Abstract
Atg5 and Atg7 have long been considered as essential molecules for autophagy. However, we found that cells lacking these molecules still form autophagic vacuoles and perform autophagic protein degradation when subjected to certain stressors. During this unconventional autophagy pathway, autophagosomes appeared to be generated in a Rab9-dependent manner by the fusion of vesicles derived from the trans-Golgi and late endosomes. Therefore, mammalian autophagy can occur via at least two different pathways; the Atg5/Atg7-dependent conventional pathway and an Atg5/Atg7-independent alternative pathway.
Keywords: alternative autophagy, Atg5, erythrocyte maturation, Golgi membrane, proteolysis
INTRODUCTION
Autophagy is a catabolic process in which cellular contents, including proteins and organelles, are degraded using autophagic vacuoles. Autophagy continuously occurs at low levels and is activated by a variety of cellular stressors, including nutrient starvation, DNA damage, the accumulation of damaged proteins, and in response to organelle damage. In many physiological and pathological contexts, autophagy functions to protect cells from such stressors.
There are at least three types of autophagy in mammals, i.e., macroautophagy, microautophagy, and chaperone-mediated autophagy. Macroautophagy is believed to be the major pathway for degrading cytoplasmic proteins and organelles (Nakatogawa et al., 2009). In this type of autophagy, subcellular constituents are enclosed by double-membrane structures called autophagosomes, and cargos are degraded by lysosomal enzymes. The second type of autophagy is microautophagy, which occurs by the direct invagination of lysosomal membranes to engulf cellular constituents (Li et al., 2012). Microautophagy can deliver entire organelles directly into lysosomes. The third type of autophagy is chaperone-mediated autophagy, in which soluble cytosolic proteins containing a specific targeting motif are delivered by the cytosolic heat shock cognate 70 chaperone to the lysosomal membranes (Kaushik and Cuervo, 2012). After docking with the lysosomal membrane receptor, the substrate proteins unfold, penetrate into the lysosomes, and are degraded. Although these three protein degradation systems are mediated by different mechanisms and different molecules, they are categorized as “autophagy” because lysosomes are involved in their proteolysis.
CHARACTERISTICS OF MACROAUTOPHAGY
Macroautophagy (hereafter referred to simply as “autophagy” unless otherwise indicated) begins from the generation of an omegasome, which is the starting point of the isolation membrane. The isolation membrane expands and its leading edges fuse together to form a double-membrane vesicle called the autophagosome, in which intracellular constituents are enclosed (Mizushima et al., 2002). By the fusion between an autophagosome and lysosome, cargos are degraded by lysosomal proteases, lipases, and DNases (Fig. 1).
Fig. 1. Hypothetical model of autophagy.
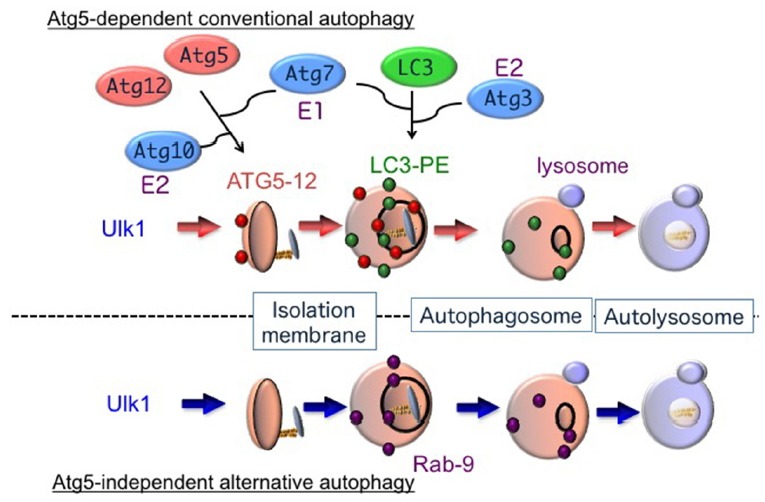
There are at least two modes of macroautophagy, i.e. conventional and alternative autophagy. Conventional autophagy requires Atg5 and Atg7, is associated with LC3 modification, and is thought to originate from the ER membrane. In contrast, alternative autophagy occurs independently of Atg5 and Atg7, as well as LC3 modification. The generation of autophagic vacuoles in alternative autophagy is mediated by the fusion of isolation membranes with vesicles derived from the trans-Golgi as well as late endosomes, in a Rab9-dependent manner.
