Abstract
For years, the use of polyhistidine tags (His-tags) has been a staple in the isolation of recombinant proteins in immobilized metal affinity chromatography experiments. Their usage has been widely beneficial in increasing protein purity from crude cell lysates. For some recombinant proteins, a consequence of His-tag addition is that it can affect protein function and stability. Functional proteins are essential in the elucidation of their biological, kinetic, structural, and thermodynamic properties. In this study, we determine the effect of N-terminal His-tags on the thermal stability of select proteins using differential scanning fluorimetry and identify that the removal of the His-tag can have both beneficial and deleterious effects on their stability.
Introduction
The production of recombinant proteins is essential for biotechnology and biomedical sciences. To be beneficial for research studies, these proteins usually have to be expressed and purified in high quantities. Once recombinant proteins are purified, they are normally stored or used under conditions that are believed to be optimal for protein stability. It is not surprising that many researchers working in the fields of biotechnology and biomedicine direct a significant amount of effort toward determining the most stable conditions for their proteins of interest.
For purification purposes, many recombinant proteins have their DNA sequence modified to add specific residues to the N- or C-terminus of the protein. These additional sequence fragments (or tags) convey specific properties to the protein. For example, a tag introduced for purification may contain a specific epitope recognized by an antibody fragment immobilized on a chromatography column.1,2 Tags that are commonly used include polyhistidine (His-tag), glutathione S-transferase, maltose-binding protein, and thioredoxin. Furthermore, protein tags may be engineered to contain a specific protease cleavage site, e.g., a site recognized by tobacco etch virus (TEV) protease. Once the tag is cleaved, the protein is typically left with just a few additional amino acids.3 Ideally, removing the tag should result in a protein that has the activity or function similar to native protein.
His-tags (typically containing six or more consecutive histidine residues) may be used for protein purification by immobilized metal affinity chromatography (IMAC), where divalent cations (usually Ni2+ or Co2+) are adhered to a solid matrix. IMAC was introduced over four decades ago and has become one of the most popular methods for recombinant protein purification.4 In some cases, the tags may not only ease protein purification but also help in protein expression, folding, and/or solubility.5
His-tags, due to their relatively small size (∼2.5 kDa), are not believed to significantly interfere with the function and structure of a majority of proteins.5,6 However, there is an increase in the number of reports showing that this tacit assumption may be false.7−10 A study by Mohanty et al. demonstrated the effects of varying the length and position of His-tags on the expression and production of a membrane protein.11 His-tags may affect the oligomeric states of proteins as well as their function.8,9,12−15 Significant reductions in the enzymatic activity of several different enzymes were observed upon the incorporation of a His-tag.8,9,12 In the case of sperm whale myoglobin, the addition of an N-terminal His-tag affected its conformational movements, which indicates that the His-tag should be taken into account during the investigation of protein structural dynamics.13 Moreover, His-tags may also have an impact on structural studies and thus the ability to crystallize proteins for X-ray diffraction studies.14,15
Over 90% of experimentally determined protein structures are of recombinant proteins,16 and a significant fraction of these proteins are purified using His-tags. In many cases, the His-tags do not interfere with the crystallization process and can even be involved in the formation of crystal contacts.14 In contrast, the B-factor, which describes the general displacement of atoms in a crystal structure, shows that the presence of a His-tag increases this value compared to structures determined without it, thus making crystallization more difficult.14 This observation also suggests that addition of a His-tag can alter protein dynamics and possibly impact protein activity or function.
In this study, differential scanning fluorimetry (DSF) was used to determine the effect of the N-terminal His-tag on protein stability. DSF, also known as thermal-shift fluorescence-based screening or Thermofluor, is a cost-effective and rapid screening method that may be used to study protein stability or ligand-binding properties.17 In this method, samples of protein are mixed with different solutions (containing, for example, ligands, buffer solutions, and salt conditions) and a fluorescent hydrophobic dye. As the mixture is gradually heated over time, the protein starts to denature and inner hydrophobic residues are exposed; the fluorescent dye binds to the exposed hydrophobic fragments, and the melting temperature (Tm) of the protein can be determined by monitoring fluorescence over time. Tm is the inflection point of the resulting fluorescence curve and serves as a proxy for the stability of a protein sample. These experiments are performed using a real-time polymerase chain reaction (RT-PCR) instrument that is compatible with small volumes and capable of reaching a broad temperature range.18 Such an approach is a low-cost method to obtain high-throughput thermal scanning results and has been proven to be very effective in the optimization of the thermal stability of proteins.19−25 Additionally, DSF can also be used as a rapid screening method for finding protein ligands and/or compounds that stabilize proteins.26−28
Several proteins with an N-terminal His-tag were studied here to monitor the impact of the tag on protein thermostability. Given the widespread use of His-tags in recombinant protein production, understanding the influence of His-tags on protein stability is central to biotechnology and biochemical engineering. To address the lack of studies on the effect of His-tags on protein stability, a diverse range of proteins were selected that differ in molecular weight and quaternary structure (Table 1). These proteins were amenable for DSF studies because they did not significantly bind the fluorescent probe, SYPRO orange, in the folded state, nor were they precipitated by the probe. The proteins express well, are soluble, have an N-terminal His-tag that can be cleaved by TEV protease, have known biochemical functions, and many have their crystal structures or homologous protein structures determined.
