast to standard statistical methods that only use correlation analysis to search for “best” features one at a time. The best ENN discovered through this process had an 85.4% true positive and true negative classification accuracy on the training data set and 84.3% accuracy on the testing data set. Signature pattern analysis (Fig V) confirmed that changes associated with brain or non-brain data sets were more complex than simply the identity of the amino acids in positions across the Nef genome. The analysis suggests that subtle adjustments of amino acid physicochemical features within multiple functional domains helps define a Nef sequence associated with HAND tissues. These slight adjustments may only be identified through modeling approaches that allow for multiple features to be used simultaneously through a nonlinear processor. Despite the finding of brain-specific Nef sequences across numerous subjects, it remains difficult to translate this information into functional viral life cycle without further assessment of such variants in biological assays.
In all three ENN experiments with 10-10-1 neural networks, the composition of the Src homology-3 (SH3) binding domain was important in the classification of brain isolates. The SH3 binding capacity by Nef is required for many other HIV protein interactions to take place (Tokarev and Guatelli 2011; Arold et al. 1997; Alvarado et al. 2014). Nef interacts with SH3 domain proteins primarily via a conserved PxxP motif in its core domain; however, other positions in Nef are also associated with SH3 binding (Fig II) (Grzesiek et al. 1996; Lee et al. 1996). One of the alignment positions involved in SH3 binding, 110G, had the a highly conserved brain amino acid signature, identified in 80% of brain sequences and in only 2% of non-brain sequences. One well-known binding partner of HIV Nef is the monocyte-specific kinase Hck (Grzesiek et al. 1996), and therefore a specific SH3-binding configuration would be consistent with infection of brain immune cells, e.g. monocyte-derived macrophages (Cornall et al. 2013).
Polarity in the Nef Loop domain and the AP-2 binding domain (a subset of amino acids in the Nef loop) was used to classify brain viruses in two of the 10-10-1 neural networks. AP-2 is a cell-specific transcription factor family consisting of five closely related proteins (α, β, γ, δ and ε) that regulate the expression of specific target genes (Damberg 2005; Eckert et al. 2005). AP-2α and β are abundant isoforms in the brain and represent binding sites for putative transcriptional factors present in the Apolipoprotein E (ApoE) promoter. ApoE has been shown to co-localize with neuropathological lesions in Alzheimer’s disease. The Nef loop has other known functions, such as trafficking and Nef internalization (Zinkernagel 1976). The CD4 binding domain is concentrated in the N-terminal flexible loop and the core domain of Nef; recruitment of the partners AP and COP I is mediated by the C-terminal loop (Geyer, Fackler, and Peterlin 2001).
The Welling scale applied to the cytokine-binding domain was found to be important the classification of brain vs. non-brain Nef sequences. This unique scale was developed based on the percentage of amino acids in known antigenic determinants of a variety of proteins compared with the percentage of the amino acids in the average composition of a protein (Welling et al. 1985). Increased antigenicity of HIV brain Nefs could associate with macrophages that produce inflammatory cytokines, which are important in driving adaptive immune responses (Koppensteiner, Brack-Werner, and Schindler 2012).
Other structural features identified through our approach (Fig VB) were within the first alpha-helix region A and two other smaller regions in the unstructured C-terminal Nef. Alpha-helix A is in the hydrophobic pocket in the core of the protein; proper positioning of the helix with a tryptophan residue at position 15 (Fig II) is thought to be necessary for positioning of Nef at membrane surfaces (Johnson et al. 2016). In our own Nef structural minimization experiments (unpublished data), we have observed that the smaller defined alpha-helices in C-terminal regions have varied potential for the formation of secondary structure which is typical for unstructured regions of the protein (Barnham et al. 1997).
A variety of subjects, tissues, and pathologies were evaluated in the study. Of the 13 patients diagnosed prior to death with neurological disease, 11 had sequences that were classified as brain. Furthermore, nef sequences from the non-brain tissues of these patients were also frequently classified as brain-like, suggesting that sequences derived from anatomical tissues outside the brain could be used a biomarker for HAND potential. An extensive phylogenetic study of the HIV env gene by Rife et al., using six CD8-depleted and twelve naturally progressing SIV-infected macaques, demonstrated early entry of virus into the brain, followed by a period of adaptation in the periphery that lead to neurotropism later in infection (Rife et al. 2016). The higher scores for non-brain sequences derived from end-stage HAD patients are consistent with these findings, although evolution in env and nef is not always comparable (Lamers et al. 2015). Notably, 13 sequences derived from PBMCs from three patients with HAD (BI, CB1, and CB3) all classified as brain. A future study could apply ZAPP to pre-cART PBMC samples combined with post-mortem tissues, in order to substantiate the hypothesis that Nef derived from PBMCs, which scores in the brain class by ZAPP, is predictive of HIV-associated neurological disease.
