Abstract
Thiolutin is a disulfide-containing antibiotic and anti-angiogenic compound produced by Streptomyces. Its biological targets are not known. We show that reduced thiolutin is a zinc-chelator that inhibits the JAB1/MPN/Mov34 (JAMM) domain-containing metalloprotease Rpn11, a de-ubiquinating enzyme of the 19S proteasome. Thiolutin also inhibits the JAMM metalloproteases Csn5, the deneddylase of the COP9 signalosome, Associated-molecule-with-the-SH3-Domain-of-STAM (AMSH), which regulates ubiquitin-dependent sorting of cell-surface receptors, and Brcc36, a K63-specific deubiquitnase of BRCC36-containing isopeptidase complex (BRISC) and BRCA1-BRCA2-containing complex (BRCC). We provide evidence that other dithiolopyrrolones also function as inhibitors of JAMM metalloproteases.
Introduction
Dithiolopyrrolones (DTPs) are bicyclic antibiotics with a disulfide bond between two ene-thioles, which are synthesized by actinomycetes and proteobacteria1. They exhibit broad-spectrum antibiotic properties, strongly inhibit adhesion of human umbilical vein endothelial cells, and S180 tumor-associated angiogenesis in mice2. DTPs are therefore potentially interesting as anti-cancer drugs, however, their biological targets are unclear and their mode of action is not known.
The antibiotic potential of the DTP thiolutin (THL) was originally attributed to its supposed ability to inhibit bacterial and fungal RNA polymerases3-5 but data accumulating over the years cast doubt on this hypothesis6-10. Moreover, THL has been indicated to affect MAPK signalling and, potentially, cellular stress responses11. Treatment of zebrafish embryos with THL led to high lethality, suggesting that it targets a major developmental process12. These mostly unexplained and partially controversial observations led us to revisit the functions of THL.
Surprisingly, we found that the reduced form of THL is an inhibitor of JAMM domain-containing proteases, a small family of Zn2+-dependent isopeptidases that cleave off conjugates of ubiquitin or nedd8. We show that reduced THL is a chelator of Zn2+-ions suggesting that it inhibits JAMM proteases by complexing the catalytic Zn2+-ion in the active center of the enzymes. The human genome encodes six characterized JAMM domain proteins, Rpn11, Csn5, AMSH, AMSH-LP, BRCC36, and MYSM1, which are involved in major aspects of protein and genome homeostasis, and MPND13, which has no known function. Rpn11 is a subunit of the 19S regulatory particle of the proteasome, and it deubiquitylates substrate proteins degraded by the 20S core14. The Rpn11 paralogue Csn5 is the corresponding subunit of the structurally related COP9 signalosome15, and it catalyzes deneddylation of cullin in cullin-RING ubiquitin ligases (CRLs)15,16. AMSH and AMSH-LP are K63-specific deubiquitinases chains required to clear cargo from endosomes17,18. BRCC36 is also a K63-specific deubiquitnase19. It is the catalytic subunit of the BRISC19 and also a subunit of the DNA damage recognition complex BRCC20,21. Finally, MYSM1 is a histone H2A deubiquitinase22.
Results
THL affects RNA PolII dependent transcript levels
To investigate the effect of THL (Fig. 1a) on transcription, we quantified the expression levels of several short-lived transcripts in Neurospora crassa, Saccharomyces cerevisiae, and in HeLa cells. The levels of the N.crassa transcripts vvd and frq dropped rapidly after incubation with a saturating amount (50 μM) of THL (Fig. 1b, c, Supplementary Results, Supplementary Fig. 1a). Similarly, the levels of the S.cerevisiae transcripts dbp2 and not3, which have a half-life of 3 min and 30 min, respectively23, decreased in the presence of THL, although the kinetics of this reduction was not fully compatible with the hypothesis that THL directly inhibits RNA polymerase II (Pol II) (Fig. 1d, Supplementary Fig. 1b). THL has been used as transcription inhibitor in bacteria and fungi but, to our knowledge, never in metazoan cells. We therefore analyzed the effect of THL on the highly unstable c-myc mRNA in HeLa cells24. The level of c-myc mRNA decreased rather slowly in the presence of THL, whereas treatment with the RNA polymerase II (Pol II) inhibitor actinomycin D led to a rapid decrease of c-myc mRNA (Fig. 1e). The data suggest that THL did not completely block transcription of c-myc.
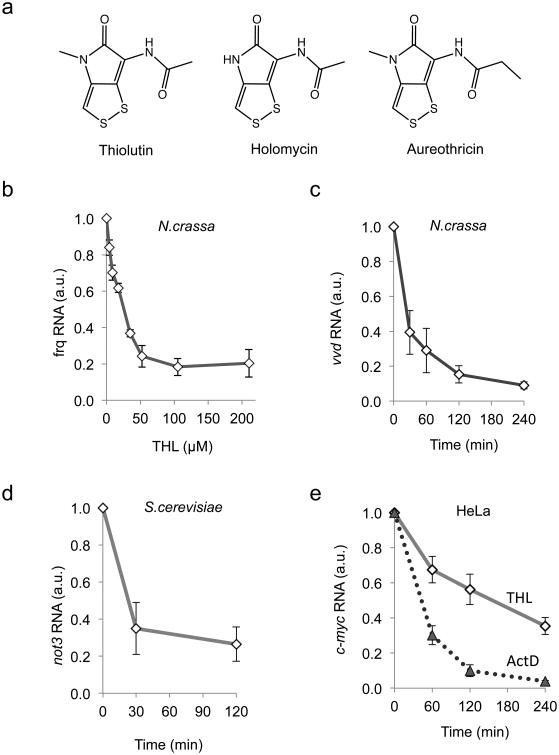
Figure 1. THL affects RNA polymerase II transcript levels.
, a: Structures of the naturally occurring dithiolopyrrolones thiolutin, holomycin and aureothricin.
b: N.crassa cultures were incubated for 4 h with the indicated concentrations of THL and frq RNA was measured.
c: N.crassa cultures were incubated in the presence 50μM THL. Samples were harvested after indicated time periods and vvd RNA was quantified by qRT-PCR and normalized to 28S rRNA.
d:S.cerevisiae cultures (OD600 = 0.5 – 0.8) were incubated in the presence of 50 μM THL and samples were harvested after the indicated time periods. Transcript levels of not3 were quantified by qRT-PCR. Data were normalized to 18S rRNA.
e: Levels of c-myc RNA in HeLa cells decrease with slower kinetics after THL treatment compared to actinomycin D. HeLa cells were incubated in the presence of 10μM THL or 1 μM ActD, respectively. Samples were harvested at indicated time points. c-myc transcript levels were quantified by qRT-PCR. Data were normalized to 18S rRNA.
b–e: Error bars indicate ±s.d. from three independent experiments measured in triplicates.
THL induces global transcriptional stress in Neurospora
To test whether THL directly inhibits eukaryotic RNA polymerases we performed in vitro run-off transcription with affinity purified Pol I-III of S.cerevisiae. None of the three yeast polymerases were inhibited by THL, whereas α-amanitin completely blocked Pol II (Fig. 2a, Supplementary Fig. 2a). These observations indicate that THL does not inhibit the catalytic activity of RNA polymerases but rather impacts on transcription via a different mechanism.
Figure 2. THL does not directly inhibit RNA polymerases.
