Abstract
The Colorado potato beetle is one of the most challenging agricultural pests to manage. It has shown a spectacular ability to adapt to a variety of solanaceaeous plants and variable climates during its global invasion, and, notably, to rapidly evolve insecticide resistance. To examine evidence of rapid evolutionary change, and to understand the genetic basis of herbivory and insecticide resistance, we tested for structural and functional genomic changes relative to other arthropod species using genome sequencing, transcriptomics, and community annotation. Two factors that might facilitate rapid evolutionary change include transposable elements, which comprise at least 17% of the genome and are rapidly evolving compared to other Coleoptera, and high levels of nucleotide diversity in rapidly growing pest populations. Adaptations to plant feeding are evident in gene expansions and differential expression of digestive enzymes in gut tissues, as well as expansions of gustatory receptors for bitter tasting. Surprisingly, the suite of genes involved in insecticide resistance is similar to other beetles. Finally, duplications in the RNAi pathway might explain why Leptinotarsa decemlineata has high sensitivity to dsRNA. The L. decemlineata genome provides opportunities to investigate a broad range of phenotypes and to develop sustainable methods to control this widely successful pest.
Introduction
The Colorado potato beetle, Leptinotarsa decemlineata Say 1824 (Coleoptera: Chrysomelidae), is widely considered one of the world’s most successful globally-invasive insect herbivores, with costs of ongoing management reaching tens of millions of dollars annually1 and projected costs if unmanaged reaching billions of dollars2. This beetle was first identified as a pest in 1859 in the Midwestern United States, after it expanded from its native host plant, Solanum rostratum (Solanaceae), onto potato (S. tuberosum)3. As testimony to the difficulty in controlling L. decemlineata, the species has the dubious honor of starting the pesticide industry, when Paris Green (copper (II) acetoarsenite) was first applied to control it in the United States in 18644. Leptinotarsa decemlineata is now widely recognized for its ability to rapidly evolve resistance to insecticides, as well as a wide range of abiotic and biotic stresses5, and for its global expansion across 16 million km2 to cover the entire Northern Hemisphere within the 20th century6. Over the course of 150 years of research, L. decemlineata has been the subject in more than 9,700 publications (according to the Web of ScienceTM Core Collection of databases) ranging from molecular to organismal biology from the fields of agriculture, entomology, molecular biology, ecology, and evolution.
In order to be successful, L. decemlineata evolved to exploit novel host plants, to inhabit colder climates at higher latitudes7–9, and to cope with a wide range of novel environmental conditions in agricultural landscapes10,11. Genetic data suggest the potato-feeding pest lineage directly descended from populations that feed on S. rostratum in the U.S. Great Plains12. This beetle subsequently expanded its range northwards, shifting its life history strategies to exploit even colder climates7,8,13, and steadily colonized potato crops despite substantial geographical barriers14. Leptinotarsa decemlineata is an excellent model for understanding pest evolution in agroecosystems because, despite its global spread, individuals disperse over short distances and populations often exhibit strong genetic differentiation15–17, providing an opportunity to track the spread of populations and the emergence of novel phenotypes. The development of genomic resources in L. decemlineata will provide an unparalleled opportunity to investigate the molecular basis of traits such as climate adaptation, herbivory, host expansion, and chemical detoxification. Perhaps most significantly, understanding its ability to evolve rapidly would be a major step towards developing sustainable methods to control this widely successful pest in agricultural settings.
Given that climate is thought to be the major factor in structuring the range limits of species18, the latitudinal expansion of L. decemlineata, spanning more than 40° latitude from Mexico to northern potato-producing countries such as Canada and Russia6, warrants further investigation. Harsh winter climates are thought to present a major barrier for insect range expansions, especially near the limits of a species’ range7,19. To successfully overwinter in temperate climates, beetles need to build up body mass, develop greater amounts of lipid storage, have a low resting metabolism, and respond to photoperiodic keys by initiating diapause20,21. Although the beetle has been in Europe for less than 100 years, local populations have demonstrating remarkably rapid evolution in life history traits linked to growth, diapause and metabolism8,13,20. Understanding the genetic basis of these traits, particularly the role of specific genes associated with metabolism, fatty acid synthesis, and diapause induction, could provide important information about the mechanism of climate adaptation.
Although Leptinotarsa decemlineata has long-served as a model for the study of host expansion and herbivory due to its rapid ability to host switch17,22, a major outstanding question is what genes and biological pathways are associated with herbivory in this species? While >35,000 species of Chrysomelidae are well-known herbivores, most species feed on one or a few host species within the same plant family23. Within Leptinotarsa, the majority of species feed on plants within Solanaceae and Asteraceae, while L. decemlineata feeds exclusively on solanaceous species24. It has achieved the broadest host range amongst its congeners (including, but not limited to: buffalobur (S. rostratum), potato (S. tuberosum), eggplant (S. melongena), silverleaf nightshade (S. elaeagnifolium), horsenettle (S. carolinense), bittersweet nightshade (S. dulcamara), tomato (S. lycopersicum), and tobacco (Nicotiana tabacum))17,22,25, and exhibits geographical variation in the use of locally abundant Solanum species26.
Another major question is what are the genes that underlie the beetle’s remarkable capacity to detoxify plant secondary compounds and are these the same biological pathways used to detoxify insecticidal compounds27? Solanaceous plants are considered highly toxic to a wide range of insect herbivore species28, because they contain steroidal alkaloids and glycoalkaloids, nitrogen-containing compounds that are toxic to a wide range of organisms, including bacteria, fungi, humans, and insects29, as well as glandular trichomes that contain additional toxic compounds30. In response to beetle feeding, potato plants upregulate pathways associated with terpenoid, alkaloid, and phenylpropanoid biosynthesis, as well as a range of protease inhibitors31. A complex of digestive cysteine proteases is known to underlie L. decemlineata’s ability to respond to potato-induced defenses32,33. There is evidence that larvae excrete34 and perhaps even sequester toxic plant-based compounds in the hemolymph35,36. Physiological mechanisms involved in detoxifying plant compounds, as well as other xenobiotics, have been proposed to underlie pesticide resistance27. To date, while cornerstone of L. decemlineata management has been the use of insecticides, the beetle has evolved resistance to over 50 compounds and all of the major classes of insecticides. Some of these chemicals have even failed to control L. decemlineata within the first year of release10, and notably, regional populations of L. decemlineata have demonstrated the ability to independently evolve resistance to pesticides and to do so at different rates37. Previous studies have identified target site mutations in resistance phenotypes and a wide range of genes involved in metabolic detoxification, including carboxylesterase genes, cytochrome P450s, and glutathione S-transferase genes38–42.
To examine evidence of rapid evolutionary change underlying L. decemlineata’s extraordinary success utilizing novel host plants, climates, and detoxifying insecticides, we evaluated structural and functional genomic changes relative to other beetle species, using whole-genome sequencing, transcriptome sequencing, and a large community-driven biocuration effort to improve predicted gene annotations. We compared the size of gene families associated with particular traits against existing available genomes from related species, particularly those sequenced by the i5k project (http://i5k.github.io), an initiative to sequence 5,000 species of Arthropods. While efforts have been made to understand the genetic basis of phenotypes in L. decemlineata (for example, pesticide resistance)32,43,44, previous work has been limited to candidate gene approaches rather than comparative genomics. Genomic data can not only illuminate the genetic architecture of a number of phenotypic traits that enable L. decemlineata to continue to be an agricultural pest, but can also be used to identify new gene targets for control measures. For example, recent efforts have been made to develop RNAi-based pesticides targeting critical metabolic pathways in L. decemlineata41,45,46. With the extensive wealth of biological knowledge and a newly-released genome, this beetle is well-positioned to be a model system for agricultural pest genomics and the study of rapid evolution.
Results and Discussion
Genome Assembly, Annotation and Assessment
A single female L. decemlineata from Long Island, NY, USA, a population known to be resistant to a wide range of insecticides47,48, was sequenced to a depth of ~140x coverage and assembled with ALLPATHS49 followed by assembly improvement with ATLAS (https://www.hgsc.bcm.edu/software/). The average coleopteran genome size is 760 Mb (ranging from 160–5,020 Mb50), while most of the beetle genome assemblies have been smaller (mean assembly size 286 Mb, range 160–710 Mb)51–55. The draft genome assembly of L. decemlineata is 1.17 Gb and consists of 24,393 scaffolds, with a N50 of 414 kb and a contig N50 of 4.9 kb. This assembly is more than twice the estimated genome size of 460 Mb56, with the presence of gaps comprising 492 Mb, or 42%, of the assembly. As this size might be driven by underlying heterozygosity, we also performed scaffolding with REDUNDANS57, which reduced the assembly size to 642 Mb, with gaps reduced to 1.3% of the assembly. However, the REDUNDANS assembly increased the contig N50 to 47.4 kb, the number of scaffolds increased to 90,205 and the N50 declined to 139 kb. By counting unique 19 bp kmers and adjusting for ploidy, we estimate the genome size as 816.9 Mb. Using just the small-insert (180 bp) 100 bp PE reads, average coverage was 24X for the ALLPATHS assembly and 26X for the REDUNDANS assembly. For all downstream analyses, the ALLPATHS assembly was used due to its increased scaffold length and reduced number of scaffolds.
The number of genes in the L. decemlineata genome predicted based on automated annotation using MAKER was 24,671 gene transcripts, with 93,782 predicted exons, which surpasses the 13,526–22,253 gene models reported in other beetle genome projects (Fig. 1)51–55. This may be in part due to fragmentation of the genome, which is known to inflate gene number estimates58. To improve our gene models, we manually annotated genes using expert opinion and additional mRNA resources (see Supplementary File for more details). A total of 1,364 genes were manually curated and merged with the unedited MAKER annotations to produce an official gene set (OGS v1.0) of 24,850 transcripts, comprised of 94,859 exons. A total of 12 models were curated as pseudogenes. A total of 1,237 putative transcription factors (TFs) were identified in the L. decemlineata predicted proteome (Fig. 2). The predicted number of TFs is similar to some beetles, such as Anoplophora glabripennis (1,397) and Hypothenemus hampei (1,148), but substantially greater than others, such as Tribolium castaneum (788), Nicrophorus vespilloides (744), and Dendroctonus ponderosae (683)51–55.
Figure 1.
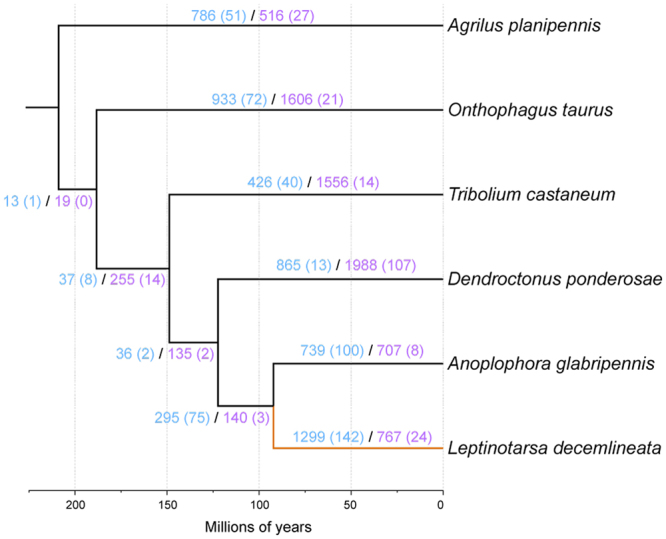
Ultrametric tree with branch lengths in millions of years for Leptinotarsa decemlineata relative to five other Coleoptera genomes. The L. decemlineata lineage is shown in orange. Branches are labeled with their length in years (top) and with the number of gene family expansions (blue) and contractions (purple) that occurred on that lineage. Rapid changes for both types are in parentheses.
Figure 2.
