Abstract
Vancomycin is a glycopeptide antibiotic used for the treatment of serious infections by Gram-positive pathogens. Vancomycin inhibits cell wall biosynthesis by targeting the d-Ala-d-Ala terminus of peptidoglycan (PG). The highly cross-linked heptapeptide aglycon structure of vancomycin is the d-Ala-d-Ala binding site. The first residue of vancomycin is N-methyl-leucine, which is crucial for the dipeptide binding. The removal of N-methyl-leucine by Edman degradation results in desleucyl-vancomycin devoid of antimicrobial activities. To investigate the function of N-methyl-leucine for the dipeptide binding in vancomycin, molecular dynamics simulations of vancomycin and three N-terminus-modified vancomycin derivatives: desleucyl-vancomycin, vancomycinNtoC, and vancomycinSar, binding to a PG unit of the sequence l-Ala-d-iso-Gln-l-Lys-d-Ala-d-Ala with an intact pentaglycine bridge structure attached to the bridge link of l-Lys were carried out. Glycopeptide–PG binding interactions were characterized by root-mean-square-deviation contour analysis of atomic positions in vancomycin and its three analogues bound to a PG unit. The overall sampling space for four glycopeptide–PG complexes shows four distinct distributions with a continuous change between the conformational spaces. The hydrogen bond analyses show that multiple hydrogen bonds between the d-Ala-d-Ala and the vancomycin aglycon structure strengthened the dipeptide binding. The simulations revealed that the removal or chemical modification of N-methyl-leucine significantly weakens the dipeptide binding to the aglycon structure and provides interesting structural insights into glycopeptide–PG binding interactions.
1. Introduction
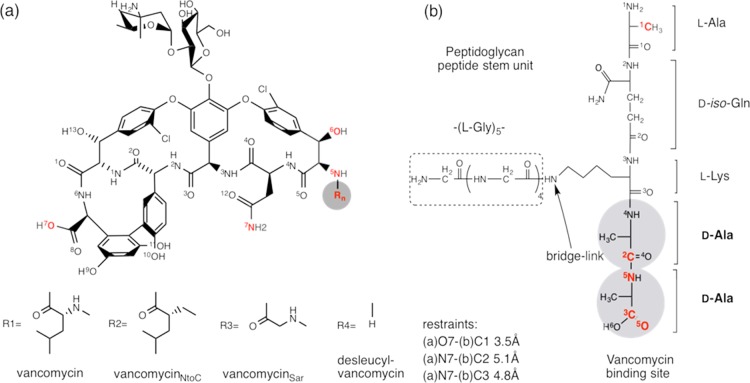
Vancomycin (Figure 1a, R1) is a glycopeptide antibiotic isolated in 1956 from the fermentation of a soil actinomycete, Amycolatopsis orientalis. Since its discovery, vancomycin has been one of the most effective therapeutic agents for the treatment of serious infections by methicillin-resistant Staphylococcus aureus.1 Vancomycin inhibits cell wall biosynthesis by binding with the d-Ala-d-Ala dipeptide of the peptidoglycan (PG)-stem unit (Figure 1b).2 The d-Ala-d-Ala dipeptide of PG binds to the aglycon structure of vancomycin, which is a highly cross-linked rigid cage-like structure formed by the phenolic ether linkages between the hydroxyphenylglycines at the second and sixth amino acid positions and between the phenolic hydroxyl residues at the fourth, fifth, and seventh positions (Figure 1a).3 The solution NMR structure of vancomycin complexed to acyl-l-Lys-d-Ala-d-Ala shows that five hydrogen bonds between the heptapeptide backbone of aglycon to the d-Ala-d-Ala dipeptide are involved in the glycopeptide–PG binding.4 In vancomycin-resistant enterococci, the d-Ala-d-Ala of the PG stem is replaced by a depsipeptide d-Ala-d-Lac, which replaces one of the hydrogen bonds with an electrostatic repulsion.5 The depsipeptide substitution reduces the vancomycin binding affinity (Kd) from micromolar for the dipeptide to millimolar, enabling vancomycin resistance in enterococcal bacteria.6
Figure 1.
Chemical structure of vancomycin, vancomycin derivatives, and PG. (a) Chemical structures of vancomycin (Rn=R1), vancomycinNtoC (Rn=R2), vancomycinSar (Rn=R3), and des-N-methylleucyl-vancomycin (Rn=R4). The key atoms are labeled with numbers shown in superscript. (b) Chemical structure of S. aureus PG-peptide stem unit without a disaccharide GlcNAc-MurNAc. A pentapeptide stem consists of the sequence l-Ala-d-iso-Gln-l-Lys-d-Ala-d-Ala, and a pentaglycine (dotted box) is attached to the ε-nitrogen side chain of l-Lys. The d-Ala-d-Ala (circles) of the PG stem is the known vancomycin-binding site. Three distance restraints were applied in the simulations: (1) “(a)O7-(b)C1 3.5 Å” which means that the distance between O7 in glycopeptide (a) and C1 in PG (b) is restrained to 3.5 Å; (2) “(a)N7-(b)C2 5.1 Å” for the restrained distance of 5.1 Å for internuclear distance between N7 in glycopeptide (a) and C2 in PG (b); and (3) “(a)N7-(b)C3 4.8 Å” for the restrained distance of 4.8 Å between N7 in (a) and C3 in (b).
To investigate vancomycin–PG interactions, glycopeptide binding to a series of peptide analogues of the bacterial cell wall were characterized using capillary electrophoresis, fluorescence correlation spectroscopy, and molecular dynamics (MD) simulations under different buffer systems.7 One of the key residues that was identified as essential for the d-Ala-d-Ala binding was N-methylleucine found at the first position of the heptapeptide-core structure in vancomycin. N-methylleucine is not part of the dipeptide binding site.4 Nevertheless, the removal of N-methylleucine by Edman degradation8 results in desleucyl-vancomycin (Figure 1a, R4) devoid of antimicrobial activity.9 Because N-methylleucine is not directly involved in the dipeptide binding, the mechanism of loss dipeptide binding by desleucyl-vancomycin remains elusive. In this study, MD simulations of glycopeptide complexed to the PG-peptide repeat unit (Figure 1b) were carried out to provide structural and dynamic insights for understanding the glycopeptide–PG interactions at atomic resolution.2 The role of the N-methylleucine was elucidated by the MD simulations of PG binding by vancomycin and N-terminus-modified vancomycin derivatives: vancomycinNtoC, vancomycinSar, and desleucyl-vancomycin. In vancomycinNtoC, the N-methyl in N-methylleucine of vancomycin is replaced with an ethyl moiety (Figure 1a, R2), and in vancomycinSar, the N-methylleucine is replaced by sarcosine (N-methylglycine) (Figure 1a, R3). Because the leucine side chain substitution by sarcosine in vancomycinSar significantly reduces the hydrophobicity, we hypothesized that the stability of the glycopeptide–PG complex will gradually transition from vancomycin, vancomycinNtoC, and vancomycinSar to desleucyl-vancomycin toward the unbound state.