Table 1. Characteristics of Proteins Studied Using DSFa.
| protein | organism | molecular weight (kDa) | no. of amino acids | oligomeric state | PDB code |
|---|---|---|---|---|---|
| β-lactamase | Staphylococcus aureus | 31.5 (29.0) | 257 | monomer | 3BLM |
| SpNadD | Streptococcus pyogenes | 27.1 (24.4) | 235 | *trimer | 1K4M* |
| VvDxr | Vibrio vulnificus | 46.1 (43.6) | 402 | dimer | 5KQO |
| mcsA | Aspergillus fumigatus | 51.0 (48.5) | 437 | dimer | 5UQO |
| MtDapB | Mycobacterium tuberculosis | 28.5 (26.0) | 245 | tetramer | 1YL7 |
| NgDapB | Neisseria gonorrhoeae | 31.0 (28.6) | 269 | tetramer** | 1YL7* |
| VvDapB | Vibrio vulnificus | 31.4 (28.9) | 269 | tetramer | 5TEM |
| Amb a 8 | Ambrosia artemisiifolia | 16.9 (14.4) | 133 | monomer | 5EM1 |
| Art v 4 | Artemisia vulgaris | 16.8 (14.4) | 133 | monomer | 5EM0 |
| Bet v 2 | Betula verrucosa | 16.9 (14.4) | 133 | monomer | 1CQA |
DapB: 4-hydroxy-tetrahydrodipicolinate reductase, Dxr: 1-deoxy-d-xylulose-5-phosphate reductoisomerase, mcsA: 2-methylcitrate synthase, NadD: nicotinic acid mononucleotide adenylyltransferase, Amb a 8, Art v 4, and Bet v 2 are allergens that belong to the profilin family. Numbers in parentheses indicate values after His-tag cleavage by TEV protease. Number of amino acids refers to a protein length without the purification tag. The purification tag in all cases has 25 amino acids and is ∼2.8 kDa. Asterisk (*) indicates a structure of homologous protein. (**) our unpublished data.
Results and Discussion
From this point forward, proteins with an attached His-tag will be referred to as His-protein and proteins where the His-tag was cleaved off by TEV protease will be referred to as the native protein name with no prefix. As previously mentioned, the proteins discussed here were produced in relatively large quantities and in all cases the cleavage of the His-tag was possible with TEV protease. Originally, our set of proteins also included Streptococcus pyogenes quinolinate phosphoribosyltransferase (SpNadC) as well as NH3-dependent NAD+ synthetase (SpNadE); however, we were not able to remove the His-tags from these proteins.29 Interestingly, although His-SpNadC and His-SpNadE were crystallized and their structures were determined, we were not able to crystallize SpNadD or His-SpNadD.29
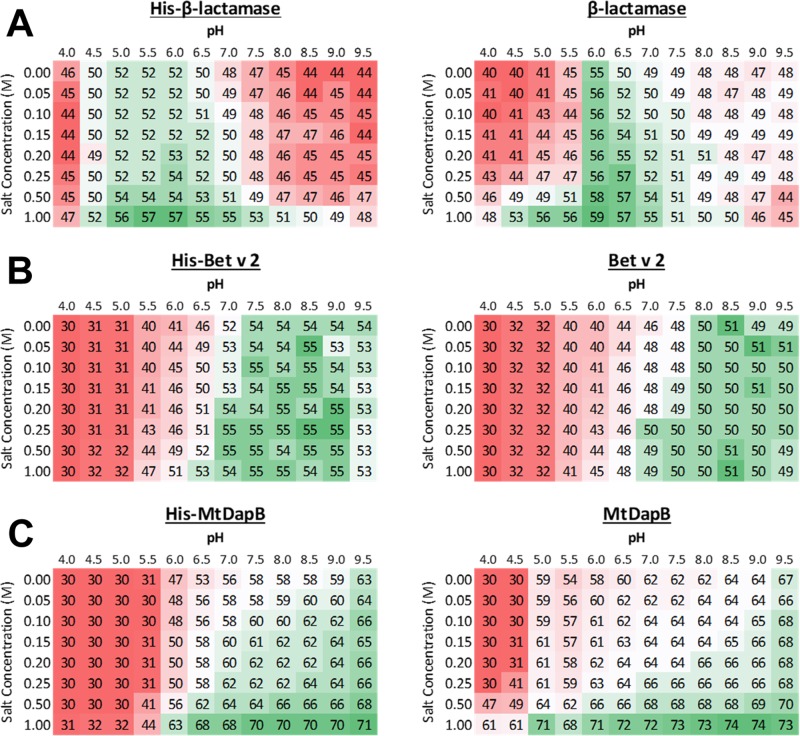
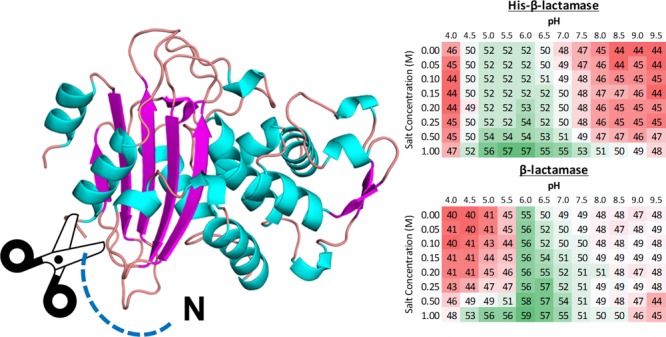
Examples of typical DSF results are shown in Figure 1, and the results for proteins not shown in Figure 1 can be found in the Supporting Information (Figure S1). The Tm of each protein is shown with a color gradient, where red, white, and green show low, average, and high melting temperatures, respectively. The color gradient is relative to each protein individually. For example, Figure 1A shows the melting temperatures for His-β-lactamase and β-lactamase. Both were found to be most stable at pH 6.0, 1.0 M salt, with β-lactamase being more stable by ∼2 °C than His-β-lactamase.
Figure 1.
Examples of screenings from DSF. A gradient of pH (4.0–9.5) and salt (0.0–1.0 M NaCl) buffers were used. Red, white, and green represent low, average, and high melting temperatures (°C), respectively. The standard deviation was typically less than 1 °C for all experiments. (A) Data for His-β-lactamase and β-lactamase. (B) Data for His-Bet v 2 and Bet v 2. (C) Data for His-MtDapB and MtDapB.