Relatively low ZAPP scores were calculated for cancer patients, suggesting that a cancer-associated Nef sequence population may exist. Therefore, a similar ZAPP workflow could be designed to identify a Nef biomarker for cancer potential in HIV-infected patients. This finding could relate to the alternative macrophage activation states in these disease processes (Lamers et al. 2012). During lymphoma pathogenesis, M2-activated macrophages produce anti-inflammatory cytokines and Nef stimulates MDMs to form conduits that selectively transfer Nef to B-cells, thus allowing the protein to bypass B-cell immunoglobulin receptors (Lamers, Fogel, et al. 2010; Xu et al. 2009; Moir and Fauci 2010). During HAD pathogenesis, M1-macrophages produce inflammatory cytokines that trigger the recruitment of further macrophages into the brain (Lamers et al. 2012).
Most subjects in the study had an unknown course of cART; however, at least four were treated until death, at which time no measurable plasma viral load was detected. These tissues were derived from donors enrolled in the National Neuralological AIDS Bank (NNAB), which clinically treats and monitors patients with neurological disease or a severe life-limiting HIV-associated comorbidity. As we reported previously (Lamers et al. 2017; Rose et al. 2016), brain tissues from these subjects were assayed for HIV using a digital droplet PCR (ddPCR) approach as well as env-nef single genome sequencing of HIV RNA and DNA. While ddPCR confirmed the presence of HIV in the brain tissues for all four of these subjects, the larger segment of HIV DNA or RNA (env-nef) was only abundantly amplifiable from subject C02*, who was diagnosed with HAND and schizophrenia over a year prior to death (Lamers et al. 2017). Three different tissue biopsies from subject C05*, whose primary pathology at death was noted as metastatic lung cancer, yielded only two env-nef brain DNA sequences. However, all of these subjects had noted brain pathology at death: C02 with neoplastic infiltrate; C04, C05, and C09 with Alzheimer’s type-2 gliosis. Interestingly C02, C09 and C05 all had sequences (brain or not) that classified as brain-like, thus again suggesting that it could be possible to use our approach to indicate HAND-risk in patients on cART using Nef sequences derived from non-brain tissues such as blood.
The best neural network did not show perfect separation between brain and non-brain classes, which should be expected as the sequences are sampled from a very complex human system, and we might not have identified all of the features required to attain perfect separation. Furthermore, previous phylogenetic studies have shown that HIV/SIV from brain tissues can move across the blood-brain barrier and infect cells in the periphery (Lamers, Salemi, et al. 2010; Rife et al. 2016). Additional re-training and testing of such nonlinear classifiers using a larger number of brain sequences, especially from diverse donors, could increase the accuracy of the system. However, with an accuracy of 84.5%, the best neural network already has high potential to identify isolates with brain-like characteristics that could be used in vivo or in vitro to better understand HIV-associated neurological comorbidities.
While it can be difficult to generate an abundance of long HIV sequences (env-nef) from most cART+ subjects, recent studies have shown that defective proviruses from patients on cART can express HIV RNA species with unusual exon combinations that usually contain Nef, even when the full HIV genome cannot be detected (Imamichi et al. 2016), thus, the Nef protein remains an attractive target for biomarker development in cART+ patients in comparison with other HIV proteins. In order to increase our understanding of the HIV Nef protein’s role in HAND, additional studies are needed to identify if Nef proteins alone are frequently within brain tissues derived from patients on cART, even when quantitative PCR, which usually targets gag RNA, are negative.
HIV Nef is a fascinating protein due to its flexible structure, multiple functions and diverse cellular targets. Furthermore, HIV Nef is relevant to neurological disease in the cART-era when considering the protein’s role in promoting survival and migration of infected brain macrophages. In this study, we show that the novel ZAPP approach has strong potential to classify brain-tropic Nef sequences, especially with the generation of additional data for training and testing.
Supplementary Material
Acknowledgments
This project was funded by the National Institutes of Health grants R01MH100984 and UM1CA181255 to Michael S. McGrath and 1U24MH100929 to Elyse J. Singer.
Footnotes
Conflict of Interest
Lamers S.L., Barbier A.E., Rodriguez C., Nolan D.J., Rose R. are employed by Bioinfoexperts LLC; Fogel GB and Liu E are employed by Natural Selection Inc.