2. Results
2.1. Root-Mean-Square Deviation (rmsd) Analysis
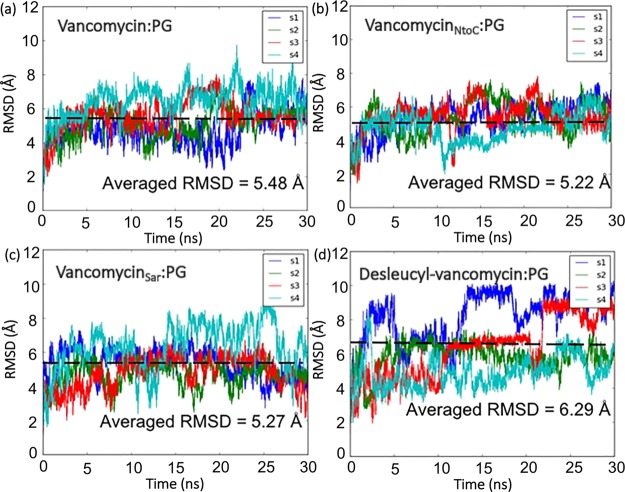
The rmsd of vancomycin (V1) and its three analogues: vancomycinNtoC (V2), vancomycinSar (V3), and desleucyl-vancomycin (V4) complexed with PG-peptide units for four independent simulations (s1 to s4) is plotted in Figure 2. The stabilities and flexibilities of the complexes of vancomycin and its analogues bound to the peptide can be estimated by rmsd values, where a small rmsd indicates high stability and low flexibility, and vice versa for a large rmsd.
Figure 2.
rmsd plots of MD simulations. The rmsd plots for the complexes of vancomycin/PG (a), vancomycinNtoC/PG (b), vancomycinSar/PG (c), and desleucyl-vancomycin/PG (d). Independent simulations for each system are labeled as s1, s2, s3, and s4. The black dashed line represents the averaged rmsd in each system.
Overall, vancomycin/PG and vancomycinNtoC/PG complexes exhibit more fluctuation patterns and a lower rmsd range compared to the vancomycinSar/PG and desleucyl-vancomycin/PG complexes, as shown in Figure 2, which suggests that the former two complexes have a lower flexibility and possibly a higher stability than the latter two. The average rmsd value for the vancomycin/PG is 5.48 Å (Figure 2a). In comparison, the average rmsd value for desleucyl-vancomycin/PG is 6.29 Å (Figure 2d), for vancomycinNtoC/PG is 5.22 Å (Figure 2b), and for vancomycinSar/PG is 5.27 Å (Figure 2c).
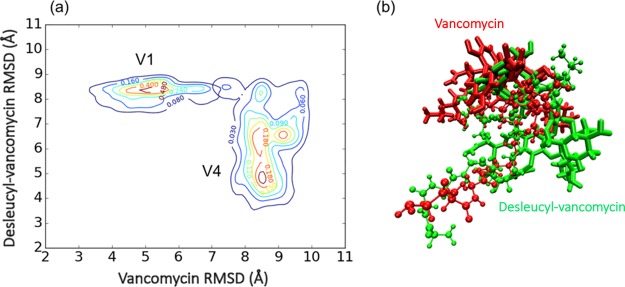
To explore and compare the conformational distribution of MD simulations of vancomycin and desleucyl-vancomycin complexes, 2D contour plots are generated to illustrate the conformational distribution of these two complexes (Figure 3). The reference structures for vancomycin/PG (red) and desleucyl-vancomycin/PG (green) complexes are shown in Figure 3b with a shared aligned peptide structure. Vancomycin and desleucyl-vancomycin complexes have significantly different conformational space, which is suggested by two completely different attraction basins on the 2D contour plot with very little overlap between the distributions (Figure 3a). In addition, the distribution for vancomycin/PG (V1) is much narrower than that of desleucyl-vancomycin/PG (V4). This is consistent with the observation that the simulations of the vancomycin/PG complex have generally lower rmsd than those of the desleucyl-vancomycin/PG complex (Figure 2). For the vancomycin/PG rmsd distribution (V1), only a single attraction basin is observed at (5.5, 8.5 Å). This is in contrast to the multiple basins observed for the rmsd distribution of the desleucyl-vancomycin/PG complex (V4) centered at (8.5, 4.5 Å), (8.5, 6.5 Å), and (9, 6.5 Å) (Figure 3a). The broad distribution with multiple basins for the desleucyl-vancomycin/PG complex supports that the PG binding by desleucyl-vancomycin is more flexible than that by vancomycin.
Figure 3.
Comparison between simulations of vancomycin and desleucyl-vancomycin. (a) Two-dimensional (2D) rmsd (Å) contour plot of the vancomycin and desleucyl-vancomycin MD simulations. The contour lines represent the density of trajectories located in the area. (b) Initial structures for MD simulations of vancomycin (red) and desleucyl-vancomycin (green) complexes as references for rmsd (Å) calculation. For each frame of simulations, two rmsd (Å) values are calculated and plotted in regard to the reference vancomycin/PG and desleucyl-vancomycin/PG complex structures.