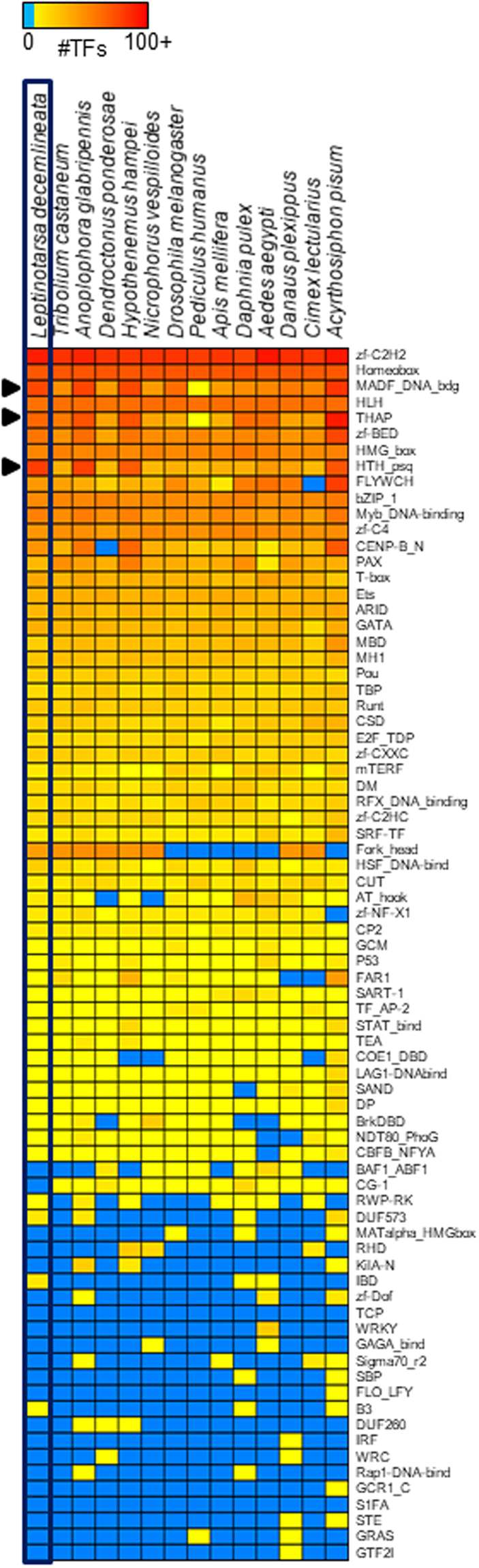
Heatmap distribution of the abundance of transcription factor families in Leptinotarsa decemlineata compared to other insects. Each entry indicates the number of TF genes for the given family per genome, based on presence of predicted DNA binding domains. Color scale is log (base 2) and the key is depicted at the top (light blue means the TF family is completely absent). Families discussed in the main text are indicated by arrows.
We assessed the completeness of both the ALLPATHS and REDUNDANS assemblies, and the OGS separately, using benchmarking sets of universal single-copy orthologs (BUSCOs) based on 35 holometabolous insect genomes59, as well as manually assessing the completeness and co-localization of the homeodomain transcription factor gene clusters in the ALLPATHS assembly. Using the reference set of 2,442 BUSCOs, the ALLPATHS genome assembly, REDUNDANS genome assembly, and OGS were 93.0%, 91.9%, and 71.8% complete, respectively. We found an additional 4.1%, 5.4%, and 17.9% of the BUSCOs present but fragmented in each dataset, respectively. For the highly conserved Hox and Iroquois Complex (Iro-C) clusters, we located and annotated complete gene models in the ALLPATHS genome assembly for all 12 expected orthologs, but these were split across six different scaffolds (Supplementary Figure 1S and Table 1S). All linked Hox genes occurred in the expected order and with the expected, shared transcriptional orientation, suggesting that the current draft assembly was correct but incomplete (see also Supplementary Figures 2S and 3S). Assuming direct concatenation of scaffolds, the Hox cluster would span a region of 3.7 Mb, similar to the estimated 3.5 Mb Hox cluster of A. glabripennis54. While otherwise highly conserved with A. glabripennis, we found a tandem duplication for Hox3/zen and an Antennapedia-class (ANTP-class) homeobox gene with no clear ortholog in other arthropods. We also assessed the ALLPATHS genome assembly for evidence of contamination using a Blobplot (Supplementary Figure 4S), which identified a small proportion of the reads (3.3%) as putative contaminants (the largest single taxonomic group represented, 3.0%, was assigned to Platyhelminthes).
Gene Annotation, Gene Family Evolution and Differential Expression
We estimated a phylogeny among six coleopteran genomes (A. glabripennis, Agrilus planipennis, D. ponderosae, L. decemlineata, Onthophagus taurus, and T. castaneum; unpublished genomes available at http://i5k.github.io/)51,52,54 using a conserved set of single copy orthologs and compared the official gene set of each species to understand how gene families evolved the branch representing Chysomelidae. Leptinotarsa decemlineata and A. glabripennis (Cerambycidae) are sister taxa (Fig. 1), as expected for members of the same superfamily Chrysomeloidea. We found 166 rapidly evolving gene families along the L. decemlineata lineage (1.4% of 11,598), 142 of which are rapid expansions and the remaining 24 rapid contractions (Table 1). Among all branches of our coleopteran phylogeny, L. decemlineata has the highest average expansion rate (0.203 genes per million years), the highest number of genes gained, and the greatest number of rapidly evolving gene families.
Table 1.
Summary of gene gain and loss events inferred after correcting for annotation and assembly error across six Coleoptera species.
| Expansions | Contractions | No Change | Average Expansion | |||||
|---|---|---|---|---|---|---|---|---|
| Families | Genes gained | Rate | Families | Genes lost | Rate | |||
| Anoplophora glabripennis | 865 (13) | 1231 | 1.42 | 1988 (107) | 3125 | 1.57 | 5850 | −0.182341 |
| Agrilus plannipennis | 739 (100) | 1991 | 2.69 | 707 (8) | 769 | 1.09 | 7257 | 0.119108 |
| Dendroctonus ponderosae | 933 (72) | 1982 | 2.12 | 1606 (21) | 1887 | 1.17 | 6164 | 0.006438 |
| Leptinotarsa decemlineata | 426 (40) | 855 | 2.01 | 1556 (48) | 2116 | 1.36 | 6721 | −0.127501 |
| Onthophagus taurus | 1299 (142) | 2952 | 2.27 | 767 (24) | 895 | 1.17 | 6637 | 0.203380 |
| Tribolium castaneum | 786 (51) | 1428 | 1.82 | 516 (27) | 909 | 1.76 | 7401 | 0.055645 |
The number of rapidly evolving families is shown in parentheses for each type of change and the rate is genes per million years.
Examination of the functional classification of rapidly evolving families in L. decemlineata (Supplementary Tables 2S and 3S) indicates that a subset of families are clearly associated with herbivory. The peptidases, comprising several gene families that play a major role in plant digestion32,60, displayed a significant expansion in genes (OrthoDB family Id: EOG8JDKNM, EOG8GTNV8, EOG8DRCSN, EOG8GTNT0, EOG8K0T47, EOG88973C, EOG854CDR, EOG8Z91BB, EOG80306V, EOG8CZF0X, EOG80P6ND,,EOG8BCH22, EOG8ZKN28, EOG8F4VSG, EOG80306V, EOG8BCH22, EOG8ZKN28, EOG8F4VSG). While olfactory receptor gene families have rapidly contracted (EOG8Q5C4Z, EOG8RZ1DX), subfamilies of odorant binding proteins and gustatory receptors have grown (see Sensory Ecology section below). The expansion of gustatory receptor subfamilies are associated with bitter receptors, likely reflecting host plant detection of nightshades (Solanaceae). Some gene families associated with plant detoxification and insecticide resistance have rapidly expanded (Glutathione S-transferases: EOG85TG3K, EOG85F05D, EOG8BCH22; UDP-glycosyltransferase: EOG8BCH22; cuticle proteins: EOG8QJV4S, EOG8DNHJQ; ABC transporters: EOG83N9TJ), whereas others have contracted (Cytochrome P450s: EOG83N9TJ). Finally, gene families associated with immune defense (fibrinogen: EOG8DNHJQ; Immunoglobulin: EOG8Q87B6, EOG8S7N55, EOG854CDX, EOG8KSRZK, EOG8CNT5M) exhibit expansions that may be linked to defense against pathogens and parasitoids that commonly attack exposed herbivores61. A substantial proportion of the rapidly evolving gene families include proteins with transposable element domains (25.3%), while other important functional groups include DNA and protein binding (including many transcription factors), nuclease activity, protein processing, and cellular transport.
Diversification of transcription factor (TF) families potentially signals greater complexity of gene regulation, including enhanced cell specificity and refined spatiotemporal signaling62. Notably, several TF families are substantially expanded in L. decemlineata, including HTH_psq (194 genes vs. a mean of only 24 across the insects shown in Fig. 2), MADF (152 vs. 54), and THAP (65 vs. 41). Two of these TF families, HTH_psq and THAP, are DNA binding domains in transposons. Of the 1,237 L. decemlineata TFs, we could infer DNA binding motifs for 189 (15%) (Supplementary Table 4S), mostly based on DNA binding specificity data from Drosophila melanogaster (124 TFs), but also from species as distant as human (45 TFs) and mouse (11 TFs). Motifs were inferred for a substantial proportion of the TFs in the largest TF families, including Homeodomain (59 of 90, 66%), bHLH (34 of 46, 74%), and Forkhead box (14 of 16, 88%). We could only infer a small number of C2H2 zinc finger motifs (21 of 439, ~5%), which is expected as these sequences evolve quickly by shuffling zinc finger arrays, resulting in largely dissimilar DNA-binding domains across metazoans63. Collectively, the almost 200 inferred DNA binding motifs for L. decemlineata TFs provide a unique resource to begin unraveling gene regulatory networks in this organism.
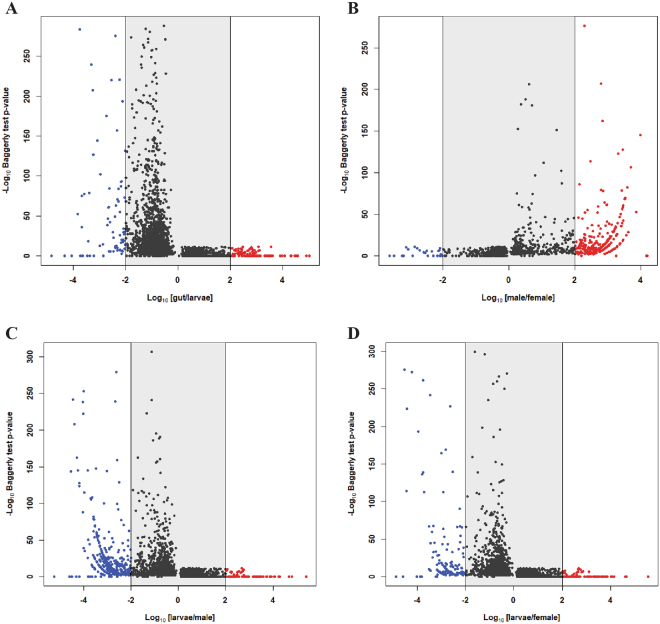
To identify genes active in mid-gut tissues, life-stages, and sex differences, we examined differential transcript expression levels using RNA sequencing data. Comparison of significantly differentially expressed genes with >100-fold change, after Bonferroni correction, indicated higher expression of digestive enzymes (proteases, peptidases, dehydrogenases and transporters) in mid-gut versus whole larval tissues, while cuticular proteins were largely expressed at lower levels (Fig. 3A, Supplementary Table 5S). Comparison of an adult male and female showed higher expression of testes and sperm related genes in males, while genes involved in egg production and sterol biosynthesis are more highly expressed in females (Fig. 3B, Supplementary Table 6S). Comparisons of larvae to both an adult male (Fig. 3C, Supplementary Table 7S) and an adult female (Fig. 3D, Supplementary Table 8S) showed higher expression of larval-specific cuticle proteins, and lower expression of odorant binding and chemosensory proteins. The adults, both drawn from a pesticide resistant population, showed higher constitutive expression of cytochrome p450 genes compared to the larval population, which is consistent with the results from previous studies of neonicotinoid resistance in this population48.
Figure 3.
Volcano plots showing statistically significant gene expression differences after Bonferroni correction in Leptinotarsa decemlineata. (A) Mid -gut tissue versus whole larvae, (B) an adult male versus an adult female, (C) an adult male versus whole larvae, and (D) an adult female versus whole larvae. Points outside the gray area indicate >100-fold-differences in expression. Blue points indicate down-regulated genes and red points indicate up-regulated genes in each contrast.