Autophagy can be classified into different types by its induction mechanism; i.e., constitutive autophagy and stimulus-dependent inducible autophagy (Table 1). The former type of autophagy occurs at low levels but continuously degrades unfolded and aged proteins to maintain cellular homeostasis. The latter type of autophagy occurs in a large scale to prevent cellular damage against various stressors. Autophagy is also classified by the type of cargo it degrades; namely, bulk autophagy and selective autophagy. Autophagy was originally considered to degrade various constituents nonselectively, in contrast to the ubiquitin-proteasome degradation system. However, increasing lines of evidence indicate the presence of a cargo-specific type of autophagy (selective autophagy) that eliminates specific organelles (Komatsu and Ichimura, 2010), including peroxisomes (pexophagy), pathogens (xenophagy), and mitochondria (mitophagy). In selective autophagy, cargos are recognized by cargo receptors and are subsequently enclosed by autophagic structures. When these classification criteria are applied, the well-studied starvation-induced type of autophagy belongs to inducible and bulk autophagy. This type of autophagy is considered to be useful for the compensation of nutrients during times of shortage. On the other hand, genotoxic stress-induced autophagy is classified into inducible and selective autophagy, which degrades damaged proteins and organelles to eliminate unfavorable constituents. Neuron-specific autophagy-deficient mice were found to develop neurodegenerative disorders, with the accumulation of ubiquitin-positive proteins in their neurons (Komatsu et al., 2005), indicating that constitutive autophagy occurs in neurons to selectively degrade ubiquitin-positive proteins. Thus, autophagy plays various essential roles in stress responses and for maintaining cellular homeostasis.
Table 1.
Classification of autophagy from the view of inducers and substrates
| Inducer | Substrate | |
|---|---|---|
|
| ||
| Bulk autophagy (non-specific degradation) | Selective autophagy (specific degradation) | |
| Constitutively working autophagy (maintain homeostasis) | Cellular metabolism | Cell protection |
| Replace old proteins/organelles with new ones | Degrade unfavorable, damaged protein and organelles | |
| Inducible autophagy (response to various stimuli) | Nutrient supply | Cellular adaptation |
| Stravation-induced | Cellular stress (DNA damage etc.) | |
| Rapamycin-treated | Differentiation | |
CLASSIFICATION OF AUTOPHAGY
Autophagy is also classified into two types based on their molecular mechanisms, i.e., the Atg5-dependent conventional type and the Atg5-independent alternative type (Fig. 1). Atg5 has long been believed to be an essential molecule for autophagy, but recently we discovered an Atg5-independent type of alternative autophagy (Nishida et al., 2009).
As described above, autophagy is defined by the formation of specific double-membrane vesicles that engulf intracellular components, and their subsequent degradation by lysosomal enzymes. This characteristic morphology is easily identified using electron microscopy (EM), and hence ultrastructural analysis has long been used to study autophagy. In particular, starvation-induced and rapamycin (a specific inhibitor of mTor)-induced autophagy has been extensively examined using EM. In contrast, ultrastructural information of cells receiving other stressors has not been fully elucidated. We thus analyzed ultrastructural features within cells after treatment with etoposide, a DNA damaging reagent, and surprisingly found the representative autophagic structures (isolation membranes, autophagosomes, and autolysosomes) not only in wild-type mouse embryonic fibroblasts (MEFs) but also in Atg5-deficient (Atg5KO) MEFs (Nishida et al., 2009). These autophagic structures were indistinguishable from those observed during starvation-induced conventional autophagy. Thus, MEFs appear to perform two distinct types of autophagy, an Atg5-dependent conventional type and an Atg5-independent alternative type.
Several lines of evidence confirmed the existence of alternative autophagy in etoposide-treated Atg5KO MEFs (Nishida et al., 2009). First, the correlative light-electron microscopy assay using Lamp2-GFP-expressing cells indicated that autolysosomes clearly contain lysosomal proteins. Second, the addition of bafilomycin A1, which inhibits the fusion between autophagosomes and lysosomes, greatly increased and decreased the formation of autophagosomes and autolysosomes, respectively. Third, bafilomycin A1 suppressed not only autolysosome generation, but also autolysosomal proteolysis.
MEMBRANE SOURCE OF ALTERNATIVE AUTOPHAGY
Morphological features are basically common to both types of autophagy, but their membrane sources are different (Table 2). Regarding conventional autophagy, the endoplasmic reticulum (ER) membrane, mitochondrial outer membrane, and mitochondria-ER contact site membrane have been reported as sources of autophagosomes (Tooze and Yoshimori, 2010). In contrast, the membranes used in alternative autophagy are thought to originate from the Golgi apparatus and late endosomes. This conclusion is based on the following five seminal observations (Nishida et al., 2009): (1) almost all autophagic vacuoles observed in alternative autophagy were localized near the Golgi apparatus, (2) Golgi ministack formation preceded autophagosome generation, (3) some isolation membranes extended from the Golgi membranes, (4) trans-Golgi proteins were observed on autophagosomes and autolysosomes, and (5) the depletion of Golgi proteins inhibited alternative autophagy but not conventional autophagy (Yamaguchi et al., 2016).