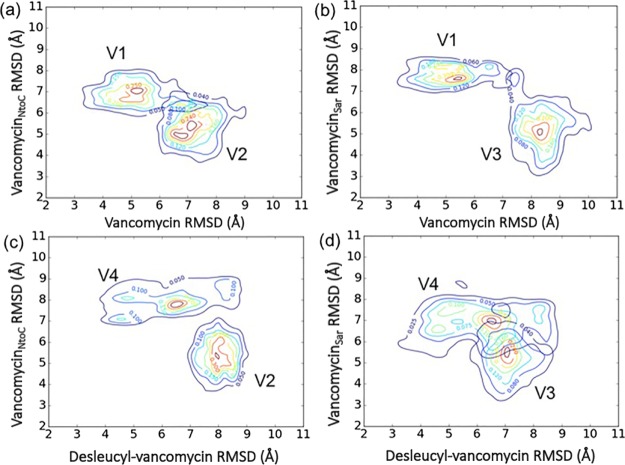
To further characterize all four glycopeptide–PG complexes, the comparative 2D rmsd distributions of vancomycin/PG (V1) and vancomycinNtoC/PG complexes (V2) are shown in Figure 4a, V1 and vancomycinSar/PG (V3) in Figure 4b, desleucyl-vancomycin/PG (V4) and V2 in Figure 4c, and V4 and V3 in Figure 4d. While the rmsd distributions for V1 and V2 show a significant overlap with attraction basins of each complex being close to each other (Figure 4a), the V1 and V3 distributions show only a minimal overlap with increased separation between the basins (Figure 4b). This indicates that the binding mode of the vancomycinNtoC/PG complex resembles more closely to that of vancomycin/PG than that of the vancomycinSar/PG complex. By contrast, the plots of vancomycinNtoC/PG (V2) and desleucyl-vancomycin/PG (V4) complex distributions show no overlap (Figure 4c). This suggests that the binding mode of vancomycinNtoC and PG resembles more closely to that of the vancomycin/PG complex than that of the desleucyl-vancomycin/PG complex. The desleucyl-vancomycin/PG complex distribution (V4), which overlaps with about one-third of the vancomycinSar/PG complex (V3) (Figure 4d), shows that these two complexes share significant conformational space.
Figure 4.
Comparative 2D rmsd (Å) contour plots of vancomycin/PG (V1) and desleucyl-vancomycin/PG (V4) complexes in regard to vancomycinNtoC/PG (V2) and vancomycinSar/PG (V3) complexes. Comparative 2D rmsd contour plots of (a) vancomycin vs vancomycinNtoC, (b) vancomycin vs vancomycinSar, (c) desleucyl-vancomycin vs vancomycinNtoC, and (d) desleucyl-vancomycin vs vancomycinSar. The rmsd (Å) values for each plot were calculated in regard to the reference structure, which is the initial structure for the MD simulation of each complex. The contour lines represent the density of trajectories that are located within the area.
2.2. Conformational Space for Glycopeptide–PG Complexes
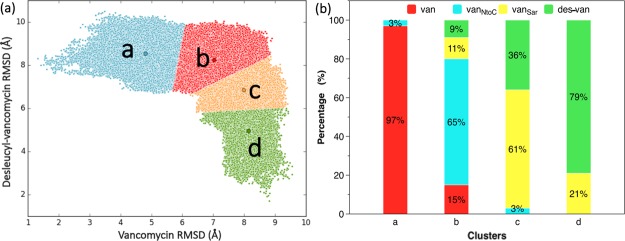
To elucidate the relationship among these four glycopeptide–PG complexes, the simulations of all four complexes are plotted on a same 2D rmsd plot using vancomycin/PG and desleucyl-vancomycin/PG complexes as the reference structures to characterize their distribution in overall conformational space. One of the advantages of this analysis is that it reveals the total conformational space for all glycopeptide–PG complexes. The overall shape of distributions is similar to the combined distribution of 2D rmsd plots shown in Figures 3 and 4. Four major clusters were identified from the overall distribution using k-means clustering analysis10 (Figure 5a). The shape and position of four clusters suggest some corresponding relations to the four glycopeptide–PG complexes.
Figure 5.
Overall sampling space and distribution of four glycopeptide–PG complexes. (a) Distribution of four glycopeptide–PG complexes in regard to vancomycin/PG and desleucyl-vancomycin/PG complexes as reference structures. The overall distribution is divided into four clusters (a, b, c, and d) using k-means clustering analysis. (b) Percentile of the contribution to each cluster in the panel (a) from simulations of each glycopeptide–PG complex: van, red (vancomycin); des-van, green (desleucyl-vancomycin); vanSar, yellow (vancomycinSar); and vanNtoC, blue (vancomycinNtoC).
The percentile contribution from the simulations of each glycopeptide–PG complex was calculated for each cluster and plotted in Figure 5b. Cluster “a” primarily corresponds to the conformational space of the vancomycin/PG complex (V1), as shown in Figures 4 and 5a. Thus, the vancomycin/PG complex distribution is the dominant contributor to cluster “a” (90%) with a minor contribution arising from the adjacent cluster “b”. In cluster “b”, the vancomycinNtoC/PG complex, which closely resembles the vancomycin/PG complex more than any other analogues, is the major contributor (65%) with minor contributions from the adjacent clusters “a” and “c” (3% to each). In cluster “c”, the vancomycinSar/PG complex distribution is the dominant contributor (61%). The contribution from cluster “d” to the complex (21%) is significantly higher than that of cluster “b” (11%). Finally, in cluster “d”, the desleucyl-vancomycin/PG complex is the dominant contributor (79%) with 21% contribution from the vancomycinSar/PG complex.
Interestingly, with an exception of vancomycin, which contributes to clusters “a” and “b” only, all three vancomycin analogues complexed with PG contribute to three clusters. For example, vancomycinNtoC contributes to clusters “a”, “b”, and “c” and vancomycinSar and desleucyl-vancomycin both contribute to clusters “b”, “c”, and “d”. This indicates that the vancomycin/PG complex has the narrowest distribution among all four complexes, and this suggests that it is also the most stable. This sequential distribution patterns exhibited by the vancomycin analogues, vancomycinNtoC and vancomycinSar (Figure 5b), are consistent with the vancomycin analogues representing the intermediate states for progressive transition in PG binding from vancomycin to desleucyl-vancomycin that results in the loss of PG-dipeptide binding.