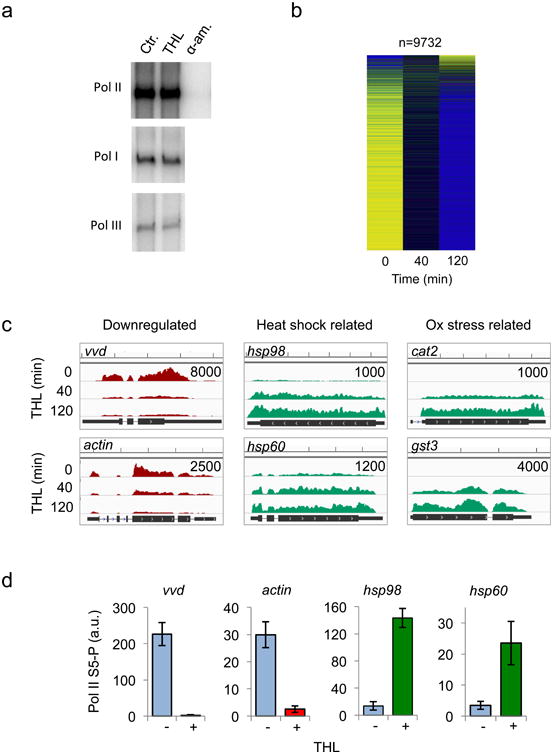
a: THL does not inhibit eukaryotic RNA polymerases in vitro. Purified yeast RNA polymerases I, II and III were pre-incubated with DMSO (Ctr.), THL and α-amanitin, when indicated. Radiolabelled transcripts were synthesized from tailed DNA templates (see Methods) and analyzed by gel electrophoresis and autoradiography.
b: THL is not a Pol II inhibitor in vivo. Heat map analysis of transcriptional profiles (RNAseq) after treatment of N.crassa with 50 μM THL for 0 min, 40 min and 120 min, respectively. Yellow: high and blue: low relative expression.
c: Examples of transcription profiles (Wig files and Browser view) of N.crassa genes up- and downregulated by treatment with 50 μM THL for the indicated time periods. The number in top right corner of each panel specifies the scale (sequence reads). Scale on the top of the panels indicates 500 bp increments.
d: Pol II S5-P occupancy (ChIP-PCR) in the presence of 50 μM THL is significantly decreased at the promoters of vvd and actin but is increased at hsp98 and hsp60. Error bars indicate ±s.d. from three independent experiments measured in triplicates.
To assess the effect of THL on transcription on a global scale we performed RNA sequencing (RNAseq) in N.crassa. The vast majority of genes were efficiently downregulated after treatment with THL for 40 or 120 min, but expression of a subset of genes increased significantly (Fig. 2b). Many of the upregulated genes are implicated in oxidative stress, heat shock, detoxification and metal binding (Fig. 2c, Supplementary Fig. 2b).
As the enzymatic activity of Pol II was not compromised by THL, we performed chromatin immunoprecipitation (ChIP) of Pol II phosphorylated at Ser5 of the C-terminal domain repeats (Pol II S5-P) to assess whether THL affects recruitment of Pol II and initiation of transcription at selected promoters25. Quantification of Pol II S5-P occupancy by ChIP-PCR correlated with the results of the RNAseq analysis: although Pol II S5-P enrichment at the promoter regions of frq, vvd, and actin decreased significantly in the presence of THL, recruitment of Pol II to hsp98 and hsp60 increased (Fig. 2d). The observation that transcription of a substantial subset of genes was induced in presence of THL demonstrates that THL is not an inhibitor of the Pol II-dependent transcription machinery per se. Rather, THL induces via an unknown mechanism a major stress response in N.crassa and triggers a dramatic global change of the transcriptome.
THL inhibits protein turnover and induces ubiquitylation
Since vvd and frq transcript levels were downregulated by THL we evaluated whether the drug could be exploited to investigate turnover of VVD and FRQ protein. When translation was blocked by cycloheximide (CHX), VVD was rapidly degraded with the expected turnover26 (Fig. 3a, Supplementary Fig. 3a). To our surprise, THL, alone or in combination with CHX, completely blocked degradation of VVD (Fig. 3a). Similarly, degradation of FRQ was blocked but resumed after a wash-out of THL (Supplementary Fig. 3a,b), indicating that the inhibition was reversible.
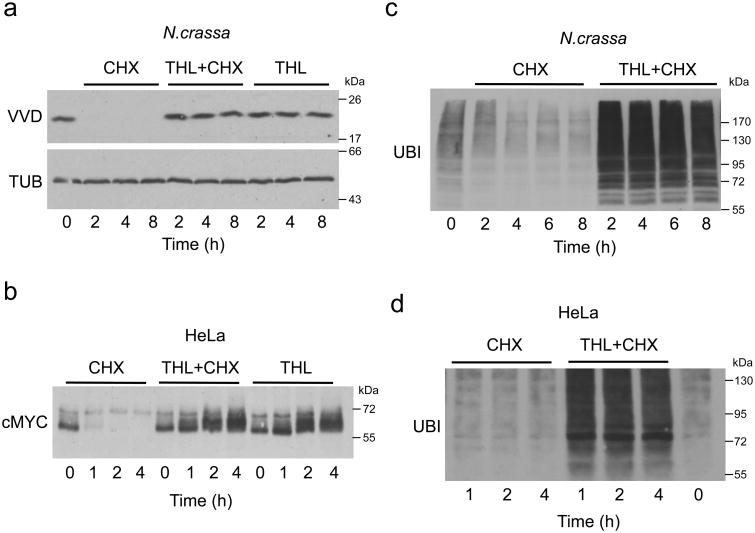
Figure 3. THL inhibits protein degradation by the proteasome.
a: Protein degradation in N.crassa is blocked by THL. Cultures were incubated with 10 μg/ml CHX, 50 μM THL or both together, respectively. Whole cell lysates were analyzed by Western blotting with VVD and β-TUB antibodies at the indicated time points.
b: THL affects protein turnover in mammalian cells. HeLa cells were incubated with 10 μg/ml CHX, 10 μM THL or both together and harvested after the indicated time periods. RIPA extracts were analyzed by Western blotting with c-MYC antibody.
c, d: THL induces accumulation of poly-ubiquitylated proteins in N.crassa (c) and HeLa (d)cells. Whole cell lysates from cells treated as indicated were analyzed by Western blotting with monoclonal Ub antibodies. Each experiment was performed three times with independent cultures. Representative western blots are shown.
To assess whether inhibition of protein degradation by THL is restricted to fungi, we analyzed turnover of c-MYC in HeLa cells (Fig. 3b, Supplementary Fig. 3c). In the presence of CHX, the level of c-MYC rapidly decreased, but THL blocked degradation of c-MYC. Furthermore, progressively phosphorylated isoforms of c-MYC accumulated. As MYC is turned over via the proteasome27, the results suggest that THL may inhibit the ubiquitin-proteasome system (UPS).
Indeed, ubiquitylated proteins accumulated in N.crassa and in HeLa cells that had been treated with THL (Fig. 3c,d, Supplementary Fig. 3b). The accumulation of ubiquitylated proteins was similar in HeLa cells treated with THL or with the proteasome inhibitor MG132 (Supplementary Fig. 3d,e). However, THL efficiently inhibited adhesion of HeLa cells as previously reported12, while MG132 did not (Supplementary Fig. 3f).
