Abstract
Autosomal dominant hypercholesterolemia (ADH) is characterized by elevated LDL-C levels leading to coronary heart disease. Four genes are implicated in ADH: LDLR, APOB, PCSK9 and APOE. Our aim was to identify new mutations in known genes, or in new genes implicated in ADH. Thirteen French families with ADH were recruited and studied by exome sequencing after exclusion, in their probands, of mutations in the LDLR, PCSK9 and APOE genes and fragments of exons 26 and 29 of APOB gene. We identified in one family a p.Arg50Gln mutation in the APOB gene, which occurs in a region not usually associated with ADH. Segregation and in-silico analysis suggested that this mutation is disease causing in the family. We identified in another family with the p.Ala3396Thr mutation of APOB, one patient with a severe phenotype carrying also a mutation in PCSK9: p.Arg96Cys. This is the first compound heterozygote reported with a mutation in APOB and PCSK9. Functional studies proved that the p.Arg96Cys mutation leads to increased LDL receptor degradation. This work shows that Next-Generation Sequencing (exome, genome or targeted sequencing) are powerful tools to find new mutations and identify compound heterozygotes, which will lead to better diagnosis and treatment of ADH.
Introduction
Hypercholesterolemia is a major risk factor for atherosclerosis and its premature cardiovascular complications. Autosomal dominant hypercholesterolemia (ADH) (MIM # 143890) is one of the most common monogenic disorders. It is caused by mutations in genes encoding key proteins involved in the LDL receptor (LDLR) endocytic and recycling pathways, causing decreased cellular uptake of LDL and increased plasma LDL-cholesterol (LDL-C) concentrations. Elevated plasma LDL-C levels give rise to tendon and skin xanthomas, arcus cornea, and vascular deposits, leading to progressive and premature atherosclerosis and coronary heart disease (CHD)1. Four genes are known to be implicated in the disease: LDLR, the gene encoding the low-density lipoprotein receptor2; APOB, which encodes the apolipoprotein B3, the ligand of the LDL receptor; PCSK9 (Proprotein convertase subtilisin kexin 9)4, which encodes a serine protease5 that plays a role in the degradation of the LDLR independently of its catalytic activity6; and APOE7. Mutations in LDL Receptor Adaptor protein 1 (LDLRAP1) gene are responsible of the recessive form of the disease8.
The prevalence of ADH is approximately estimated to 1/217 in Northern Europe9,10 but varies geographically from a region to another and is higher in some populations due to a founder effect11. Recent studies showed that ADH is under-diagnosed and undertreated in the general population12 and the probability of identifying a genetic component in individuals with hypercholesterolemia increases when LDL-C levels increased10.
The respective contribution of each known gene to ADH slightly varies from one country to another. A study in a French cohort, which included probands and families with hypercholesterolemia recruited through the French Research Network for ADH from several regions of France, showed that the LDLR gene is implicated in 73.9% of the cases, while mutations in APOB and PCSK9 are responsible of 6.6% and 0.7% of the cases respectively. Nevertheless, in 18.8% of the ADH probands studied, no mutation was found13.
Genetic diagnosis of ADH was generally performed by direct DNA sequencing or a combination of direct sequencing with the multiplex ligation-dependent probe amplification (MLPA) to detect large insertion or deletion mutations in genes known to be implicated in the disease. Concerning the APOB gene, the p.Arg3527Gln, also known as APOB3527 or APOB3500, is the first and most common ADH-related mutation in APOB reported in late 1980s3. It is responsible alone for more than 95% of Familial Defective apolipoprotein B cases (FDB)14 (MIM# 144010). A few other mutations leading to hypercholesterolemia were described in the following years and were all located in a specific region of the APOB gene. Therefore, this gene was not classically entirely studied in ADH by Sanger sequencing and routinely only a fragment of exon 26 and another of exon 29 are analyzed, covering the regions where the functional mutations causing hypercholesterolemia have been described15,16.
Targeted next-generation sequencing (NGS) panel is now currently used to screen for ADH-causing mutations10,17, while exome sequencing (targeted sequencing of all protein-coding regions of the genome) is used to identify mutations in new genes. This approach, now slowly substituted by whole genome sequencing, has emerged as an effective tool for gene discovery in families with suspected monogenic disorders18.
The aim of our study was to investigate the genetic causes of ADH in French probands. After the exclusion in 127 ADH probands, of mutations in the LDLR, APOE and PCSK9 genes and fragments of exons 26 and 29 of APOB, thirteen ADH families were recruited. Two or three affected members of each family were studied by exome sequencing in order to identify new mutations in known genes or in new genes, which will eventually lead to a better understanding of the mechanism of this disease.
Results
A double heterozygous patient with a mutation in APOB and PCSK9 in HC138 family
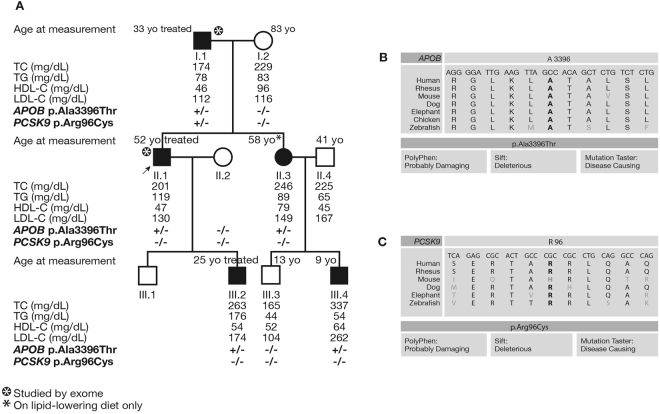
Exome sequencing was used to investigate the genetic cause of ADH in the HC138 family using DNA from patients I.1 and II.1 (Fig. 1). The proband II.1 had a history of arcus cornea with high levels of total-cholesterol (201 mg/dL) and LDL-C (130 mg/dL) despite treatment by rosuvastatin 10 mg and Ezetimibe. His father I.1 suffered from myocardial infarction at the age of 50. He also presented arcus cornea and had a very high level of total-cholesterol reaching 440 mg/dL before he started statin treatment. The familial autosomal dominant transmission was confirmed by the recruitment of other members of the family. Details of their clinical measurements are given in Fig. 1A.