Effect of pH and His-Tag on Protein Stability
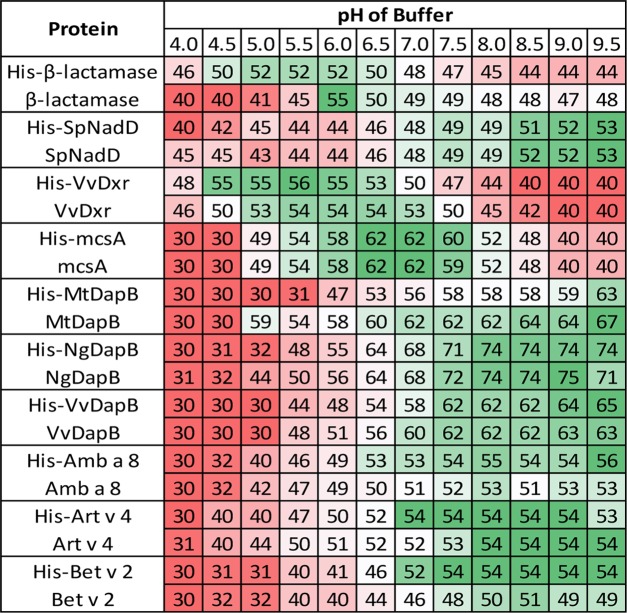
Figure 2 shows DSF results for all proteins in the presence of sodium chloride along with varied pH, whereas Table 2 summarizes the highest melting temperatures and best buffering conditions for proteins studied. β-Lactamase was most stable in the pH range 6.0–7.0 and without the His-tag. SpNadD was most stable in the pH range 8.5–9.5, and the His-tag did not affect stability based on pH. VvDxr was most stable in the pH range 5.5–6.5 with His-VvDxr being slightly more stable under these conditions than VvDxr. McsA was most stable in the pH range 6.0–7.0 and the His-tag did not significantly impact stability. MtDapB was most stable in the pH range 8.5–9.5, and His-MtDapB was less stable in this same range by ∼4 °C. Furthermore, His-MtDapB was very unstable in the pH range 5.0–6.0, whereas MtDapB showed much higher stability (∼20 °C). NgDapB was the most stable in the pH range 8.0–9.0, and the His-tag did not impact stability except at lower pH values (5.0–6.0), where His-NgDapB was less stable. Amb a 8 was most stable in the pH range 8.5–9.5, and the His-tag slightly increased its stability. Art v 4 was most stable in the pH range 8.0–9.5, and the His-tag did not significantly impact stability. Bet v 2 was most stable in the pH range 8.5–9.5, and the His-tag increased its stability by ∼4 °C.
Figure 2.
Effect of pH and His-tag on protein stability. Only melting temperatures observed from solutions with no salt are shown. Red, white, and green represent low, average, and high melting temperatures, respectively. All temperatures are in degree Celsius.
Table 2. Summary of Highest Melting Temperatures and Best Buffering Conditions for Proteins Studieda.
| protein | pI | highest Tm (°C) | best pH stability | best NaCl stability for pH (M) | least stable pH (lowest Tm) |
|---|---|---|---|---|---|
| β-lactamase | 9.4 (9.5) | 57 (59) | 5.0–6.5 (6.0–8.0) | 1.0 (1.0) | 4.0, 8.0–9.5 (4.0–5.0) |
| SpNadD | 5.5 (5.2) | 58 (59) | 8.5–9.5 (8.5–9.5) | 1.0 (1.0) | 4.0–5.5 (4.0–5.5) |
| VvDxr | 5.1 (5.4) | 62 (62) | 4.5–6.5 (5.0–7.5) | 1.0 (1.0) | 9.0–9.5 (9.5) |
| mcsA | 6.7 (6.9) | 64 (64) | 6.0–8.5 (6.0–8.5) | 1.0 (1.0) | 4.0–4.5 (4.0–4.5) |
| MtDapB | 5.8 (5.5) | 71 (74) | 7.0–9.5 (6.5–9.5) | 1.0 (1.0) | 4.0–5.5 (4.0–4.5) |
| NgDapB | 6.0 (5.9) | 76 (76) | 7.5–9.5 (7.5–9.5) | no significant impact | 4.0–5.0 (4.0–4.5) |
| VvDapB | 5.5 (5.2) | 71 (71) | 7.5–9.5 (7.5–9.5) | 1.0 (1.0) | 4.0–5.0 (4.0–5.0) |
| Amb a 8 | 5.4 (4.8) | 56 (53) | 6.5–9.5 (6.5–9.5) | 0 (0) | 4.0–4.5 (4.0–4.5) |
| Art v 4 | 5.3 (4.7) | 54 (54) | 6.5–9.5 (6.5–9.5) | no significant impact | 4.0 (4.0) |
| Bet v 2 | 5.6 (5.0) | 55 (50) | 7.0–9.5 (7.0–9.5) | no significant impact | 4.0–5.0 (4.0–5.0) |
Numbers in parentheses indicate values after His-tag cleavage by TEV protease. The pH values are given in ranges. Best NaCl stability for pH indicates the salt concentration that gave the highest thermal stability for the best pH range; no significant impact means that protein stability was not affected by salt concentration.
Analysis of Table 2 shows that the biggest shift in the value of the calculated pI (decrease by 0.6) is observed for profilins, which is expected as these proteins are the smallest among those studied here, and the His-tag corresponds to a significant fraction of the polypeptide chain (Table 1). The results presented in Table 2 also show that for most of the studied proteins (SpNadD, MtDapB, NgDapB, VvDapB, Amb a 8, Arv v 4, and Bet v 2) the regions of the best stability correspond to solutions with pH values significantly higher than the calculated pI value. At the same time, these proteins are least stable in solutions that have pH values close to or lower than the calculated pI. In the case of β-lactamase, the lowest melting temperatures are observed in solutions with pH close to or slightly lower than the calculated pI. In addition, β-lactamase is most stable in solutions with pH values significantly lower than pI values, but not under very acidic conditions. VvDxr and mcsA behave in a completely different manner as the pH values for solutions in which they display their best thermal stability overlap with calculated pIs.