References
- Aboderin AA. An empirical hydrophobicity scale for a-aminoacids and some of its applications. Int J Biochem. 1971;2:537–44. [Google Scholar]
- Abraham DJ, Leo AJ. Extension of the fragment method to calculate amino acid zwitterion and side chain partition coefficients. Proteins. 1987;2:130–52. doi: 10.1002/prot.340020207. [DOI] [PubMed] [Google Scholar]
- Acharjee S, Branton WG, Vivithanaporn P, Maingat F, Paul AM, Dickie P, Baker GB, Power C. HIV-1 Nef expression in microglia disrupts dopaminergic and immune functions with associated mania-like behaviors. Brain Behav Immun. 2014;40:74–84. doi: 10.1016/j.bbi.2014.02.016. [DOI] [PubMed] [Google Scholar]
- Alvarado JJ, Tarafdar S, Yeh JI, Smithgall TE. Interaction with the Src homology (SH3–SH2) region of the Src-family kinase Hck structures the HIV-1 Nef dimer for kinase activation and effector recruitment. J Biol Chem. 2014;289:28539–53. doi: 10.1074/jbc.M114.600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaspec, Inc. pK and pI Values of Amino Acids. 2013 http://www.anaspec.com/html/pK_n_pl_Values_of_AminoAcids.html.
- Annunziata P. Blood-brain barrier changes during invasion of the central nervous system by HIV-1. Old and new insights into the mechanism. J Neurol. 2003;250:901–6. doi: 10.1007/s00415-003-1159-0. [DOI] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69:1789–99. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arold S, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997;5:1361–72. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- Arold ST, Baur AS. Dynamic Nef and Nef dynamics: how structure could explain the complex activities of this small HIV protein. Trends Biochem Sci. 2001;26:356–63. doi: 10.1016/s0968-0004(01)01846-1. [DOI] [PubMed] [Google Scholar]
- Barnham KJ, Monks SA, Hinds MG, Azad AA, Norton RS. Solution structure of a polypeptide from the N terminus of the HIV protein Nef. Biochemistry. 1997;36:5970–80. doi: 10.1021/bi9629945. [DOI] [PubMed] [Google Scholar]
- Bell JE. The neuropathology of adult HIV infection. Rev Neurol (Paris) 1998;154:816–29. [PubMed] [Google Scholar]
- Bentham M, Mazaleyrat S, Harris M. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J Gen Virol. 2006;87:563–71. doi: 10.1099/vir.0.81200-0. [DOI] [PubMed] [Google Scholar]
- Bergonzini V, Calistri A, Salata C, Del Vecchio C, Sartori E, Parolin C, Palu G. Nef and cell signaling transduction: a possible involvement in the pathogenesis of human immunodeficiency virus-associated dementia. J Neurovirol. 2009;15:238–48. doi: 10.1080/13550280902939748. [DOI] [PubMed] [Google Scholar]
- Bhaskaran K, Mussini C, Antinori A, Walker AS, Dorrucci M, Sabin C, Phillips A, Porter K, Cascade Collaboration Changes in the incidence and predictors of human immunodeficiency virus-associated dementia in the era of highly active antiretroviral therapy. Ann Neurol. 2008;63:213–21. doi: 10.1002/ana.21225. [DOI] [PubMed] [Google Scholar]
- Black SD, Mould DR. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Analytical biochemistry. 1991;193:72–82. doi: 10.1016/0003-2697(91)90045-u. [DOI] [PubMed] [Google Scholar]
- Browne CA, Bennett HP, Solomon S. The isolation of peptides by high-performance liquid chromatography using predicted elution positions. Analytical biochemistry. 1982;124:201–8. doi: 10.1016/0003-2697(82)90238-x. [DOI] [PubMed] [Google Scholar]
- Budka H. The definition of HIV-specific neuropathology. Acta Pathol Jpn. 1991;41:182–91. doi: 10.1111/j.1440-1827.1991.tb01645.x. [DOI] [PubMed] [Google Scholar]
- Budka H, Costanzi G, Cristina S, Lechi A, Parravicini C, Trabattoni R, Vago L. Brain pathology induced by infection with the human immunodeficiency virus (HIV). A histological, immunocytochemical, and electron microscopical study of 100 autopsy cases. Acta Neuropathol. 1987;75:185–98. doi: 10.1007/BF00687080. [DOI] [PubMed] [Google Scholar]
- Budka H, Wiley CA, Kleihues P, Artigas J, Asbury AK, Cho ES, Cornblath DR, Dal Canto MC, DeGirolami U, Dickson D, et al. HIV-associated disease of the nervous system: review of nomenclature and proposal for neuropathology-based terminology. Brain Pathol. 1991;1:143–52. doi: 10.1111/j.1750-3639.1991.tb00653.x. [DOI] [PubMed] [Google Scholar]
- Bull HB, Breese K. Surface tension of amino acid solutions: a hydrophobicity scale of the amino acid residues. Archives of biochemistry and biophysics. 1974;161:665–70. doi: 10.1016/0003-9861(74)90352-x. [DOI] [PubMed] [Google Scholar]