Fungi are only poorly sensitive to MG132 due to efficient removal of the drug by efflux pumps such as PDR5 in S.cerevisiae28. In the MG132-hypersensitive yeast strain pdr5Δ THL and MG132 led to comparable levels of ubiquitinated-protein accumulation (Supplementary Fig. 3g). Collectively, our data support the notion that THL inhibits the UPS. Interestingly, the DTP aureothricin (see Fig. 1a) also induced accumulation of ubiquitylated proteins in N.crassa (Supplementary Fig. 3h) suggesting that the UPS might be a common target of DTPs.
We then synthesized and assayed a pyrrolone derivative, PYR2604 (1) (Supplementary Note), lacking the disulfide of THL (Supplementary Fig. 4a). PYR2604 did neither affect expression of frq and hsp30 RNA nor did it induce accumulation of ubiquitylated proteins (Supplementary Fig. 4b and c) suggesting that the disulfide of THL is required for its activity.
THL inhibits Rpn11 of the 19S proteasomal subunit
The 26S proteasome is composed of a 20S proteolytic core particle and a 19S regulatory particle that de-ubiquitylates substrate proteins and funnels them into the proteolytic core14. THL did not inhibit the trypsin-, chymotrypsin-, and caspase-like activities of the proteolytic core (Supplementary Fig. 5a,b). Thus, we reasoned that the drug might inhibit a step upstream, including binding, unfolding, and de-ubiquitylation of substrate proteins by the 19S cap.
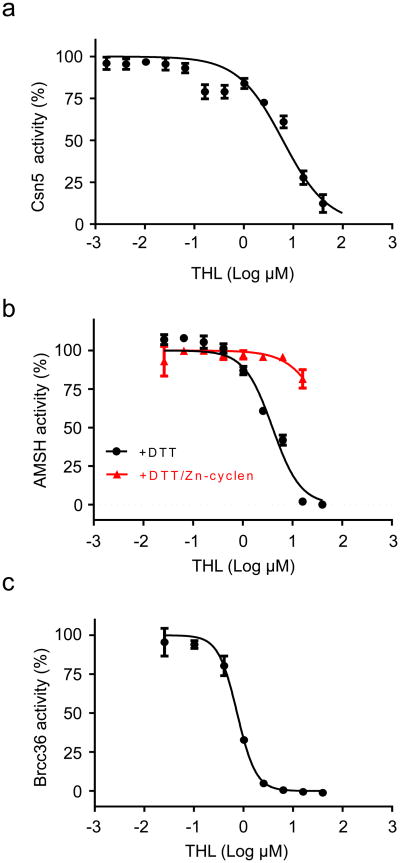
We therefore tested the effect of THL on purified 26S proteasome and observed that it blocked removal of ubiquitin from a substrate designed to measure activity of the 19S de-ubiquitinase, Rpn11 (IC50 = 0.53 μM) (Fig. 4a, black curve)29. Moreover, THL blocked cleavage of di-ubiquitin by purified Rpn11–RPN8 heterodimer (Fig. 4b), strongly implicating Rpn11 as the direct target. Holomycin, a natural methyl derivative of THL (see Fig. 1a), inhibited 19S de-ubiquitinase activity even more efficiently (IC50 = 0.18 μM) than THL (Fig. 4c), suggesting that Rpn11 is a common target of DTPs.
Figure 4. THL is an inhibitor of Rpn11.
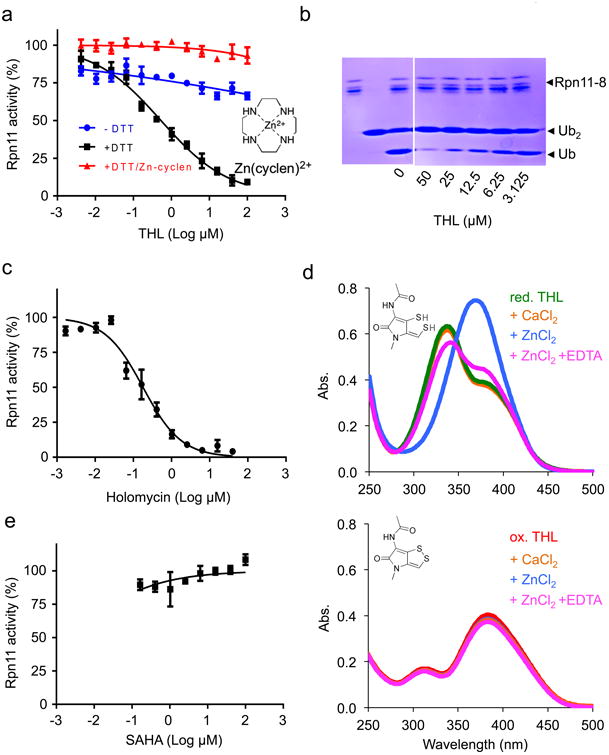
a: Titration curves for inhibition of Rpn11 by THL (IC50 = 0.53 μM ± 0.05 μM (s.d.)) in the absence and presence of 1 mM DTT (blue and black, respectively) and in the presence of 1 mM DTT and 0,1 mM Zn(cyclen)2+ (red). Chemical structure of Zn(cyclen)2+ is shown.
b: THL directly inhibits Rpn11 activity. Purified Rpn11•Rpn8 was incubated with K48 linked di-ubiquitin in the presence of different concentrations of THL, and reactions were stopped by 2× SDS sample buffer and analyzed by SDS-PAGE with Coomassie Blue staining. Shown is the representative gel image of two independent experiments.
c: Titration curve for inhibition of Rpn11 by holomycin (IC50 = 0.18 μM ± 0.03 (s.d.)).
d: Reduced THL binds Zn2+- ions. Absorption spectra of 40 μM reduced THL (upper panel) and oxidized THL (lower panel) in absence of metal ions (green and red curve, respectively), in presence of 500 μM CaCl2 (orange curves), in presence of 40 μM ZnCl2 (blue curves), and in presence of 40 μM ZnCl2 and 0.2 mM EDTA (magenta curves). Experiment was performed with β-mercaptoethanol (20 mM) as reducing agent. Identical results were obtained when repeating experiment with TCEP (0.1 mM) or DTT (1 mM) for reduction of THL.
e: Titration curve showing that the metal chelating HDAC inhibitor SAHA does not inhibit Rpn11. a,c,e: Error bars represent s.d., n = 4 wells. Shown is one representative of three independent experiments.