Transposable Elements
Transposable elements (TEs) are ubiquitous mobile elements within most eukaryotic genomes and play critical roles in both genome architecture and the generation of genetic variation64. Through insertional mutagenesis and recombination, TEs are a major contributor to the generation of novel mutations (50–80% of all mutation events within the genome of D. melanogaster)65–67, and are increasingly thought to generate much of the genetic diversity that contributes to rapid evolution68–70. In addition to finding that genes with TE domains comprise 25% of the rapidly evolving gene families within L. decemlineata, we found that at least 17% of the genome consists of TEs (Supplementary Table 9S). This is substantially greater than the 6% found in T. castaneum51, but less than some Lepidoptera (35% in Bombyx mori and 25% in Heliconius melpomene)71,72. LINEs were the largest TE class, comprising ~10% of the genome, while SINEs were not detected. Curation of the TE models with intact protein domains resulted in 334 current models of potentially active TEs, meaning that these TEs are capable of transposition and excision. Within the group of active TEs, we found 191 LINEs, 99 DNA elements, 38 LTRs, and 5 Helitrons. Given that TEs have been associated with the ability of species to rapidly adapt to novel selection pressures73–75, particularly via alterations of gene expression patterns in neighboring genomic regions, we scanned gene rich regions (1 kb neighborhood size) for active TE elements. Genes with active neighboring TEs have functions that include transport, protein digestion, diapause, and metabolic detoxification (Supplementary Table 10S). Because TE elements have been implicated in conferring insecticide resistance in other insects76, future work should investigate the role of these TE insertions on rapid evolutionary changes within pest populations of L. decemlineata.
Population Genetic Variation and Invasion History
To understand the propensity for L. decemlineata pest populations to rapidly evolve across a range of environmental conditions, we examined geographical patterns of genomic variability and the evolutionary history of L. decemlineata (to see the genomic analysis of environmental adaptation, refer to the Diapause and Environmental Stress section of Supplementary File). High levels of standing variation are one mechanism for rapid evolutionary change77. We identified 1.34 million biallelic single nucleotide polymorphisms (SNPs) from pooled RNAseq datasets, or roughly 1 variable site for every 22 base pairs of coding DNA. This rate of polymorphism is exceptionally high when compared to vertebrates (e.g. ~1 per kb in humans, or ~1 per 500 bp in chickens)78,79, and is 8-fold higher than other beetles (1 in 168 for D. ponderosae and 1 in 176 bp for O. taurus)52,80 and 2 to 5-fold higher than some dipterans (1 in 54 bp for D. melanogaster and 1 in 125 bp for Anopheles gambiae)78,81. It is likely that these values simply scale with effective population size, although the dipterans, with the largest known population sizes, have reduced variation due to widespread selective sweeps and genetic bottlenecks82.
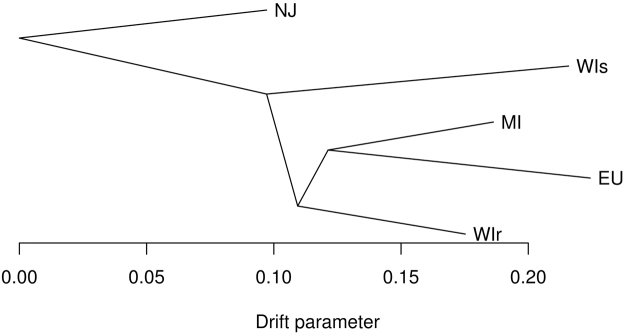
Evolutionary relationships and the amount of genetic drift among Midwestern USA, Northeastern USA, and European L. decemlineata populations were estimated based on genome-wide allele frequency differences using a population graph. A substantial amount of local genetic structure and high genetic drift is evident among all populations, although both the reference lab strain from New Jersey and European populations appear to have undergone more substantial drift, suggestive of strong inbreeding (Fig. 4). Population genetic divergence (FST) values (Supplementary Table 11S) range from 0.035 (among Wisconsin populations) to 0.182 (Wisconsin compared to Europe). The allele frequency spectrum was calculated for populations in Wisconsin, Michigan, and Europe to estimate the population genetic parameter θ, or the product of the mutation rate and the ancestral effective population size, and the ratio of contemporary to ancestral population size in models that allowed for single or multiple episodes of population size change. Estimates of ancestral θ are much higher for Wisconsin (θ = 12595) and Michigan (θ = 93956) than Europe (θ = 3.07; Supplementary Table 12S), providing support for a single introduction into Europe following a large genetic bottleneck15. In all three populations, a model of population size growth is supported, in agreement with historical accounts of the beetles expanding from the Great Plains into the Midwestern U.S. and Europe3,15, but the dynamics of each population appear independent, with the population from Michigan apparently undergoing a very recent decline in contemporary population size (the ratio of contemporary to ancient population size is 0.066, compared to 3.3 and 2.1 in Wisconsin and Europe, respectively).
Figure 4.
Population genetic relationships and relative rates of genetic drift among Leptinotarsa decemlineata pest populations based on single nucleotide polymorphism data. Population codes: NJ- New Jersey lab strain, WIs- imidacloprid susceptible population from Arlington, Wisconsin, WIr- imidacloprid resistant population from Hancock, Wisconsin, MI- imidacloprid resistant population from Michigan, and EU- European samples combined from Italy and Russia.
Sensory Ecology and Host Plant Detection
To interact with their environment, insects have evolved neurosensory organs to detect environmental signals, including tactile, auditory, chemical and visual cues83. We examined neural receptors, olfactory genes, and light sensory (opsin) genes to understand the sensory ecology and host-plant specializations of L. decemlineata.
We found high sequence similarity in the neuroreceptors of L. decemlineata compared to other Coleoptera. The transient receptor potential (TRP) channels are permeable transmembrane proteins that respond to temperature, touch, pain, osmolarity, pheromones, taste, hearing, smell and visual cues of insects84,85. In most insect genomes, there are typically 13–14 TRP genes located in insect stretch receptor cells and several are targeted by commercial insecticides86. We found 12 TRP genes present in the L. decemlineata genome, including the two TRPs (Nanchung and Inactive) that are targeted by commercial insecticides, representing a complete set of one-to-one orthologs with T. castaneum. Similarly, the 20 known amine neurotransmitter receptors in T. castaneum are present as one-to-one orthologs in L. decemlineata87,88. Amine receptors are G-protein-coupled receptors that interact with biogenic amines, such as octopamine, dopamine and serotonin. These neuroactive substances regulate behavioral and physiological traits in animals by acting as neurotransmitters, neuromodulators and neurohormones in the central and peripheral nervous systems89.
The majority of phytophagous insects are restricted to feeding on several plant species within a genus, or at least restricted to a particular plant family90. Thus, to find their host plants within heterogeneous landscapes, insect herbivores detect volatile organic compounds through olfaction, which utilizes several families of chemosensory gene families, such as the odorant binding proteins (OBPs), odorant receptors (ORs), gustatory receptors (GRs), and ionotropic receptors (IRs)91. OBPs directly bind with volatile organic compounds emitted from host plants and transport the ligands across the sensillar lymph to activate the membrane-bound ORs in the dendrites of the olfactory sensory neurons92. The ORs and GRs are 7-transmembrane proteins related within a superfamily93 of ligand-gated ion channels94. The ionotropic receptors are related to ionotropic glutamate receptors and function in both smell and taste95. These four gene families are commonly large in insect genomes, consisting of tens to hundreds of members.
We compared the number of genes found in L. decemlineata in the four chemosensory gene families to T. castaneum and A. glabripennis (Table 2, as well as Supplementary Tables 13S–16S for details). While the OBP family is slightly enlarged, the three receptor families are considerably smaller in L. decemlineata than in either A. glabripennis or T. castaneum, consistent with the specialization of this beetle on one genus of plants. However, each beetle species exhibits species-specific gene subfamily expansions (Supplementary Figures 5S–8S); in particular, some lineages of GRs related to bitter taste are expanded in L. decemlineata relative to A. glabripennis and other beetles. Among the OBPs, we identified a major L. decemlineata-specific expansion of proteins belonging to the Minus-C class (OBPs that have lost two of their six conserved cysteine residues) that appear unrelated to the ‘traditional’ Minus-C subfamily in Coleoptera, indicating that coleopteran OBPs have lost cysteines on at least two occasions.
Table 2.
Numbers of putatively functional proteins in four chemosensory families in Leptinotarsa decemlineata and other beetle species.
| Species | Odorant Binding Proteins (OBPs) | Olfactory Receptors (ORs) | Gustatory Receptors (GRs) | Ionotropic Receptors (IRs) |
|---|---|---|---|---|
| Leptinotarsa decemlineata | 58 | 75 | 144 | 27 |
| Anoplophora glabripennis | 52 | 120 | 234 | 72 |
| Tribolium castaneum | 49 | 264 | 219 | 72 |
To understand the visual acuity of L. decemlineata, we examined the G-protein-coupled transmembrane receptor opsin gene family. We found five opsins, three of which are members of rhabdomeric opsin (R-opsin) subfamilies expressed in the retina of insects51,96. Specifically, the L. decemlineata genome contains one member of the long wavelength-sensitive R-opsin and two short wavelength UV-sensitive R-opsins. The latter were found to be closely linked in a range of less than 20,000 bp, suggestive of recent tandem gene duplication. Overall, the recovered repertoire of retinally-expressed opsins in L. decemlineata97 is consistent with the beetle’s attraction to yellow light and to the yellow flowers of its ancestral host plant, S. rostratum98,99, and is consistent with the beetle’s sensitivity in the UV- and LW-range100. In addition, we found a member of the Rh7 R-opsin subfamily, which is broadly conserved in insects including other beetle species (A. glabripennis), although it is missing from T. castaneum. Finally, L. decemlineata has a single ortholog of the c-opsin subfamily shared with T. castaneum, which is absent in A. glabripennis and has an unclear role in photoreception101.
Host Plant Utilization
Protein digestion
Insect herbivores are fundamentally limited by nitrogen availability102, and thus need to efficiently break down plant proteins in order to survive and develop on host plants103. Leptinotarsa decemlineata has serine and cysteine digestive peptidases (coined “intestains”)32,104,105, as well as aspartic and metallo peptidases33, for protein digestion. For the vast majority of plant-eating beetles (the infraorder Cucujiformia, which includes Chrysomelidae), cysteine peptidases contribute most strongly to proteolytic activity in the gut105,106. In response to herbivory, plants produce a wide range of proteinase inhibitors to prevent insect herbivores from digesting plant proteins103,105,107. Coleopteran peptidases are differentially susceptible to plant peptidase inhibitors, and our annotation results suggest that gene duplication and selection for inhibitor insensitive genotypes may have contributed to the success of leaf-feeding beetles (Chrysomelidae) on different plants. We found that gene expansion of cysteine cathepsins from the C1 family in L. decemlineata correlates with the acquisition of greater digestive function by this group of peptidases, which is supported by gene expression activity of these genes in mid-gut tissue (Fig. 3A, Fig. 5). The gene expansion may be explained by an evolutionary arms race between insects and plants that favors insects with a variety of digestive peptidases in order to resist plant peptidase inhibitors108,109 and allows for functional specialization110.
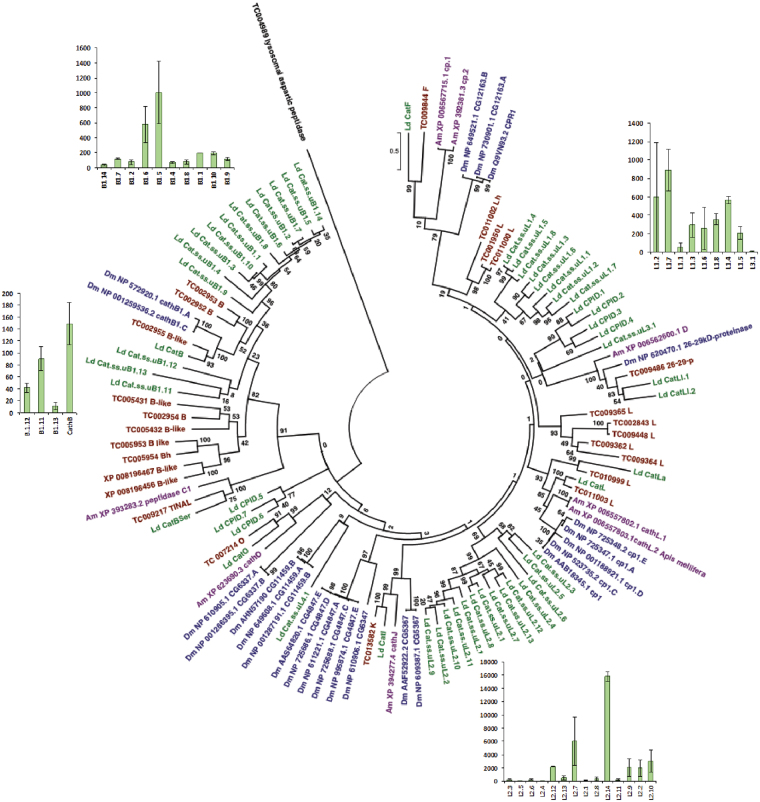
Figure 5.