- Chompre G, Cruz E, Maldonado L, Rivera-Amill V, Porter JT, Noel RJ., Jr Astrocytic expression of HIV-1 Nef impairs spatial and recognition memory. Neurobiol Dis. 2013;49:128–36. doi: 10.1016/j.nbd.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Advances in enzymology and related areas of molecular biology. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cornall A, Mak J, Greenway A, Tachedjian G. HIV-1 infection of T cells and macrophages are differentially modulated by virion-associated Hck: a Nef-dependent phenomenon. Viruses. 2013;5:2235–52. doi: 10.3390/v5092235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan R, Whittaker RG. Hydrophobicity indices for amino acid residues as determined by high-performance liquid chromatography. Peptide research. 1990;3:75–80. [PubMed] [Google Scholar]
- Damberg M. Transcription factor AP-2 and monoaminergic functions in the central nervous system. J Neural Transm (Vienna) 2005;112:1281–96. doi: 10.1007/s00702-005-0325-1. [DOI] [PubMed] [Google Scholar]
- Darby NJ, Creighton TE. Protein structure: In focus. Oxford University Press; 1993. [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. Atlas of Protein Sequence and Structure 1978 [Google Scholar]
- Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein engineering. 1987;1:289–94. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–42. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Everall I, Vaida F, Khanlou N, Lazzaretto D, Achim C, Letendre S, Moore D, Ellis R, Cherner M, Gelman B, Morgello S, Singer E, Grant I, Masliah E. Cliniconeuropathologic correlates of human immunodeficiency virus in the era of antiretroviral therapy. Journal of neurovirology. 2009;15:360–70. doi: 10.3109/13550280903131915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchere JL, Pliska VE. Hydrophobic parameters-pi of amino-acid side-chains from the partitioning of N-acetyl-amino-acid amides. Eur J Med Chem. 1983;18:369–75. [Google Scholar]
- Fiala M, Rhodes RH, Shapshak P, Nagano I, Martinez-Maza O, Diagne A, Baldwin G, Graves M. Regulation of HIV-1 infection in astrocytes: expression of Nef, TNF-alpha and IL-6 is enhanced in coculture of astrocytes with macrophages. J Neurovirol. 1996;2:158–66. doi: 10.3109/13550289609146878. [DOI] [PubMed] [Google Scholar]
- Fogel DB, Fogel LJ, Porto VW. Evolving Neural Networks. Biological Cybernetics. 1990;63:487–93. [Google Scholar]
- Fogel GB. Computational intelligence approaches for pattern discovery in biological systems. Briefings in bioinformatics. 2008;9:307–16. doi: 10.1093/bib/bbn021. [DOI] [PubMed] [Google Scholar]
- Fogel GB, Enoch SL, Salemi M, Lamers SL, McGrath MS. Evolved Neural Networks for HIV-1 Co-receptor Identification. IEEE World Congress on Computational Intelligence; Beijing, China. 2014. [Google Scholar]
- Fogel GB, Lamers SL, Liu ES, Salemi M, McGrath MS. Identification of dual-tropic HIV-1 using evolved neural networks. Biosystems. 2015;137 doi: 10.1016/j.biosystems.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. Humana Press; 2005. [DOI] [PubMed] [Google Scholar]
- Geyer M, Fackler OT, Peterlin BM. Structure–function relationships in HIV-1 Nef. EMBO Rep. 2001;2:580–5. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione Y, Turk G. Nef performance in macrophages: the master orchestrator of viral persistence and spread. Curr HIV Res. 2011;9:505–13. doi: 10.2174/157016211798842080. [DOI] [PubMed] [Google Scholar]
- Glass JD, Fedor H, Wesselingh SL, McArthur JC. Immunocytochemical quantitation of human immunodeficiency virus in the brain: correlations with dementia. Ann Neurol. 1995;38:755–62. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez MP, O’Connell O, Lin R, Sullivan WM, Bell J, Simmonds P, Clapham PR. Independent evolution of macrophage-tropism and increased charge between HIV-1 R5 envelopes present in brain and immune tissue. Retrovirology. 2012;9:20. doi: 10.1186/1742-4690-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Gray LR, Gabuzda D, Cowley D, Ellett A, Chiavaroli L, Wesselingh SL, Churchill MJ, Gorry PR. CD4 and MHC class 1 down-modulation activities of nef alleles from brain- and lymphoid tissue-derived primary HIV-1 isolates. Journal of neurovirology. 2011a;17:82–91. doi: 10.1007/s13365-010-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu JS, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol. 1996;3:340–5. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- Gupta MK, Kaminski R, Mullen B, Gordon J, Burdo TH, Cheung JY, Feldman AM, Madesh M, Khalili K. HIV-1 Nef-induced cardiotoxicity through dysregulation of autophagy. Sci Rep. 2017;7:8572. doi: 10.1038/s41598-017-08736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy HR. Amino acid side-chain partition energies and distribution of residues in soluble proteins. Biophysical journal. 