Table 2.
Comparison between conventional and alternative autophagy
| Conventional type | Alternative type | |
|---|---|---|
| Morphology | Autophagosome/Autolysosome | Autophagosome/Autolysosome |
| Function | Organelle/Protein Digestion | Organelle/Protein Digestion |
| Phylogenetic Conservation | Yeast to Mammals | Yeast to Mammals |
| Atg5/Atg7 Requirement | Yes | No |
| LC3 Modifications | Yes | No |
| P62 digestion | Yes | No |
| Membrane Source | ER, mitochondria | Golgi membrane |
Evidence from various sources suggests that there are two types of biological membranes: the thin type (8.5 nm), such as the membranes of the ER and mitochondria, and the thick type (10 nm), such as the membranes of lysosomes, endosomes, and the trans-Golgi. These two membrane types do not fuse. In alternative autophagy, trans-Golgi-derived thick membranes and lysosomal thick membranes fuse to generate autophagosomes.
Initial step of autophagy
Several of the core molecules required for autophagosome maturation are different between the two types of mammalian autophagy (Table 2). However, both types of autophagy utilize several of the same molecules at the initial step. One such molecule is Unc-51-like kinase 1 (Ulk1), a homologue of yeast Atg1. Ulk1 is a serine/threonine kinase that forms the Ulk1 complex together with Fip200, Atg13, and Atg101 (Wong et al., 2013). In healthy conditions, Ulk1 is phosphorylated and inactivated by mammalian target of rapamycin complex 1 and AMP-activated protein kinase at different serine/threonine residues (Egan et al., 2011; Kim et al., 2011; Shang et al., 2011). In the case of starvation-induced conventional autophagy, Ulk1 is dephosphorylated by protein phosphatase 2A and subsequently translocates to pre-autophagosomal membranes (Wong et al., 2015), which are the initial platforms of the isolation membrane. We demonstrated that during DNA damage-induced conventional autophagy, Ulk1 dephosphorylation at Ser637 by Mg2+/Mn2+ dependent 1D, a protein phosphatase, is essential (Torii et al., 2016). This Ulk1 dephosphorylation facilitates Ulk1 puncta formation, Ulk1 kinase function (assessed by Atg13 phosphorylation), DFCP1 puncta formation, and the subsequent progression of autophagy. Because point mutations at Ser637 almost completely inhibited the downstream events, this serine residue was concluded to be crucial for genotoxic stress-induced conventional autophagy (Torii et al., 2016).
The Ulk1 protein is also known to be crucial for alternative autophagy. The addition of etoposide induced the accumulation and phosphorylation of Ulk1 in Atg5-deficient MEFs. Furthermore, no autophagic membranes were observed within Ulk1-silenced Atg5-deficient MEFs in response to etoposide. Similar results were also observed when Fip200, another component of the Ulk1 complex, was silenced. Thus, Ulk1 is required for the initial step of both conventional and alternative autophagy (Nishida et al., 2009), and the mechanism for Ulk1 activation during alternative autophagy remains to be elucidated.
CORE MACHINERY OF AUTOPHAGY
After Ulk1 complex activation, vesicle nucleation occurs via activation of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex, including PtdIns3K and Beclin 1, in both types of autophagy. However, the molecules required for the expansion and closure of isolation membranes are completely different. In conventional autophagy, these steps occur via two ubiquitin-like conjugation pathways, namely, the Atg5–Atg12 pathway and the microtubule-associated protein 1 light chain 3 (LC3) pathway (Mizushima et al., 2002). Atg7 is required for the conjugation of Atg12 to Atg5 as an E1-like enzyme. Conjugation of phosphatidylethanolamine to LC3 is mediated by the actions of Atg3 and the Atg5–Atg12 complex, as E2-like and E3-like enzymes, respectively. This event is coupled with the translocation of LC3 from the cytosol to the isolation membrane, and hence this translocation is considered to be a reliable marker of autophagy. In the final step, UV radiation resistance-associated gene (UVRAG) and the PtdIns3K complex, excluding Atg14L, facilitate autophagosome–lysosome fusion. Syntaxin 17 is also required for this fusion (Itakura et al., 2012).