Phylogenetic relationships of the cysteine peptidase gene family in Leptinotarsa decemlineata compared to model insects. Species abbreviations are: L. decemlineata (Ld, green color), Drosophila melanogaster (Dm, blue color), Apis mellifera (Am, purple color), and Tribolium castaneum (Tc, red color). Mid-gut gene expression (TPM) of highly expressed L. decemlineata cysteine peptidases is shown as bar graphs across three replicate treatments.
Cysteine peptidases of the C1 family were represented by more than 50 genes separated into four groups with different structure and functional characteristics (Supplementary Table 18S): cathepsin L subfamily, cathepsin B subfamily, TINAL-like genes, and cysteine peptidase inhibitor domains (CPIDs). Cathepsin L subfamily cysteine peptidases are endopeptidases111 that can be distinguished by the cathepsin propeptide inhibitor domain I29 (pfam08246)112,113. Within the cathepsin L subfamily, we found sequences that were similar to classical cathepsin L, cathepsin F, and cathepsin O. However, there were 28 additional predicted peptidases of this subfamily that could not be assigned to any of the “classical” cathepsin types, and most of these were grouped into two gene expansions (uL1 and uL2) according to their phylogenetic and structural characteristics. Cathepsin B subfamily cysteine peptidases are distinguished by the specific peptidase family C1 propeptide domain (pfam08127). Within the cathepsin B subfamily, there was one gene corresponding to typical cathepsin B peptidases and 14 cathepsin B-like genes. According to the structure of the occluding loop, only the typical cathepsin B may have typical endo- and exopeptidase activities, while a large proportion of cathepsin B-like peptidases presumably possesses only endopeptidase activity due to the absence of a His-His active subsite in the occluding loop, which is responsible for exopeptidase activity111. Only one gene corresponding to a TINAL-like-protein was present, which has a domain similar to cathepsin B in the C-terminus, but lacks peptidase activity due to the replacement of the active site Cys residue with Ser114. Cysteine peptidase inhibitor domain (CPID) genes encode the I29 domain of cysteine peptidases without the mature peptidase domain. Within the CPID group, there were seven short inhibitor genes that lack the enzymatic portion of the protein. A similar trend of “stand-alone inhibitors” has been observed in other insects, such as B. mori115. These CPID genes may be involved in the regulation of cysteine peptidases. We note that we found multiple fragments of cysteine peptidase genes, suggesting that the current list of L. decemlineata genes may be incomplete. Comparison of these findings with previous data on L. decemlineata cysteine peptidases116 demonstrates that intestains correspond to several peptidase genes from the uL1 and uL2 groups (Supplementary Table 18S). These data, as well as literature for Tenebrionidae beetles117, suggest that intensive gene expansion is typical for peptidases that are involved in digestion.
We also found a high number of digestion-related serine peptidase genes in the L. decemlineata genome (Supplementary Table 19S), but they contribute only a small proportion of the beetle’s total gut proteolytic activity32. Of the 31 identified serine peptidase genes and fragments, we annotated 16 as trypsin-like peptidases and 15 as chymotrypsin-like peptidases. For four chymotrypsin-like and one trypsin-like peptidase, we identified only short fragments. All complete (and near-complete) sequences have distinctive S1A peptidase subfamily motifs, a conserved catalytic triad, conserved sequence residues such as the “CWC” sequence and cysteines that form disulfide bonds in the chymotrypsin protease fold. The number of serine peptidases was higher than expected based upon the number of previously identified EST clones32, but lower than the number of chymotrypsin and trypsin genes in the T. castaneum genome.
Carbohydrate digestion
Carbohydrates are the other category of essential nutrients for L. decemlineata. The enzymes that assemble and degrade oligo- and polysaccharides, collectively termed Carbohydrate active enzymes (CAZy), are categorized into five major classes: glycoside hydrolases (GH), polysaccharide lyases, carbohydrate esterases (CE), glycosyltransferases (GT) and various auxiliary oxidative enzymes118. Due to the many different roles of carbohydrates, the CAZy family profile of an organism can provide insight into “glycobiological potential” and, in particular, mechanisms of carbon acquisition119. We identified 182 GHs assigned to 25 families, 181 GTs assigned to 41 families, and two CEs assigned to two families in L. decemlineata; additionally, 99 carbohydrate-binding modules (which are non-catalytic modules associated with the above enzyme classes) were present and assigned to 9 families (Supplementary Table 20S; the list of CAZy genes is presented in Supplementary Table 21S). We found that L. decemlineata has three families of genes associated with plant cell wall carbohydrate digestion (GH28, GH45 and GH48) that commonly contain enzymes that target pectin (GH28) and cellulose (GH45 and GH48), the major structural components of leaves120. We found evidence of massive gene duplications in the GH28 family (14 genes) and GH45 family (11 genes, plus one additional splicing variant), whereas GH48 is represented by only three genes in the genome121. Overall, the genome of L. decemlineata shows a CAZy profile adapted to metabolize pectin and cellulose contained in leaf cell walls. The absence of specific members of the families GH43 (α-L-arabinofuranosidases) and GH78 (α-L-rhamnosidases) suggests that L. decemlineata can break down homogalacturonan, but not substituted galacturonans such as rhamnogalacturonan I or II120. The acquisition of these plant cell wall degrading enzymes has been linked to horizontal transfer in the leaf beetles and other phytophagous beetles120, with strong phylogenetic evidence supporting the transfer of GH28 genes from a fungal donor (Pezizomycotina) in L. decemlineata, as well as in the beetles D. ponderosae and Hypothenemus hampei, but a novel fungal donor in the more closely related cerambycid beetle A. glabripennis54 and a bacterial donor in the weevil Callosobruchus maculatus120.
Insecticide Resistance
To understand the functional genomic properties of insecticide resistance, we examined genes important to neuromuscular target site sensitivity, tissue penetration, and prominent gene families involved in Phase I, II, and III metabolic detoxification of xenobiotics122. These include the cation-gated nicotinic acetylcholine receptors (nAChRs), the γ-amino butyric acid (GABA)-gated anion channels and the histamine-gated chloride channels (HisCls), cuticular proteins, cytochrome P450 monooxygenases (CYPs), and the Glutathione S-transferases (GSTs).
Many of the major classes of insecticides (organochlorides, organophosphates, carbamates, pyrethroids, neonicotinoids and ryanoids) disrupt the nervous system (particularly ion channel signaling), causing paralysis and death123. Resistance to insecticides can come from point mutations that reduce the affinity of insecticidal toxins to ligand-gated ion superfamily genes124. The cys-loop ligand-gated ion channel gene superfamily is comprised of receptors involved in mediating synaptic ion flow during neurotransmission125. A total of 22 cys-loop ligand-gated ion channels were identified in the L. decemlineata genome in numbers similar to those observed in other insects126, including 12 nAChRs, three GABA receptors, and two HisCls (Supplementary Figure 9S). The GABA-gated chloride channel homolog of the Resistance to dieldrin (Rdl) gene of D. melanogaster was examined due to its role in resistance to dieldrin and other cyclodienes in Diptera127. The coding sequence is organized into 10 exons (compared to nine in D. melanogaster) on a single scaffold, with duplications of the third and sixth exon (Supplementary Figure 10S). Alternative splicing of these two exons encodes for four different polypeptides in D. melanogaster128,129, and as the splice junctions are present in L. decemlineata, we expect the same diversity of Rdl. The point mutations in the transmembrane regions TM2 and TM3 of Rdl are known to cause insecticide resistance in Diptera124,127, but were not observed in L. decemlineata.
Cuticle genes have been implicated in imidacloprid resistant L. decemlineata130 and at least one has been shown to have phenotypic effects on resistance traits following RNAi knockdown131. A total of 163 putative cuticle protein genes were identified and assigned to one of seven families (CPR, CPAP1, CPAP3, CPF, CPCFC, CPLCG, and TWDL) (Supplemental Figure 11S and Table 22S). Similar to other insects, the CPR family, with the RR-1 (soft cuticle), RR-2 (hard cuticle), and unclassifiable types, constituted the largest group of cuticle protein genes (132) in the L. decemlineata genome. While the number of genes in L. decemlineata is slightly higher than in T. castaneum (110), it is similar to D. melanogaster (137)132. Numbers in the CPAP1, CPAP3, CPF, and TWDL families were similar to other insects, and notably no genes with the conserved sequences for CPLCA were detected in L. decemlineata, although they are found in other Coleoptera.
A total of 89 CYP (P450) genes were identified in the L. decemlineata genome, an overall decrease relative to T. castaneum (143 genes). Due to their role in insecticide resistance in L. decemlineata and other insects38,48,133, we examined the CYP6 and CYP12 families in particular. Relative to T. castaneum, we observed reductions in the CYP6BQ, CYP4BN, and CYP4Q subfamilies. However, five new subfamilies (CYP6BJ, CYP6BU, CYP6E, CYP6F and CYP6K) were identified in L. decemlineata that were absent in T. castaneum, and the CYP12 family contains three genes as opposed to one gene in T. castaneum (CYP12h1). We found several additional CYP genes not present in T. castaneum, including CYP413A1, CYP421A1, CYP4V2, CYP12J and CYP12J4. Genes in CYP4, CYP6, and CYP9 are known to be involved in detoxification of plant allelochemicals as well as resistance to pesticides through their constitutive overexpression and/or inducible expression in imidacloprid resistant L. decemlineata48,130.
GSTs have been implicated in resistance to organophosphate, organochlorine and pyrethroid insecticides134 and are responsive to insecticide treatments in L. decemlineata130,135. A total of 27 GSTs were present in the L. decemlineata genome, and while they represent an expansion relative to A. glabripennis, all have corresponding homologs in T. castaneum. The cytosolic GSTs include the epsilon (11 genes), delta (5 genes), omega (4 genes), theta (2 genes) and sigma (3 genes) families, while two GSTs are microsomal (Supplementary Figure 12S). Several GST-like genes present in the L. decemlineata genome represent the Z class previously identified using transcriptome data135.
Pest Control via the RNAi Pathway
RNA interference (RNAi) is the process by which small non-coding RNAs trigger sequence-specific gene silencing136, and is important in protecting against viruses and mobile genetic elements, as well as regulating gene expression during cellular development137. The application of exogenous double stranded RNA (dsRNA) has been exploited as a tool to suppress gene expression for functional genetic studies138,139 and for pest control41,140.
We annotated a total of 49 genes associated with RNA interference, most of them were found on a single scaffold. All genes from the core RNAi machinery (from all three major RNAi classes) were present in L. decemlineata, including fifteen genes encoding components of the RNA Induced Silencing Complex (RISC) and genes known to be involved in double-stranded RNA uptake, transport, and degradation (Supplementary Table 22S). A complete gene model was annotated for R2D2, an essential component of the siRNA pathway that interacts with dicer-2 to load siRNAs into the RISC, and not previously detected in the transcriptome of the L. decemlineata mid-gut141. The core components of the small interfering RNA (siRNA) pathway were duplicated, including dicer-2, an RNase III enzyme that cleaves dsRNAs and pre-miRNAs into siRNAs and miRNAs respectively142,143. The dicer-2a and dicer-2b CDS have 60% nucleotide identity to each other, and 56% and 54% identity to the T. castaneum dicer-2 homolog, respectively. The argonaute-2 gene, which plays a key role in RISC by binding small non-coding RNAs, was also duplicated. A detailed analysis of these genes will be necessary to determine if the duplications provide functional redundancy. The duplication of genes in the siRNA pathway may play a role in the high sensitivity of L. decemlineata to RNAi knockdown144 and could benefit future efforts to develop RNAi as a pest management technology.