2.3. Interaction between PG and Vancomycin Analogues
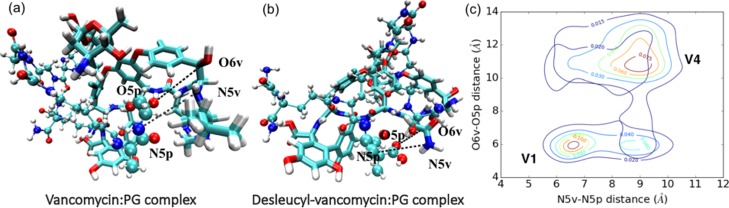
A crucial insight into the molecular interaction for vancomycin and desleucyl-vancomycin with PG-peptide stem units is obtained from the detailed analysis of key atomic pairs found in the model structures. Model structures of vancomycin and desleucyl-vancomycin bound to the PG representing the centers of attraction basins in contour plots of these two simulations are illustrated in Figure 6a,b, respectively. The distribution of two pairs of key atomic distances, between N5 from desleucyl-vancomycin and N5 in d-Ala-d-Ala of PG (N5v−N5p) and between O6 from desleucyl-vancomycin and O5 in d-Ala-d-Ala of PG (O6v–O5p), from the simulations is co-plotted for both vancomycin/PG (V1) and desleucyl-vancomycin/PG complexes (V4) in Figure 6c. The N5v–N5p and O6v–O5p distances represent key interactions between the aglycon structure of vancomycin and the bound d-Ala-d-Ala dipeptide of PG. The distributions of V1 and V4 do not co-localize on the 2D contour plot, indicating that vancomycin and desleucyl-vancomycin have an intrinsic difference in their interactions with d-Ala-d-Ala. The vancomycin/PG complex (V1) has a narrower distribution for N5v–N5p and O6v–O5p distances centered around (6.5, 6 Å) than desleucyl-vancomycin/PG (V4) centered around (9, 11 Å). The result from pair distance distribution simulations is consistent with the evidence that the conformational space distribution of vancomycin interaction with the d-Ala-d-Ala moiety of PG is narrower and significantly stronger (Figure 5b) than that of desleucyl-vancomycin. This provides molecular insights into the loss of antimicrobial activity in desleucyl-vancomycin as evidenced in the model structures shown in Figure 6. While vancomycin adopts a binding pocket conformation that is favorable for binding the d-Ala-d-Ala moiety of PG (Figure 6a), the glycon structure of desleucyl-vancomycin adopts an extended conformation that is not amenable for the dipeptide binding (Figure 6b).
Figure 6.
Key interaction between vancomycin and PG represented as atomic pair distance distributions. Model structures of vancomycin/PG (a) and desleucyl-vancomycin/PG (b) complexes. In (a,b), the stick structures represent (desleucyl-) vancomycin, and ball and stick structures represent PG units with the d-Ala-d-Ala part represented by a larger sphere. Color codes of ball and stick structures: oxygen (red), nitrogen (blue), hydrogen (white), carbon (cyan), and chlorine (green). The nomenclatures N5v and O6v refer to the amide nitrogen and the oxygen on the carbonyl carbon at the fifth residue in glycopeptides, respectively. N5p and O5p refer to the amide nitrogen and the oxygen from the d-Ala (fifth amino acid) in PG, respectively. (c) Contour plots of atomic pair distance distributions for the vancomycin/PG complex (V1) and desleucyl-vancomycin/PG complex (V4).
2.4. Hydrogen Bonds, Entropy, and Binding Free Energy
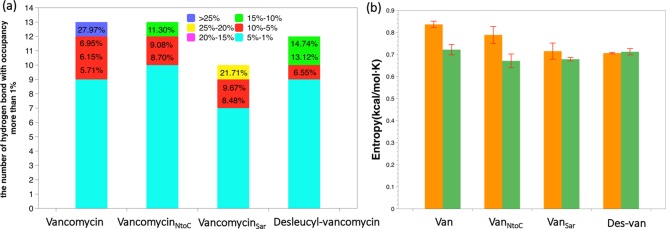
The total number of hydrogen bonds (H-bonds) in each of four analogues/PG complexes was estimated using VMD 1.9.2 with the donor–acceptor distance defined as within 3 Å with the angle cutoff as 20° within a linear (180°) configuration. Only the H-bonds with greater than 1% occupancy (stable for more than 1% of the simulations) were considered for the analysis. The total number of H-bonds formed during the simulations of four glycopeptide–PG complexes ranged from 10 to 13 (Figure 7a). Our simulations show that the vancomycin/PG complex had the largest number of H-bonds (five H-bonds) among all four complexes. The five H-bonds in the vancomycin/PG complex showed high occupancy (up to 27.97%) (Table 1), consistent with multiple H-bonds stabilizing the binding of the d-Ala-d-Ala to the aglycon of vancomycin. By contrast, the desleucyl-vancomycin/PG complex had the least number of H-bonds (one H-bond) with a relatively low partial occupancy (up to 14.74%) (Table 2). The specific hydrogen bonds with occupancy higher than 3.00% are listed in the Tables 1 and 2.
Figure 7.
(a) Number of hydrogen bonds found in the simulations of vancomycin, vancomycinNtoC, vancomycinSar, and desleucyl-vancomycin binding with PG. Only the hydrogen bonds that have more than 1% occupancy (being stable for more than 1% of the simulations for each complex) are considered in the analysis. (b) Configurational entropies of Van (vancomycin), VanNtoC (vancomycinNtoC), VanSar (vancomycinSar), and Des-van (desleucyl-vancomycin) (orange bar) and PG in four complexes (green bar). Error bars represent the standard deviation from four independent simulations.
Table 1. Atom Pairs of Hydrogen Bonds in the Vancomycin/PG Complex.
| donora | acceptora | occupancya (%) |
|---|---|---|
| Van-Side-N6 | Pep-Side-O2 | 27.97 |
| Pep-Side-O6 | Van-Side-O3 | 6.95 |
| Van-Side-N6 | Pep-Side-O3 | 6.15 |
| Van-Side-N6 | Pep-Side-O1 | 5.71 |
| Van-Side-N3 | Pep-Side-O5 | 4.87 |
| Van-Side-O7 | Pep-Side-N1 | 3.54 |
The donors, acceptors, and occupancy (>3.00%) and 59951 frames. “Van-Side” or “Pep-Side” represent where the atoms come from, van-side is for vancomycin, and pep-side is for peptide units.
