Abstract
Anti-biofilm assay guided fractionation of the marine sponge Stylissa massa revealed the butanol soluble fraction that was possessing the inhibitory activity toward the biofilm formation of bacterium E. coli. Chromatographic separation of the bioactive fraction resulted in the isolation of 32 bromopyrrole alkaloids, including six new alkaloids, named stylisines A–F (1–6). The structures of new alkaloids were established by comprehensive analyses of the two-dimensional (2D) NMR (COSY, HMQC, and HMBC) and the high resolution electrospray ionization mass spectroscopy (HRESIMS) data, while the absolute configurations were determined by the X-ray diffraction and the electronic circular dichroism (ECD) data. Bioassay results indicated that phakellin-based alkaloids, including dibromoisophakellin and dibromophakellin, significantly reduced the biofilm formation of the bacterium E. coli. Present work provided a group of new natural scaffolds for the inhibitory effects against the biofilm formation of E. coli.
Keywords: marine sponge, Stylissa massa, antimicrobial activity, antibiofilm, structure elucidation
1. Introduction
Planktonic bacteria attached onto solid surfaces, creating a complex community of bacteria to form a biofilm, which protects bacteria against antibiotics and causes many recalcitrant infections, as well as resistance to antibiotics [1,2,3,4]. Biofilms account for >80% of human bacterial infections, and a biofilm matrix resists antibiotics at concentrations over 1000 times higher than conventionally used doses [5,6,7]. In nature, most bacteria are likely to form surface-attached biofilm communities as a survival strategy. Pathogenic biofilms pose a challenge because they enhance resistance not only to conventional antibiotics, but also to host defenses and external stresses. Thus, they are difficult to control in medical and industrial settings. So far, no specific biofilm inhibitor is commercially available, while bacterial biofilms remain a frontier of microbiology as they exhibit important tolerance and resistance in many aspects. Currently, an explosive amount of biofilm research is being conducted to discover novel compounds that are capable of inhibiting biofilms, without allowing bacteria to develop drug resistance. Marine benthic invertebrates have been proved a rich source of bioactive compounds with multiple ecological functions. Among the sessile organisms, sponges are the marine sessile filter feeders that are exposed to large amounts of bacteria and also viruses in the surrounding seawater. Because bacteria play a key role in microfilm formation [8], it becomes vital for sponges to regulate bacterial epibiosis on their surfaces. One epibacterial defense strategy for sponge-symbiont community is to produce bioactive secondary metabolites in order to outcompete other bacteria in the water column from causing infection or biofouling [9,10,11,12,13,14]. Among the sponge-derived active metabolites, pyrrole-imidazole alkaloids are a class of structurally unique natural products with a range of ecological and significant pharmacological bioactivities [15,16,17]. For instance, oroidin and stevensine were demonstrated to possess antifeedant properties, while oroidin and its analogues have been extensively studied as biofilm inhibitors [18,19,20,21]. In our research program for the discovery of bioactive natural products that are derived from marine sponges, HPLC-DAD-MS data and bioassay guided examination of the sponge extracts revealed that the n-BuOH extracts of the sponge Stylissa massa contain a profile of brominated alkaloids, and possess an inhibitory effect against the biofilm formation of E. coli. Chromatographic separation of the alkaloid-containing extracts resulted in the isolation of 32 bromopyrrole alkaloids, including six new compounds, namely stylisines A–F (1–6) (Figure 1). The antibacterial and anti-biofilm effects of the isolated alkaloids were evaluated.
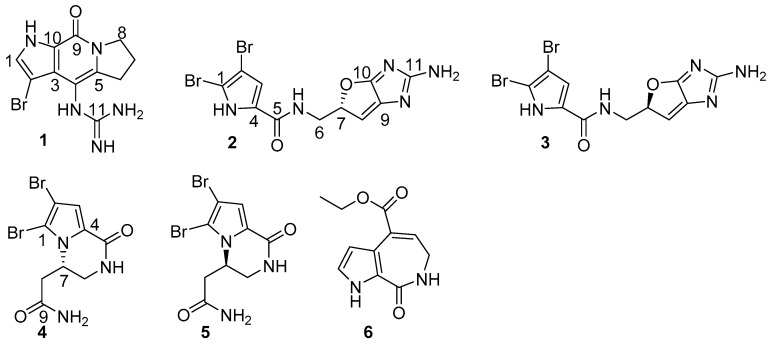
Figure 1.