1985;47:61–70. doi: 10.1016/S0006-3495(85)83877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Clifford DB, Franklin DR, Jr, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–96. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G, Gras G, Khan KA, Abbas W. Macrophage signaling in HIV-1 infection. Retrovirology. 2010;7:34. doi: 10.1186/1742-4690-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann HJ, Hadge D. On the theoretical prediction of protein antigenic determinants from amino acid sequences. Biomedica biochimica acta. 1987;46:855–66. [PubMed] [Google Scholar]
- Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981;78:3824–8. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’Doherty U, Paxinos EE, Fauci AS, Lane HC. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A. 2016;113:8783–8. doi: 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J. Surface and inside volumes in globular proteins. Nature. 1979;277:491–2. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- Jia X, Singh R, Homann S, Yang H, Guatelli J, Xiong Y. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat Struct Mol Biol. 2012;19:701–6. doi: 10.1038/nsmb.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AL, Dirk BS, Coutu M, Haeryfar SM, Arts EJ, Finzi A, Dikeakos JD. A Highly Conserved Residue in HIV-1 Nef Alpha Helix 2 Modulates Protein Expression. mSphere. 2016;1 doi: 10.1128/mSphere.00288-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DD. Amino acid properties and side-chain orientation in proteins: a cross correlation appraoch. Journal of theoretical biology. 1975;50:167–83. doi: 10.1016/0022-5193(75)90031-4. [DOI] [PubMed] [Google Scholar]
- Jung J, Byeon IJ, Ahn J, Gronenborn AM. Structure, dynamics, and Hck interaction of full-length HIV-1 Nef. Proteins. 2011;79:1609–22. doi: 10.1002/prot.22986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestler HW, 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–62. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- Khan MB, Lang MJ, Huang MB, Raymond A, Bond VC, Shiramizu B, Powell MD. Nef exosomes isolated from the plasma of individuals with HIV-associated dementia (HAD) can induce Abeta(1–42) secretion in SH-SY5Y neural cells. J Neurovirol. 2016;22:179–90. doi: 10.1007/s13365-015-0383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koedel U, Kohleisen B, Sporer B, Lahrtz F, Ovod V, Fontana A, Erfle V, Pfister HW. HIV type 1 Nef protein is a viral factor for leukocyte recruitment into the central nervous system. J Immunol. 1999;163:1237–45. [PubMed] [Google Scholar]
- Koppensteiner R, Brack-Werner H, Schindler M. Macrophages and their relevance in Human Immunodeficiency Virus Type I infection. Retrovirology. 2012;9:82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo LS, Baugh LL, Denial SJ, Watkins RL, Liu M, Garcia JV, Foster JL. Overlapping effector interfaces define the multiple functions of the HIV-1 Nef polyproline helix. Retrovirology. 2012;9:47. doi: 10.1186/1742-4690-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lamers SL, Fogel GB, Huysentruyt LC, McGrath MS. HIV-1 nef protein visits B-cells via macrophage nanotubes: a mechanism for AIDS-related lymphoma pathogenesis? Curr HIV Res. 2010;8:638–40. doi: 10.2174/157016210794088209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Fogel GB, Liu ES, Nolan DJ, Salemi M, Barbier AE, Rose R, Singer EJ, McGrath MS. Predicted coreceptor usage at end-stage HIV disease in tissues derived from subjects on antiretroviral therapy with an undetectable plasma viral load. Infect Genet Evol. 2017;51:194–97. doi: 10.1016/j.meegid.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Fogel GB, Liu ES, Salemi M, McGrath MS. On the Physicochemical and Structural Modifications Associated with HIV-1 Subtype B Tropism Transition. AIDS Res Hum Retroviruses. 2016;32:829–40. doi: 10.1089/aid.2015.0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Fogel GB, Singer EJ, Salemi M, Nolan DJ, Huysentruyt LC, McGrath MS. HIV-1 Nef in macrophage-mediated disease pathogenesis. International reviews of immunology. 2012;31:432–50. doi: 10.3109/08830185.2012.737073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Gray RR, Salemi M, Huysentruyt LC, McGrath MS. HIV-1 phylogenetic analysis shows HIV-1 transits through the meninges to brain and peripheral tissues. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2011;11:31–7. doi: 10.1016/j.meegid.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Nolan DJ, Rife BD, Fogel GB, McGrath MS, Burdo TH, Autissier P, Williams KC, Goodenow MM, Salemi M. Tracking the Emergence of Host-Specific Simian Immunodeficiency Virus env and nef Populations Reveals nef Early Adaptation and Convergent Evolution in Brain of Naturally Progressing Rhesus Macaques. J Virol. 2015;89:8484–96. doi: 10.1128/JVI.01010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers AF, Poon SL, McGrath MS. HIV-1 nef protein structures associated with brain infection and dementia pathogenesis. PLoS One. 2011;6:e16659. doi: 10.1371/journal.pone.0016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Rose R, Maidji E, Agsalda-Garcia M, Nolan DJ, Fogel GB, Salemi M, Garcia DL, Bracci P, Yong W, Commins D, Said J, Khanlou N, Hinkin CH, Valdes Sueiras M, Mathisen G, Donovan S, Shirimizu B, Stoddart CA, McGrath MS, Singer EJ. HIV DNA is frequently present within pathologic tissues evaluated at autopsy from cART-treated patients with undetectable viral load. J Virol. 2016 doi: 10.1128/JVI.00674-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Salemi M, Galligan DC, Morris A, Gray R, Fogel G, Zhao L, McGrath MS. Human immunodeficiency virus-1 evolutionary patterns associated with pathogenic processes in the brain. J Neurovirol. 2010;16:230–41. doi: 10.3109/13550281003735709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Salemi M, McGrath MS, Fogel GB. Prediction of R5, X4, and R5X4 HIV-1 coreceptor usage with evolved neural networks. IEEE/ACM Trans Comput Biol Bioinform. 2008;5:291–300. doi: 10.1109/TCBB.2007.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–42. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17:4277–85. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Lee P, Ng HL, Yang OO. Immune selection in vitro reveals human immunodeficiency virus type 1 Nef sequence motifs important for its immune evasion function in vivo. J Virol. 2012;86:7126–35. doi: 10.1128/JVI.00878-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifson S, Sander C. Antiparallel and parallel beta-strands differ in amino acid residue preferences. Nature. 1979;282:109–11. doi: 10.1038/282109a0. [DOI] [PubMed] [Google Scholar]
- Manavalan P, Ponnuswamy PK. Hydrophobic character of amino acid residues in globular proteins. Nature. 1978;275:673–4. doi: 10.1038/275673a0. [DOI] [PubMed] [Google Scholar]
- Masliah E, Heaton RK, Marcotte TD, Ellis RJ, Wiley CA, Mallory M, Achim CL, McCutchan JA, Nelson JA, Atkinson JH, Grant I. Dendritic injury is a pathological substrate for human immunodeficiency virus-related cognitive disorders. HNRC Group The HIV Neurobehavioral Research Center. Ann Neurol. 1997;42:963–72. doi: 10.1002/ana.410420618. [DOI] [PubMed] [Google Scholar]
- McArthur JC, McDermott MP, McClernon D, Hillaire CSt, Conant K, Marder K, Schifitto G, Selnes OA, Sacktor N, Stern Y, Albert SM, Kieburtz K, deMarcaida JA, Cohen B, Epstein LG. Attenuated central nervous system infection in advanced HIV/AIDS with combination antiretroviral therapy. Archives of neurology. 2004;61:1687–96. doi: 10.1001/archneur.61.11.1687. [DOI] [PubMed] [Google Scholar]
- McCaldon P, Argos P. Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide sequences. Proteins. 1988;4:99–122. doi: 10.1002/prot.340040204. [DOI] [PubMed] [Google Scholar]
- Meek JL. Prediction of peptide retention times in high-pressure liquid chromatography on the basis of amino acid composition. Proc Natl Acad Sci U S A. 1980;77:1632–6. doi: 10.1073/pnas.77.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milich B, Margolin L, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–34. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa S, Jernigan RL. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256:623–44. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- Mohana Rao JK, Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochimica et biophysica acta. 1986;869:197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Moir S, Fauci AS. Nef, macrophages and B cells: a highway for evasion. Immunol Cell Biol. 2010;88:1–2. doi: 10.1038/icb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordelet E, Kissa K, Cressant A, Gray F, Ozden S, Vidal C, Charneau P, Granon S. Histopathological and cognitive defects induced by Nef in the brain. FASEB J. 2004;18:1851–61. doi: 10.1096/fj.04-2308com. [DOI] [PubMed] [Google Scholar]