It has been suggested that oxidized DTPs are pro-drugs, and that the reduced dithiol forms are the active antibiotics1. Given that the cytoplasm is a reducing environment, our in vitro assays were performed in the presence of DTT, which reduced THL to its dithiol form (Supplementary Fig. 6a). We next tested the inhibitory activity of oxidized versus reduced THL on purified 26S and found that only reduced THL inhibited deubiquitylation of substrates by 19S (Fig. 4a). Rpn11 belongs to the group of JAMM (JAB1/MPN/Mov34) domain-containing metalloproteases, which have a Zn2+-ion in their active center. To elucidate whether reduced THL acts on Rpn11 by chelating the Zn2+-ion we analyzed the effect of reduced THL on RNP11 activity in the presence of an excess of Zn(cyclen)2+, a Zn2+ coordination complex that retains the ability to interact with metal-binding groups. Addition of Zn(cyclen)2+ in excess over THL abrogated inhibition of Rpn11 (Fig. 4a), suggesting that THL inhibits Rpn11 by binding of Zn2+. Indeed, the absorption spectrum of reduced but not oxidized THL shifted in presence of Zn2+ and was reversible by addition of EDTA (Fig. 4d). In contrast, Ca2+ did not affect the absorption spectrum of reduced or oxidized THL (Fig. 4d). Similarly, reduced but not oxidized THL competed with fluoZin™-1, a fluorescent indicator of Zn2+ (Supplementary Fig. 6b), indicating that THL is a chelator of Zn2+. However, Rpn11 was not inhibited by the metal-chelator SAHA, which is a potent inhibitor of histone deacetylases (HDACs) (Fig. 4e). Futhermore, in a screen of 96 metal binding compounds Rpn11 was inhibited by only 3 chelators30. Hence, Rpn11 is not generally inhibited by chelating reagents.
THL is an inhibitor of JAMM proteases
A structural paralogue of the proteasomal 19S cap is the COP9 signalosome (CSN), which regulates the ubiquitin–proteasome pathway by inactivating cullin-RING ubiquitin ligases (CRLs)15,16. The subunit Csn5 is the paralogue of Rpn11 and catalyzes deneddylation of cullin15. We found that THL can inhibit Csn5 with an IC50 of 6.2 μM (Fig. 5a). In addition, THL could inhibit the JAMM protease family AMSH, which cleaves K63-linked ubiquitin chains of endosomal proteins17,18. AMSH was inhibited by THL with an IC50 that was ∼8-fold higher (4 μM) than that measured for Rpn11, and inhibition was reversible by Zn(cyclen)2+ (Fig. 5b). Rpn11, Csn5 and AMSH (and AMSH-like protein31) are conserved between mammals and fungi. The human genome encodes three additional JAMM-family members, BRCC36, MYSM1, and MPND. BRCC36 is a K63-specific deubiquitinase of BRISC19 and BRCC20,21. THL inhibited de-ubiquitylation of substrate by BRISC (Fig. 5c). MYSM1 is a histone H2A deubiquitinase22, which we have not assayed and MPND has no known function.
Figure 5. THL inhibits JAMM proteases.
a: Titration curve for inhibition of Csn5 by THL (IC50 = 6.16 μM± 1.88 (s.d.)).
b: Titration curves for inhibition of AMSH by THL (IC50 = 3.96μM± 0.59 (s.d.)) in the absence (black) and presence (red) of Zn(cyclen)2+.
c: Titration curve for inhibition of Brcc36 by THL (IC50 = 0,79 μM± 0.03 (s.d.)).
a–c: Error bars represent s.d., n = 4 wells. Shown is one representative of three independent experiments.
In addition to JAMM proteases THL also inhibited the bacterial metalloproteinase thermolysin with an IC50 of about 2.5 μM (Supplementary Fig. 7a,b), suggesting that it has the potential to inhibit other metalloproteinases. The Rpn11 inhibitor quinoline-8-thiol (8TQ)30,32 also inhibited thermolysin (Supplementary Fig. 7b). However, THL did not reduce the overall concentration of intracellular Zn2+ in HEK293T cells (Supplementary Fig. 7c). THL did not inhibit de-ubiquitylation of substrates by the UPS5, a cystein isopeptidase that contains a Zn2+-finger domain participating in substrate recognition33, and THL did not inhibit the cystein isopeptidase UPS25 (Supplementary Fig. 7d). Together the data indicate that THL inhibits Rpn11 via chelation of its Zn2+- ion.
To assess some aspects of the structure-activity relationship of DTPs, we tested synthetic derivatives of holomycin carrying various substitutions in place of its methyl group (2-15) (Supplementary Note). All derivatives were active in cell culture and inhibited Rpn11, Csn5 and AMSH in in vitro activity assays (Supplementary Table). Yet, none of the derivatives exhibited a substantial increase in affinity. Interestingly, however, benzoyl-holothin did not induce detachment of cells at concentrations when the UPS was inhibited (Supplementary Fig. 8). This finding suggests that the mechanism of inhibition of cell adhesion by DTPs might be distinct from the mechanism of UPS inhibition. The data indicate that modification of holomycin at the methyl position does not enhance the potency of the drug. However, given that such modifications did not interfere with drug uptake or UPS inhibition, the methyl position could be derivatized to make affinity reagents or labelled probes.
Finally, as THL needs to be reduced to its dithiol form to become active, we asked whether treatment with reduced THL may enhance its antibiotic properties. Surprisingly, pre-treatment of THL with DTT nearly completely abolished its antibiotic effect on N.crassa. Specifically, ubiquitylated proteins did not accumulate, transcription of frq was not inhibited and hsp30 was not induced upon treatment with reduced THL (Supplementary Fig. 9a,b). The data suggest that cells may not take up THL in its reduced form.
Impact of THL on transcription in HeLa cells
Having established that THL is an inhibitor of Rpn11 and other JAMM proteases we asked whether it affects transcription in HeLa cells. Since most mammalian mRNAs are stable for several hours we quantified elongating Pol II rather than transcript levels. In addition to THL we analyzed the transcriptional impact of the Rpn11 inhibitor 8TQ30,32. Treatment of HeLa cells with 5 μM and 50 μM THL or 8TQ did not significantly affect transcription (Pol II Ser2 occupancy) of actin while transcription of c-myc was attenuated (Supplementary Fig. 10a, b). In contrast, transcription of hsp70 was induced by THL and to a somewhat lesser extent also by 8TQ (Supplementary Fig. 10c). Thus, the data indicate that the Rpn11 inhibitors do not inhibit Pol II transcription in mammalian cells. Rather, both inhibitors appear to trigger a stress response that affects transcription in a gene-specific manner. Whether this indirect impact on transcription is a pleiotropic consequence of inhibition of Rpn11 or triggered by inhibition an unknown target of THL and 8TQ cannot be decided.
Discussion
We showed here that the reduced forms of THL and other DTPs act as Zn2+ chelators to inhibit the JAMM metalloproteases Rpn11, Csn5 AMSH, and Brcc36. In vivo THL may target other metal-dependent enzymes since it inhibited the metalloproteinase thermolysin in vitro. However, many metalloproteinases including matrix-metalloproteinases (MMPs), ADAM-metalloproteinases, and angiotensin-converting enzyme (ACE) are, like bacterial thermolysin, secreted enzymes that act in the extracellular space. Since oxidized THL is an inactive cell permeable prodrug that becomes activated but apparently membrane-impermeable when reduced within the cell, these enzymes may not be targeted by THL in living cells or in vivo.
The effect of THL on transcription in Neurospora and HeLa cells was indirect since transcription of a substantial subset of genes was not affected or even upregulated. Thus, THL did not inhibit the Pol II dependent transcription machinery per se but triggered a stress response that impacted on transcription in a gene-specific manner. Hence, THL should not be used as a general transcription inhibitor. Corresponding results were obtained with 8QT. Thus, both inhibitors targeted Rpn11 and JAMM proteases, and at high concentration potentially other intracellular metalloproteases/enzymes. It is not clear whether the indirect effect of THL and 8TQ on transcription is related to inhibition of Rpn11 or other targets.