Conclusion
The whole-genome sequence of L. decemlineata, provides novel insights into one of the most diverse animal taxa, Chrysomelidae. It is amongst the largest beetle genomes sequenced to date, with a minimum assembly size of 640 Mb (ranging up to 1.17 Gb) and 24,740 genes. The genome size is driven in part by a large number of transposable element families, which comprise at least 17% of the genome and appear to be rapidly expanding relative to other beetles. Population genetic analyses suggest high levels of nucleotide diversity, local geographic structure, and evidence of recent population growth, which helps to explain how L. decemlineata rapidly evolves to exploit novel host plants, climate space, and overcome a range of pest management practices (including a large and diverse number of insecticides). Digestive enzymes, in particular the cysteine peptidases and carbohydrate-active enzymes, show evidence of gene expansion and elevated expression in gut tissues, suggesting the diversity of the genes is a key trait in the beetle’s phytophagous lifestyle. Additionally, expansions of the gustatory receptor subfamily for bitter tasting might be a key adaptation to exploiting hosts in the nightshade family, Solanaceae, while expansions of novel subfamilies of CYP and GST proteins are consistent with rapid, lineage-specific turnover of genes implicated in L. decemlineata’s capacity for insecticide resistance. Finally, L. decemlineata has interesting duplications in RNAi genes that might increase its sensitivity to RNAi and provide a promising new avenue for pesticide development. The L. decemlineata genome promises new opportunities to investigate the ecology, evolution, and management of this species, and to leverage genomic technologies in developing sustainable methods of pest control.
Materials and Methods
Genome Characteristics and Sampling of DNA and RNA
Previous cytological work determined that L. decemlineata is diploid and consists of 34 autosomes plus an XO system in males, or an XX system in females145. Twelve chromosomes are submetacentric, while three are acrocentric and two are metacentric, although one chromosome is heteromorphic (acrocentric and/or metacentric) in pest populations146. The genome size has been estimated with Feulgen densitometry at 0.46 pg, or approximately 460 Mb47. To generate a reference genome sequence, DNA was obtained from a single adult female, sampled from an imidacloprid resistant strain developed from insects collected from a potato field in Long Island, NY. Additionally, whole-body RNA was extracted for one male and one female from the same imidacloprid resistant strain. Raw RNAseq reads for 8 different populations were obtained from previous experiments: two Wisconsin populations (PRJNA297027)130, a Michigan population (PRJNA400685), a lab strain originating from a New Jersey field population (PRJNA275431), and three samples from European populations (PRJNA79581 and PRJNA236637)43,121. All RNAseq data came from pooled populations or were combined into a population sample from individual reads. In addition, RNA samples of an adult male and female from the same New Jersey population were sequenced separately using Illumina HiSeq 2000 as 100 bp paired end reads (deposited in the GenBank/EMBL/DDBJ database, PRJNA275662), and three samples from the mid-gut of 4th-instar larvae were sequenced using SOLiD 5500 Genetic Analyzer as 50 bp single end reads (PRJNA400633).
Genome Sequencing, Assembly, Annotation and Assessment
Four Illumina sequencing libraries were prepared, with insert sizes of 180 bp, 500 bp, 3 kb, and 8 kb, and sequenced with 100 bp paired-end reads on the Illumina HiSeq 2000 platform at estimated 40x coverage, except for the 8kb library, which was sequenced at estimated 20x coverage. ALLPATHS-LG v3521849 was used to assemble reads. Two approaches were used to scaffold contigs and close gaps in the genome assembly. The reference genome used in downstream analyses was generated with ATLAS-LINK v1.0 and ATLAS GAP-FILL v2.2 (https://www.hgsc.bcm.edu/software/). In the second approach, REDUNDANS was used57, as it is optimized to deal with heterozygous samples. The raw sequence data and L. decemlineata genome have been deposited in the GenBank/EMBL/DDBJ database (Bioproject accession PRJNA171749, Genome assembly GCA_000500325.1, Sequence Read Archive accessions: SRX396450- SRX396453). This data can be visualized, along with gene models and supporting data, at the i5k Workspace@NAL: https://i5k.nal.usda.gov/Leptinotarsa_decemlineata and https://apollo.nal.usda.gov/lepdec/jbrowse/ 147. We estimated the size of our genome using a kmer distribution plot in JELLYFISH148, where we mapped the 100 bp paired-end reads from the 180 bp insert library, used a 19 bp kmer distribution plot, and corrected for ploidy.
Automated gene prediction and annotation were performed using MAKER v2.0149, using RNAseq evidence and arthropod protein databases. The MAKER predictions formed the basis of the first official gene set (OGS v0.5.3). To improve the structural and functional annotation of genes, these gene predictions were manually and collaboratively edited using the interactive curation software Apollo150. For a given gene family, known insect genes were obtained from model species, especially T. castaneum51 and D. melanogaster151, and the nucleotide or amino acid sequences were used in BLASTx or tBLASTn152,153 to search the L. decemlineata OGS v0.5.3 or genome assembly, respectively, on the i5k Workspace@NAL. All available evidence (AUGUSTUS, SNAP, RNA data, etc.), including additional RNAseq data not used in the MAKER predictions, were used to inspect and modify gene predictions. Changes were tracked to ensure quality control. Gene models were inspected for quality, incorrect splits and merges, internal stop codons, and gff3 formatting errors, and finally merged with the MAKER-predicted gene set to produce the official gene set (OGS v1.0; merging scripts are available upon request). For focal gene families (e.g. peptidase genes, odorant and gustatory receptors, RNAi genes, etc.), details on how genes were identified and assigned names based on functional predictions or evolutionary relationship to known reference genes are provided in the Supplementary Material (Supplementary File).
To assess the quality of our genome assemblies, we used BUSCO v2.059 to determine the completeness of each genome assembly and the official gene set (OGS v1.0), separately. We benchmarked our data against 35 insect species in the Endopterygota obd9 database, which consists of 2,442 single-copy orthologs (BUSCOs). Secondly, we annotated and examined the genomic architecture of the Hox and Iroquois Complex gene clusters. For this, tBLASTn searches were performed against the genome using orthologous Hox gene protein sequences from T. castaneum (Tcas3.0) and A. glabripennis. Provisional L. decemlineata models were refined, and potential gene duplications were identified, via iterative and reciprocal BLAST and by manual inspection and correction of protein alignments generated with ClustalW2154, using RNAseq expression evidence when available. Finally, we used BlobTools v1.0155 to assess the assembled genome for possible contamination by generating a Blobplot. This plot integrates guanine and cytosine (GC) content of sequences, read coverage and sequence similarity via blast searches to assess genome contamination. Putative contaminants were identified using the combined results (‘bestsumorder’ command) of BLASTn against NCBI’s nt database156 and DIAMOND BLAST157 against the UniProt reference database158, and read coverage was assessed by mapping the 100 bp paired-end reads from the 180 bp insert library to the ALLPATHS genome with the Burrows-Wheeler Aligner (bwa-mem) version 0.7.5a159. As these sequences are nested within the genome scaffolds, and could represent horizontally transferred DNA, we left the scaffolds intact and provide a supplementary dataset of the contaminant sequences.
Gene Family Evolution
In order to identify rapidly evolving gene families along the L. decemlineata lineage, we obtained ~38,000 ortho-groups from 72 Arthropod species as part of the i5k pilot project (Thomas et al. in prep) from OrthoDB version 8160. For each ortho-group, we took only those genes present in the order Coleoptera, which was represented by the following six species: A. glabripennis, A. planipennis, D. ponderosae, L. decemlineata, O. taurus, and T. castaneum (http://i5k.github.io/)51,52,54. Finally, in order to make accurate inferences of ancestral states, families that were present in only one of the six species were removed. This resulted in a final count of 11,598 gene families that, among these six species, form the comparative framework that allowed us to examine rapidly evolving gene families in the L. decemlineata lineage.
Aside from the gene family count data, an ultrametric phylogeny is also required to estimate gene gain and loss rates. To make the tree, we considered only gene families that were single copy in all six species and that had another arthropod species also represented with a single copy as an outgroup. Outgroup species were ranked based on the number of families in which they were also single copy along with the coleopteran species, and the highest ranking outgroup available was chosen for each family. For instance, Pediculus humanus was the most common outgroup species. For any gene family, we chose P. humanus as the outgroup if it was also single copy. If it was not, we chose the next highest ranking species as the outgroup for that family. This process resulted in 3,932 single copy orthologs that we subsequently aligned with PASTA161. We used RAxML162 with the PROTGAMMAJTTF model to make gene trees from the alignments and ASTRAL163 to make the species tree. ASTRAL does not give branch lengths on its trees, a necessity for gene family analysis, so the species tree was again given to RAxML along with a concatenated alignment of all one-to-one orthologs for branch length estimation. Finally, to generate an ultrametric species tree with branch lengths in millions of years (my) we used the software r8s164, with a calibration range based on age estimates of a crown Coleopteran fossil at 208.5–411 my165. This calibration point itself was estimated in a similar fashion in a larger phylogenetic analysis of all 72 Arthropod species (Thomas et al. in prep).
With the gene family data and ultrametric phylogeny as the input data, gene gain and loss rates (λ) were estimated with CAFE v3.0166. CAFE is able to estimate the amount of assembly and annotation error (ε) present in the input data using a distribution across the observed gene family counts and a pseudo-likelihood search, and then is able to correct for this error and obtain a more accurate estimate of λ. Our analysis had an ε value of about 0.02, which implies that 3% of gene families have observed counts that are not equal to their true counts. After correcting for this error rate, λ = 0.0010 is on par with those previously those found for other Arthropod orders (Thomas et al. in prep). Using the estimated λ value, CAFE infers ancestral gene counts and calculates p-values across the tree for each family to assess the significance of any gene family changes along a given branch. Those branches with low p-values are considered rapidly evolving.
Gene Expression Analysis
RNAseq analyses were conducted to (1) establish male, female, and larva-enriched gene sets and (2) identify specific genes that are enriched within the digestive tract (mid-gut) compared to entire larva. RNAseq datasets were trimmed with CLC Genomics v.9 (Qiagen) and quality was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Each dataset was mapped to the predicted gene set (OGS v1.0) using CLC Genomics. Reads were mapped with >90% similarity over 60% of length, with two mismatches allowed. The number of reads were corrected to reads per million mapped to allow comparison between RNAseq datasets that having varying coverage. Transcripts per million (TPM) was used as a proxy for gene expression and fold changes were determined as the TPM in one sample relative to the TPM of another dataset167. The Baggerly’s test (t-type test statistic) followed by Bonferroni correction was used to identify genes with significant enrichment in a specific sample168. Statistical values for Bonferroni correction were reported as the number of genes x α value. This stringent statistical analysis was used as only a single replicate was available for each treatment. Enriched genes were removed, and mapping and expression analyses were repeated to ensure low expressed genes were not missed. Genes were identified by BLASTx searching against the NCBI non-redundant protein databases for arthropods with an expectation value (E-value) <0.001.
Transposable Elements
We investigated the identities (family membership), diversity, and genomic distribution of active transposable elements within L. decemlineata in order to understand their contribution to genome structure and to determine their potential positional effect on genes of interest (focusing on a TE neighborhood size of 1 kb). To identify TEs and analyze their distribution within the genome, we developed three repeat databases using: (1) RepeatMasker169, which uses the library repeats within Repbase (http://www.girinst.org/repbase/), (2) the program RepeatModeler170, which identifies de-novo repeat elements, and (3) literature searches to identify beetle transposons that were not found within Repbase. The three databases were used within RepeatMasker to determine the overall TE content in the genome.
To eliminate false positives and examine the genome neighborhood surrounding active TEs, all TE candidate models were translated in 6 frames and scanned for protein domains from the Pfam and CDD database (using the software transeq from Emboss, hmmer3 and rps-blast). The protein domain annotations were manually curated in order to remove: (a) clear false positives, (b) old highly degraded copies of TEs without identifiable coding potential, and (c) the correct annotation when improper labels were given. The TE models that contained protein domains were mapped onto the genome and used for our neighborhood analysis: we extracted the 1 kb flanking regions for each gene and scanned these regions for TEs with intact protein coding domains.