Figure 1.
Pedigree of family HC138 with status of the p.Ala3396Thr mutation of APOB and the p.Arg96Cys of PCSK9 for each patient. (A) Lipid levels are given when available in mg/dL with the age at clinical measurement. The −/− indicates the absence of the mutation while +/− indicates the heterozygous carriers. The asterisk shows the patients studied by exome sequencing. (B–C) Conservation of the alanine at position 3396 of APOB and the arginine at position 96 of PCSK9 between different species. In-silico prediction analysis of both mutations using Polyphen, Sift, and Mutation taster tools.
Exome sequencing of the two patients allowed the identification of the c.10186 G > A mutation in exon 26 of the APOB gene, which led to the substitution p.Ala3396Thr as previously described19. This mutation was then confirmed by Sanger sequencing. It segregated with the disease in the family with a penetrance of 100% and with no phenocopy. It was not reported in the different databases: Exome Variant Server (evs.gs.washington.edu/EVS/), dbSNP (ncbi.nlm.nih.gov/SNP/), and gnomAD browser (gnomad.broadinstitute.org/). The in-silico bioinformatics tools predicted that it is disease causing, and showed a high conservation of the alanine at position 3396 among different species (Fig. 1B).
Furthermore, exome sequencing analysis showed in patient I.1 a variation in exon 2 of PCSK9: c.286 C > T leading to the p.Arg96Cys substitution. This variant was confirmed by Sanger sequencing. It was not carried by any other member of the family (Fig. 1A). The p.Arg96Cys (rs185392267) is reported with a very low frequency of 2.166e-5 (only 6 times over 277054 in gnomAD browser). The arginine at position 96 is highly conserved among species and its replacement by a cysteine is predicted to be pathologic by the in silico prediction tools (Fig. 1C).
Characterization of the p.Arg96Cys PCSK9 mutation
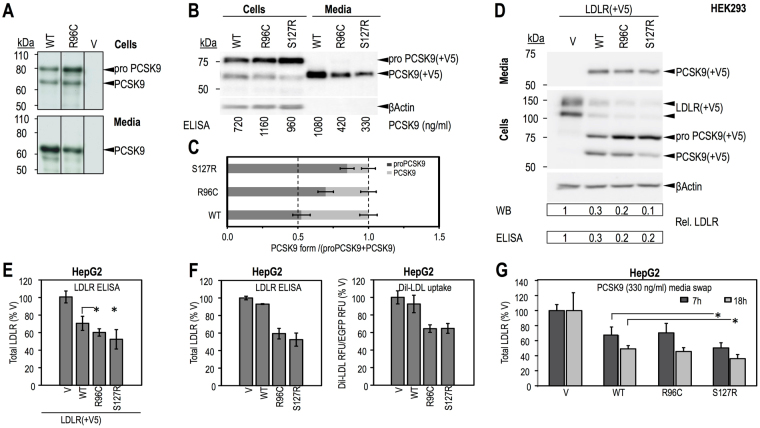
Functional analyses were performed to study the effect of the p.Arg96Cys PCSK9 mutation on PCSK9 maturation and on PCSK9-mediated LDLR degradation. PCSK9-WT and p.Ser127Arg (S127R), a well-characterized gain-of-function (GOF) mutation, were used as controls. Cell-based functional characterizations of wild-type (WT) PCSK9 and mutants R96C and S127R in HEK293 and HepG2 cells are shown in Fig. 2. Biosynthetic analysis (Fig. 2A), Western Blot (WB) (Fig. 2B) and Elisa (Fig. 2C) analyses for PCSK9 showed that the p.Arg96Cys mutation results in increased cellular levels of total PCSK9 (~60%), reduced processing of proPCSK9 zymogen to PCSK9 (by ~30%), and decreased PCSK9 secretion (by ~60%) relative to WT protein. Overexpression of PCSK9-R96C results in significantly increased LDLR degradation compared to PCSK9-WT. This was confirmed by WB and by Elisa assays for total overexpressed LDLR after V5-tagged LDLR was co-transfected in HEK293 cells with V5-tagged PCSK9-WT (WT), PCSK9-R96C (R96C), PCSK9-S127R (S127R) or empty vector (V), as control (Fig. 2D), or in HepG2 cells (Fig. 2E). Similar results were obtained for endogenous LDLR in HepG2 cells after overexpression of WT and mutants PCSK9 (Fig. 2F). Measurement of Dil-LDL uptake under these conditions (Fig. 2F, right panel), showed that R96C PCSK9, like the GOF S127R, was significantly more potent than WT PCSK9 in reducing LDL uptake. However, when added extracellularly to HepG2 cells for short or long times, PCSK9-R96C degraded endogenous LDLR to the same extent as PCSK9-WT (Fig. 2G).
Figure 2.
Cell-based functional characterization of PCSK9 wild-type (WT) and p.Arg96Cys (R96C). HEK293 cells were transiently transfected with V5-tagged PCSK9-WT (WT), PCSK9-R96C (R96C) or empty vector (V) (A), or gain of function PCSK9-S127R (S127R) (positive control) (B and C) and analyzed for PCSK9 cellular expression, zymogen processing and secretion of mature protein. (A) Biosynthetic analysis; 48 h post-transfection cells were pulsed-labeled with [35S]Met/Cys for 3 h, followed by anti-V5 immunoprecipitation, SDS-PAGE and autoradiography. (B) WB analysis using anti V5-HRP. Levels of total cellular and secreted PCSK9 were quantified by ELISA and the concentrations are listed. The bands corresponding to proPCSK9 and PCSK9 in (B) were quantified, their values normalized to β−Actin and ratios of the value of each form to the sum of the two forms were graphed in (C). V5-tagged LDLR was co-transfected with V5-tagged PCSK9-WT, PCSK9-R96C, PCSK9-S127R or empty vector, as control, in HEK293 cells (D) or HepG2 cells (E). Transfected HEK293 cells were analyzed by WB using anti V5-HRP (D). Total cellular levels of LDLR were quantified from the WB and by ELISA for HEK293 cells (D) or by ELISA only for HepG2 cells (E) and were normalized to values of the control. (F) HepG2 cells were transiently transfected with V5-tagged PCSK9-WT, PCSK9-R96C, PCSK9-S127R or empty vector, as control, and the total cellular endogenous levels of LDLR were quantified by ELISA (left). Dil-LDL uptake (right) was measured over 2 h. Measured values are reported as % control. (G) HepG2 cells were incubated for 7 h or 18 h with conditioned media from HEK293 cells (produced, illustrated and quantified in B): no PCSK9 control-media (V) or PCSK9-media (330 ng/ml), WT, R96C or S127R, and analyzed by ELISA for total cellular LDLR. Data are representative of two independent experiments performed at least in duplicate, with the exception of (F), where LDLR-ELISA was completed for one experiment performed in triplicate, while Dil-LDL uptake was measured in one experiment performed in 16 independent replicates per condition. Quantifications are averages ± SD. *p < 0.05; **p < 0.01; ***p < 0.001 (t-test). Full-length blots of Fig. 2A,B and D are presented in Supplementary Figure 2, where only the lanes selected in Fig. 2 are labeled.