Table 2. Atom Pairs of Hydrogen Bonds in the Desleucyl Vancomycin/PG Complexa.
| donor | acceptor | occupancy (%) |
|---|---|---|
| Des-van-Side-N6 | Pep-Side-O2 | 14.74 |
| Pep-Side-N4 | Des-van-Side-O3 | 13.12 |
| Des-van-Side-O11 | Pep-Side-O3 | 6.55 |
The donors, acceptors, and occupancy (>3.00%) and 59951 frames. “Des-van-Side” or “Pep-Side” represent where the atoms come from, “Des-van-side” is for desleucyl-vancomycin, and “Des-pep-side” is for peptide units.
We also calculated the configurational entropies of the four complexes (Figure 7b and Table 3). For the comparison, the configurational entropy for each of the vancomycin analogue and PG was calculated separately. Interestingly, vancomycin has the largest configurational entropy (836.6 cal/mol·K) among four analogues with desleucyl-vancomycin as the least (706.4 cal/mol·K). Although vancomycinNtoC has greater number of atoms than vancomycin, it has smaller entropy than vancomycin. The calculated entropies show the decreasing trend from vancomycin, vancomycinNtoC, and vancomycinSar to desleucyl-vancomycin. A similar trend is also observed for the normalized entropies, where the calculated entropies are divided by the number of atoms or by the atomic mass of each analogue (Table 3), revealing intrinsic properties of these analogues. The calculated entropies in Table 3 are for the glycopeptides only without PG binding, whereas the calculated rmsd values in Figures 2 through 5 are for the glycopeptide–PG complexes. We observed that vancomycin with the highest entropy when bound to PG results in a vancomycin/PG complex with the lowest rmsd. Likewise, the desleucyl-vancomycin with the lowest entropy when bound to PG results in a desleucyl-vancomycin/PG complex with the highest rmsd. Hence, the low entropy of a glycopeptide does not necessarily correlate with the low rmsd in glycopeptide–PG complexes and vice versa.
Table 3. Configurational Entropies of Glycopeptide–PG Complexesa.
| vancomycin | vancomycinNtoC | vancomcyinSar | desleucyl-vancomycin | |
|---|---|---|---|---|
| number of atoms | 282 | 283 | 270 | 260 |
| molar mass of molecules | 1450 | 1449 | 1394 | 1322 |
| configurational entropyb | 836.6 | 788.9 | 715.1 | 706.4 |
| normalization entropyb (divided by the number of atoms) | 2.97 | 2.79 | 2.65 | 2.72 |
| normalization entropyb (divided by atomic mass) | 0.577 | 0.544 | 0.512 | 0.534 |
The first row is the number of atoms from vancomycin, vancomycinNtoC, vancomycinSar, and desleucyl vancomycin.
The unit of configurational entropy is calorie/mol·K.
The binding free energy was calculated using the molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) method to further compare the stability of vancomycin and desleucyl-vancomycin binding with PG. The use of distance restraints during the simulations prevented the accurate calculation of the binding free energy; however, all simulations were subjected to the same distance restraints, and thus the effects of distance restraints on the binding free energy are likely to be comparable for all complexes. The estimated binding free energies of vancomycin/PG and desleucyl-vancomycin/PG complexes are −37.84 ± 2.16 and −11.17 ± 16.07 kcal/mol, respectively. Hence, PG binding to vancomycin is significantly more favorable than to desleucyl-vancomycin by a ΔΔG of approximately −27 kcal/mol. Despite the error in the binding free energy calculation associated with large variance in trajectories, a significant difference in ΔΔG is consistent with the stable vancomycin/PG complex compared to the desleucyl-vancomycin/PG complex.
3. Discussions
The X-ray crystal11,12 and solution NMR structures of vancomycin and related glycopeptides, with an exception of ristocetin,13,14 are found as dimers or oligomers. Hence, drug dimerization is thought to play an important role in glycopeptide mode of action.15,16 However, the in situ characterization of disaccharide-modified glycopeptides, including oritavancin that readily forms drug dimers in solution, when complexed to intact whole cells and isolated cell walls of S. aureus are found as monomers without dimerization.17−24 Glycopeptide antibiotics binding to PG in cell walls as a monomer is due to complex multivalent interactions among the cell wall glycans with drug sugar disaccharide and glycopeptide interactions with the non-d-Ala-d-Ala segment of the PG-stem structure.25 To characterize the monomeric glycopeptide–PG interactions, MD simulations of vancomycin and its three N-terminus-modified vancomycin derivatives bound to a PG-stem unit, l-Ala-d-iso-Gln-l-Lys-d-Ala-d-Ala, with a (Gly)5 bridge attached, were carried out.
The first amino acid in type I glycopeptide antibiotics, which include vancomycin, chloroeremomycin, and eremomycin, is N-methyl-leucine. N-methyl-leucine is essential for the dipeptide binding, where the removal by Edman degradation results in a hexapeptide26 with 100-fold reduction in dipeptide binding affinity and the loss of antimicrobial activities.27 Accordingly, our simulations show that the desleucyl-vancomycin/PG complex in general is less ordered than the vancomycin/PG complex. For example, the average rmsd value for desleucyl-vancomycin/PG simulation was significantly larger and had much broader rmsd distribution than that for vancomycin/PG simulation (Figure 2). This indicated that the desleucyl-vancomycin had greater conformational space with more flexible PG binding than vancomycin. Moreover, rmsd distributions in the 2D contour plot of desleucyl-vancomycin and vancomycin did not share a common space, indicating that desleucyl-vancomycin has a distinct binding structure. The desleucyl-vancomycin/PG complex, in comparison to the vancomycin/PG complex, showed reduced binding energy of approximately 27 kcal/mol (Figure 3). The key atomic distance distributions of selected atoms in the bound dipeptide to the aglycon structure of vancomycin and desleucyl-vancomycin (Figure 6c) further reveal that the d-Ala-d-Ala moiety of PG has a much tighter binding mode with vancomycin than desleucyl-vancomycin. The superimposed model structures of vancomycin and desleucyl-vancomycin bound to PG (Figure 6b) show that the binding cleft of desleucyl-vancomycin exhibits an extended conformation.