Population Genetic Variation and Demographic Analysis
Population genetic diversity of pooled RNAseq samples was used to examine genetic structure of pest populations and past population demography. For Wisconsin, Michigan and the lab strains from New Jersey, we aligned the RNAseq data to the genomic scaffolds, using Bowtie2 version 2.1.0171 to index the genome and generate aligned SAM files. We used bwa150 to align the RNAseq from the three populations from Europe. SAMtools/BCFtools version 0.1.19172 was used to produce BAM and VCF files. All calls were filtered with VCFtools version 0.1.11173 using a minimum quality score of 30 and minimum depth of 10. All indels were removed from this study. Population specific VCF files were sorted and merged using VCFtools, and the allele counts were extracted for each SNP. These allele frequency data were then used to infer population splits and relative rates of genetic drift using Treemix version 1.12174. We ran Treemix with SNPs in groups of 1000, choosing to root the tree with the Wisconsin population. In addition, for each pair of populations, we estimated the average genetic divergence by using F-statistics (FST) in VCFtools to calculate the ratio of among- to within-population genetic divergence across SNP loci.
To infer patterns of demographic change in the Midwestern USA (Wisconsin and Michigan) and European populations, the genome-wide allele frequency spectrum was used in dadi version 1.6.3175 to infer demographic parameters under several alternative models of population history. The history of L. decemlineata as a pest is relatively well-documented. The introduction of L. decemlineata into Europe in 1914176 is thought to have involved a strong bottleneck15 followed by rapid expansion. Similarly, an outbreak of L. decemlineata in Nebraska in 1859 is thought to have preceded population expansion into the Midwest reaching Wisconsin in 18653. For each population, a constant-size model, a two-epoch model of instantaneous population size change at a time point τ, a bottle-growth model of instantaneous size change followed by exponential growth, and a three-epoch model with a population size change of fixed duration followed by exponential growth, was fit to infer θ, the product of the ancestral effective population size and mutation rate, and relative population size changes.
Availability of data and material
All data generated or analyzed during this study have been made publicly available (see Methods for NCBI accession numbers), or included in this published article and its supplementary information. The genome assembly and official gene sets can be accessed at: https://data.nal.usda.gov/dataset/leptinotarsa-decemlineata-genome-assembly-10_5667, https://data.nal.usda.gov/dataset/leptinotarsa-decemlineata-genome-annotations-v053_5668 and https://data.nal.usda.gov/dataset/leptinotarsa-decemlineata-official-gene-set-v11.
Electronic supplementary material
Acknowledgements
We sincerely thank the sequencing, assembly and annotation teams at the Baylor College of Medicine Human Genome Sequencing Center for their efforts. We would like to acknowledge the following funding sources: sequencing, assembly and automated annotation was supported by NIH grant NHGRI U54 HG003273 to RAG; the UVM Agricultural Experiment Station Hatch grant to YHC (VT-H02010); the NIH postdoctoral training grant to RFM (K12 GM000708); MMT’s work with Apollo was supported by NIH grants (5R01GM080203 from NIGMS, and 5R01HG004483 from NHGRI) and by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy (contract No. DE-AC02-05CH11231); the National Science Centre (2012/07/D/NZ2/04286) and Ministry of Science and Higher Education scholarship to AM. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author Contributions
M.N.A., A.B., J.H.B., K.C., A.K.C., O.C., E.M.D., E.N.E., P.E., M.F., I.G.-R., C.G., A.G., M.G., B.H., E.C.J., J.W.J., M.K., S.A.K., A.K., F.L., V.L., X.M., A.M., N.J.M., R.F.M,. B.O., S.R.P., K.A.P., Y.P., L.C.P., M.P., É.R., J.P.R., H.M.R., A.J.R., V.M.R.-A., G.S., A.S.T., I.M.V.J., A.D.Y., G.D.Y., and J.-S.Y. contributed to the manual annotation effort, data analysis, and data interpretation, in addition to reading and approving the final manuscript. S.D.S. and Y.H.C. coordinated the project and drafted the manuscript. S.R. and R.A.G. coordinated genome sequencing, assembly and automated annotation at the Baylor College of Medicine Human Genome Sequencing Center. M.F.P., C.C., M.M.T., and M.-J.M.C. coordinated the biocuration of the genome. G.W.C.T. generated the phylogeny and conducted the gene family analysis. M.T.W. conducted the transcription factor analysis. A.G., K.G., M.P., and S.Z. contributed additional RNAseq data. J.B.B. coordinated the RNAseq data analysis. K.B., A.M., and Y.H.C. conducted the transposable element analysis. S.D.S. and J.C. conducted the population genetics analysis.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20154-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grafius E. Economic impact of insecticide resistance in the Colorado potato beetle (Coleoptera: Chrysomelidae) on the Michigan potato industry. J. Econ. Entomol. 1997;90:1144–1151. doi: 10.1093/jee/90.5.1144. [DOI] [Google Scholar]
- 2.Skryabin KDoRussia. and Eastern Europe need GM plants? Nat. Biotechnol. 2010;27:593–595. doi: 10.1016/j.nbt.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Walsh BD. The new potato bug and its natural history. Practical Entomol. 1865;1:1–4. [Google Scholar]
- 4.Gauthier, N. L., Hofmaster, R. N. & Semel, M. History of Colorado potato beetle control in Advances in potato pest management (ed. Lashomb, J. H. & Casagrande, R.) 13–33 (Hutchinson Ross Publishing Co., 1981).
- 5.Hare JD. Ecology and management of the Colorado potato beetle. Annu. Rev. Entomol. 1990;41:81–100. doi: 10.1146/annurev.en.35.010190.000501. [DOI] [Google Scholar]
- 6.Weber, D. C. Colorado potato beetle, Leptinotarsa decemlineata (Say) (Coleoptera: Chrysomelidae) in Encyclopedia of entomology (ed. Capinera, J. L.) 1008–1013 (Springer Netherlands, 2008).
- 7.Lyytinen A, Boman S, Grapputo A, Lindström L, Mappes J. Cold tolerance during larval development: effects on the thermal distribution limits of Leptinotarsa decemlineata. Entomol. Exp. Appl. 2009;133:92–99. doi: 10.1111/j.1570-7458.2009.00908.x. [DOI] [Google Scholar]
- 8.Hiiesaar K, et al. Factors affecting development and overwintering of second generation Colorado potato beetle (Coleoptera: Chrysomelidae) in Estonia in 2010. Acta Agric. Scand. Sect. B - Soil Plant Sci. 2013;63:506–515. [Google Scholar]
- 9.Hiiesaar K, et al. Phenology and overwintering of the Colorado potato beetle Leptinotarsa decemlineata Say in 2008–2015 in Estonia. Acta Agric. Scand. Sect. B — Soil Plant Sci. 2016;66:1–8. [Google Scholar]
- 10.Alyokhin A, Baker M, Mota-Sanchez D, Dively G, Grafius E. Colorado potato beetle resistance to insecticides. Am J Potato Res. 2008;85:395–413. doi: 10.1007/s12230-008-9052-0. [DOI] [Google Scholar]
- 11.Sparks TC, Nauen R. IRAC: Mode of action classification and insecticide resistance management. Pestic. Biochem. Physiol. 2015;121:122–128. doi: 10.1016/j.pestbp.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Izzo, V., Chen, Y. H., Schoville, S. D., Wang, C. & Hawthorne, D. J. Origin of pest lineages of the Colorado potato beetle, Leptinotarsa decemlineata. bioRxiv 156612, 10.1101/156612 (2017).
- 13.Lehmann P, Lyytinen A, Piiroinen S, Lindström L. Northward range expansion requires synchronization of both overwintering behaviour and physiology with photoperiod in the invasive Colorado potato beetle (Leptinotarsa decemlineata) Oecologia. 2014;176:57–68. doi: 10.1007/s00442-014-3009-4. [DOI] [PubMed] [Google Scholar]
- 14.Liu N, Li Y, Zhang R. Invasion of Colorado potato beetle, Leptinotarsa decemlineata, in China: dispersal, occurrence, and economic impact. Entomol. Exp. Appl. 2012;143:207–217. doi: 10.1111/j.1570-7458.2012.01259.x. [DOI] [Google Scholar]
- 15.Grapputo A, Boman S, Lindström L, Lyytinen A, Mappes J. The voyage of an invasive species across continents: genetic diversity of North American and European Colorado potato beetle populations. Mol. Ecol. 2005;14:4207–4219. doi: 10.1111/j.1365-294X.2005.02740.x. [DOI] [PubMed] [Google Scholar]
- 16.Azeredo-Espin A, Schroder R, Huettel M, Sheppard W. Mitochondrial DNA variation in geographic populations of Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera; Chrysomelidae) Experientia. 1991;47:483–485. doi: 10.1007/BF01959950. [DOI] [Google Scholar]
- 17.Lu W, Kennedy GG, Gould F. Genetic analysis of larval survival and larval growth of two populations of Leptinotarsa decemlineata on tomato. Entomol. Exp. Appl. 2001;99:143–155. doi: 10.1046/j.1570-7458.2001.00812.x. [DOI] [Google Scholar]
- 18.Gaston KJ. The how and why of biodiversity. Nature. 2003;421:900–901. doi: 10.1038/421900a. [DOI] [PubMed] [Google Scholar]
- 19.Turnock WJ, Fields PG. Winter climates and cold hardiness in terrestrial insects. Eur. J. Entomol. 2005;102:561–576. doi: 10.14411/eje.2005.081. [DOI] [Google Scholar]
- 20.Piiroinen S, Ketola T, Lyytinen A, Lindström L. Energy use, diapause behaviour and northern range expansion potential in the invasive Colorado potato beetle. Funct. Ecol. 2011;25:527–536. doi: 10.1111/j.1365-2435.2010.01804.x. [DOI] [Google Scholar]
- 21.Lehmann P, Lyytinen A, Piiroinen S, Lindström L. Latitudinal differences in diapause related photoperiodic responses of European Colorado potato beetles (Leptinotarsa decemlineata) Evol. Ecol. 2015;29:269–282. doi: 10.1007/s10682-015-9755-x. [DOI] [Google Scholar]
- 22.Hsiao TH. Host plant adaptations among geographic populations of the Colorado potato beetle. Entomol. Exp. Appl. 1978;24:237–247. doi: 10.1111/j.1570-7458.1978.tb02804.x. [DOI] [Google Scholar]
- 23.Jolivet, P. Food habits and food selection of Chrysomelidae. Bionomic and evolutionary perspectives in Biology of the chrysomelidae (ed. Jolivet, P., Petitpierre, E. & Hsiao, T. H.) 1–24 (Kluwer Acad. Press, 1988).
- 24.Hsiao, T. H. Host specificity, seasonality and bionomics of Leptinotarsa beetles in Biology of the chrysomelidae (ed. Jolivet, P., Petitpierre, E. & Hsiao, T. H.) 581–599 (Kluwer Acad. Press, 1988).
- 25.Hare JD, Kennedy GG. Genetic variation in plant-insect associations: survival of Leptinotarsa decemlineata populations on Solanum carolinense. Evolution. 1986;40:1031–1043. doi: 10.1111/j.1558-5646.1986.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 26.Jacques, R. L. Jr. & Fasulo, T. R. Colorado potato beetle, Leptinotarsa decemlineata (Say) and False potato beetle, Leptinotarsa juncta (Germar) (Insecta: Coleoptera: Chrysomelidae). Univ. Florida IFAS Ext. EENY146, 1–5 (2000).