Structures of stylisines A–F (1–6).
2. Results
The sponge tissue was extracted with 95% alcohol, and the crude extract was partitioned between 40% MeOH-H2O and CH2Cl2 to remove lipids. The water soluble extract was then partitioned between H2O and n-BuOH. The 1H NMR spectrum in association with the LC-ESIMS data of n-BuOH extract characterized a profile of brominated alkaloids. Chromatographic separation, including the semi-preparative HPLC purification, conduced the isolation of 32 bromopyrrole derivatives, including six new alkaloids (Figure 1).
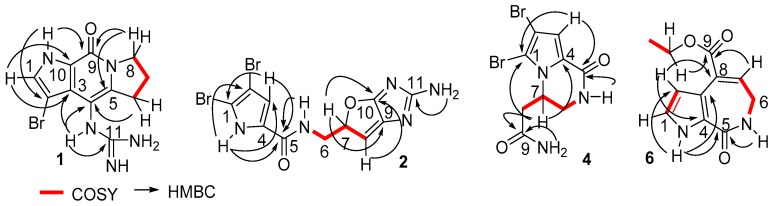
Stylisine A (1) was obtained as a light yellow amorphous solid. Its molecular formula was determined as C11H13N5OBr on the basis of the high resolution electrospray ionization mass spectroscopy (HRESIMS) and NMR data. The pseudomolecular ion peaks at m/z 310 and 312, with a ratio of 1:1 in the ESIMS spectrum also supported a bromine atom in the molecule. The 13C NMR and distortionless enhancement by polarization transfer (DEPT) spectra exhibited 11 carbon resonances, which were classified into three methylenes, one methine, and seven quaternary carbons. The COSY in association with the HMBC correlations established a tetrahydropyrroloindolizinone nucleus, as evident from the COSY relationships for an alkyl spin system from H2-6 (δH 2.87, 2.99) to H2-8 (δH 3.98, 4.10), in addition to the HMBC correlations from the pyrrole proton H-1 (δH 7.46, brs) to C-3 (δC 123.1) and C-10 (δC 126.4), NH (δH 12.54, brs, pyrrole) to C-2 (δC 86.8), C-3, and C-9 (δC 152.6), and between H2-8/C-9 and H2-6/C-4 (δC 102.8). The remaining moiety involved a quaternary carbon at δC 157.5 (C-11) and three nitrogen atoms, which was characteristic of a guanidine unit that was substituted at C-4 due to the HMBC correlations from a NH (9.16, s) to C-3, C-4, C-5 (δC 142.2), and C-11 (Figure 2). In addition, the significantly shielded carbon C-2 (δC 86.8) was attributed to a bromine atom substitution.
Figure 2.
Key COSY and HMBC correlations of 1, 2, 4 and 6.