- Navia BD, Jordan BA, Price RW. The AIDS dementia complex: I. Clinical features. Ann Neurol. 1986;19:517–24. doi: 10.1002/ana.410190602. [DOI] [PubMed] [Google Scholar]
- Neumann M, Felber BK, Kleinschmidt A, Froese B, Erfle V, Pavlakis GN, Brack-Werner R. Restriction of human immunodeficiency virus type 1 production in a human astrocytoma cell line is associated with a cellular block in Rev function. J Virol. 1995;69:2159–67. doi: 10.1128/jvi.69.4.2159-2167.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetta E, Percario Z, Fiorucci G, Mattia G, Schiavoni I, Dennis C, Jager J, Harris M, Romeo G, Affabris E, Federico M. HIV-1 Nef induces the release of inflammatory factors from human monocyte/macrophages: involvement of Nef endocytotic signals and NF-kappa B activation. J Immunol. 2003;170:1716–27. doi: 10.4049/jimmunol.170.4.1716. [DOI] [PubMed] [Google Scholar]
- Olivieri KC, Agopian KA, Mukerji J, Gabuzda D. Evidence for adaptive evolution at the divergence between lymphoid and brain HIV-1 nef genes. AIDS Res Hum Retroviruses. 2010;26:495–500. doi: 10.1089/aid.2009.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholser ED, Coleman GD, Bennett JL, Casaday RJ, Zink MC, Barber SA, Clements JE. Expression of simian immunodeficiency virus (SIV) nef in astrocytes during acute and terminal infection and requirement of nef for optimal replication of neurovirulent SIV in vitro. J Virol. 2003;77:6855–66. doi: 10.1128/JVI.77.12.6855-6866.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JM, Guo D, Hodges RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–32. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- Percario ZA, Ali M, Mangino G, Affabris E. Nef, the shuttling molecular adaptor of HIV, influences the cytokine network. Cytokine Growth Factor Rev. 2015;26:159–73. doi: 10.1016/j.cytogfr.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Porto VW, Fogel DB, Fogel LBJ. Alternative Neural Network Training Methods. IEEE Expert. 1995;10:16–22. [Google Scholar]
- Pumarola-Sune T, Navia BA, Cordon-Cardo C, Cho ES, Price RW. HIV antigen in the brains of patients with the AIDS dementia complex. Ann Neurol. 1987;21:490–6. doi: 10.1002/ana.410210513. [DOI] [PubMed] [Google Scholar]
- Ranki A, Nyberg M, Ovod V, Haltia M, Elovaara I, Raininko R, Haapasalo H, Krohn K. Abundant expression of HIV Nef and Rev proteins in brain astrocytes in vivo is associated with dementia. AIDS. 1995;9:1001–8. doi: 10.1097/00002030-199509000-00004. [DOI] [PubMed] [Google Scholar]
- Raymond AD, Diaz P, Chevelon S, Agudelo M, Yndart-Arias A, Ding H, Kaushik A, Jayant RD, Nikkhah-Moshaie R, Roy U, Pilakka-Kanthikeel S, Nair MP. Microglia-derived HIV Nef+ exosome impairment of the blood-brain barrier is treatable by nanomedicine-based delivery of Nef peptides. J Neurovirol. 2016;22:129–39. doi: 10.1007/s13365-015-0397-0. [DOI] [PubMed] [Google Scholar]
- Rife BD, Nolan DJ, Lamers SL, Autissier P, Burdo T, Williams KC, Salemi M. Evolution of Neuroadaptation in the Periphery and Purifying Selection in the Brain Contribute to Compartmentalization of Simian Immunodeficiency Virus (SIV) in the Brains of Rhesus Macaques with SIV-Associated Encephalitis. J Virol. 2016;90:6112–26. doi: 10.1128/JVI.00137-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229:834–8. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- Rose R, Lamers SL, Nolan DJ, Maidji E, Faria NR, Pybus OG, Dollar JJ, Maruniak SA, McAvoy AC, Salemi M, Stoddart C, Singer E, McGrath MS. HIV maintains an evolving and dispersed population among multiple tissues during suppressive cART with periods of rapid expansion corresponding to the onset of cancer. J Virol. 2016 doi: 10.1128/JVI.00684-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman MA. Hydrophilicity of polar amino acid side-chains is markedly reduced by flanking peptide bonds. J Mol Biol. 1988;200:513–22. doi: 10.1016/0022-2836(88)90540-2. [DOI] [PubMed] [Google Scholar]
- Salemi M, Lamers SL, Huysentruyt LC, Galligan D, Gray RR, Morris A, McGrath MS. Distinct patterns of HIV-1 evolution within metastatic tissues in patients with non-Hodgkins lymphoma. PLoS One. 2009;4:e8153. doi: 10.1371/journal.pone.0008153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sami Saribas A, Cicalese S, Ahooyi TM, Khalili K, Amini S, Sariyer IK. HIV-1 Nef is released in extracellular vesicles derived from astrocytes: evidence for Nef-mediated neurotoxicity. Cell Death Dis. 2017;8:e2542. doi: 10.1038/cddis.2016.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Khalili K, Sariyer IK. Dysregulation of autophagy by HIV-1 Nef in human astrocytes. Cell Cycle. 2015;14:2899–904. doi: 10.1080/15384101.2015.1069927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Guillemin GJ, Pemberton L, Kerr S, Nath A, Smythe GA, Brew BJ. Quinolinic acid is produced by macrophages stimulated by platelet activating factor, Nef and Tat. Journal of neurovirology. 2001;7:56–60. doi: 10.1080/135502801300069692. [DOI] [PubMed] [Google Scholar]