Effect of Salt Concentration and His-Tag on Protein Stability
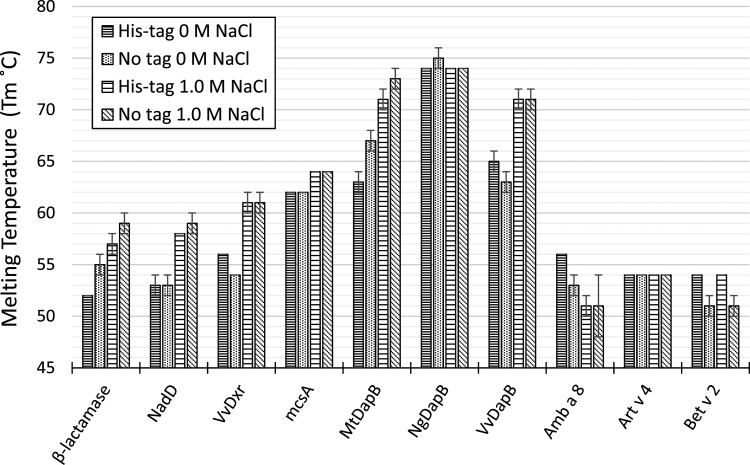
Results comparing protein stability with no salt and 1.0 M salt are shown in Figure 3. Each salt condition shown is respective to the pH in which a protein was most stable (Figure 2). His-tag lowered the stabilities of β-lactamase and MtDapB for both no salt and 1.0 M salt; had little or no effect on the stabilities of SpNadD, mcsA, NgDapB, VvDapB, and Art v 4 for both no salt and 1.0 M salt; and increased the stabilities of VvDxr, Amb a 8, and Bet v 2 in the presence of no salt only. His-β-lactamase Tm increased by ∼5 °C, His-SpNadD by ∼5 °C, His-VvDxr by ∼5 °C, His-mcsA by ∼2 °C, His-MtDapB by ∼8 °C, and His-VvDapB by ∼6 °C in the presence of 1.0 M salt compared to no salt. On the other hand, Tm for β-lactamase increased by ∼4 °C, SpNadD by ∼6 °C, VvDxr by ∼7 °C, mcsA by ∼2 °C, MtDapB by ∼6 °C, and VvDapB by ∼8 °C, exhibiting a slight change in the trend as compared to the His-tagged proteins under the same conditions. Overall, increasing salt concentration increased the stability of most proteins except NgDapB, Art v 4, Amb a 8, and Bet v 2; NgDapB and Art v 4 were not affected by salt concentration, whereas Amb a 8 and Bet v 2 were destabilized by increasing salt concentration.
Figure 3.
Effect of salt concentration and His-tag on protein stability. Melting temperatures are shown for solutions containing no salt and 1.0 M salt. The pH of each solution was the pH that displayed the best thermal stability for each respective protein.
Quaternary Structure and Protein Stability
The most stable proteins (MtDapB, NgDapB, and VvDapB) form tetrameric assemblies, whereas the least stable proteins are monomeric (β-lactamase, Amb a 8, Art v 4, Bet v 2). Dimeric proteins (VvDxr and mcsA) show intermediate stability. However, the trimeric SpNadD also displays low temperature stability. Taking into account the limited number of proteins studied, as well as the small variety of the oligomeric structures represented, it is not possible to establish any meaningful trend correlating the thermal stability with the quaternary structure.
Homologous Proteins
Among the ten proteins studied here, six of them belong to two protein families: the profilins and DapB proteins. Amb a 8, Art v 4, and Bet v 2 have over 75% of identical residues. DapB proteins are less similar in terms of the sequence with 53% of sequence identity between VvDapB and NgDapB and approximately 30% of sequence identity between MtDapB and both NgDapB and VvDapB. Despite the differences in amino acid composition, proteins in both groups have similar calculated pIs and surprisingly very similar stabilities. Although the similarities in the highest Tm values are not so pronounced, these proteins have very similar profiles of stability when pH of the solution is taken into consideration (Figures 2, 3, and Table 2).
The varied pH and NaCl concentration results showed that the profilins (Amb a 8, Art v 4, and Bet v 2) were found to be least stable with an average Tm around 50–54 °C (Figures 1 and S1). They all are unstable under conditions with pH below 5.5. NgDapB, MtDapB, and VvDapB were found to have the highest stability with Tm above 70 °C under the same conditions (Figures 1 and S1). Although NgDapB displays similar thermal stability at all NaCl concentrations, MtDapB and VvDapB are more stable at higher NaCl concentrations. These experiments indicate that pH of the solution has a more significant impact on protein thermal stability than salt concentration. All proteins analyzed here were stable over a relatively wide range of pH. However, decreased stability in acidic solutions (≤pH 5.0) was seen for most of the proteins, which may be due to the fact that most of the proteins studied here have a calculated isoelectric point around 5–6 (Table 2). These proteins showed an overall trend of being more stable in higher salt concentrations with the exception of Amb a 8, which was slightly destabilized as salt concentration increased.
Limitations of DSF
It was observed that DSF, as the method, has a limited application for proteins that bind to large hydrophobic ligands (like SYPRO orange), have high disulfide bond content, and/or have high thermal stability.15 For example, we were not able to determine thermal stability of Ara h 8 (PR-10-related peanut allergen with a large ligand(s) binding cavity),30 Cor a 8 (nonspecific lipid-transfer protein and hazelnut allergen),31 Act d 5 (kiwi fruit allergen with unknown function),32 and ragweed allergens Amb a 9 and Amb a 10 (calcium-binding proteins) using DSF. Information on the stability (and thermostability) of the allergens may be especially useful in the case of the food allergens as these proteins very often undergo thermal processing.
Our results on the thermal stabilities of Amb a 8, Art v 4, and Bet v 2 (inhaled allergens originating from pollens) clearly show that indeed these proteins are relatively unstable.33 The presence of the His-tag has no significant impact on the thermal stabilities of Amb a 8 and Art v 4, but it slightly increases the stability of Bet v 2. This observation is consistent with the high sequence identity between Amb a 8 and Art v 4 (∼89%) and lower sequence identity between Bet v 2 and the two other profilins (∼76%). Nonetheless, all these allergens have very similar thermal stability, and it is somewhat surprising that the addition of a relatively large purification tag to a small protein does not have a more pronounced effect.
Furthermore, it should be noted that our studies were performed with a purification tag that is 25 amino acids, where 4 linker residues (SGSG) are retained after tag removal. The length of the tag and linker, the amino acid composition, and their sequence order can have potential implications on the protein’s thermal stability. However, in the scope of the experiments performed here, different tag characteristics and their impact on protein thermal stability were not investigated.