Vertebrates but not fungi express the JAMM-domain containing histone H2A deubiquitinase MYSM122, which is likely also a target of THL. Ubiquitylation of H2A is associated with transcriptional silencing34-36. A knockout of Mysm1 in mice affects p53 function but has little affect on general transcription37. Mysm1 deficient mice show increased embryonic lethality, growth retardation, and defects in hematopoesis38-40. Hence, the THL-dependent inhibition of angiogenesis in mice2 and the THL-induced lethality of zebrafish embryos12 may be related to inhibition of MYSM1.
As JAMM proteases are key players in protein homeostasis, they might be promising drug targets in cancer therapy41. Cancer cells harbor numerous genomic mutations and aberrations that may perturb protein homeostasis. Therefore, cancer cells are thought to be particularly dependent on the UPS14. Indeed, inhibitors of the core proteasome such as bortezomib or carfilzomib are successful in treatment of multiple myeloma and mantle cell lymphoma, which both exhibit high secretory activity14,42,43. However, these inhibitors are not successful in the treatment of solid tumors14, which may be more difficult to reach and differ less from healthy tissues in their dependence on the UPS. Hence, novel UPS inhibitors with distinct specificity, pharmacodynamics and kinetics are needed. As DTPs are not chemically related to bortezomib or carfilzomib and target distinct components of the UPS or the lysosomal system, they merit further analysis for potential antitumor activity. We, therefore, suggest that a broad variety of natural and synthetic DTP analogues, that are available or can be synthesized1, be tested for their specificity towards Rpn11, Csn5, AMSH,Brcc36 and potentially MYSM1, and their pharmacokinetic properties in vivo.
Online Methods
N.crassa strains and culture conditions
Conidial suspensions in 1M Sorbitol were prepared from samples grown on standard solid growth medium (2.2% Agar, 0.3% D+ Glucose monohydrate, 0.17% L-arginine, 1× Vogel's medium and 0.1% biotin). Liquid culture medium contained 2% Glucose, 2% Arginine and 1× Vogel's medium. Cycloheximid (CHX) was added to a final concentration of 10 μg/ml. Thiolutin (THL) and MG132 dissolved in DMSO were added to final concentrations as indicated.
Protein extraction and analysis of N.crassa
Extraction of N.crassa protein was performed as described44. Protein concentrations were measured with NanoDrop® (PeqLab) and 400 μg protein was loaded unless stated otherwise. Western blotting was performed as described45,46. Enhanced chemiluminescence signals were detected with X-ray films (Fuji RX). Antibodies used against FRQ and VVD were previously described26,46. Mouse monoclonal anti-β-tubulin (WA3) was used at 1:1000 in 5% milk TBS. The Ubiquitin antibody (Ub P4D1 sc-8017) was purchased from Santa Cruz Biotechnology and used following the Technical Services recommendation.
Quantitative Real Time PCR of N.crassa RNA
Total mRNA from ground frozen mycelia was prepared using peqGOLD TriFAST™ (PeqLab) and reverse transcription was performed using the QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer's instructions. Transcript levels were analyzed by quantitative real time PCR as described previously26. Sequences of primers and probes used for qRT-PCR were: frq: F: ttg taa tga aag gtg tcc gaa ggt, R: gga gga aga agc gga aaa cg, Probe: 6-FAM - acc tcc caa tct ccg aac tcg cct g - TAMRA; vvd: F: acg tca tgc gct ctg att ctg, R: aaa agc ttc cga ggc gta ca, Probe: 6-FAM - cga cct gaa gca aaa aga cac gcc a - TAMRA; 28s rRNA: F: cct gtt cga gcg tca ttt ca, R: agc ccg cca ctg att ttg, 6-FAM - cca tca agc tct gct tgc gtt ggg - TAMRA; hsp30: F: cac tcg cga ggt tca agg, R: tcg aac ttg gga ctg aag gt, Probe: UPL #23 (UPL = universal probe library; Roche Applied Science).
RNA PolII ChIP in Neurospora
The RNA polymerase II ChIP was performed as described25. Briefly, light grown cultures were incubated for 40 min in the presence of DMSO and 50 μM THL, respectively. Samples were cross-linked for 10 min with 1% formaldehyde and subjected to ChIP with antibodies against Pol II Ser5-P25. Precipitated DNA was quantified by qRT–PCR with promoter specific probes: actin: F: tatcatgggcacaagctgat, R: cttacccattgtcgatgacg, Probe: 6-FAM - tgcgttcccgccaacagaag - TAMRA; vvd: F: ttagcctttgccagttaggg, R: tgttggtagggtattatgatggttt, Probe: UPL #41; hsp98: F: cattttgctggtccttgctc, R: cccgttcatctggcagtc, Probe: UPL #14; hsp60: F: gcgttgaccagagcttcc, R: ggatcgtaccttgtgagcaaa, Probe: UPL #3.
RNA PolII ChIP in HeLa cells
HeLa cells were incubated 40 min in DMEM containing DMSO, THL or 8TQ (8-quinolinethiol, Sigma-Aldrich) and then immediately cross-linked in 1% formaldehyde for 10 min. Scraped cells were washed twice with cold PBS and recollected by centrifugation at 2000g for 5 min at 4°C. The cellular pellet was resuspended in 1 ml IP buffer (150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1% Triton X-100, 50 mM Tris-HCl, pH 7.5) and washed twice with IP buffer by centrifugation at 12.000g for 1 min at 4°C. Pelleted nuclei were sonicated in 300 μl IP buffer supplemented with 0.8% SDS (30 s on/30 s off cycles for 40 min) using a Bioruptor (Diagenode Inc.). The chromatin (equivalent of 5×105 cells) was then incubated overnight at 4°C with 4 μl affinity purified anti-Pol II Ser2-P antibody25 and precipitated with salmon sperm DNA blocked protein A-sepharose beads. Beads were washed 5 times with IP buffer and co-precipitated DNA was recovered by boiling for 10 min in 10% Chelex slurry (Bio-Rad) and treatment with Proteinase K (150 μg/ml, NEB) at 55°C for 30 min. Proteinase K and beads were removed by boiling at 95°C for 10 min and subsequent centrifugation 12,000g for 10 min at 4°C. DNA-containing supernatants were analyzed by qPCR using Maxima SYBR Green kit (ThermoFisher), and values were normalized to percentage of input. Primer sequences used were: β-actin (ACTB)47: F: ccttgagtgggggtgtagtg, R: cccctaccccaacttgactt; c-myc (MYC)47: F: tggctgcttgtgagtacagg, R: aactggcttcttcccaggag; HSP70 (HSPA1A)48: F: gagacggccggaggcgtgat, R: cggagggatgccgctcagct.
HeLa cell culture conditions
HeLa cells (ATCC CCL-2.2) were cultivated in Dulbecco's Modified Eagle Medium (PAA laboratories) supplemented with 10% (v/v) FCS and 100 μg/ml Streptomycin/Penicillin. CHX was added to a final concentration of 25 μg/ml. THL and MG 132 were dissolved in DMSO and added to final concentrations as indicated.
HeLa cell adhesion measurement
24-well plates were densely seeded with 200.000 HeLa cells. After 24 h THL or MG132 was added to the indicated final concentrations (from a dilution series of THL stocks to keep added volume constant). After 2 h the supernatant was removed and the detached cells were counted with a Neubauer chamber. For Suppl. Fig. 5 cell adhesion was assessed by microscopic observations on the morphological changes. Images were taken under 20× magnification using a Molecular Devices ImageXpress Micro (Molecular Devices).