A new mutation p.Arg50Gln in APOB, causing ADH
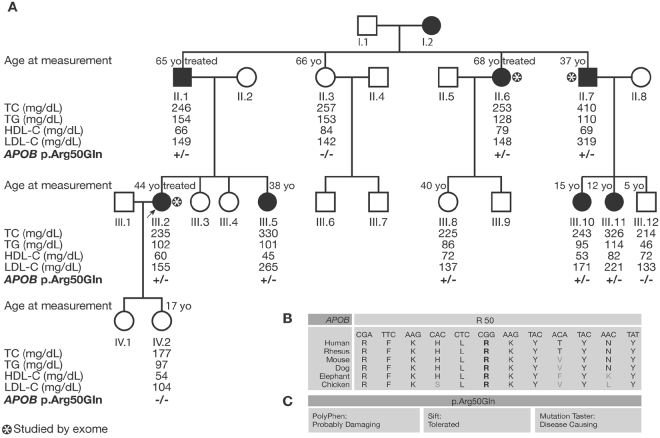
Furthermore, exome sequencing was performed in the HC706 family using DNA from II.6, II.7 and III.2 affected individuals (with asterisk in Fig. 3). The female proband (III.2) presented high levels of total cholesterol (235 mg/dL) and LDL-C (155 mg/dL) at the age of 44 even after treatment with rosuvastatin 20 mg and Ezetimibe. Family history indicated that her 65 years-old father (II.1) had a level of total cholesterol of 300 mg/dL before treatment, and kept high levels of total cholesterol (246 mg/dL) and LDL-C (149 mg/dL) at 65 years old under treatment. Interestingly, a paternal aunt (II.6) also had a history of elevated level of total cholesterol (315 mg/dL before treatment) that reached 253 mg/dL on statin treatment with a LDL-C level of 148 mg/dL at the age of 68. Additionally, a paternal uncle (II.7) presented a level of total cholesterol of 410 mg/dL and LDL-C level of 319 mg/dL at the age of 37 before starting statin treatment. Recruitment of other family members and further investigations confirmed the autosomal dominant trait. Lipid measurements of the different members of the family before treatment (when available) or after are provided in Fig. 3A.
Figure 3.
Segregation analysis of the p.Arg50Gln mutation of APOB in family HC706: (A) Lipid levels are given when available in mg/dL with the age of the patients at clinical measurement. The −/− indicates absence of the mutation while +/− indicates heterozygous carriers. The asterisk shows the patients studied by exome sequencing. (B) The arginine at position 50 of APOB is a conserved amino acid between different species. (C) This mutation is predicted to be probably damaging by Polyphen, tolerated by Sift, and disease causing by mutation taster when studied in-silico.
A substitution c.149 G > A in exon 3 in the APOB gene causing a missense variant p.Arg50Gln was identified in the three affected members studied by exome sequencing. Sanger sequencing was used to validate the variation and showed that the variation segregated with the disease in the family with a penetrance of 91% and absence of phenocopy. Only one case of incomplete penetrance was detected over the eleven studied members of the family for whom DNA were available: individual III.8 who carries the variation but does not present high lipid levels as depicted in Fig. 3. This variant is not reported in the databases. The arginine residue at position 50 is a conserved amino acid in different species (Fig. 3B). The p.Arg50Gln variant is predicted to be damaging by bioinformatics tools such as Sift, mutation taster and Polyphen-2 as shown in Fig. 3C.
Subsequently, we analyzed by Sanger sequencing the DNA of 127 French probands with hypercholesterolemia in whom we excluded the known mutations in LDLR, APOE and PCSK9 genes and fragments of exons 26 and 29 of APOB, to look for the p.Arg50Gln variation in the APOB gene. None of those probands carried this variation.