In Most Cases, the Presence of His-Tag Decreased Thermal Stability
Out of the ten proteins evaluated in this study, protein thermal stability results for four (Amb a 8, Art v 4, mcsA, and SpNadD) were not significantly affected by cleavage of the His-tag. For the remaining six proteins, a change in stability was observed that was related to the presence or absence of the His-tag. His-Bet v 2 was the only protein observed to be more stable than its native counterpart, Bet v 2, in the majority of pH and salt concentrations tested (Figures 2 and 3). In the case of MtDapB, the cleavage of the His-tag resulted in a more tolerable pH range; both constructs are stable in pH 6.0, although MtDapB is significantly more stable at pH 5.0 than His-MtDapB (about a 24 °C Tm difference). A similar result is seen for VvDapB at pH 5.5 in which the Tm difference is at least 8 °C (Figure 3). An increase in thermal stability at lower pH is also observed for NgDapB. His-β-lactamase and His-VvDxr are more stable at lower pH and less stable at higher pH than β-lactamase and VvDxr, respectively. However, this effect is significantly more pronounced for β-lactamase. Bet v 2 was the only recombinant protein that had increased stability with a His-tag. This protein was more stable when the His-tag was intact, which may be indicative of high entropy residues at the N-terminus of the protein. It was also demonstrated that the presence of poly-l-proline peptides or urea may affect the thermal stability of profilin proteins.33 Binding of the proline-rich peptides may be affected by the presence of additional residues at the N-terminus of profilin as this part of the protein participates in the peptide binding.
The most significant changes in thermal stability were observed for β-lactamase and MtDapB. Various changes in the overall trend were seen between the native and His-tag versions of the proteins. In the case of β-lactamase, removal of His-tag resulted in a two-way change. First, the pH range in which the protein is stable significantly shifted toward more basic conditions. Second, notable changes were recorded in Tm in accordance with this shift. For instance, the protein Tm increased under conditions of pH 7.0–9.5 by an average of 3 °C, whereas an average decrease of 7 °C was seen under low-pH (4.0–6.0) conditions. In the case of MtDapB, His-tag removal caused the protein to be stable over a wider pH range, specifically under conditions of pH 5.0 and 5.5; the Tm increased from 30 and 31 to 59 and 54 °C, respectively.
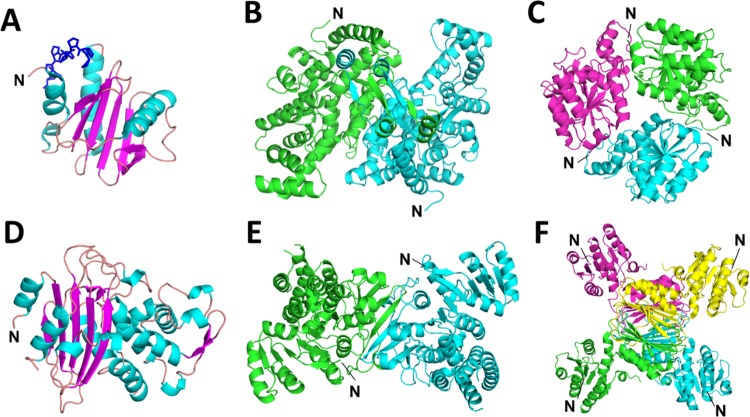
Most of the proteins mentioned herein have had their crystal structures determined (Figure 4). For all of the proteins, the His-tag is solvent-exposed and thus susceptible to TEV protease cleavage. Despite this fact, removal of the His-tag most often resulted in increased thermal stability. Although changes observed in Tm were not drastic between His-tagged and native proteins, it is apparent that the presence of a His-tag can contribute to the thermal instability of the protein and that its removal may prove beneficial when designing in vitro experiments and optimal conditions for storage and/or structural biology.34
Figure 4.
Cartoon representations of proteins analyzed in this study with marked positions of their N-termini. (A) Amb a 8 (PDB code: 5EV0) in complex with polyproline (blue). Both Arv 4 and Bet v 2 have the same overall fold as Amb a 8. (B) mcsA (PDB code: 5UQO). (C) Homolog of SpNadD (NadD from Escherichia coli (E. coli); PDB code: 1K4M). (D) β-Lactamase from Staphylococcus aureus (PDB code: 3BLM). (E) VvDxr (PDB code: 5KQO). (F) MtDapB (PDB code: 1YL7). Both NgDapB and VvDapB have the same overall fold and quaternary structure as MtDapB.
Information on protein thermal stability is not only important for designing experiments related to functional and characterization studies of protein, but it may be also used during protein purification.35 For example, in a case of a protein with high thermal stability, one may include a step involving heating of a sample to remove less thermally stable impurities. These impurities, like native proteins from E. coli, are quite often co-purified with IMAC.36,37 The same procedure could also be done while varying the pH and salt concentration to improve isolation. Furthermore, it has been recently shown that DSF and information on thermal stability of proteins may be used to guide the development of protocols for protein refolding.38
Thermal stability testing could also be used for the determination of optimal conditions for techniques that require minimal buffer conditions to lessen background noise from experimental results. For example, isothermal titration calorimetry tends to provide more accurate results when buffer/salt concentrations are at their lowest.39 DSF thermal stability testing can be utilized to give an indication of how long a protein will remain stable at a particular temperature within minimally buffered solutions. In summary, the use of affinity tags, like the polyhistidine tag, provides a highly effective means for isolating various proteins. However, it is important to consider the possibility that the introduction of even a relatively small purification tag may alter the behavior of a protein of interest.