The peptide core of vancomycin is highly cross-linked to form a rigid structure. By contrast, the side chain of Asn and N-methyl-Leu is highly flexible in the absence of d-Ala-d-Ala binding. This flexibility is crucial for the dipeptide binding as these side chains are thought to function as “flaps” to swing into the ligand-binding site as a surrogate in the absence of a ligand.11 The partial occupancy of the binding cleft by the Asn and N-methyl-Leu side chains is thought to prevent the hydration of the aglycon structure. In the presence of a ligand, the side chains of Asn and N-methyl-Leu facilitate the desolvation of the binding cleft necessary for the d-Ala-d-Ala binding.12 Following the d-Ala-d-Ala binding, the side chains of Asn and N-methyl-Leu become rigid.28 Our rmsd distribution analysis shows that the replacement of N-methyl-Leu by a shortened side-chain length significantly diminished PG binding and its stability. The order of the most stable to the least stable glycopeptide–PG complexes are: vancomycin, vancomycinNtoC, vancomycinSar, and desleucyl-vancomycin. In the case of vancomycinSar, replacing the positively charged N-methyl-leucine with an ethyl moiety in the aglycon structure interferes with the binding to the carboxyl terminus of the PG dipeptide.28 The calculated glycopeptide stability correlated with the overall strength of hydrogen bonding interactions with the highest for the vancomycin/PG complex and the least for the desleucyl-vancomycin/PG complex (Figure 7a). The changes in entropy, despite the shared hydrogen bonds, indicated that the highest entropy, corresponding to the lowest free energy, could be the crucial determinant for ligand binding. We anticipate that the glycopeptide–PG interactions are not local but involves multiple interactions between the aglycon structure and the non-d-Ala-d-Ala segment of the PG-stem structure that are likely to be cooperative and critical for the overall effectiveness of PG binding by the glycopeptide antibiotics. Our study provides a new approach to characterize the complex interactions between the PG and glycopeptide antibiotics that will facilitate the design and development of novel antibiotics against the emerging multidrug-resistant Gram-positive pathogens.
4. Conclusions
In this study, we built simplified simulation models to study the molecular mechanism of vancomycin as an antibiotic through binding with bacterial cell wall structures. The Edman degradation of vancomycin cleaves the first residue from the aglycon structure, resulting in desleucyl-vancomycin with a damaged binding pocket and devoid of any antimicrobial activities. To reveal the atomic details of vancomycin and cell wall peptide structure interactions, two vancomycin derivatives with chemical modification of the first residue in the aglycon structure were built as intermediate analogues between vancomycin and desleucyl-vancomycin and referred to as vancomycinNtoC and vancomycinSar. Using MD simulations, we show that the binding in vancomycin/PG is the tightest among all four analogues. The rmsd distribution analyses revealed the continuous conformational distribution among these four analogues from vancomycin to desleucyl-vancomycin. Through atomic distance analyses, it is also suggested that the d-Ala-d-Ala segment of the PG-stem structure binds more favorably with vancomycin than its desleucyl analogue. The calculated entropies and binding free energies of four complexes also displayed a consistent trend from the most favorable binding of vancomycin against the cell wall peptide to the least favorable binding of desleucyl-vancomycin. Overall, the MD simulations provided a new approach to provide insights for the development of novel glycopeptide antibiotics with improved antimicrobial activities against the evolving glycopeptide resistance in pathogens.
5. Computational Methods
5.1. MD Simulations
The structures of vancomycin and its derivatives (Figure 1) complexed with the PG-peptide repeat unit were based on the computational models from a previous study25,29 and optimized to the B3LYP/6-311G(d,p) level of theory using the Gaussian09 program package.30 The CHARMM General Force Field (CGenFF) for the simulation system was generated using the online server ParamChem (https://cgenff.paramchem.org/).31 Atomic charges of the simulation system were taken from the Gaussian calculations. All systems were solvated in a water box using a TIP3P model32 with the addition of sodium and chlorine as charge-balancing ions. Simulation box sizes, number of ions, and the ionic strength are listed in Table 4. The following equation was used to calculate the ionic strength based on the simulation box size and number of ions
| 1 |
where i is the ion identification number and z represents the charge of the ion.
Table 4. Ionic Strengths of Vancomycin/PG, VancomycinNtoC/PG, VancomycinSar/PG, and Desleucyl Vancomycin/PG Complexes.
| vancomycin | vancomycinNtoC | vancomcyinSar | desleucyl-vancomycin | |
|---|---|---|---|---|
| ionic strength (mol·L–1) | 84.9 | 84.8 | 85.2 | 71.0 |
| number of ions | 4Cl–, 6Na+ | 4Cl–, 6Na+ | 4Cl–, 6Na+ | 4Cl–, 6Na+ |
| box size (Å) | 46.1 | 46.1 | 46.0 | 48.9 |
The simulation boxes were subjected to 200 steps of the steepest descent energy minimization and then further energy was minimized using the adopted basis Newton–Raphson method until the total gradient of the system was lower than 0.03 kcal/(mol·Å). Subsequently, the minimized simulation systems were subjected to 24 ps of the MD simulations at a temperature of 300 K as the equilibrium. Then, the MD simulation of the system was run for 30 ns via an isothermal–isobaric ensemble at 300 K and 1 atm. The time step for MD simulations is 2 fs, with all the bonds associated with hydrogen being fixed during the simulation. The cutoff distance for the nonbonded interaction is 12 Å and using a Nosé–Hoover thermostat33,34 to keep the temperature of the system at 300 K for 30 ns. The leapfrog Verlet scheme was used for the integration of the atomic velocities and coordinates in simulations. All simulations used periodic boundary conditions, and electrostatic interactions were modeled using the particle mesh Ewald method.35 Three atomic pair distance restraints were applied during the simulations based on solid-state NMR experiments.36,37 All simulations were carried out using a MM simulation program, CHARMM version 40b1.38
5.2. Root-Mean-Square Deviation (rmsd)
The rmsd is used to measure the difference of the conformation for each snapshot of the MD simulations from the reference structure. For a molecular structure represented by a Cartesian coordinate vector ri (i = 1 to N) of N atoms, the rmsd is calculated as follows
| 2 |
The Cartesian coordinate vector ri0 is the ith atom in the reference structure. The transformation matrix U is defined as the best-fit alignment between the vancomycin/PG complex structures along trajectories in respect of the reference structure.
5.3. Cross-Correlation Matrix
The correlation of motion between all atomic pairs in each simulation was measured through a cross-correlation matrix. The element Cij of the cross-correlation matrix C, which measures the correlation between the movement of atoms i and j in the simulation, is defined as
| 3 |
where ri and rj are Cartesian coordinate vectors from the least-square fitted structures in MD simulation trajectories. It should be noted that the least-square fitting of the MD trajectory effectively project out the translational and rotational motions of the vancomycin and its analogues complexed with the peptide. The normalized matrix elements Cij have their values ranging between −1 and 1. A positive Cij value associates with an overall positive correlation between atoms i and j, and a negative value corresponds to a negative correlation.