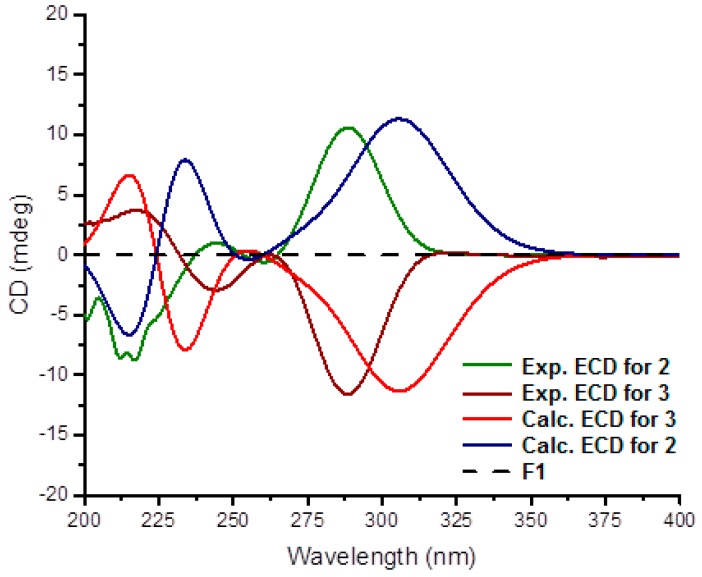
Stylisine B (2) was obtained as a yellow amorphous solid. Its ESIMS data presented the ion peaks at m/z 401.9, 403.9, and 405.9 [M + H]+ with the ratio of 1:2:1, indicating two bromine atoms in the molecule, while the HRESIMS and NMR data provided a molecular formula of C11H10N5O2Br2. Analyses of the two-dimensional (2D) NMR data revealed that compound 2 consists of two units of substructures. The first unit was determined to be a pyrrole-2-carboxamide, based on the HMBC correlations from H-3 (δH 6.95, s) to C-1 (δC 105.2) and a carbonyl carbon C-5 (δC 159.5), and from NH (δH 12.68, s, pyrrole) to C-2 (δC 98.4), C-3 (δC 113.5), and C-5. The second unit was assigned to a furoimidazole nucleus, according to additional HMBC correlations from H-7 (δH 4.37, m) to C-9 (δC 128.6) and C-10 (δC 153.5), and H-8 (δH 5.76, s) to C-7 (δC 74.0) and C-10. An amine group to be positioned at C-11 (δC 159.5) was ascribed by the HMBC correlation between NH2 (δH 9.23, s) and C-11. The connection of two units across a methylene group to form an amide was clarified by the COSY relationships from H2-6 (δH 3.43, m) to an amide NH (δH 8.15, t, J = 6.0 Hz) and H-7, along with the HMBC correlations from C-5 to the amide proton and H2-6. The significantly shielded quaternary carbons C-1 and C-2 were indicative of the substitution of bromine atoms at C-1 and C-2, respectively [22]. The absolute configuration at C-7 was determined on the basis of the comparison of the experimental and calculated electronic circular dichroism (ECD) data. The experimental ECD spectrum of 2 exhibited the positive Cotton effects (CE) at 240 and 280 nm, and negative CE at 230 nm, which were in accordance with the calculated ECD data for the model compound of 2 with 7S configuration, using the time-dependent density functional theory (TDDFT) method (Figure 3) [23,24]. Thus, C-7 of 2 was in agreement with S configuration.
Figure 3.
Experimental and calculated electronic circular dichroism (ECD) data for 2 and 3 in MeOH.
The planer structure of stylisine C (3) was the same as that of 2, as determined by the duplicated NMR (Table 1) and MS data. However, the specific rotation of 2 ([α] +30.8, MeOH) showed the opposite phase to that of 3 ([α] − 31.2, MeOH), suggesting 3 to be a stereoisomer of 2. This assumption was confirmed by the opposite Cotton effects of 3 in comparison with those of 2 in the ECD spectra (Figure 3). The comparable Cotton effects of the experimental ECD of 3 and the calculated ECD for the 7R isomer of 2, indicated the absolute configuration of 3 to be 7R.
Table 1.
1H and 13C NMR data of 1–6 in dimethyl sulfoxide (DMSO-d6).