- Snider WD, Simpson DM, Nielsen S, Gold JW, Metroka CE, Posner JB. Neurological complications of acquired immune deficiency syndrome: analysis of 50 patients. Ann Neurol. 1983;14:403–18. doi: 10.1002/ana.410140404. [DOI] [PubMed] [Google Scholar]
- Strickland SL, Gray RR, Lamers SL, Burdo TH, Huenink E, Nolan DJ, Nowlin B, Alvarez X, Midkiff CC, Goodenow MM, Williams K, Salemi M. Significant genetic heterogeneity of the SIVmac251 viral swarm derived from different sources. AIDS Res Hum Retroviruses. 2011;27:1327–32. doi: 10.1089/aid.2011.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet RM, Eisenberg D. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J Mol Biol. 1983;171:479–88. doi: 10.1016/0022-2836(83)90041-4. [DOI] [PubMed] [Google Scholar]
- Tanford C. Contribution of hydrophobic interactions to the stability of the globular conformation of proteins. J Am Chem Soc. 1962;84 [Google Scholar]
- Tokarev A, Guatelli J. Misdirection of membrane trafficking by HIV-1 Vpu and Nef: Keys to viral virulence and persistence. Cell Logist. 2011;1:90–102. doi: 10.4161/cl.1.3.16708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays. Journal of virological methods. 2007;146:61–73. doi: 10.1016/j.jviromet.2007.06.003. [DOI] [PubMed] [Google Scholar]
- van Marle G, Henry S, Todoruk T, Sullivan A, Silva C, Rourke SB, Holden J, McArthur JC, Gill MJ, Power C. Human immunodeficiency virus type 1 Nef protein mediates neural cell death: a neurotoxic role for IP-10. Virology. 2004;329:302–18. doi: 10.1016/j.virol.2004.08.024. [DOI] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- Welling GW, Weijer WJ, van der Zee R, Welling-Wester S. Prediction of sequential antigenic regions in proteins. FEBS letters. 1985;188:215–8. doi: 10.1016/0014-5793(85)80374-4. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods in molecular biology. 1999;112:531–52. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Williams K, Burdo TH. Monocyte mobilization, activation markers, and unique macrophage populations in the brain: observations from SIV infected monkeys are informative with regard to pathogenic mechanisms of HIV infection in humans. Journal of neuroimmune pharmacology: the official journal of the Society on NeuroImmune Pharmacology. 2012;7:363–71. doi: 10.1007/s11481-011-9330-3. [DOI] [PubMed] [Google Scholar]
- Williams KC, Corey S, Westmoreland SV, Pauley D, Knight H, deBakker C, Alvarez X, Lackner AA. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. The Journal of experimental medicine. 2001;193:905–15. doi: 10.1084/jem.193.8.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KJ, Honegger A, Stotzel RP, Hughes GJ. The behaviour of peptides on reverse-phase supports during high-pressure liquid chromatography. The Biochemical journal. 1981;199:31–41. doi: 10.1042/bj1990031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R, Andersson L, Cullis PM, Southgate CC. Affinities of amino acid side chains for solvent water. Biochemistry. 1981;20:849–55. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- Xu W, Santini PA, Sullivan JS, He B, Shan M, Ball SC, Dyer WB, Ketas TJ, Chadburn A, Cohen-Gould L, Knowles DM, Chiu A, Sanders RW, Chen K, Cerutti A. HIV-1 evades virus-specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long-range intercellular conduits. Nat Immunol. 2009;10:1008–17. doi: 10.1038/ni.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, London E. An amino acid “transmembrane tendency” scale that approaches the theoretical limit to accuracy for prediction of transmembrane helices: relationship to biological hydrophobicity. Protein science: a publication of the Protein Society. 2006;15:1987–2001. doi: 10.1110/ps.062286306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JM, Eliezer N, Simha R. The characterization of amino acid sequences in proteins by statistical methods. Journal of theoretical biology. 1968;21:170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- Zinkernagel RM. Virus-specific T-cell-mediated cytotoxicity across the H-2 barrier to virus-altered alloantigen. Nature. 1976;261:139–41. doi: 10.1038/261139a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