5.4. Configurational Entropy
Entropy was estimated for the simulation systems using quasi-harmonic approximations based on MD simulations. Quasi-harmonic analysis was carried out through the inversion of the cross-correlation matrix C
| 4 |
In eq 4, Fij is the element of the force constant matrix F describing the quasi-harmonic potential,39kB is the Boltzmann constant, and T is the temperature.
Configurational entropy Sconfig of the simulation system could be calculated using the vibration frequency ω of the molecule with N atoms
| 5 |
ℏ is the reduced Planck constant.
The vibration frequency ω in the quasi-harmonic model of the molecule on the effective quasi-harmonic potential can be calculated through the solution of the secular equation
| 6 |
where M is the mass matrix of the molecule.
5.5. MM/PBSA Binding Free Energy Calculation
Total free energy of binding ΔGbinding was computed using the MM/PBSA method. This method uses a thermodynamic cycle to calculate the free energy of binding for vancomycin and its analogues against the PG peptide. The free energies of binding are computed using the equation
| 7 |
where ΔGbindingsol is the total free energy of binding in solution and ΔGcomplex, ΔGvansol, and ΔGPG are free energies in solution of the complex, vancomycin/analogues, and PG, respectively. The free energy in solution of each entity (ΔGsol) is calculated by the following equations
| 8 |
| 9 |
| 10 |
where ΔGgas is the free energy in gas phase and ΔGsolvation is the solvation energy. ΔGgas is the sum of the internal energy (Einternal), van der Waals (EvdW) and Coulombic (Eelectrostatic) interactions, as well as entropic contributions (ΔS). The internal energy includes bond stretching, bond angle, and torsional contributions to the total MM energies. The solvation energy ΔGsolvation includes polar (ΔGPB) and nonpolar (ΔGnonpolar) contributions.
Acknowledgments
This work was partially supported by the Edward R. Biehl Graduate Fellowship (H.Z.), SMU Dean’s Research Council research grant (P.T.). Computational time was provided by Southern Methodist University’s Center for Scientific Computation and Texas Advanced Computing Center (TACC) at the University of Texas at Austin. S.J.K. was supported by the National Institutes of Health under grant number GM116130.
The authors declare no competing financial interest.
References
- Wright G. D.; Walsh C. T. d-Alanyl-d-alanine ligases and the molecular mechanism of vancomycin resistance. Acc. Chem. Res. 1992, 25, 468–473. 10.1021/ar00022a006. [DOI] [Google Scholar]
- Gokhale R. S.; Tsuji S. Y.; Cane D. E.; Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science 1999, 284, 482–485. 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- Nagarajan R. Structure-activity relationships of vancomycin-type glycopeptide antibiotics. J. Antibiot. 1993, 46, 1181–1195. 10.7164/antibiotics.46.1181. [DOI] [PubMed] [Google Scholar]
- Barna J. C. J.; Williams D. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 1984, 38, 339–357. 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- Arthur M.; Molinas C.; Bugg T. D.; Wright G. D.; Walsh C. T.; Courvalin P. Evidence for in vivo incorporation of D-lactate into peptidoglycan precursors of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 1992, 36, 867–869. 10.1128/aac.36.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg T. D. H.; Wright G. D.; Dutka-Malen S.; Arthur M.; Courvalin P.; Walsh C. T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 1991, 30, 10408–10415. 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- Xing B.; Jiang T.; Wu X.; Liew R.; Zhou J.; Zhang D.; Yeow E. K. L. Molecular interactions between glycopeptide vancomycin and bacterial cell wall peptide analogues. Chem.—Eur. J. 2011, 17, 14170–14177. 10.1002/chem.201102195. [DOI] [PubMed] [Google Scholar]
- Bischoff D.; Pelzer S.; Holtzel A.; Nicholson G. J.; Stockert S.; Wohlleben W.; Jung G.; Sussmuth R. D. The Biosynthesis of Vancomycin-Type Glycopeptide Antibiotics—New Insights into the Cyclization Steps. Angew. Chem., Int. Ed. 2001, 40, 1693–1696. . [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M.; Jones P. G.; Kennard O.; Williams D. H.; Smith G. A. Structure of vancomycin and its complex with acetyl-D-alanyl-D-alanine. Nature 1978, 271, 223–225. 10.1038/271223a0. [DOI] [PubMed] [Google Scholar]
- Lloyd S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. 10.1109/tit.1982.1056489. [DOI] [Google Scholar]
- Schäfer M.; Schneider T. R.; Sheldrick G. M. Crystal structure of vancomycin. Structure 1996, 4, 1509–1515. 10.1016/s0969-2126(96)00156-6. [DOI] [PubMed] [Google Scholar]
- Loll P. J.; Bevivino A. E.; Korty B. D.; Axelsen P. H. Simultaneous recognition of a carboxylate-containing ligand and an intramolecular surrogate ligand in the crystal structure of an asymmetric vancomycin dimer. J. Am. Chem. Soc. 1997, 119, 1516–1522. 10.1021/ja963566p. [DOI] [Google Scholar]
- Batta G.; Sztaricskai F.; Kövér K. E.; Rüdel C.; Berdnikova T. F. An NMR study of eremomycin and its derivatives. Full 1H and 13C assignment, motional behavior, dimerization and complexation with Ac-D-Ala-D-Ala. J. Antibiot. 1991, 44, 1208–1221. 10.7164/antibiotics.44.1208. [DOI] [PubMed] [Google Scholar]
- Groves P.; Searle M. S.; Mackay J. P.; Williams D. H. The structure of an asymmetric dimer relevant to the mode of action of the glycopeptide antibiotics. Structure 1994, 2, 747–754. 10.1016/s0969-2126(94)00075-1. [DOI] [PubMed] [Google Scholar]
- Cristofaro M. F.; Beauregard D. A.; Yan H.; Osborn N. J.; Williams D. H. Cooperativity between non-polar and ionic forces in the binding of bacterial cell wall analogues by vancomycin in aqueous solution. J. Antibiot. 1995, 48, 805–810. 10.7164/antibiotics.48.805. [DOI] [PubMed] [Google Scholar]