| 1 | 2/3 | 4/5 | 6 | |||||
|---|---|---|---|---|---|---|---|---|
| 1 | 7.46, d (2.8) | 127.38, CH | 105.20, C | 105.67, C | 6.98, dd (2.4, 2.8) | 121.52, CH | ||
| 2 | 86.79, C | 98.36, C | 99.24, C | 6.52, dd (2.0, 2.4) | 109.32, CH | |||
| 3 | 123.14, C | 6.95, s | 113.46,CH | 6.84, s | 113.86, CH | 119.98, C | ||
| 4 | 102.79, C | 128.36, C | 125.81, C | 126.22, C | ||||
| 5 | 142.18, C | 159.51, C | 157.56, C | 163.29, C | ||||
| 6 | 2.99, m; 2.87, m | 28.42, CH2 | 3.43, m | 42.36, CH2 | 3.77, dd (2.0, 13.20) 3.34, dd (5.2, 13.2) |
42.06, CH2 | 3.48, dd (4.8, 7.2) | 37.60, CH2 |
| 7 | 2.15, m | 21.56, CH2 | 4.37, m | 74.01, CH | 4.64, m | 50.59, CH | 6.84, t (7.2) | 132.54, CH |
| 8 | 4.10, m; 3.98, m | 48.27, CH2 | 5.76, brs | 115.41, CH | 2.63, dd (11.2, 12.8) 2.26, dd (1.5, 12.8) |
36.16, CH2 | 131.59, C | |
| 9 | 152.56, C | 128.58, C | 170.25, C | 165.98, C | ||||
| 10 | 126.39, C | 153.49, C | 4.21, q (7.0) | 60.56, CH2 | ||||
| 11 | 157.51, C | 159.50, C | 1.26, t (7.0) | 14.08, CH3 | ||||
| NH | 12.54, d (2.8) | 12.68, s | 7.87, brd (5.2) | 11.85, dd (2.0, 2.8) | ||||
| NH | 8.15, t (5.0) | 7.73, t (4.8) | ||||||
| NH (CN3H4) | 9.16, s | |||||||
| NH2 (CN3H4) | 7.48, br | 9.23, brs | ||||||
| NH (CN3H4) | 7.24, br | |||||||
| CONH2 | 7.13, brs; 7.54, brs |
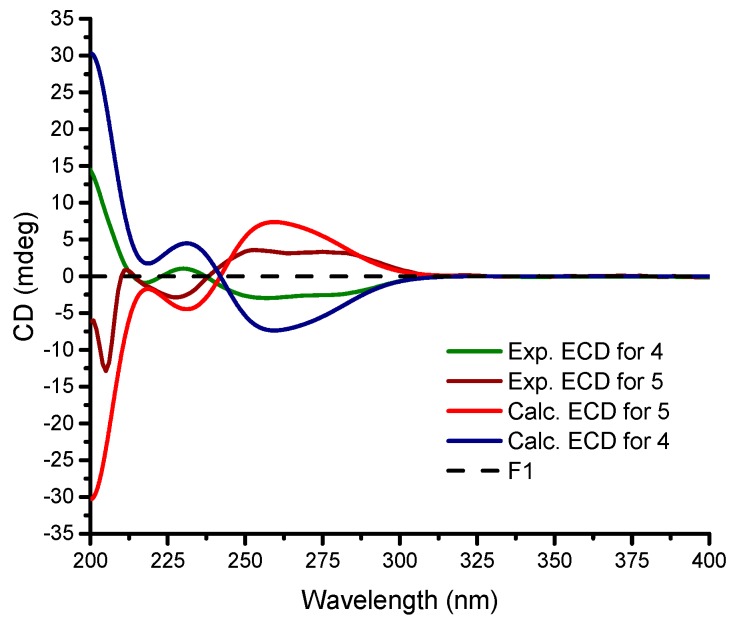
Stylisine D (4) was obtained as yellow amorphous solid. Its HRESIMS and NMR data established a molecular formula of C9H10N3O2Br2, containing two bromine atoms. A comparison of the NMR data (Table 1) indicated that 4 shared the partial structure (1,2-dibromopyrrole-4-carboxamide) of 2. The COSY relationships established an alkyl spin system from H2-6 (δH 3.34, 3.77) to H2-8 (δH 2.26, 2.63), while H2-6 extended the COSY correlation to an NH proton (δH 7.87, t). The HMBC correlations from a carbonyl carbon C-9 (δC 170.3) to H2-8, H-7 (δH 4.64), and NH2 (δH 7.13, 7.54) allowed for the linkage of a terminal amide to C-8 (δC 36.2). The connection of C-7 (δC 50.6) to the nitrogen atom of the pyrrole ring was deduced by the HMBC correlations from H-7 to C-1 (δC 105.7) and C-4 (δC 125.8). In addition, the HMBC correlations from NH (δH 7.87) to C-4 and the carbonyl carbon C-5 (δC 157.6) and between H-3 (δH 6.84, s) and C-5, resulted in the formation of a δ-lactam. The experimental ECD data showed positive CE at 228 nm and negative CE at 260 nm, which were in accordance with the calculated ECD data for a model compound of 4 with 7S configuration (Figure 4).