Discussion
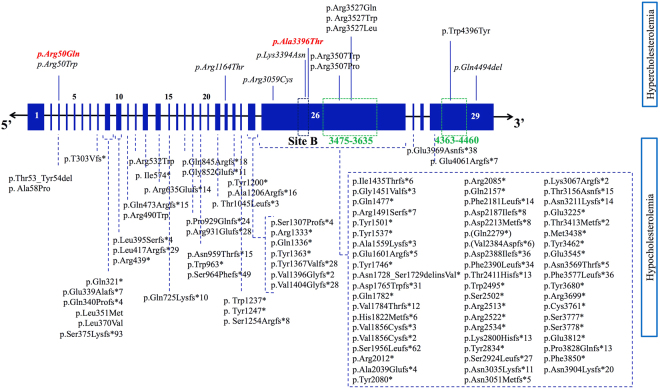
In this study, we show that new sequencing technologies like exome sequencing are powerful tools to find new mutations in genes already known to be implicated in ADH, APOB in particular, especially because the Sanger sequencing strategies classically performed in ADH genetic diagnosis do not target the entire region of APOB. Indeed, we identified by exome sequencing in 2 unrelated French ADH families, 2 new mutations in the APOB gene, p.Arg50Gln and p.Ala3396Thr which are not located in the regions of exons 26 and 29 routinely investigated using Sanger sequencing when searching for APOB mutations implicated in ADH. In fact, direct Sanger sequencing has been classically targeting only these particular regions due to the size of the apoB (4536 amino acids) and because the previously described functional ADH mutations are located in these specific regions of the gene15,16 (Fig. 4). Although numerous other mutations of APOB were reported in its entire coding-region, these mutations are known to cause familial hypocholesterolemia (hypobetalipoproteinemia or FHBL) and are mostly nonsense, frameshift, or splicing variants that lead to various C-terminally truncated apoB species20 (Fig. 4). As for FDB, the most common mutation associated with an ADH phenotype is the p.Arg3527Gln carried by approximately 0.1% of Northern Europeans and US Caucasians. In addition, the p.Arg3527Trp variant is known to make a significant contribution to familial hypercholesterolemia among East Asians21. Few APOB mutations were proven to be pathogenic in ADH: p.Arg3507Trp, p.Arg3527Gln, and p.Trp4396Tyr15,22. Despite the fact that these mutations are not directly involved in the binding to the LDLR segment that concerns residues 3386 to 339623 (Fig. 4), they might destabilize the apoB–LDL receptor interaction by altering some critical residues that are crucial for apoB–LDL receptor affinity24, which result in a defective receptor-binding23,25. Interestingly, the p.Ala3396Thr mutation that we identified recently19 occurs in exon 26 of APOB and is located in the LDLR binding site B of apoB (Fig. 4). Consequently, this mutation might lead to a defective apoB-LDLR binding and an abnormal LDL internalization. Its good segregation with the disease in the family along with its localization in an important region for the normal apoB function are strong evidences for its pathogenic effect.
Figure 4.
Different mutations reported in APOB, causing hypercholesterolemia (FDB) and hypocholesterolemia (FHBL). The APOB gene is constituted of 29 exons. The binding site for the LDL receptor originally described as site B is formed primarily by residues 3386–3396 (anciently known by 3359–3369). Regions 3475–3635 and 4363–4460 are the ones we classically sequence when looking for APOB mutations in ADH. Mutations causing familial hypocholesterolemia (FHBL) are distributed on the entire coding-region, and they are mostly nonsense, frameshift, or splicing variants. Few mutations causing hypercholesterolemia (FDB) are described in a particular region of APOB, the p.Arg3527Gln mutation being the most frequent one. Some others have been recently reported to cause ADH outside the classical regions of APOB, shown in italic; the p.Arg50Gln and p.Ala3396Thr are detailed in this article and highlighted in bold.
Add to that, the p.Arg50Gln mutation is new and occurs in exon 3 of APOB, a region not routinely analyzed in ADH. Several arguments are in favor of a mutation responsible of the ADH phenotype in this family: (1) it showed good segregation with the disease in the family, (2) in-silico prediction tools are in favor of its deleterious effect at the protein level, (3) this variant was never reported before, and thus its rare incidence is in favor of a mutation rather than a polymorphism and (4) exome sequencing revealed no other interesting genetic event that could be responsible for the ADH phenotype in this family. Furthermore, another ADH causing mutation at the same position, the p.Arg50Trp, has been reported in another ADH family26 and was confirmed by exome sequencing in the UK10K project27. Thomas et al. showed that the p.Arg50Trp variant of APOB accumulates in the circulation of affected carriers, which suggest its defective hepatic uptake. In fact, exon 3 forms a small part of the first domain (βα1), which is predicted to direct hepatic assembly of lipoprotein molecules, as well as to affect the interaction of LDL particles with lipases and macrophage scavenger receptors. Nevertheless, detailed information on the specific role of exon 3 in these processes is not available26.
Furthermore, a recent study identified 2 novel APOB mutations, p.Arg3059Cys and p.Lys3394Asn, both associated with a significant decrease in binding to the LDL receptor, despite having a low penetrance when studying the co-segregation in the families16. Another study proved that p.Arg1164Thr and p.Gln4494del of APOB presented a 40% decrease in internalization in lymphocytes and HepG2 cells, very similar to APOB352715 even though they didn’t show complete penetrance. These results suggest that APOB can carry more ADH causing mutations outside of the classically studied regions, which means that all regions of the APOB should be investigated when diagnosing ADH. The arginine at position 50 seems to be crucial for a normal cholesterol metabolism since 2 different mutations causing ADH have been identified at this same position. Interestingly, a recent study suggested that PCSK9 could bind to amino acid sequences within the N-terminal region of apoB28. Whether or not APOB mutations in this region might affect its interaction with other proteins such as PCSK9 for instance are still to be uncovered.
Besides the p.Ala3396Thr identified in the HC138 family, the analyses of the variants observed in one of the members of this family in whom exome sequencing has been performed, allowed the identification of a new mutation in exon 2 of PCSK9: p.Arg96Cys. This patient carrying the 2 mutations in APOB and PCSK9 showed a severe phenotype with a maximal level of 440 mg/dL of total cholesterol before statin treatment. He also suffered from a myocardial infarction at the age of 50. The p.Arg96Cys mutation of PCSK9 has been recently reported to be responsible alone for the ADH phenotype in 3 patients from Denmark14, with mean untreated values of total cholesterol of 271.5 ± 46.0 mg/dL and LDL-C of 191.4 ± 34.4 mg/dL. Interestingly, 2 of those 3 patients presented coronary artery disease (CAD)14. Our functional studies performed herein demonstrated that the p.Arg96Cys mutation is a new GOF mutation of PCSK9, which alone may be responsible for the ADH phenotype. Although synthesized in cells at a higher level than the WT protein, the p.Arg96Cys is less secreted compared to WT. When overexpressed in cells, this GOF mutant degrades the LDLR to a higher extent than PCSK9-WT. The latter activity of PCSK9-R96C correlated with a decreased Dil-LDL uptake in HepG2 cells, an effect similar to that observed with the PCSK9-S127R GOF mutant. Interestingly, when added extracellularly to HepG2 cells, PCSK9-R96C degraded endogenous LDLR to the same extent as PCSK9-WT, whereas the GOF PCSK9-S127R displayed a significantly higher activity toward the LDLR. Collectively, our cell-based data suggest that PCSK9-R96C is a GOF mutant that better interacts with LDLR intracellularly (intracellular pathway)29, but similarly at the cell surface (extracellular pathway). In the liver, the extracellular pathway is predominant, as wild type PCSK9 acts mostly extracellularly on hepatocytes. However, the presence of a mutation on one allele of the PCSK9 gene could result in a shift of the prevalence of one pathway over the other, as was also reported for the LDLR-R410S mutation30. Thus, genetic evidence together with the cell-based functional characterization of PCSK9-R96C compared to WT and S127R are strong enough to conclude that the p.Arg96Cys is a GOF mutation of PCSK9, which alone could cause ADH, and aggravate the phenotype when carried together with another ADH causing mutation.