HeLa cell protein extraction
Cells were grown in a 6 well plate to 100 % confluency. Cells were washed twice with cold PBS, resuspended (scraped off) in 1 ml PBS, and transferred into a 1.5 ml tube. The cells were collected by centrifugation at 500 × g, 3 min, 4°C and the pelleted cells were lysed with 100 μl RIPA buffer (50 mM Tris/HCl, pH 7.4,150 mM NaCl, 1% Triton X-100, 0.25% Na-deoxycholate, 3 mM EDTA supplemented with complete protease inhibitors and PhosSTOP® (Roche)). The suspension was vortexed at the maximum setting for 10 s with subsequent sonification for 10 min at 4°C. After centrifugation (20000 × g, 10 min, 4°C), the supernatant was collected (cell lysate). Protein concentration of cell lysate was determined with Bradford-Reagent 5× (Bio-Rad).c-Myc polyclonal antibody (rabbit) was raised against the C-terminal peptide NH2-CRREQLKHKLEQLRNS-CONH2 (Pineda), affinity purified and used for western blots 1:500 in 5% milk TBS.
Quantitative Real Time PCR of HeLa RNA
Cells were grown in a 6 well plate to 100 % confluency, and RNA was isolated using RNeasy kit (Qiagen). cDNA was synthesized with QuantiTect Reverse Transcription Kit. Sequences of primers and probes used for qRT-PCR were:c-myc: F:caccagcagcgactctga, R: gatccagactctgaccttttgc, Probe: UPL #34; 18S rRNA: F: ttgactcaacacgggaaacc, R: cgctccaccaactaagaacg, Probe: UPL#77.
Yeast strain and culture conditions
Cells of the yeast strains DS1-2b (Y2197; MAT, his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52) and pdr5Δ∷kanMX in ESM356-1 (MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63; gift from M. Knop, ZMBH Heidelberg)were cultured in YPD medium at 30°C49. CHX was added to a final concentration of 20 μg/ml. THL and MG132 were added as indicated.
Protein extraction and analysis of yeast cells
Denaturing extracts of yeast cells were prepared as described50. The cell pellet was resuspended in 40 μl 0.1 M NaOH and 14 μl 4×Leammli sample buffer and incubated at 95 °C for 5 min. Aliquots corresponding to 0.5 OD600 of cells were analyzed by SDS–PAGE.
In vitro transcription using tailed templates
In vitro transcription using tailed templates (10 nM) was performed as described51. In brief, THL or control reactants were preincubated with the respective polymerase. Transcription was carried out for 30 min at 30°C in reaction buffer (20 mM Hepes pH 8.0, 10 mM MgCl2, 5 mM EGTA, 0.05 mM EDTA, 2.5 mM DTT) with 0.5 mM of each ATP, UTP and GTP, and 24 μM CTP and 1.25 μM α-32P-CTP (total volume 20 μl). Reactions were stopped by addition of Proteinase K (15 min, 37°C) and RNA was extracted by ethanol precipitation. Transcripts were separated on a 6% denaturing polyacrylamide gel and visualized10. Signals were quantified using Multi Gauge (Fuji).
Quantitative Real Time PCR of Yeast RNA
cDNA was synthesized using the QuantiTect Reverse Transcription Kit and qRT-PCR was performed as described above. Sequences of primers and probes: dbp2: F: gtagaggtggttacggtggtg, R: tcaatagtttgaacgacctctgtta, Probe: UPL#83; not3: F:aaaggtctcgtgatatgtgtgaaa, R: ccaagggagatggtggatta, Probe: UPL#64; 18S rRNA: F: cgatggaagtttgaggcaat, R: cctctcggccaaggttaga, Probe: UPL#36.
RNAseq mapping
Raw reads were mapped to N.crassa genome (NC10) using Bowtie52, where parameters were set to allow maximum 3 mismatches and suppress alignments which mapped to more than one location.
Mapping summary:
| Time Point (min.) | Total reads | Uniquely mapped | Mapping percentage |
| 0 | 20,363,638 | 17,766,411 | 87.25% |
| 40 | 18,075,145 | 15,991,479 | 88.47% |
| 120 | 18,426,954 | 15,695,743 | 85.18% |
Gene expression quantification and normalization: For RNA-seq, gene expression was quantified by the number of reads that fall into exons. Number of reads mapping to the supercontig 8 (ribosomal loci) was used to compute the normalization factor. This is based on the assumption that reads mapping to ribosomal RNA are not affected by THL on the time scale of the experiment. Based on their mean sequence reads the lower quartile of genes were considered as not expressed and discarded. Up-regulated genes: The normalized data was sorted based on the regression coefficient between time in THL and log expression value; the fitting was carried out using lm function in R. Up-regulated genes were defined as those, which have a positive coefficient, and show monotonous increase of sequence reads. Using these criteria 267 genes were detected to be up-regulated.
In vitro DUB activity assay
Human USP5 and USP25 were expressed in E.coli and purified as described53,54.4 nM USP25 or USP5 were preincubated in SAB buffer (20 mM Hepes, pH 7.3, 110 mM KOAc, 2 mM Mg(OAc)2, 1.0 mM EGTA, 1 mM DTT, 0.05% Tween 20, 0.2 mg/ml Ovalbumin and 1 mg/ml each of leupeptin, aprotinin and pepstatin) with either DMSO, 50 μM THL or 50 μM PR619 inhibitor (Sigma-Aldrich, SML0430) for 30 min at 30°C. Deubiquitination reaction was started by addition of 1 μM tetra-Ubiquitin K48 linked chains (BostonBiochem, UC-210B). After 10 or 30 min, reactions were stopped by addition of 2×SDS sample buffer + 1 mM DTT. Samples were analyzed by Western blot using mouse anti-Ub antibody (Santa Cruz, sc-8017).
20S proteasome assay Kit, SDS-Activation Format (BostonBiochem, USA)
Purified 20S proteasome (1.4 nM) was preincubated at 37°C with reaction buffer (25mM Hepes pH 7.6, 0.5mM EDTA and 0.03% SDS). The different substrates (S1 Suc-Leu-Leu-Val-Tyr-AMC in DMSO; BostonBiochem, S2 Suc-Leu-Tyr-AMC in DMSO; BostonBiochem, S3 Bz-Val-Gly-Arg-AMC in DMSO; ENZO Life Sciences and S4 Z-Leu-Leu-Glu-AMC in DMF; ENZO Life Sciences) were added to a final concentration of 10 μM and the hydrolytic activity of the 20S proteasome (rate of AMC release) was measured at 20 min intervals. The reaction was monitored at λEm380 and λEx460 nm using an EnSpire plate reader (Perkin Elmer).
THL spectra
THL spectra were recorded between 250 and 500 nm using an NP80 nanophotometer (Implen). 500μl reactions containing 40 μM THL in 20 mM Tris pH 7.5 were supplemented either with reducing agent DTT (1 mM), TCEP (100μM) or β-mercaptoethanol (20 mM), respectively and spectra were recorded after 3 mins of incubation. CaCl2 was added at a final concentration of 500 μM and ZnCl2 at a final concentration of 40 μM. Spectra were recorded after 1 min of incubation. Where indicated EDTA was added to the reaction at a final concentration of 200 μM and spectra were measured after 1 min of incubation.