Experimental Procedures
Cloning
Genes encoding all proteins in this study were synthesized by ATUM, Inc. (Newark CA) (http://www.atum.bio/), codon-optimized for expression in E. coli, and inserted into the plasmid pJExpress411. This plasmid contains an inducible T7 promoter with isopropyl β-d-1-thiogalactopyranoside (IPTG) and kanamycin resistance. Each plasmid was transformed into E. coli BL21 (DE3) by heat shock.40 Upon transformation, correct gene sequences were verified by DNA sequencing from Eton Bioscience (Research Triangle Park, NC) using T7 promoter and T7 terminator primers. All proteins used in this study were designed to contain an N-terminal purification tag (MHHHHHHSSGVDLGTENLYFQ↓SGSG). The purification tag includes a TEV-cleavage site (cut site marked with an arrow), and its sequence was designed based on the His-tag coded by vector pMCSG7.41
Protein Expression and Purification
For expression and purification, E. coli cells were grown to an OD600 of 0.6–0.8 at 37 °C, induced with IPTG at a working concentration of 0.4–1.0 mM, and shaken at 16 °C overnight. Cell cultures were centrifuged, and cell pellets were resuspended in lysis buffer (50 mM Tris, pH 7.5, 500 mM sodium chloride, 10 mM imidazole, 2% glycerol, and 20 mM β-mercaptoethanol (β-ME)) supplemented with Pierce protease inhibitors following the manufacturer’s protocol and lysed by sonication; NgDapB and β-lactamase were resuspended in lysis buffer with no β-ME and no glycerol. A pH of 6.5 was used for β-lactamase (this applies to all of the buffers for NgDapB and β-lactamase purification mentioned herein). The homogenate was then centrifuged, and the supernatant was separated from the insoluble cellular debris. The crude extract was loaded onto Ni-NTA (nitrilotriacetic acid, Qiagen or Pierce) IMAC resin previously equilibrated with wash buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 30 mM imidazole, 2% glycerol, and 20 mM β-ME). After washing, proteins were eluted with elution buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 250 mM imidazole, 2% glycerol, and 20 mM β-ME). The fractions containing protein were then pooled and dialyzed in 10 mM Tris, pH 7.5, 150 mM NaCl, and 5.0 mM β-ME using SnakeSkin dialysis tubing (ThermoFisher, Grand Island, NY) with 3 or 10 kDa molecular weight cutoff dependent on protein size.
His-tag cleavage was achieved by supplementing purified protein (1–2 mg/mL) with TEV protease in a 1:100 (w/w) protease-to-protein ratio and then dialyzing overnight at 4 °C. After incubation with TEV protease, the solution was passed through a Ni-NTA column equilibrated in dialysis buffer to collect non-tagged protein and concentrated using Amicon centrifugal filters (EMD Millipore, Billerica, MA) with 3 or 10 kDa molecular weight cutoff dependent on protein size. Protein concentration was determined by the Bradford method.42 After concentration, all protein samples were further purified using a Superdex 200 PG gel filtration column (equilibrated in dialysis buffer) attached to an ÄKTA Pure liquid chromatography system (GE Healthcare). Protein purity was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and all were isolated with an average purity of 90% or better. All proteins selected for this study were expressed with yields ranging from 5 to 40 mg/L of E. coli culture
Differential Scanning Fluorimetry
Using a MASTERBLOCK 96 deep well plate (Greiner; Hampton Research, Aliso Viejo, CA), solutions were prepared as an in-house pH and salinity screen. All conditions contained working concentrations of 50 mM buffer with a pH range 4–9.5 (in 0.5 unit increments) and a salt range (sodium chloride) 0–1.0 M (no salt, 0.05, 0.10, 0.15, 0.20, 0.25, 0.50, and 1.0 M); although a salt gradient was used, residual salt from purification buffers was present in all protein samples, which slightly alters the working concentration of salt under each condition. Buffers used to accommodate the pH ranges were acetate (pH 4.0–5.0), bis-Tris (pH 5.5–6.5), Tris (pH 7.0–8.0), and N-cyclohexyl-2-aminoethanesulfonic acid (pH 8.5–9.5). SYPRO orange dye (ThermoFisher, Waltham, MA) was diluted 1:1000 in 1.0 mL of 1–2 mg/mL protein. This solution was mixed under each screen condition 1:1 for a 20 μL of total reaction volume in a Bio-Rad Hardshell 96-well RT-PCR plate (Hercules, CA). The plates were sealed using Microseal PCR plate sealing film (Bio-Rad, Hercules, CA), and stability was measured in a Bio-Rad CFX96 RT-PCR instrument. Each experiment was performed in triplicate. An experimental protocol was created to increase the temperature of the 96-well plate from 30 to 90 °C in 2 °C increments after 1.0 min intervals. Upon conclusion of the experiment, Bio-Rad CFX Manager software was used to determine the inflection point of melting curves (Tm).
Various Computational Calculations
Figures were prepared with Pymol.43 The pI values were calculated with ProtParam.44
Acknowledgments
The authors acknowledge the Biomedical Engineering Program and thank Dr. Silke Henrich for access to their instrumentation. This work was partially supported by grants from National Institute of Allergy and Infectious Diseases (R01AI120987 and R01AI077653).
Glossary
Abbreviations
- His-tag
polyhistidine
- IMAC
immobilized metal affinity chromatography
- TEV
tobacco etch virus
- DSF
differential scanning fluorimetry
- DapB
4-hydroxytetrahydrodipicolinate reductase
- Dxr
1-deoxy-d-xylulose-5-phosphate reductoisomerase
- mcsA
2-methylcitrate synthase
- NadD
nicotinic acid mononucleotide adenylyltransferase
- IPTG
β-d-1-thiogalactopyranoside
- β-ME
β-mercaptoethanol
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b01598.