Consequently, exome sequencing helped us identify, in a patient with severe ADH, a mutation in APOB together with a new mutation in PCSK9. To the best of our knowledge, this is the first report of an ADH patient carrying a mutation in both APOB and PCSK9 genes concomitantly. Unfortunately, parents or other family members were not available for further investigations to rule out the possibility of a de-novo mutation that might have occurred in the PCSK9 gene in this patient, and that it was not transmitted to his children. These findings demonstrate that exome sequencing can help in the diagnosis and the identification of compound heterozygotes in ADH. In fact, some studies described patients and families with both LDLR/APOB31–36, or LDLR/PCSK9 mutations37,38. However, the clinical characteristics of double-heterozygous ADH patients are underreported and the diagnosis of double-heterozygous ADH can be easily missed39. Thus other APOB/PCSK9 double heterozygous might exist. It is noteworthy that the molecular identification of double heterozygosity is very important for family screening and adequate ADH diagnosis and treatment of all affected carriers of the family. Furthermore, the other genetic event occurring in another gene might explain differences in phenotypes and would be important for revealing the cause of phenocopies when studying familial segregation. This underscores the necessity of fully screening the LDLR, APOB, PCSK9 and APOE genes in all patients.
In summary, our work shows the importance of next generation sequencing technologies such as exome sequencing in identifying new mutations in the genes known to be implicated in ADH, and in revealing double heterozygous mutations, which improves familial screening and genetic counseling, as well as understanding the transmission of the disease and its severity in different members of the same family. The investigation for ADH mutations should include the entire coding regions of LDLR, PCSK9, APOE and APOB, with a particular attention to the region of the arginine at position 50 of the APOB. This will improve diagnosis and treatment of the disease and prevent its cardiovascular complications. Physicians, clinical biochemists and health professionals worldwide should join their effort to fight against this disease by offering the most adequate screening and diagnosis of individuals and families with ADH. Thus, genetic studies are of great importance in these extremely high-risk individuals and families. They can help implementing the most effective strategies to prevent and treat ADH, and might lead to new therapeutic class of lipid lowering drugs like it was the case with the discovery of PCSK940.
Methods
Patients and Families
Probands and families from different cities in France were recruited by The French National Research Network on Hypercholesterolemia based on the inclusion criteria previously described13: LDL-C levels above the 95th percentile when compared with a sex and age-matched French population (STANISLAS cohort, B. Herbeth, G. Siest & S Visvikis-Siest41, personal communication), with normal levels of triglycerides and HDL-C, with an autosomal dominant transmission of hypercholesterolemia in the family. The study was performed in accordance with French bioethics regulations and all subjects gave informed consent. This study was conducted as part of trial # 05-07-06 approved by French Consultative Committee for the Protection of Person in Biomedical Research (CCPPRBs) Paris, Necker.
Sanger Sequencing and MLPA
In all subjects, genes were studied sequentially: first, the p.Arg3527Gln mutation of the APOB (NM_000384.2) was looked for as previously described42 and then regions 3475–3635 and 4363–4460 of APOB were analyzed by Sanger sequencing. The promoters and the 18 exons of LDLR (NM_000527.4), as well as close flanking intronic sequences were amplified and sequenced. If no mutation was found, the search for a deletion/duplication of one or several LDLR’ exon(s) was performed with SALSA MLPA kit (P062) and data were analyzed with Coffalyser software (MRC-Holland). Finally, if no deletion/duplication was discovered, the 12 exons of PCSK9 (NM_174936.3) and the 4 exons of APOE (NM_000041.3) as well as the flanking intronic regions were sequenced. Primer sequences and annealing temperatures are available upon request. Electrophoregrams were analyzed using Gensearch®, or CodonCode Alligner®.
Exome Sequencing
The family members analyzed by exome sequencing where chosen based on DNA availability for at least 2 affected members of the family and presence of informed consent allowing for genetic studies with prioritization of phenotypic extremes when possible. These selected samples underwent exome sequencing at the Broad Institute after that the institutional review board and all participating sites approved the study protocols and all individuals who were selected for sequencing provided informed consent as previously described19.
Analysis of Exome Sequencing Data
In order to find the causal mutation out of thousands of different variations obtained by the exome sequencing, we used a comprehensive framework for prioritizing variants, which is commonly used in the analysis of exome sequencing studies43. Starting with the total number of variants shared by individuals from the family, we excluded variants conflicting with the ADH inheritance pattern and common variants that have a frequency of ≥ 1% in the general population, by examining different databases such as dbSNP, 1000Genome, Exac and gnomAD browsers. Then, we excluded silent and non-genic variants which do not alter protein sequence since most Mendelian syndromes are caused by coding or splice site mutations that alter the protein sequence. The remaining single nucleotide variants and short insertions or deletions were considered candidates. The variants were subsequently analyzed separately by exploring those whose genes have physical protein-protein interaction with the 4 known genes of ADH, or share the same biological pathways. When an interesting variation is found, familial segregation was studied to demonstrate its possible co-segregation with the phenotype in the family.
In silico Analyses
The frequency of the variations found by sequencing was estimated using different databases: Exome Variant Server (evs.gs.washington.edu/EVS/), dbSNP (ncbi.nlm.nih.gov/SNP/), and gnomAD browser (gnomad.broadinstitute.org/).