- 27.Alyokhin A, Chen YH. Adaptation to toxic hosts as a factor in the evolution of insecticide resistance. Curr. Opin. Insect Sci. 2017;21:33–38. doi: 10.1016/j.cois.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Cárdenas PD, et al. The bitter side of the nightshades: Genomics drives discovery in Solanaceae steroidal alkaloid metabolism. Phytochemistry. 2015;113:24–32. doi: 10.1016/j.phytochem.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Milner SE, et al. Bioactivities of glycoalkaloids and their aglycones from Solanum species. J. Agric. Food Chem. 2011;59:3454–3484. doi: 10.1021/jf200439q. [DOI] [PubMed] [Google Scholar]
- 30.Dimock MB, Tingey WM. Host acceptance behaviour of Colorado potato beetle larvae influenced by potato glandular trichomes. Physiol. Entomol. 1988;13:399–406. doi: 10.1111/j.1365-3032.1988.tb01123.x. [DOI] [Google Scholar]
- 31.Lawrence SD, Novak NG, Ju CJ-T, Cooke JEK. Examining the molecular interaction between potato (Solanum tuberosum) and Colorado potato beetle Leptinotarsa decemlineata. Botany. 2008;86:1080–1091. doi: 10.1139/B08-074. [DOI] [Google Scholar]
- 32.Petek M, et al. A complex of genes involved in adaptation of Leptinotarsa decemlineata larvae to induced potato defense. Arch. Insect Biochem. Physiol. 2012;79:153–181. doi: 10.1002/arch.21017. [DOI] [PubMed] [Google Scholar]
- 33.Novillo C, Castañera P, Ortego F. Characterization and distribution of chymotrypsin-like and other digestive proteases in Colorado potato beetle larvae. Arch. Insect Biochem. Physiol. 1997;36:181–201. doi: 10.1002/(SICI)1520-6327(1997)36:3<181::AID-ARCH3>3.0.CO;2-X. [DOI] [Google Scholar]
- 34.Armer CA. Colorado potato beetle toxins revisited: Evidence the beetle does not sequester host plant glycoalkaloids. J. Chem. Ecol. 2004;30:883–888. doi: 10.1023/B:JOEC.0000028495.26931.c7. [DOI] [PubMed] [Google Scholar]
- 35.Deroe C, Pasteels JM. Defensive mechanisms against predation in the Colorado beetle (Leptinotarsa decemlineata, Say) Arch. Biol. Sci. 1977;88:289–304. [Google Scholar]
- 36.Hsiao TH, Fraenkel G. Properties of Leptinotarsin: a toxic hemolymph protein from the Colorado potato beetle. Toxicon. 1969;7:119–130. doi: 10.1016/0041-0101(69)90074-9. [DOI] [PubMed] [Google Scholar]
- 37.Forgash, A. J. Insecticide resistance in the Colorado potato beetle in Proceedings of the symposium on the Colorado potato beetle, XVII International Congress of Entomology, August 1984 (ed. Ferro, D. N. &Voss, R. H.). 33–52 (Massachusetts Agricultural Experimental Station, 1985).
- 38.Wan P-J, et al. Identification of cytochrome P450 monooxygenase genes and their expression profiles in cyhalothrin-treated Colorado potato beetle, Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 2013;107:360–368. doi: 10.1016/j.pestbp.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Pelletier Y, Goyer C. Identification of potential detoxification enzyme genes in Leptinotarsa decemlineata (Say) and study of their expression in insects reared on different plants. J. Plant Sci. 2008;88:621–629. [Google Scholar]
- 40.Lü F-G, et al. Identification of carboxylesterase genes and their expression profiles in the Colorado potato beetle Leptinotarsa decemlineata treated with fipronil and cyhalothrin. Pestic. Biochem. Physiol. 2015;122:86–95. doi: 10.1016/j.pestbp.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 41.Zhu F, Xu J, Palli R, Ferguson J, Palli SR. Ingested RNA interference for managing the populations of the Colorado potato beetle. Leptinotarsa decemlineata. Pest Manag. Sci. 2011;67:175–182. doi: 10.1002/ps.2048. [DOI] [PubMed] [Google Scholar]
- 42.Clements J, Schoville S, Peterson N, Lan Q, Groves RL. Characterizing molecular mechanisms of imidacloprid resistance in select populations of Leptinotarsa decemlineata in the Central Sands region of Wisconsin. PLoS One. 2016;11:e0147844. doi: 10.1371/journal.pone.0147844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar A, et al. Sequencing, de novo assembly and annotation of the Colorado potato beetle, Leptinotarsa decemlineata, transcriptome. PLoS One. 2014;9:e86012. doi: 10.1371/journal.pone.0086012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinkevich FD, et al. Multiple evolutionary origins of knockdown resistance (kdr) in pyrethroid-resistant Colorado potato beetle. Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 2012;104:192–200. doi: 10.1016/j.pestbp.2012.08.001. [DOI] [Google Scholar]
- 45.Zhou LT, et al. RNA interference of a putative S-adenosyl-L-homocysteine hydrolase gene affects larval performance in Leptinotarsa decemlineata (Say) J. Insect Physiol. 2013;59:1049–1056. doi: 10.1016/j.jinsphys.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, et al. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science. 2015;347:991–994. doi: 10.1126/science.1261680. [DOI] [PubMed] [Google Scholar]
- 47.Mota-Sanchez D, Hollingworth RM, Grafius EJ, Moyer DD. Resistance and cross-resistance to neonicotinoid insecticides and spinosad in the Colorado potato beetle, Leptinotarsa decemlineata (Say) (Coleoptera: Chrysomelidae) Pest Manag. Sci. 2006;62:30–37. doi: 10.1002/ps.1120. [DOI] [PubMed] [Google Scholar]
- 48.Zhu F, Moural TW, Nelson DR, Palli SR. A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple cytochrome P450s. Sci. Rep. 2016;6:20421. doi: 10.1038/srep20421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butler J, et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–20. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gregory, T. R. Animal genome size database. http://www.genomesize.com (2017).
- 51. Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature452, 949–55 (2008). [DOI] [PubMed]
- 52.Keeling CI, et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013;14:R27. doi: 10.1186/gb-2013-14-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cunningham CB, et al. The genome and methylome of a beetle with complex social behavior, Nicrophorus vespilloides (Coleoptera: Silphidae) Genome Biol. Evol. 2015;7:3383–3396. doi: 10.1093/gbe/evv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKenna DD, et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 2016;17:227. doi: 10.1186/s13059-016-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vega FE, et al. Draft genome of the most devastating insect pest of coffee worldwide: the coffee berry borer, Hypothenemus hampei. Sci. Rep. 2015;5:12525. doi: 10.1038/srep12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petitpierre E, Segarra C, Juan C. Genome size and chromosomal evolution in leaf beetles (Coleoptera, Chrysomelidae) Hereditas. 1993;6:1–6. [Google Scholar]
- 57.Pryszcz LP, Gabaldón T. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 2016;44:e113–e113. doi: 10.1093/nar/gkw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denton JF, et al. Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput. Biol. 2014;10:e1003998. doi: 10.1371/journal.pcbi.1003998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 60.Gruden K, et al. The cysteine protease activity of Colorado potato beetle (Leptinotarsa decemlineata Say) guts, which is insensitive to potato protease inhibitors, is inhibited by thyroglobulin type-1 domain inhibitors. Insect Biochem. Mol. Biol. 1998;28:549–560. doi: 10.1016/S0965-1748(98)00051-4. [DOI] [PubMed] [Google Scholar]
- 61.Smilanich AM, Dyer LA, Chambers JQ, Bowers MD. Immunological cost of chemical defence and the evolution of herbivore diet breadth. Ecol. Lett. 2009;12:612–621. doi: 10.1111/j.1461-0248.2009.01309.x. [DOI] [PubMed] [Google Scholar]
- 62.Vidal NM, Grazziotin AL, Iyer LM, Aravind L, Venancio TM. Transcription factors, chromatin proteins and the diversification of Hemiptera. Insect Biochem. Mol. Biol. 2016;69:1–13. doi: 10.1016/j.ibmb.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Najafabadi HS, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotech. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 64.Chénais B, Caruso A, Hiard S, Casse N. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene. 2012;509:7–15. doi: 10.1016/j.gene.2012.07.042. [DOI] [PubMed] [Google Scholar]
- 65.Tu, Z. Insect transposable elements in Insect molecular biology and biochemistry (ed. Gilbert, L. I.) 57–89 (Academic Press, 2012).
- 66.Biémont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 67.González J, Petrov DA. The adaptive role of transposable elements in the Drosophila genome. Gene. 2009;448:124–133. doi: 10.1016/j.gene.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 71.Lavoie CA, Platt RN, Novick PA, Counterman BA, Ray DA. Transposable element evolution in Heliconius suggests genome diversity within Lepidoptera. Mob DNA. 2013;4:21. doi: 10.1186/1759-8753-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1046–1057. doi: 10.1016/j.ibmb.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 73.Schrader L, et al. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat. Commun. 2014;5:5495. doi: 10.1038/ncomms6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.González J, Karasov TL, Messer PW, Petrov DA. Genome-wide patterns of adaptation to temperate environments associated with transposable elements in. Drosophila. PLoS Genet. 2010;6:e1000905. doi: 10.1371/journal.pgen.1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cridland JM, Thornton KR, Long AD. Gene expression variation in Drosophila melanogaster due to rare transposable element insertion alleles of large effect. Genetics. 2015;199:85–93. doi: 10.1534/genetics.114.170837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daborn PJ, et al. A single P450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- 77.Messer PW, Petrov DA. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 2013;28:659–669. doi: 10.1016/j.tree.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aquadro CF, DuMont VB, Reed FA. Genome-wide variation in the human and fruitfly: a comparison. Curr. Opin. Genet. Dev. 2001;11:627–634. doi: 10.1016/S0959-437X(00)00245-8. [DOI] [PubMed] [Google Scholar]
- 79.International Chicken Polymorphism Map Consortium. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature432, 717–722 (2004). [DOI] [PMC free article] [PubMed]
- 80.Choi J-H, et al. Gene discovery in the horned beetle Onthophagus taurus. BMC Genomics. 2010;11:703. doi: 10.1186/1471-2164-11-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morlais I, Ponçon N, Simard F, Cohuet A, Fontenille D. Intraspecific nucleotide variation in Anopheles gambiae: new insights into the biology of malaria vectors. Am. J. Trop. Med. Hyg. 2004;71:795–802. [PubMed] [Google Scholar]
- 82.Campos JL, Zhao L, Charlesworth B. Estimating the parameters of background selection and selective sweeps in Drosophila in the presence of gene conversion. Proc. Natl Acad. Sci. USA. 2017;114:E4762–E4771. doi: 10.1073/pnas.1619434114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dangles O, Irschick D, Chittka L, Casas J. Variability in sensory ecology: expanding the bridge between physiology and evolutionary biology. Q. Rev. Biol. 2009;84:51–74. doi: 10.1086/596463. [DOI] [PubMed] [Google Scholar]
- 84.Matsuura H, Sokabe T, Kohno K, Tominaga M, Kadowaki T. Evolutionary conservation and changes in insect TRP channels. BMC Evol Biol. 2009;9:228. doi: 10.1186/1471-2148-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Venkatachalam K, Montell C. TRP channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nesterov A, et al. TRP channels in insect stretch receptors as insecticide targets. Neuron. 2015;86:665–671. doi: 10.1016/j.neuron.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 87.Hauser F, et al. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front. Neuroendocrinol. 2008;29:142–165. doi: 10.1016/j.yfrne.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 88.Nishi Y, Sasaki K, Miyatake T. Biogenic amines, caffeine and tonic immobility in Tribolium castaneum. J. Insect Physiol. 2010;56:622–628. doi: 10.1016/j.jinsphys.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 89.Roeder T. Biochemistry and molecular biology of receptors for biogenic amines in locusts. Microsc. Res. Tech. 2002;56:237–247. doi: 10.1002/jemt.10027. [DOI] [PubMed] [Google Scholar]
- 90.Schoonhoven, L., Van Loon, J. J. A. & Dicke, M. Insect–Plant Biology 2nd edn (Oxford University Press, 2005).
- 91.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 92.Pelosi P, Zhou J-J, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sato K, Tanaka K, Touhara K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. USA. 2011;108:11680–11685. doi: 10.1073/pnas.1019622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rytz R, Croset V, Benton R. Ionotropic Receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 2013;43:888–897. doi: 10.1016/j.ibmb.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 96.Jackowska M, et al. Genomic and gene regulatory signatures of cryptozoic adaptation: Loss of blue sensitive photoreceptors through expansion of long wavelength-opsin expression in the red flour beetle Tribolium castaneum. Front. Zool. 2007;4:24. doi: 10.1186/1742-9994-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharkey CR, et al. Overcoming the loss of blue sensitivity through opsin duplication in the largest animal group, beetles. Sci. Rep. 2017;7:8. doi: 10.1038/s41598-017-00061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Izzo VM, Mercer N, Armstrong J, Chen YH. Variation in host usage among geographic populations of Leptinotarsa decemlineata, the Colorado potato beetle. J. Pest. Sci. 2014;87:597–608. doi: 10.1007/s10340-014-0578-2. [DOI] [Google Scholar]
- 99.Otálora-Luna F, Dickens JC. Multimodal stimulation of Colorado potato beetle reveals modulation of pheromone response by yellow light. PLoS One. 2011;6:e20990. doi: 10.1371/journal.pone.0020990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Briscoe AD, Chittka L. The evolution of color vision in insects. Annu. Rev. Entomol. 2001;46:471–510. doi: 10.1146/annurev.ento.46.1.471. [DOI] [PubMed] [Google Scholar]
- 101.Leung NY, Montell C. Unconventional roles of opsins. Annu. Rev. Cell. Dev. Biol. 2017;33:241–264. doi: 10.1146/annurev-cellbio-100616-060432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Price, P. W., Denno, R. F., Eubanks, M. D., Finke, D. L. & Kaplan, I. Insect ecology: behavior, populations and communities (Cambridge University Press, 2011).