Examples of screening using DSF for the following proteins: Amb a 8, Art v 4, mcsA, NgDapB, SpNadD, VvDapB, and VvDxr (PDF)
Author Contributions
∥ W.T.B. and C.R.S. equally contributed to the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Hopp T. P.; Prickett K. S.; Price V. L.; Libby R. T.; March C. J.; Cerretti D. P.; Urdal D. L.; Conlon P. J. A short polypeptide marker sequence useful for recombinant protein identification and purification. Nat. Biotechnol. 1988, 6, 1204–1210. 10.1038/nbt1088-1204. [DOI] [Google Scholar]
- Munro S.; Pelham H. R. Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70. EMBO J. 1984, 3, 3087–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust R. B.; Tozser J.; Fox J. D.; Anderson D. E.; Cherry S.; Copeland T. D.; Waugh D. S. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001, 14, 993–1000. 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Porath J.; Carlsson J.; Olsson I.; Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 1975, 258, 598–599. 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- Gräslund S.; Nordlund P.; Weigelt J.; Hallberg B. M.; Bray J.; Gileadi O.; Knapp S.; Oppermann U.; Arrowsmith C.; Hui R.; Ming J.; dhe-Paganon S.; Park H. W.; Savchenko A.; Yee A.; Edwards A.; Vincentelli R.; Cambillau C.; Kim R.; Kim S. H.; Rao Z.; Shi Y.; Terwilliger T. C.; Kim C. Y.; Hung L. W.; Waldo G. S.; Peleg Y.; Albeck S.; Unger T.; Dym O.; Prilusky J.; Sussman J. L.; Stevens R. C.; Lesley S. A.; Wilson I. A.; Joachimiak A.; Collart F.; Dementieva I.; Donnelly M. I.; Eschenfeldt W. H.; Kim Y.; Stols L.; Wu R.; Zhou M.; Burley S. K.; Emtage J. S.; Sauder J. M.; Thompson D.; Bain K.; Luz J.; Gheyi T.; Zhang F.; Atwell S.; Almo S. C.; Bonanno J. B.; Fiser A.; Swaminathan S.; Studier F. W.; Chance M. R.; Sali A.; Acton T. B.; Xiao R.; Zhao L.; Ma L. C.; Hunt J. F.; Tong L.; Cunningham K.; Inouye M.; Anderson S.; Janjua H.; Shastry R.; Ho C. K.; Wang D.; Wang H.; Jiang M.; Montelione G. T.; Stuart D. I.; Owens R. J.; Daenke S.; Schutz A.; Heinemann U.; Yokoyama S.; Bussow K.; Gunsalus K. C. Protein production and purification. Nat. Methods 2008, 5, 135–146. 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M.; Forsberg G.; Moks T.; Hartmanis M.; Nilsson B. Fusion proteins in biotechnology. Curr. Opin. Biotechnol. 1992, 3, 363–369. 10.1016/0958-1669(92)90164-E. [DOI] [PubMed] [Google Scholar]
- Ledent P.; Duez C.; Vanhove M.; Lejeune A.; Fonze E.; Charlier P.; Rhazi-Filali F.; Thamm I.; Guillaume G.; Samyn B.; Devreese B.; Van Beeumen J.; Lamotte-Brasseur J.; Frere J. M. Unexpected influence of a C-terminal-fused His-tag on the processing of an enzyme and on the kinetic and folding parameters. FEBS Lett. 1997, 413, 194–196. 10.1016/S0014-5793(97)00908-3. [DOI] [PubMed] [Google Scholar]
- Majorek K. A.; Kuhn M. L.; Chruszcz M.; Anderson W. F.; Minor W. Double trouble-Buffer selection and His-tag presence may be responsible for nonreproducibility of biomedical experiments. Protein Sci. 2014, 23, 1359–1368. 10.1002/pro.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek A.; Pietrow O.; Filipkowski P.; Synowiecki J. Effects of the polyhistidine tag on kinetics and other properties of trehalose synthase from Deinococcus geothermalis. Acta Biochim. Pol. 2013, 60, 163–166. [PubMed] [Google Scholar]
- Sabaty M.; Grosse S.; Adryanczyk G.; Boiry S.; Biaso F.; Arnoux P.; Pignol D. Detrimental effect of the 6 His C-terminal tag on YedY enzymatic activity and influence of the TAT signal sequence on YedY synthesis. BMC Biochem. 2013, 14, 28. 10.1186/1471-2091-14-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A. K.; Wiener M. C. Membrane protein expression and production: effects of polyhistidine tag length and position. Protein Expression Purif. 2004, 33, 311–325. 10.1016/j.pep.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Araújo A. P. U.; Oliva G.; Henrique-Silva F.; Garratt R. C.; Caceres O.; Beltramini L. M. Influence of the histidine tail on the structure and activity of recombinant chlorocatechol 1,2-dioxygenase. Biochem. Biophys. Res. Commun. 2000, 272, 480–484. 10.1006/bbrc.2000.2802. [DOI] [PubMed] [Google Scholar]
- Thielges M. C.; Chung J. K.; Axup J. Y.; Fayer M. D. Influence of histidine tag attachment on picosecond protein dynamics. Biochemistry 2011, 50, 5799–5805. 10.1021/bi2003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M.; Johnson D. H.; McDonald H.; Brouillette C.; Delucas L. J. His-tag impact on structure. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2007, 63, 295–301. 10.1107/S0907444906052024. [DOI] [PubMed] [Google Scholar]
- Deller M. C.; Kong L.; Rupp B. Protein stability: a crystallographer’s perspective. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2016, 72, 72–95. 10.1107/S2053230X15024619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda Z. S. The use of recombinant methods and molecular engineering in protein crystallization. Methods 2004, 34, 354–363. 10.1016/j.ymeth.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Pantoliano M. W.; Petrella E. C.; Kwasnoski J. D.; Lobanov V. S.; Myslik J.; Graf E.; Carver T.; Asel E.; Springer B. A.; Lane P.; Salemme F. R. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screening 2001, 6, 429–440. 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- Lavinder J. J.; Hari S. B.; Sullivan B. J.; Magliery T. J. High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J. Am. Chem. Soc. 2009, 131, 3794–3795. 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F.; Orru R.; Bonivento D.; Chiarelli L. R.; Mattevi A. ThermoFAD, a Thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding. FEBS J. 2009, 276, 2833–2840. 10.1111/j.1742-4658.2009.07006.x. [DOI] [PubMed] [Google Scholar]
- Kean J.; Cleverley R. M.; O’Ryan L.; Ford R. C.; Prince S. M.; Derrick J. P. Characterization of a CorA Mg2+ transport channel from Methanococcus jannaschii using a Thermofluor-based stability assay. Mol. Membr. Biol. 2008, 25, 653–663. 10.1080/09687680802541169. [DOI] [PubMed] [Google Scholar]