- Jia Z.; O’Mara M. L.; Zuegg J.; Cooper M. A.; Mark A. E. Vancomycin: ligand recognition, dimerization and super-complex formation. FEBS J. 2013, 280, 1294–1307. 10.1111/febs.12121. [DOI] [PubMed] [Google Scholar]
- Kim S. J.; Cegelski L.; Studelska D. R.; O’Connor R. D.; Mehta A. K.; Schaefer J. Rotational-echo double resonance characterization of vancomycin binding sites in Staphylococcus aureus. Biochemistry 2002, 41, 6967–6977. 10.1021/bi0121407. [DOI] [PubMed] [Google Scholar]
- Kim S. J.; Cegelski L.; Preobrazhenskaya M.; Schaefer J. Structures of Staphylococcus aureus cell-wall complexes with vancomycin, eremomycin, and chloroeremomycin derivatives by 13C{19F} and 15N{19F} rotational-echo double resonance. Biochemistry 2006, 45, 5235–5250. 10.1021/bi052660s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J.; Cegelski L.; Stueber D.; Singh M.; Dietrich E.; Tanaka K. S.; Parr T. R.; Far A. R.; Schaefer J. Oritavancin exhibits dual mode of action to inhibit cell-wall biosynthesis in Staphylococcus aureus. J. Mol. Biol. 2008, 377, 281–293. 10.1016/j.jmb.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J.; Matsuoka S.; Patti G. J.; Schaefer J. Vancomycin derivative with damaged D-Ala-D-Ala binding cleft binds to cross-linked peptidoglycan in the cell wall of Staphylococcus aureus. Biochemistry 2008, 47, 3822–3831. 10.1021/bi702232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J.; Schaefer J. Hydrophobic side-chain length determines activity and conformational heterogeneity of a vancomycin derivative bound to the cell wall of Staphylococcus aureus. Biochemistry 2008, 47, 10155–10161. 10.1021/bi800838c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J.; Singh M.; Schaefer J. Oritavancin binds to isolated protoplast membranes but not intact protoplasts of Staphylococcus aureus. J. Mol. Biol. 2009, 391, 414–425. 10.1016/j.jmb.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J.; Tanaka K. S. E.; Dietrich E.; Far A. R.; Schaefer J. Locations of the hydrophobic side chains of lipoglycopeptides bound to the peptidoglycan of Staphylococcus aureus. Biochemistry 2013, 52, 3405–3414. 10.1021/bi400054p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J.; Singh M.; Sharif S.; Schaefer J. Desleucyl-oritavancin with a damaged d-Ala-d-Ala binding site inhibits the transpeptidation step of cell-wall biosynthesis in whole cells of Staphylococcus aureus. Biochemistry 2017, 56, 1529–1535. 10.1021/acs.biochem.6b01125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.; Zhou H.; Preobrazhenskaya M.; Tao P.; Kim S. J. The carboxyl terminus of eremomycin facilitates binding to the non-d-Ala-d-Ala segment of the peptidoglycan pentapeptide stem. Biochemistry 2016, 55, 3383–3391. 10.1021/acs.biochem.6b00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth P. M.; Williams D. H. Preparation and conformational analysis of vancomycin hexapeptide and aglucovancomycin hexapeptide. J. Chem. Soc., Perkin Trans. 1 1989, 2335–2339. 10.1039/p19890002335. [DOI] [Google Scholar]
- Allen N. E.; LeTourneau D. L.; Hobbs J. N. Jr.; Thompson R. C. Hexapeptide derivatives of glycopeptide antibiotics: tools for mechanism of action studies. Antimicrob. Agents Chemother. 2002, 46, 2344–2348. 10.1128/aac.46.8.2344-2348.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z.; O’Mara M. L.; Zuegg J.; Cooper M. A.; Mark A. E. Vancomycin: ligand recognition, dimerization and super-complex formation. FEBS J. 2013, 280, 1294–1307. 10.1111/febs.12121. [DOI] [PubMed] [Google Scholar]
- Jusuf S.; Axelsen P. H. Synchronized conformational fluctuations and binding site desolvation during molecular recognition. Biochemistry 2004, 43, 15446–15452. 10.1021/bi048357t. [DOI] [PubMed] [Google Scholar]
- Frisch M.; Trucks G.; Schlegel H. B.; Scuseria G.; Robb M.; Cheeseman J.; Scalmani G.; Barone V.; Mennucci B.; Petersson G.. Gaussian 09, revision a. 02, Gaussian. Inc.: Wallingford, CT, 2009, 200.
- Vanommeslaeghe K.; Hatcher E.; Acharya C.; Kundu S.; Zhong S.; Shim J.; Darian E.; Guvench O.; Lopes P.; Vorobyov I.; Mackerell A. D. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Evans D. J.; Holian B. L. The nose–hoover thermostat. J. Chem. Phys. 1985, 83, 4069–4074. 10.1063/1.449071. [DOI] [Google Scholar]
- Tobias D. J.; Martyna G. J.; Klein M. L. Molecular dynamics simulations of a protein in the canonical ensemble. J. Phys. Chem. 1993, 97, 12959–12966. 10.1021/j100151a052. [DOI] [Google Scholar]
- Brooks B. R.; Bruccoleri R. E.; Olafson B. D.; States D. J.; Swaminathan S.; Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. 10.1002/jcc.540040211. [DOI] [Google Scholar]
- Singh M.; Kim S. J.; Sharif S.; Preobrazhenskaya M.; Schaefer J. REDOR constraints on the peptidoglycan lattice architecture of Staphylococcus aureus and its FemA mutant. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 363–368. 10.1016/j.bbamem.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J.; Chang J.; Singh M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 350–362. 10.1016/j.bbamem.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T. A. The representation of van der Waals (vdW) interactions in molecular mechanics force fields: potential form, combination rules, and vdW parameters. J. Am. Chem. Soc. 1992, 114, 7827–7843. 10.1021/ja00046a032. [DOI] [Google Scholar]
- Levy R. M.; Srinivasan A. R.; Olson W. K.; McCammon J. A. Quasi-harmonic Method for Studying Very Low Frequency Modes in Proteins. Biopolymers 1984, 23, 1099–1112. 10.1002/bip.360230610. [DOI] [PubMed] [Google Scholar]