Zn binding assay using fluorescent zinc indicator FluoZin™-1
Reactions were performed in a final volume of 100μl containing 50 μM FluoZin™-1 (ThermoFisher, F24180) and 50 μM ZnCl2 in20 mM Tris pH 7.5, 5% DMSO. THL was added at the indicated concentrations in the presence and absence of 2.5 mM β-mercaptoethanol. FluoZin™-1 fluorescence (Ex/Em: 495/515nm) was measured after 5 mins of incubation in black 96-well plates using an EnSpire multilabel plate reader with a monochromator.
Thermolysin inhibition assay
100 ng of overexpressed and purified FLAG-tagged Neurospora FREQUENCY was incubated in 100 μl reactions with 30 ng of thermolysin (Sigma-Aldrich, P1512) in a buffer containing 50 mM Hepes pH 7.4, 140 mM NaCl, 10% Glycerol and 5% DMSO in the presence and absence of the indicated concentrations of THL or 8TQ. Reducing agent TCEP was added at a final concentration of 500 μM where indicated. Samples were incubated at 37 °C for 1 h (Supplementary Figure 7a) or for 3, 6, 10, 20 and 40 min, respectively (Supplementary Figure 7b) and reactions were stopped by adding SDS sample buffer and boiling at 95 °C for 5 minutes. Western blots were decorated with FLAG antibodies. Densitometric quantification of western blots was done using Adobe® Photoshop.
Protein expression and purification of Rpn11•Rpn8 dimer
pETDuet-1 expression vector for Rpn11•Rpn8 dimer was kindly provided by Dr. Andreas Martin from University of California, Berkeley. We expressed and purified Zn2+ bound Rpn11•Rpn8 dimer based on the protocol previously described55.
Rpn11•Rpn8 di-Ub cleavage assay
Di-ubiquitin cleavage assay was performed as previously described55. In brief, 5 μM Rpn11•Rpn8 dimer was incubated with DiUbK48 in the presence of different concentrations of THL at 30°C for 2 hours. The reaction buffer contains 40 mM HEPES, pH7.5, 100 mM NaCl, 100 mM KCl and 5% glycerol. The reactions were stopped with 2× SDS sample buffer and analyzed by SDS-PAGE with coomassie blue staining.
In vitro Rpn11 activity assay
Rpn11 activity was measured with synthetic model substrate32 and human 26S proteasome (Enzo Life Sciences) by fluorescence polarization at 30°C using a PHERAstar plate reader (BMGlabtech) with excitation at 480 nm and emission at 520 nm. Titration was done in low-volume 384 well solid black plates (Molecular Devices) in quadruplicate. 100 μM Zn(cyclen)2+ was used for the experiments performed with Zn(cyclen)2+. Collected data was normalized to DMSO control and fitted to a dose-response curve to determine the IC50 value.
In vitro Csn5 activity assay
Csn5 activity was measured using the purified CSN holoenzyme and the fluorescent substrate OGNedd8–SCFSkp2, in which a unique cysteine engineered into Nedd8 at the N-terminus was labeled with Oregon Green 488, after which the product was conjugated to SCFSkp2 as previously described56. Incubation of the substrate with CSN resulted in decreased fluorescence polarization due to cleavage of labeled Nedd8 from the substrate complex. The assay was performed in buffer containing 25 mM Tris–HCl pH 7.6, 50 mM NaCl, 1 mM DTT, 0.01% Triton X-100, 1% glycerol, 25 mM trehalose and 15 μg/ml ovalbumin. Polarization data was collected with a PHERAstar plate reader with excitation (480 nm) and emission (520 nm) filter sets using black low-volume 384-well plates. Reaction rates were calculated from the linear phase of the reaction progression curves and these raw numbers were normalized to the DMSO control and fitted to a dose–response curve to determine the IC50 value.
In vitro AMSH activity assay
AMSH activity was measured as described32. In brief, titration was performed with 10 nM purified AMSH (E-548B, Boston Biochem) and 80 nM di-ubiquitin substrate DiUbK63TAMRA (UF-310, Boston Biochem) in the presence of different concentration of compound. TAMRA fluorescence intensity was monitored using a PHERAstar plate reader with excitation at 540 nm and emission at 590 nm. Reactions were performed in black low-volume 384 well plates and analyzed as described above.
In vitro BRCC36 activity assay
The BRISC complex (kindly provided by Dr Elton Zeqiraj, University of Leeds) consists of four different protein including BRCC36, KIAA0157, BRCC45 and MERIT40. The complex was produced by co-expression in insect cells and purified to greater than 95% purity57. BRCC36 activity was measured using 1nM purified BRISC complex and 80 nM di-ubiquitin substrate DiUbK63TAMRA (UF-310, Boston Biochem). Reactions were recorded and analyzed as described above.
Imaging of cellular zinc
Imaging experiments were performed on a Nikon Ti-E wide-field fluorescence microscope equipped with Nikon elements software, Ti-E perfect focus system, an iXon3 EMCCD camera (Andor), mercury arc lamp, and YFP FRET (434/16 excitation, 458 dichroic, 535/20 emission), CFP (434/16 excitation, 458 dichroic, 470/24 emission), and YFP (495/10 excitation, 515 dichroic, 535/20 emission) filter sets. Images were collected every minute using a 40× air objective (NA 0.95), 100 ms exposure time, 5.1× conversion gain, and a neutral density filter with 12.5% light transmission. Zn2+ sensor constructs were transfected into HEK293T cells with Lipofectamine 3000 (Thermo Fisher Scientific). Cells were imaged 24-48 hours post-transfection. Cells were maintained at 37°C and 5% CO2 in a LiveCell™ environment chamber (Pathology Devices) during the experiment. Cells were imaged in phosphate-free HEPES-buffered HBSS, pH = 7.4 to prevent zinc precipitation. All data were analyzed in MATLAB (Mathworks).
Data Availability
The RNAseq raw data evaluated for this study have been deposited as Sequence Read Archives (SRA) at NCBI under BioProject PRJNA347916.
Supplementary Material
Acknowledgments
We thank TJ Simpson (Univ Bristol) for thiomarinol, Seth Cohen (UCSD) for Zn(cyclen)2+,Dr. Elton Zeqiraj (University of Leeds) for BRISC complex and Daniela Marzoll for purified recombinant FLAG-FRQ.The work was supported by SFB1036 grants to M.B. and F.M., DFG grant DI1874/1 to A.D. and NIH grant CA164803 to R.J.D. F.M. and M.B are Investigators of Cellnetworks. R.J.D. is an Investigator of and was supported by the Howard Hughes Medical Institute.
Footnotes
Author contributions: A.D. and M.B. designed the research and wrote the manuscript;
J.L. Y.Z. and R.J.D. designed, performed, and interpreted the Rpn11/Csn5/AMSH/Brcc36 experiments.
W.J.W. and A.A.B. synthesized the holothin derivatives.
K.P.C and A.E.P designed and performed the measurements of intracellular zinc.
N.S.-V. and F.M. designed and performed the USP5 and USP25 experiments;
P.M, S.O. and H.T. designed and performed the Pol I – III in vitro transcription assays.
L.L., I.A.C., A.D. and T.S. performed the Neurospora experiments. L.L. and N.H. performed the RNA-seq analysis. L.L. performed the yeast experiments. A.S. and L.L. performed the HeLa experiments.