- Nettleship J. E.; Brown J.; Groves M. R.; Geerlof A. Methods for protein characterization by mass spectrometry, thermal shift (ThermoFluor) assay, and multiangle or static light scattering. Methods Mol. Biol. 2008, 426, 299–318. 10.1007/978-1-60327-058-8_19. [DOI] [PubMed] [Google Scholar]
- Ericsson U. B.; Hallberg B. M.; Detitta G. T.; Dekker N.; Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 2006, 357, 289–298. 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Cummings M. D.; Farnum M. A.; Nelen M. I. Universal screening methods and applications of ThermoFluor. J. Biomol. Screening 2006, 11, 854–863. 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- Matulis D.; Kranz J. K.; Salemme F. R.; Todd M. J. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 2005, 44, 5258–5266. 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- Carver T. E.; Bordeau B.; Cummings M. D.; Petrella E. C.; Pucci M. J.; Zawadzke L. E.; Dougherty B. A.; Tredup J. A.; Bryson J. W.; Yanchunas J. Jr.; Doyle M. L.; Witmer M. R.; Nelen M. I.; DesJarlais R. L.; Jaeger E. P.; Devine H.; Asel E. D.; Springer B. A.; Bone R.; Salemme F. R.; Todd M. J. Decrypting the biochemical function of an essential gene from Streptococcus pneumoniae using ThermoFluor technology. J. Biol. Chem. 2005, 280, 11704–11712. 10.1074/jbc.M413278200. [DOI] [PubMed] [Google Scholar]
- Mezzasalma T. M.; Kranz J. K.; Chan W.; Struble G. T.; Schalk-Hihi C.; Deckman I. C.; Springer B. A.; Todd M. J. Enhancing recombinant protein quality and yield by protein stability profiling. J. Biomol. Screening 2007, 12, 418–428. 10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]
- Fan J.; Heng J.; Dai S.; Shaw N.; Zhou B.; Huang B.; He Z.; Wang Y.; Jiang T.; Li X.; Liu Z.; Wang X.; Zhang X. C. An efficient strategy for high throughput screening of recombinant integral membrane protein expression and stability. Protein Expression Purif. 2011, 78, 6–13. 10.1016/j.pep.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Malik K.; Matejtschuk P.; Thelwell C.; Burns C. J. Differential scanning fluorimetry: rapid screening of formulations that promote the stability of reference preparations. J. Pharm. Biomed. Anal. 2013, 77, 163–166. 10.1016/j.jpba.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Booth W. T.; Morris T. L.; Mysona D. P.; Shah M. J.; Taylor L. K.; Karlin T. W.; Clary K.; Majorek K. A.; Offermann L. R.; Chruszcz M. Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3-dependent NAD+ synthetase. FEBS J. 2017, 284, 2425–2441. 10.1111/febs.14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt B. K.; Offermann L. R.; McBride J. K.; Majorek K. A.; Maleki S. J.; Chruszcz M. Structure and function of the peanut panallergen Ara h 8. J. Biol. Chem. 2013, 288, 36890–36901. 10.1074/jbc.M113.517797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann L. R.; Bublin M.; Perdue M. L.; Pfeifer S.; Dubiela P.; Borowski T.; Chruszcz M.; Hoffmann-Sommergruber K. Structural and functional characterization of the hazelnut allergen cor a 8. J. Agric. Food Chem. 2015, 63, 9150–9158. 10.1021/acs.jafc.5b03534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann L. R.; Giangrieco I.; Perdue M. L.; Zuzzi S.; Santoro M.; Tamburrini M.; Cosgrove D. J.; Mari A.; Ciardiello M. A.; Chruszcz M. Elusive structural, functional, and immunological features of act d 5, the green kiwifruit kiwellin. J. Agric. Food Chem. 2015, 63, 6567–6576. 10.1021/acs.jafc.5b02159. [DOI] [PubMed] [Google Scholar]
- Offermann L. R.; Schlachter C. R.; Perdue M. L.; Majorek K. A.; He J. Z.; Booth W. T.; Garrett J.; Kowal K.; Chruszcz M. Structural, functional, and immunological characterization of profilin panallergens amb a 8, art v 4, and Bet v 2. J. Biol. Chem. 2016, 291, 15447–15459. 10.1074/jbc.M116.733659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos S. P.; Bandeiras T. M.; Pinto A. F.; Teixeira M.; Carrondo M. A.; Romao C. V. Thermofluor-based optimization strategy for the stabilization and crystallization of Campylobacter jejuni desulforubrerythrin. Protein Expression Purif. 2012, 81, 193–200. 10.1016/j.pep.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Boivin S.; Kozak S.; Meijers R. Optimization of protein purification and characterization using Thermofluor screens. Protein Expression Purif. 2013, 91, 192–206. 10.1016/j.pep.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Bolanos-Garcia V. M.; Davies O. R. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim. Biophys. Acta 2006, 1760, 1304–1313. 10.1016/j.bbagen.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Niedzialkowska E.; Gasiorowska O.; Handing K. B.; Majorek K. A.; Porebski P. J.; Shabalin I. G.; Zasadzinska E.; Cymborowski M.; Minor W. Protein purification and crystallization artifacts: the tale usually not told. Protein Sci. 2016, 25, 720–733. 10.1002/pro.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biter A. B.; de la Pena A. H.; Thapar R.; Lin J. Z.; Phillips K. J. DSF guided refolding as a novel method of protein production. Sci. Rep. 2016, 6, 18906 10.1038/srep18906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff M. R. Jr.; Grubbs J.; Howell E. E. Isothermal titration calorimetry for measuring macromolecule-ligand affinity. J. Visualized Exp. 2011, 55, e2796 10.3791/2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Die I. M.; Bergmans H. E.; Hoekstra W. P. Transformation in Escherichia coli: studies on the role of the heat shock in induction of competence. J. Gen. Microbiol. 1983, 129, 663–670. 10.1099/00221287-129-3-663. [DOI] [PubMed] [Google Scholar]
- Stols L.; Gu M.; Dieckman L.; Raffen R.; Collart F. R.; Donnelly M. I. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expression Purif. 2002, 25, 8–15. 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- DeLano W. S.The PyMOL Molecular Graphics System; Schrödinger, LLC, 2002.
- Gasteiger E.; Gattiker A.; Hoogland C.; Ivanyi I.; Appel R. D.; Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.