The causal effect of each new molecular event was estimated with in silico prediction of protein function tools: Polyphen (genetics.bwh.harvard.edu/pph), SIFT (sift.jcvi.org), MutationTaster (mutationtaster.org) using Alamut Visual version 2.7.1.
cDNAs, cell culture and transfections
The cDNAs encoding human LDLR30 and human PCSK9 and its mutants44 were cloned in pIRES2-EGFP (Clontech Labs), a bicistronic plasmid allowing the independent expression of a fluorescent EGFP from an internal ribosome entry site (IRES) and C-terminally V5-tagged wild-type (WT) human PCSK9 or its p.Arg96Cys (R96C) mutant (identity confirmed by DNA sequencing) under the control of a CMV promoter. The WT PCSK9 and the GOF PCSK9-S127R44 served as control. HEK293 (human embryonic kidney-derived epithelial) and HepG2 (human hepatocellular carcinoma) cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco’s modified Eagle medium (DMEM) (HEK293 cells) or in Eagle minimal essential medium (EMEM) (HepG2 cells) supplemented with 10% (v/v) fetal bovine serum (FBS) (Invitrogen) and were maintained at 37 °C under 5% CO2. HEK293 cells were seeded in poly-L-lysine (50 µg/mL) coated 12-well plates (3.5 × 105 cells/well) or 10 cm plates (4 × 106 cells/well) for PCSK9 media production and the following day transfected using jetPRIME (PolyPlus) and a total of 0.5 µg of cDNA or 6 µg of cDNA, respectively. 24 h post-transfection, the culturing medium was changed to serum-free and the cells were treated according to each experiment. Alternatively, for media swap experiments, HepG2 cells (3.5 × 105 cells/well) were seeded in 12-well cell culture plates. 24 h later cells were starved for 24 h in serum-free media and following were incubated for 7 h or 18 h with 24 h serum-free conditioned media of HEK293 cells overexpressing human PCSK9 (WT, S127R or R96C) (see above). The concentrations of the secreted PCSK9 into the media were measured using an in-house ELISA assay as previously described45. For Dil-LDL uptake experiments and LDLR ELISA measurements, HepG2 cells were plated in 96-well plates (0.2 × 105 cells/well) (CellBind black plate with clear bottom, Corning; Cat # 3340) or 12-well plates (2 × 105 cells/well), respectively. Transfections were performed at the time of seeding with 0.125 µg of cDNA (96-well plate) or 1 µg of cDNA (12-well plate) and using FuGENE® HD (Promega). 24 h post-seeding and -transfection, the cells were starved for 24 h in serum-free media and treated according to each experiment.
Biosynthetic Analyses
48 h post-transfection, HEK293 cells were washed and pulse-labeled for 3 h with 250 µCi/ml [35S]Met/Cys (PerkinElmer Life Sciences)44. Following 2 washes with ice-cold PBS, the cells were lysed in modified radioimmune precipitation assay buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5; 1% Nonidet P-40; 0.5% sodium deoxycholate; 0.1% SDS) and protease inhibitor mixture (Roche Applied Science). Cell lysates and media were immunoprecipitated with monoclonal V5-Ab (1:500; Invitrogen) and immunoprecipitates were resolved by SDS-PAGE (8% Tris-Tricine gels) followed by autoradiography (2 h at −80 °C).
Human LDLR ELISA in cell lysates
Following the treatments specific to each experiment, HEK293 or HepG2 cells were washed twice with ice-cold PBS and lysed on ice with ice-cold, non-denaturing cell lysis buffer (20 mM Tris-HCl, pH 8, 137 mM NaCl, 2 mM Na2EDTA, 1% NP-40, 10% glycerol, 4% protease inhibitor cocktail without EDTA) for 40 min, with gentle rotation. Cell lysates were cleared by centrifuging for 12 min at 15,000xg at 4 °C. The supernatants corresponding to the non-denatured cell lysates were saved and subjected to measurement of total human LDLR protein levels (human LDLR DuoSet ELISA Development kit, DY218; R&D Systems) and of total protein (Bio-Rad DC Protein assay), following the manufacturers’ protocol. The optical densities of the colored products were determined using a SpectraMax i3 plate reader (Molecular Devices). All measured LDLR concentrations (pg/mL) were corrected for total protein concentration (mg/mL). Corrected LDLR content (pg LDLR/ml total protein) is reported as % vector (pIRES-EGFP) control.
Western Blotting
Following the incubation times and treatments specific to each experiment, cultured cells were washed and lysed and the lysates cleared, as described above. Thirty to fifty micrograms of protein were separated on 8% Tris glycine SDS-PAGE gels and transferred to a PVDF membrane. Western blotting was performed for human LDLR-V5 and human PCSK9-V5 (anti-V5-HRP, 1:5000; R96125; Invitrogen) and for β-actin (rabbit anti-β-actin, 1:5000; A2066; Sigma). After incubation with the appropriate secondary antibodies, if required, the membranes were revealed using Clarity Western ECL Substrate (Bio-Rad), imaged with a GelDoc XR+ instrument (Bio-Rad) and the bands of interest quantified using ImageLab 5.2.1 software (Bio-Rad).
Dil-LDL uptake
48 h post-seeding and -transfection and after a 24 h starvation in serum-free medium, HepG2 cells were incubated for 2 h at 37 °C with 6 μg/ml Dil-LDL (Alfa Aesar; Cat # J675330) (20 μl of 36 μg/ml Dil-LDL were added to each well containing 100 μl of conditioned media). Each condition was prepared in 16 replicates. At the end of the 2 h incubation, the media was removed and the cells washed 3 times (200 μl/well) with ice-cold D-PBS no ions (Wisent). After the final wash was removed, 100 μl of PBS was added to each well and the plate was scanned (bottom read) on a SpectraMax i3 plate reader (Molecular Devices). For each well, raw Dil-LDL uptake was measured as the average fluorescence intensity (RFU) (ex: 534 nm/em: 572 nm) of 21 points equally distributed in a fill pattern. Dil-LDL uptake in each well was corrected for the total number of transfected cells (average EGFP fluorescence intensity; ex: 488 nm/ em: 513 nm) and reported as the fluorescence ratio of Dil-LDL/EGFP. Corrected Dil-LDL uptake is reported as % vector (pIRES-EGFP) control and was obtained from 16 wells.