Figure 4.
Experimental and calculated ECD data for 4 and 5 in MeOH.
The identical NMR data of stylisine E (5) and 4 (Table 1), in association with the NMR and the HRESIMS data, assigned the same planar structure of both 5 and 4. The distinction was attributed to the absolute configuration at C-7, which was determined to be 7R, according to the Cotton effects of 5 showing opposite phases to those of 4, and to be similar with those calculated for 7R isomer of 4 (Figure 4).
Stylisine F (6) was obtained as a yellow amorphous solid. Its molecular formula was established as C11H12N2O3 on the basis of the HRESIMS and NMR data, requiring seven degrees of unsaturation. The 13C NMR and DEPT spectra exhibited 11 carbon resonances, of which, eight sp2 carbons and three sp3 carbons were defined. The COSY correlations from H-1 (δH 6.98) to H-2 (δH 6.52) and NH (δH 11.85), in association with the HMBC correlations from H-1 and H-2 to the quaternary carbons C-3 (δC 120.0) and C-4 (δC 126.2), disclosed a 3,4-disubstituted pyrrole ring. The HMBC correlations from NH to the carbons in the pyrrole ring and a carbonyl carbon C-5 (δC 163.3) ascertained a carbonyl group at C-4. A spin system coupling H2-6 (δH 3.48, dd) with NH (δH 7.73, t) and H-7 (δH 6.84, t) in the COSY spectrum, in addition to the HMBC correlations from H2-6 to C-5, and the olefinic carbons C-7 (δC 132.5) and C-8 (δC 131.6) and between H-7 and C-3, established a pyrroloazepine nucleus. Additional HMBC correlations from H-7 and the ethoxy methylene protons (δH 4.21, t) to the carbonyl carbon C-9 (δC 166.0) confirmed an ethoxycarbonyl group at C-8. Compound 6 was suggested to be an artifact derived during the process of the EtOH extraction, while 8-carboxylic analogue was considered to be the natural origin of 6.
In addition, 26 known alkaloids including pyrrole-2-carbamide type derivatives (1,2-dibromopyrrole-4-carbamide (7) [25], 2-bromopyrrole-4-carbamide (8), 1,2-dibromopyrrole-4-ethylate (9) [26], oroidin (10) [27], hymenidin (11) [28], dispacamide 1 (12) [29], keramadine (13) [30], laughine (14) [31], taurodispacamide A (15) [32]); aldizine type analogues (aldizine (16), 1-bromoaldizine (17) [33], 10Z-hymenialdisine (18), 10E-hymenialdisine (19) [34], hymenin (20) [35], stevensine (21) [36], (10E)-3-bromohymenialdisine (22), (10Z)-3-bromohymenialdisine (23) [37] and (10Z)-debromohymenialdisine (24)); phakellin type analogues (dibromoisophakellin (25), dibromophakellin (26) [38], monobromophakellin (27), monobromoisophakellin (28)); mamzacidin type analogues (manzacidins B-C (29, 30) [39], N-methylmanzacidin C (31) [40]); in addition to longamide B (32) (Supporting Information, Figure S41) [41], were identified by the comprehensive analyses and the comparison of one-dimensional (1D) and 2D NMR, and HRESIMS data, in association with the specific rotations. The absolute configurations of N-methylmanzacidin C were confirmed by the X-ray diffraction of a single crystal using the Flack parameter for the first time (Figure 5).
Figure 5.
Plot of the X-ray diffraction of N-methylmanzacidin C.
Biogenetically, pyrrole-2-carbamide and brominated pyrrole-2-carbamides, such as 7 and 8, were postulated to be the precursors to derive the isolated alkaloids [42,43,44]. Condensation of 8 with homoarginine yielded laughine (14), while the latter compound underwent dehydrogenation and cyclization to generate 1. Condensation of 7 with 3-amino-l-(2-aminoimidazolyl)-prop-l-ene generated oroidin (10), which followed oxygenation and olefinic migration to afford dispacamide 1 (12). Heterocyclization of 12 through the formation of an ether bridge to derive the stereoisomers 2 and 3. Compounds 4 and 5 were considered to be generated from 2 and 3 by ring rearrangement and oxygenation (Scheme 1).