For alternative autophagy, neither the Atg5–Atg12 nor the LC3 conjugation pathway is required. Thus, it is unknown as to how extension and closure of autophagic membranes are accomplished during alternative autophagy without these two ubiquitin-like systems. Detailed morphological analysis has, however, yielded some clues. As described above, autophagic membranes originate from the trans-Golgi. Subsequent expansion and closure occur via fusion between trans-Golgi-derived membranes and endosomal vesicular membranes, which was shown by the colocalization of mannose-6-phosphate receptors (a trans-Golgi/late endosomal marker) and syntaxin 7 (a late endosomal marker) with lamp2-positive vacuoles in etoposide-treated Atg5-deficient MEFs (Nishida et al., 2009). Formation of autophagic vacuoles by trans-Golgi/endosomal fusion is also supported by studies showing the requirement for Rab9, a GTPase essential for the trafficking of proteins from late endosomes to trans-Golgi membranes. First, GFP-Rab9 was found to colocalize with lamp2-positive autolysosomes in etoposide-treated Atg5-deficient MEFs, and this colocalization was increased by the transfection of GFP-Rab9Q66L, a constitutively active Rab9 mutant, and reduced by GFP-Rab9S21N, a GDP-preferring dominant-negative Rab9 mutant. Moreover, Rab9 silencing by a targeted siRNA reduced the number of autophagic vacuoles but induced the accumulation of isolation membranes. Numerous isolation membranes are normally generated in cells by their exposure to etoposide, so siRab9 did not merely slow down the progression of autophagy but rather inhibited autophagosome maturation. Therefore, Rab9-mediated extension and closure of isolation membranes in the alternative autophagy pathway presumably replaces Atg5/Atg7/LC3 in conventional autophagy (Fig. 1).
BIOLOGICAL ROLES OF ALTERNATIVE AUTOPHAGY
A variety of potential physiological functions of conventional autophagy have been demonstrated by the analysis of systemic and tissue-specific Atg-gene knockout mice, including resistance to early neonatal starvation, clearing of neuronal protein aggregates, and maintenance of cardiac function (Mizushima and Levine, 2010). On the other hand, research on the biological roles of alternative autophagy has just been started. The first identified biological role is mitochondrial elimination during erythrocyte maturation. Erythroblasts have nuclei and other organelles. At the final stage of erythrocyte maturation, erythroblasts lose their nuclei to become reticulocytes, and reticulocytes transform into erythrocytes by the elimination of their organelles, including mitochondria (Fig. 2A). The elimination of mitochondria is considered to occur via autophagy, because ultrastructural studies have detected autophagic structures engulfing the mitochondria. Interestingly, however, erythrocyte maturation proceeds normally in Atg5-deficient embryos. Ultrastructural analysis also demonstrated that even in Atg5-deficient embryos, autophagic vacuoles in reticulocytes normally engulfed and digested mitochondria, indicating that mitochondrial elimination is not mediated by conventional autophagy in reticulocytes. In contrast, a large number of mitochondria remained in Ulk1-deficient reticulocytes due to the lack of mitophagy (Honda et al., 2014; Kundu et al., 2008). Ulk1 is the initiator of conventional and alternative autophagy, but mainly functions in alternative autophagy in erythroid cells. Therefore, mitochondrial elimination mediated by alternative autophagy is crucial for erythrocyte maturation (Fig. 2B). Interestingly, conventional autophagy is also activated in reticulocytes to digest other organelles, including ribosomes and the ER. Therefore, conventional and alternative autophagy can occur in the same cell to digest different organelles.
Fig. 2. Involvement of alternative autophagy in mitochondrial clearance during erythrocyte maturation.
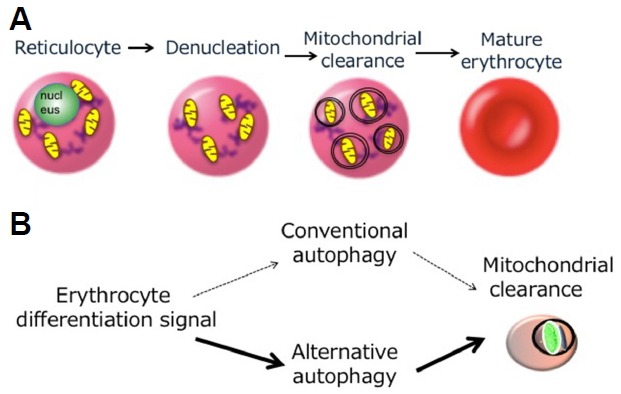
(A) The final stage of red blood cell maturation. During erythrocyte maturation, erythroblasts lose their nuclei to become reticulocytes, and reticulocytes transform into erythrocytes by the elimination of their mitochondria. Autophagy is involved in the latter process. (B) Mechanism of mitochondrial elimination during erythrocyte maturation. Mitochondrial elimination mainly occurs via Ulk1-dependent alternative autophagy and only partially via Atg5-dependent conventional autophagy.