Electronic supplementary material
Acknowledgements
We would like to thank Pr Sekar Kathiresan (Broad Institute of MIT and Harvard University, Cambridge, MA, USA; Cardiology Division, Department of Medicine, Massachusetts General Hospital and Harvard Medical School, Boston, MA USA) for his help in the exome sequencing experiments and analysis. S.K. was supported by NIH R01 HL107816. This work was supported by a grant from Leducq Foundation [FLQ # 13 CVD 03] through the Transatlantic Networks of Excellence in Cardiovascular Research program (‘The function and regulation of PCSK9: a novel modulator of LDLR activity’); and Institut National de la Santé et de la Recherche Médicale (INSERM); and Conseil de la Recherche de l’Université Saint-Joseph, Beirut, Lebanon; and Lebanese National Council for Scientific Research (CNRS-L); and Canadian Institutes of Health Research (CIHR) Foundation grant (NGS; # 148363); and Canada Research Chair (NGS; # 950–231335); This work was also supported by the national project CHOPIN (CHolesterol Personalized Innovation) granted by the Agence Nationale de la Recherche (ANR-16-RHUS-0007) and coordinated by the Centre Hospitalo-Univesitaire (CHU) de Nantes. S Elbitar and Y Ghaleb are supported by grants from Lebanese National Council for Scientific Research (CNRS-L); and Council of Research of Saint-Joseph University of Beirut, Lebanon and Institut Français du Liban.
Author Contributions
C.B. and M.AF. conceived and designed the study; S.EB., JP.R., M.AF. performed the molecular analyses; G.P., N.S. performed the exome sequencing; M.V., P.EK. and Y.G. participated in the validation of the molecular results; V.C. and E.B. performed the clinical assessments; N.S. conceived and designed biosynthetic analyses; D.SR., J.H. and A.BDO. performed the Western blot and in vitro analysis; S.EB., M.AF., C.B. and N.S. analyzed the data and wrote the article. All authors reviewed and approved the present article.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20281-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abifadel M, et al. Living the PCSK9 adventure: from the identification of a new gene in familial hypercholesterolemia towards a potential new class of anticholesterol drugs. Curr. Atheroscler. Rep. 2014;16:439. doi: 10.1007/s11883-014-0439-8. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein JL, Brown MS. Familial hypercholesterolemia: pathogenesis of a receptor disease. Johns Hopkins Med. J. 1978;143:8–16. [PubMed] [Google Scholar]
- 3.Innerarity TL, et al. Familial defective apolipoprotein B-100: low density lipoproteins with abnormal receptor binding. Proc. Natl. Acad. Sci. USA. 1987;84:6919–6923. doi: 10.1073/pnas.84.19.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 5.Seidah NG, et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 2003;100:928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNutt MC, Lagace TA, Horton JD. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J. Biol. Chem. 2007;282:20799–20803. doi: 10.1074/jbc.C700095200. [DOI] [PubMed] [Google Scholar]
- 7.Marduel M, et al. Description of a large family with autosomal dominant hypercholesterolemia associated with the APOE p.Leu167del mutation. Hum. Mutat. 2013;34:83–87. doi: 10.1002/humu.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia CK, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–1398. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- 9.Benn M, Watts GF, Tybjærg-Hansen A, Nordestgaard BG. Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur. Heart J. 2016;37:1384–1394. doi: 10.1093/eurheartj/ehw028. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, et al. Polygenic Versus Monogenic Causes of Hypercholesterolemia Ascertained Clinically. Arterioscler. Thromb. Vasc. Biol. 2016;36:2439–2445. doi: 10.1161/ATVBAHA.116.308027. [DOI] [PubMed] [Google Scholar]
- 11.Sjouke B, et al. Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: prevalence, genotype-phenotype relationship, and clinical outcome. Eur. Heart J. 2015;36:560–565. doi: 10.1093/eurheartj/ehu058. [DOI] [PubMed] [Google Scholar]
- 12.Nordestgaard BG, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur. Heart J. 2013;34:3478–3490a. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marduel M, et al. Molecular spectrum of autosomal dominant hypercholesterolemia in France. Hum. Mutat. 2010;31:E1811–1824. doi: 10.1002/humu.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins PN, et al. Characterization of Autosomal Dominant Hypercholesterolemia Caused by PCSK9 Gain of Function Mutations and Its Specific Treatment With Alirocumab, a PCSK9 Monoclonal Antibody. Circ. Cardiovasc. Genet. 2015;8:823–831. doi: 10.1161/CIRCGENETICS.115.001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves AC, Etxebarria A, Soutar AK, Martin C. & Bourbon, M. Novel functional APOB mutations outside LDL-binding region causing familial hypercholesterolaemia. Hum. Mol. Genet. 2014;23:1817–1828. doi: 10.1093/hmg/ddt573. [DOI] [PubMed] [Google Scholar]
- 16.Motazacker MM, et al. Advances in genetics show the need for extending screening strategies for autosomal dominant hypercholesterolaemia. Eur. Heart J. 2012;33:1360–1366. doi: 10.1093/eurheartj/ehs010. [DOI] [PubMed] [Google Scholar]
- 17.Maglio C, et al. Genetic diagnosis of familial hypercholesterolaemia by targeted next-generation sequencing. J. Intern. Med. 2014;276:396–403. doi: 10.1111/joim.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 19.Stitziel NO, et al. Exome sequencing in suspected monogenic dyslipidemias. Circ. Cardiovasc. Genet. 2015;8:343–350. doi: 10.1161/CIRCGENETICS.114.000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schonfeld G. Familial hypobetalipoproteinemia: a review. J. Lipid Res. 2003;44:878–883. doi: 10.1194/jlr.R300002-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Andersen LH, Miserez AR, Ahmad Z, Andersen RL. Familial defective apolipoprotein B-100: A review. J. Clin. Lipidol. 2016;10:1297–1302. doi: 10.1016/j.jacl.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Rabès JP, et al. R3531C mutation in the apolipoprotein B gene is not sufficient to cause hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2000;20:E76–82. doi: 10.1161/01.ATV.20.10.e76. [DOI] [PubMed] [Google Scholar]