R.J.D. implicated THL as an Rpn11 inhibitor and edited the manuscript.
Competing financial interests: R.J.D. is a founder, shareholder, member of the SAB, and consultant of Cleave Biosciences, which is developing drugs that target protein homeostasis in cancer. The remaining authors declare no competing financial interests.
Additional information: Any supplementary information, chemical compound information and source data are available in the online version of the paper.
References
- 1.Li B, Wever WJ, Walsh CT, Bowers AA. Dithiolopyrrolones: biosynthesis, synthesis, and activity of a unique class of disulfide-containing antibiotics. Nat Prod Rep. 2014;31:905–23. doi: 10.1039/c3np70106a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minamiguchi K, et al. Thiolutin, an inhibitor of HUVEC adhesion to vitronectin, reduces paxillin in HUVECs and suppresses tumor cell-induced angiogenesis. International journal of cancer Journal international du cancer. 2001;93:307–16. doi: 10.1002/ijc.1321. [DOI] [PubMed] [Google Scholar]
- 3.Tipper DJ. Inhibition of yeast ribonucleic acid polymerases by thiolutin. Journal of bacteriology. 1973;116:245–56. doi: 10.1128/jb.116.1.245-256.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimenez A, Tipper DJ, Davies J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae. Antimicrobial agents and chemotherapy. 1973;3:729–38. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khachatourians GG, Tipper DJ. Inhibition of messenger ribonucleic acid synthesis in Escherichia coli by thiolutin. Journal of bacteriology. 1974;119:795–804. doi: 10.1128/jb.119.3.795-804.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Molecular and cellular biology. 2004;24:5534–47. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelechano V, Perez-Ortin JE. The transcriptional inhibitor thiolutin blocks mRNA degradation in yeast. Yeast. 2008;25:85–92. doi: 10.1002/yea.1548. [DOI] [PubMed] [Google Scholar]
- 8.Sivasubramanian N, Jayaraman R. Thiolutin resistant mutants of Escherichia coli are they RNA chain initiation mutants? Molecular & general genetics : MGG. 1976;145:89–96. doi: 10.1007/BF00331562. [DOI] [PubMed] [Google Scholar]
- 9.Joshi A, Verma M, Chakravorty M. Thiolutin-resistant mutants of Salmonella typhimurium. Antimicrobial agents and chemotherapy. 1982;22:541–7. doi: 10.1128/aac.22.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roza J, Blanco MG, Hardisson C, Salas JA. Self-resistance in actinomycetes producing inhibitors of RNA polymerase. J Antibiot (Tokyo) 1986;39:609–12. doi: 10.7164/antibiotics.39.609. [DOI] [PubMed] [Google Scholar]
- 11.Dai S, et al. Comprehensive characterization of heat shock protein 27 phosphorylation in human endothelial cells stimulated by the microbial dithiole thiolutin. Journal of proteome research. 2008;7:4384–95. doi: 10.1021/pr800376w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia Y, et al. Thiolutin inhibits endothelial cell adhesion by perturbing Hsp27 interactions with components of the actin and intermediate filament cytoskeleton. Cell stress & chaperones. 2010;15:165–81. doi: 10.1007/s12192-009-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 14.Deshaies RJ. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 2014;12:94. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–86. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 17.Agromayor M, Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J Biol Chem. 2006;281:23083–91. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- 18.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–92. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper EM, et al. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 2009;28:621–31. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu P, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–21. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–24. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dani C, et al. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984;81:7046–50. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cesbron F, Oehler M, Ha N, Sancar G, Brunner M. Transcriptional refractoriness is dependent on core promoter architecture. Nat Commun. 2015;6:6753. doi: 10.1038/ncomms7753. [DOI] [PubMed] [Google Scholar]
- 26.Malzahn E, Ciprianidis S, Kaldi K, Schafmeier T, Brunner M. Photoadaptation in Neurospora by Competitive Interaction of Activating and Inhibitory LOV Domains. Cell. 2010;142:762–772. doi: 10.1016/j.cell.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Dai MS, Jin Y, Gallegos JR, Lu H. Balance of Yin and Yang: ubiquitylation-mediated regulation of p53 and c-Myc. Neoplasia. 2006;8:630–44. doi: 10.1593/neo.06334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins GA, Gomez TA, Deshaies RJ, Tansey WP. Combined chemical and genetic approach to inhibit proteolysis by the proteasome. Yeast. 2010;27:965–74. doi: 10.1002/yea.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 30.Perez C, et al. Discovery of an Inhibitor of the Proteasome Subunit Rpn11. J Med Chem. 2017;60:1343–1361. doi: 10.1021/acs.jmedchem.6b01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato Y, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–62. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 32.Li J, et al. Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonnet J, Romier C, Tora L, Devys D. Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem Sci. 2008;33:369–75. doi: 10.1016/j.tibs.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 35.Zhou W, Wang X, Rosenfeld MG. Histone H2A ubiquitination in transcriptional regulation and DNA damage repair. Int J Biochem Cell Biol. 2009;41:12–5. doi: 10.1016/j.biocel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Belle JI, et al. p53 mediates loss of hematopoietic stem cell function and lymphopenia in Mysm1 deficiency. Blood. 2015;125:2344–8. doi: 10.1182/blood-2014-05-574111. [DOI] [PubMed] [Google Scholar]
- 38.Jiang XX, et al. Control of B cell development by the histone H2A deubiquitinase MYSM1. Immunity. 2011;35:883–96. doi: 10.1016/j.immuni.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nijnik A, et al. The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood. 2012;119:1370–9. doi: 10.1182/blood-2011-05-352666. [DOI] [PubMed] [Google Scholar]
- 40.Nandakumar V, Chou Y, Zang L, Huang XF, Chen SY. Epigenetic control of natural killer cell maturation by histone H2A deubiquitinase, MYSM1. Proc Natl Acad Sci U S A. 2013;110:E3927–36. doi: 10.1073/pnas.1308888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Hideshima T, et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci U S A. 2005;102:8567–72. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson PG, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352:2487–98. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
Online Methods references
- 44.Diernfellner AC, Schafmeier T, Merrow MW, Brunner M. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes & development. 2005;19:1968–73. doi: 10.1101/gad.345905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schafmeier T, et al. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–46. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 46.Gorl M, et al. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. The EMBO journal. 2001;20:7074–84. doi: 10.1093/emboj/20.24.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson L, Muniz L, West S. 3′rmation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014;8:342–56. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vera M, et al. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. Elife. 2014;3:e03164. doi: 10.7554/eLife.03164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics - A Laboratory Course Manual. Vol. 198. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- 50.Knop M, et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–72. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 51.Merkl P, et al. Binding of the termination factor Nsi1 to its cognate DNA site is sufficient to terminate RNA polymerase I transcription in vitro and to induce termination in vivo. Mol Cell Biol. 2014;34:3817–27. doi: 10.1128/MCB.00395-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz S, et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012;13:930–8. doi: 10.1038/embor.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–9. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 55.Worden EJ, Padovani C, Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol. 2014;21:220–7. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 56.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeqiraj E, et al. Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function. Mol Cell. 2015;59:970–83. doi: 10.1016/j.molcel.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq raw data evaluated for this study have been deposited as Sequence Read Archives (SRA) at NCBI under BioProject PRJNA347916.