Recently, we showed that alternative autophagy also occurs when the membrane trafficking from the Golgi to the PM is disrupted (Yamaguchi et al., 2016). This alternative autophagy is utilized for the degradation of undelivered cargo molecules. One example is the degradation of (pro)insulin granules in glucose-deprived β-cells. When β-cells are subjected to glucose deprivation, crinophagy (Orci et al., 1984) and starvation-induced nascent granule degradation (SINGD) (Goginashvili et al., 2015) are the main pathways for the degradation of unused (pro)insulin. These pathways degrade old and fresh insulin granules, respectively, via their direct fusion with lysosomes. Alternative autophagy also plays a subsidiary function. Interestingly, despite crinophagy/SINGD are not macroautophagy, they are largely suppressed in the β-cells lacking conventional autophagy. In such cells, alternative autophagy mainly contributed to degrade insulin granules. The alternative autophagy also plays a role in mitochondrial elimination from dedifferentiating iPS cells (Ma et al., 2015) and in the protection of intestinal epithelial cells from inflammatory bowel disease (Ra et al., 2016). A deeper understanding of the physiological and pathological relevance of alternative autophagy should emerge from analyses of knockout mice with targeted deletions of genes specific to the alternative autophagic pathway.
CONCLUSION
In this review, we described two distinct autophagic pathways, namely, conventional and alternative autophagy, and compared their nature. The presence of at least two mechanistically distinct forms of autophagy in mammalian cells underscores autophagy as a highly adaptable cellular stress response. Further elucidation of the biological roles of autophagy will require a more complete understanding of (1) the molecular mechanisms of alternative autophagy, (2) the unique functional roles of these two pathways in vivo, and (3) the contribution of each pathway to pathology, particularly of diseases associated with the accumulation of misfolded proteins or damaged organelles.
REFERENCES
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goginashvili A, Zhang Z, Erbs E, Spiegelhalter C, Kessler P, Mihlan M, Pasquier A, Krupina K, Schieber N, Cinque L, et al. Insulin granules. Insulin secretory granules control autophagy in pancreatic β cells. Science. 2015;347:878–882. doi: 10.1126/science.aaa2628. [DOI] [PubMed] [Google Scholar]
- Honda S, Arakawa S, Nishida Y, Yamaguchi H, Ishii E, Shimizu S. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat Commun. 2014;5:4004. doi: 10.1038/ncomms5004. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE Syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells. 2010;15:923–933. doi: 10.1111/j.1365-2443.2010.01433.x. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Li J, Xu Y, Yu C, Xu T, Wang H, Liu K, Cao N, Nie BM, Zhu SY, et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol. 2015;17:1379–1387. doi: 10.1038/ncb3256. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome Formation in Mammalian Cells Tracing of autophagosome formation with mammalian Apg proteins Initial step of autophagosome formation. Cell. 2002;429:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms : lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:1–10. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Amherdt M, Yanaihara C, Yanaihara N, Halban P, Renold AE, Perrelet A. Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B-cell. J Cell Biol. 1984;98:222–228. doi: 10.1083/jcb.98.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra EA, Lee TA, Kim SW, Park A, Choi HJ, Jang I, Kang S, Cheon JH, Cho JW, Lee JE, et al. TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nature Commun. 2016;7 doi: 10.1038/ncomms11726. Article number: 11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci USA. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- Torii S, Yoshida T, Arakawa S, Honda S, Nakanishi A, Shimizu S. Identification of protein phosphatase 1D magnesium-dependent delta isoform as an essential Ulk1 phosphatase for genotoxic stress-induced autophagy. EMBO R. 2016;11:1552–1564. doi: 10.15252/embr.201642565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PM, Feng Y, Wang J, Shi R, Jiang X. Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A. Nat Commun. 2015;6:8048. doi: 10.1038/ncomms9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Arakawa S, Kanaseki T, Miyatsuka T, Fujitani Y, Watada H, Tsujimoto H, Shimizu S. Golgi membrane-associated degradation pathway in yeast and mammals. EMBO J. 2016;35:1991–2007. doi: 10.15252/embj.201593191. [DOI] [PMC free article] [PubMed] [Google Scholar]