- 23.Boren J, et al. Identification of the low density lipoprotein receptor-binding site in apolipoprotein B100 and the modulation of its binding activity by the carboxyl terminus in familial defective apo-B100. J. Clin. Invest. 1998;101:1084–1093. doi: 10.1172/JCI1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miserez AR, Keller U. Differences in the phenotypic characteristics of subjects with familial defective apolipoprotein B-100 and familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 1995;15:1719–1729. doi: 10.1161/01.ATV.15.10.1719. [DOI] [PubMed] [Google Scholar]
- 25.Borén J, Ekström U, Agren B, Nilsson-Ehle P, Innerarity TL. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J. Biol. Chem. 2001;276:9214–9218. doi: 10.1074/jbc.M008890200. [DOI] [PubMed] [Google Scholar]
- 26.Thomas ERA, et al. Identification and biochemical analysis of a novel APOB mutation that causes autosomal dominant hypercholesterolemia. Mol. Genet. Genomic Med. 2013;1:155–161. doi: 10.1002/mgg3.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futema M, et al. Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J. Med. Genet. 2014;51:537–544. doi: 10.1136/jmedgenet-2014-102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H, et al. Proprotein convertase subtilisin/kexin type 9 interacts with apolipoprotein B and prevents its intracellular degradation, irrespective of the low-density lipoprotein receptor. Arterioscler. Thromb. Vasc. Biol. 2012;32:1585–1595. doi: 10.1161/ATVBAHA.112.250043. [DOI] [PubMed] [Google Scholar]
- 29.Poirier S, et al. Dissection of the endogenous cellular pathways of PCSK9-induced low density lipoprotein receptor degradation: evidence for an intracellular route. J. Biol. Chem. 2009;284:28856–28864. doi: 10.1074/jbc.M109.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susan-Resiga D, et al. The Proprotein Convertase Subtilisin/Kexin Type 9-resistant R410S Low Density Lipoprotein Receptor Mutation: A Novel Mechanism Causing Familial Hypercholesterolemia. J. Biol. Chem. 2017;292:1573–1590. doi: 10.1074/jbc.M116.769430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauh G, et al. Identification of a heterozygous compound individual with familial hypercholesterolemia and familial defective apolipoprotein B-100. Klin. Wochenschr. 1991;69:320–324. doi: 10.1007/BF01644767. [DOI] [PubMed] [Google Scholar]
- 32.Benlian P, et al. Phenotypic expression in double heterozygotes for familial hypercholesterolemia and familial defective apolipoprotein B-100. Hum. Mutat. 1996;7:340–345. doi: 10.1002/(SICI)1098-1004(1996)7:4<340::AID-HUMU8>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.deCampo A, Schallmoser K, Schmidt H, Toplak H, Kostner GM. A novel splice-site mutation in intron 7 causes more severe hypercholesterolemia than a combined FH-FDB defect. Atherosclerosis. 2001;157:524–525. doi: 10.1016/S0021-9150(01)00535-4. [DOI] [PubMed] [Google Scholar]
- 34.Rubinsztein DC, et al. Characterization of six patients who are double heterozygotes for familial hypercholesterolemia and familial defective apo B-100. Arterioscler. Thromb. J. Vasc. Biol. 1993;13:1076–1081. doi: 10.1161/01.ATV.13.7.1076. [DOI] [PubMed] [Google Scholar]
- 35.Tai ES, et al. Compound heterozygous familial hypercholesterolemia and familial defective apolipoprotein B-100 produce exaggerated hypercholesterolemia. Clin. Chem. 2001;47:438–443. [PubMed] [Google Scholar]
- 36.Taylor A, et al. A double heterozygote for familial hypercholesterolaemia and familial defective apolipoprotein B-100. Ann. Clin. Biochem. 2010;47:487–490. doi: 10.1258/acb.2010.010089. [DOI] [PubMed] [Google Scholar]
- 37.Pisciotta L, et al. Additive effect of mutations in LDLR and PCSK9 genes on the phenotype of familial hypercholesterolemia. Atherosclerosis. 2006;186:433–440. doi: 10.1016/j.atherosclerosis.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Bertolini S, et al. Spectrum of mutations and phenotypic expression in patients with autosomal dominant hypercholesterolemia identified in Italy. Atherosclerosis. 2013;227:342–348. doi: 10.1016/j.atherosclerosis.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Sjouke B, et al. Double-heterozygous autosomal dominant hypercholesterolemia: Clinical characterization of an underreported disease. J. Clin. Lipidol. 2016;10:1462–1469. doi: 10.1016/j.jacl.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Elbitar S, et al. Proprotein convertase subtilisin / kexin 9 (PCSK9) inhibitors and the future of dyslipidemia therapy: an updated patent review (2011–2015) Expert Opin. Ther. Pat. 2016;26:1377–1392. doi: 10.1080/13543776.2016.1206080. [DOI] [PubMed] [Google Scholar]
- 41.Siest G, et al. Objectives, design and recruitment of a familial and longitudinal cohort for studying gene-environment interactions in the field of cardiovascular risk: the Stanislas cohort. Clin. Chem. Lab. Med. 1998;36:35–42. doi: 10.1515/CCLM.1998.007. [DOI] [PubMed] [Google Scholar]
- 42.Rabès JP, et al. Familial ligand-defective apolipoprotein B-100: simultaneous detection of the ARG3500– > GLN and ARG3531– > CYS mutations in a French population. Hum. Mutat. 1997;10:160–163. doi: 10.1002/(SICI)1098-1004(1997)10:2<160::AID-HUMU8>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 43.Li M-X, Gui H-S, Kwan JSH, Bao S-Y, Sham PC. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40:e53. doi: 10.1093/nar/gkr1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjannet S, et al. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 2004;279:48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 45.Dubuc G, et al. A new method for measurement of total plasma PCSK9: clinical applications. J. Lipid Res. 2010;51:140–149. doi: 10.1194/jlr.M900273-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.