Scheme 1.
Biogenetic postulation to derive 1–5.
Hymenidin, taurodiscapamide A, and oroidin have been reported to display significant growth inhibition against the Gram positive bacteria Staphylococcus aureus and Bacillus subtilis, and the Gram negative bacteria Escherichia coli and Pseudomonas aeruginosa, as well as the fungus C. albicans [22]. Oroidin was also reported to inhibit the growth of Rhodospirillum salexigens, a marine bacterium that is known to form biofilms [19]. 1,2-Dibromopyrrole-4-carbamide promoted larval metamorphosis of the ascidian Ciona savignyi [25]. In addition, oroidin-based derivatives incorporating a 2-aminoimidazole motif served as molecules with a unique scaffold to inhibit and disperse bacterial biofilms [45,46].
The literature data stimulated us to examine the isolated alkaloids individually for growth inhibition against pathogenic bacteria, including the Gram positive bacteria S. aureus ATCC 25923, S. haemolyticus ATCC 29970, and B. subtilis ATCC 11562, and the Gram negative bacteria E. coli ATCC 25922, Xanthomanes vesicatoria ATCC 11633, P. lachrymans ATCC 11921, Agrobaterium tumefaciens No. 8 ATCC 11158, Ralstonia solanacearum ATCC 11696. Unexpectedly, most compounds showed weak or no activity against the panel of bacteria with MIC ≥ 128 μg/mL (Supporting Information, Table S1), with the exception of oroidin that showed MIC values ranging from 32–128 μg/mL.
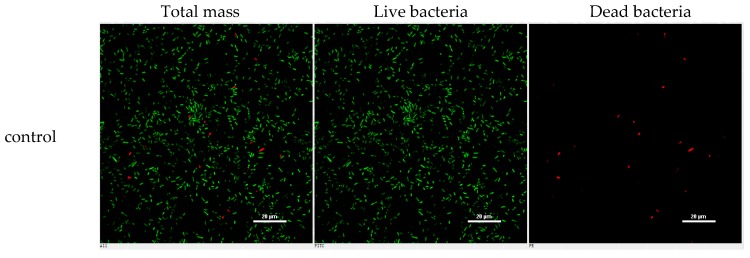
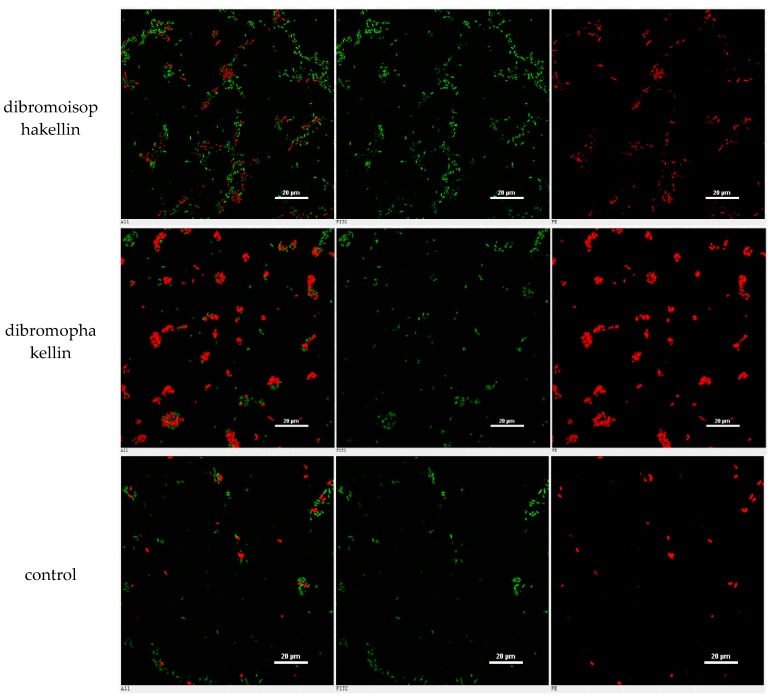
However, the initial bioassay indicated that the n-BuOH extract (an extract of total alkaloids) exerted inhibitory effects against the biofilm formation of E. coli. The bioassay guided fractionation of n-BuOH extract revealed three fractions (F2 to F4) to be active (Table 2), while the most active fraction was attributed to F2, in which dibromoisophakellin and dibromophakellin were isolated as the main components. Both of the compounds exerted significant inhibition against the biofilm formation of E. coli with IC50 value of 50.9 and 31.3 μg/mL, respectively. In order to understand the bioactivities of the relative analogues, we extended the anti-biofilm assay of remaining alkaloids. However, those alkaloid derivatives showed weak or no inhibition toward the biofilm formation (Table 3). Preliminary analyses of the structure-activity relationship indicated that phakellin-type scaffold with an aminoimidazole ring is essential to induce the inhibitory activity, while analogues with dibromo substitution showed more active than those with monobromo substitution. In parallel, confocal laser scanning microscopy (CLSM) was used to detect the ability of