- 103.Martinez M, et al. Phytocystatins: defense proteins against phytophagous insects and Acari. Int. J. Mol. Sci. 2016;17:1747. doi: 10.3390/ijms17101747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wolfson JL, Murdock LL. Suppression of larval Colorado potato beetle growth and development by digestive proteinase inhibitors. Entomol. Exp. Appl. 1987;44:235–240. doi: 10.1111/j.1570-7458.1987.tb00550.x. [DOI] [Google Scholar]
- 105.Bolter CJ, Jongsma MA. Colorado potato beetles (Leptinotarsa decemlineata) adapt to proteinase inhibitors induced in potato leaves by methyl jasmonate. J. Insect Physiol. 1995;41:1071–1078. doi: 10.1016/0022-1910(95)00073-4. [DOI] [Google Scholar]
- 106.Murdock LL, et al. Cysteine digestive proteinases in Coleoptera. Biochem. Physiol. 1987;87:783–787. [Google Scholar]
- 107.Chye M-L, Sin S-F, Xu Z-F, Yeung EC. Serine proteinase inhibitor proteins: Exogenous and endogenous functions. Vitr. Cell. Dev. Biol.-Plant. 2006;42:100–108. doi: 10.1079/IVP2005741. [DOI] [Google Scholar]
- 108.Gruden K, Popovič T, Cimerman N, Križaj I, Štrukelj B. Diverse enzymatic specificities of digestive proteases, “intestains”, enable Colorado potato beetle larvae to counteract the potato defence mechanism. Biol. Chem. 2003;384:305–310. doi: 10.1515/BC.2003.034. [DOI] [PubMed] [Google Scholar]
- 109.Gruden K, et al. Molecular basis of Colorado potato beetle adaptation to potato plant defence at the level of digestive cysteine proteinases. Insect Biochem. Mol. Biol. 2004;34:365–375. doi: 10.1016/j.ibmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 110.Goptar IA, et al. Cysteine digestive peptidases function as post-glutamine cleaving enzymes in tenebrionid stored-product pests. Comp. Biochem. Physiol. Part B. 2012;161:148–154. doi: 10.1016/j.cbpb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 111.Turk V, et al. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta - Proteins Proteomics. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2013;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wex T, et al. TIN-ag-RP, a novel catalytically inactive cathepsin B-related protein with EGF domains, is predominantly expressed in vascular smooth muscle cells. Biochemistry. 2001;40:1350–1357. doi: 10.1021/bi002266o. [DOI] [PubMed] [Google Scholar]
- 115.Yamamoto Y, Kurata M, Watabe S, Murakami R, Takahashi SY. Novel cysteine proteinase inhibitors homologous to the proregions of cysteine proteinases. Curr. Protein Pept. Sci. 2002;3:231–238. doi: 10.2174/1389203024605331. [DOI] [PubMed] [Google Scholar]
- 116.Sainsbury F, Rhéaume A-J, Goulet M-C, Vorster J, Michaud D. Discrimination of differentially inhibited cysteine proteases by activity-based profiling using cystatin variants with tailored specificities. J. Proteome Res. 2012;11:5983–5993. doi: 10.1021/pr300699n. [DOI] [PubMed] [Google Scholar]
- 117.Martynov AG, Elpidina EN, Perkin L, Oppert B. Functional analysis of C1 family cysteine peptidases in the larval gut of Тenebrio molitor and Tribolium castaneum. BMC Genomics. 2015;16:75. doi: 10.1186/s12864-015-1306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels. 2013;6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scully ED, Hoover K, Carlson JE, Tien M, Geib SM. Mid-gut transcriptome profiling of Anoplophora glabripennis, a lignocellulose degrading cerambycid beetle. BMC Genomics. 2013;14:850. doi: 10.1186/1471-2164-14-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kirsch R, et al. Horizontal gene transfer and functional diversification of plant cell wall degrading polygalacturonases: key events in the evolution of herbivory in beetles. Insect Biochem. Mol. Biol. 2014;52:33–50. doi: 10.1016/j.ibmb.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 121.Pauchet Y, Wilkinson P, Chauhan R, Ffrench-Constant RH. Diversity of beetle genes encoding novel plant cell wall degrading enzymes. PLoS One. 2010;5:e15635. doi: 10.1371/journal.pone.0015635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meyer UA. Overview of enzymes of drug metabolism. J. Pharmacokinet. Pharmacodyn. 1996;24:449–459. doi: 10.1007/BF02353473. [DOI] [PubMed] [Google Scholar]
- 123.Bloomquist JR. Chloride channels as tools for developing selective insecticides. Arch. Insect Biochem. Physiol. 2003;54:145–156. doi: 10.1002/arch.10112. [DOI] [PubMed] [Google Scholar]
- 124.Le Goff G, Hamon A, Bergé J-B, Amichot M. Resistance to fipronil in Drosophila simulans: influence of two point mutations in the RDL GABA receptor subunit. J. Neurochem. 2005;92:1295–1305. doi: 10.1111/j.1471-4159.2004.02922.x. [DOI] [PubMed] [Google Scholar]
- 125.Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel gene superfamily of the nematode, Caenorhabditis elegans. Invert. Neurosci. 2008;8:41–47. doi: 10.1007/s10158-008-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jones, A. K. & Sattelle, D. B. Diversity of insect nicotinic acetylcholine receptor subunits in Insect nicotinic acetylcholine receptors. (ed. Thany, S. H.) 25–43 (Springer, 2010). [DOI] [PubMed]
- 127.Ffrench-Constant RH, Mortlock DP, Shaffer CD, MacIntyre RJ, Roush RT. Molecular cloning and transformation of cyclodiene resistance in Drosophila: an invertebrate gamma-aminobutyric acid subtype A receptor locus. Proc. Natl. Acad. Sci. USA. 1991;88:7209–7213. doi: 10.1073/pnas.88.16.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ffrench-Constant RH, Rocheleau TA. Drosophila γ-aminobutyric acid receptor gene Rdl shows extensive alternative splicing. J. Neurochem. 1993;60:2323–2326. doi: 10.1111/j.1471-4159.1993.tb03523.x. [DOI] [PubMed] [Google Scholar]
- 129.Buckingham SD, Biggin PC, Sattelle BM, Brown LA, Sattelle DB. Insect GABA receptors: splicing, editing, and targeting by antiparasitics and insecticides. Mol. Pharmacol. 2005;68:942–951. doi: 10.1124/mol.105.015313. [DOI] [PubMed] [Google Scholar]
- 130.Clements J, Schoville S, Peterson N, Lan Q, Groves RL. Characterizing molecular mechanisms of Imidacloprid resistance in select populations of Leptinotarsa decemlineata in the Central Sands Region of Wisconsin. PLoS One. 2016;11:e0147844. doi: 10.1371/journal.pone.0147844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clements J, et al. RNA interference of three up-regulated transcripts associated with insecticide resistance in an imidacloprid resistant population of Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 2017;135:35–40. doi: 10.1016/j.pestbp.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 132.Willis JH. Structural cuticular proteins from arthropods: Annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol. 2010;40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McDonnell CM, et al. Evolutionary toxicogenomics: diversification of the Cyp12d1 and Cyp12d3 Genes in Drosophila species. J. Mol. Evol. 2012;74:281–96. doi: 10.1007/s00239-012-9506-3. [DOI] [PubMed] [Google Scholar]
- 134.Hu F, Dou W, Wang J-J, Jia F-X, Wang J-J. Multiple glutathione S-transferase genes: identification and expression in oriental fruit fly, Bactrocera dorsalis. Pest Manag. Sci. 2014;70:295–303. doi: 10.1002/ps.3558. [DOI] [PubMed] [Google Scholar]
- 135.Han J-B, Li G-Q, Wan P-J, Zhu T-T, Meng Q-W. Identification of glutathione S-transferase genes in Leptinotarsa decemlineata and their expression patterns under stress of three insecticides. Pestic. Biochem. Physiol. 2016;133:26–34. doi: 10.1016/j.pestbp.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 136.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 137.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 138.Revuelta L, et al. Contribution of ldace1 gene to acetylcholinesterase activity in Colorado potato beetle. Insect Biochem. Mol. Biol. 2011;41:795–803. doi: 10.1016/j.ibmb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 139.Wan P-J, Fu K-Y, Lü F-G, Guo W-C, Li G-Q. Knockdown of a putative alanine aminotransferase gene affects amino acid content and flight capacity in the Colorado potato beetle Leptinotarsa decemlineata. Amino Acids. 2015;47:1445–1454. doi: 10.1007/s00726-015-1978-1. [DOI] [PubMed] [Google Scholar]
- 140.Baum JA, et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007;25:1322. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 141.Swevers L, et al. Colorado potato beetle (Coleoptera) gut transcriptome analysis: expression of RNA interference-related genes. Insect Mol. Biol. 2013;22:668–684. doi: 10.1111/imb.12054. [DOI] [PubMed] [Google Scholar]
- 142.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 143.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/S0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 144.Yoon J-S, Shukla JN, Gong ZJ, Mogilicherla K, Palli SR. RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: Identification of key contributors. Insect Biochem. Mol. Biol. 2016;78:78–88. doi: 10.1016/j.ibmb.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 145.Guénin H-A, Scherler M. La formule chromosomiale du doryphore Leptinotarsa decemlineata Stål. Rev. Suisse Zool. 1951;58:359–370. [Google Scholar]
- 146.Hsiao TH, Hsiao C. Chromosomal analysis of Leptinotarsa and Labidomera species. Genetica. 1983;60:139–150. doi: 10.1007/BF00127500. [DOI] [Google Scholar]
- 147.Poelchau M, et al. The i5k Workspace@NAL—enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 2015;43:D714–D719. doi: 10.1093/nar/gku983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee E, et al. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013;14:R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 152.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 154.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 155.Laetsch DR, Blaxter ML. BlobTools: Interrogation of genome assemblies. F1000Research. 2017;6:1287. doi: 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- 156.National Center for Biotechnology Information Nucleotide. Nucleotide (nt) database. Available from: https://www.ncbi.nlm.nih.gov/nucelotide/(accessed January 05, 2018).
- 157.Buchfink B, Xie C, Huson D. Fast and sensitive protein alignment using DIAMOND. Nature Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 158. The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed]
- 159.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kriventseva EV, et al. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015;43:D250–D256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mirarab S, et al. PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J. Comput. Biol. 2015;22:377–386. doi: 10.1089/cmb.2014.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:2010–2011. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mirarab S, Warnow T. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics. 2015;31:i44–i52. doi: 10.1093/bioinformatics/btv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 165.Wolfe JM, Daley AC, Legg DA, Edgecombe GD. Fossil calibrations for the arthropod Tree of Life. Earth-Sci. Rev. 2016;160:43–110. doi: 10.1016/j.earscirev.2016.06.008. [DOI] [Google Scholar]
- 166.Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 167.Rosendale AJ, Romick-Rosendale LE, Watanabe M, Dunlevy ME, Benoit JB. Mechanistic underpinnings of dehydration stress in the American dog tick revealed through RNA-seq and metabolomics. J. Exp. Biol. 2016;219:1808–1819. doi: 10.1242/jeb.137315. [DOI] [PubMed] [Google Scholar]
- 168.Baggerly KA, Deng L, Morris JS, Aldaz CM. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics. 2003;19:1477–1483. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
- 169.Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0.
- 170.Smit, A. & Hubley, R. RepeatModeler Open-1.0.
- 171.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jacques, R. L. Jr. The potato beetles (No. 3) (Taylor & Francis, 1988).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.