dibromophakellin and dibromoisophakellin to interfere with the biofilm formation of E. coli. In this experiment, the biofilm-embedded cells were LIVE/DEAD stained. The viable bacteria were stained with intact membranes fluorescing green (SYTO9) to distinguish the dead bacteria that were stained with damaged membrane fluorescing red (propidium iodine) [47,48]. As shown in Figure 6, the number of the viable bacteria of E. coli in the biofilm and the total biofilm thickness were greatly reduced after the treatment with dibromoisophakellin and dibromophakellin individually at a dose of 100 μg/mL in comparison with the compact and condensed biofilm control. These images provided additional evidence to support the inhibitory effects of both compounds against the biofilm formation. In parallel experiment, the untreated biofilms of E. coli in the vehicle group (DMSO) showed an inhomogeneous distribution with visible cell clusters and clearly rod shaped, whereas the E. coli cells treated by dibromoisophakellin and dibromophakellin changed their cell morphology to a more spherical shape, and tended to be aggregated.
Table 2.
Inhibitory effects of fractions against E. coli biofilm formation.
| Sample | E. coli Biofilm Formation Inhibition Rate (%) a |
|---|---|
| Positive control b | 98.4 |
| n-BuOH fraction | 7.7 |
| F1 | na |
| F2 | 84.4 |
| F3 | 26.4 |
| F4 | 18.2 |
| F5 | na |
a Measured in a dose of 200 μg/mL. b Positive control: chloramphenicol. Measured in a dose of 100 μg/mL.
Table 3.
Inhibitory effects of alkaloids against E. coli biofilm formation.
| Sample | E. coli Biofilm Formation Inhibition Rate (%) a | IC50 (μg/mL) |
|---|---|---|
| Positive control | 98.4 ± 0.8 | |
| 12 | 10.4 ± 1.7 | >100 |
| 25 | 72.1 ± 2.8 | 50.9 |
| 26 | 76.5 ± 1.1 | 31.3 |
| 27 | 25.9 ± 2.4 | >100 |
a Measured in a dose of 100 μg/mL. Positive control: chloramphenicol.
Figure 6.
Microscopy images of dibromoisophakellin and dibromophakellin showing inhibition toward Escherichia coli biofilms by LIVE/DEAD-stained after 24 h.
A–C = DMSO control, D–F = treated with dibromoisophakellin (100 μg/mL), G–I = treated with dibromophakellin (100 μg/mL), and J–L = treated with chloramphenicol (20 μg/mL). Viable bacteria are visible in green and dead bacteria in red. Left panels (A, D, G, J) for integrated red and green flourescent channels, middle panels (B, E, H, K) for green (live bacteria) and right panels (C, F, I, L) for red (dead bacteria).
3. Discussion
To the best of our knowledge, this study uncovered the largest numbers of brominated alkaloids with diverse scaffolds that were derived from marine sponge S. massa, implying more complexity of ecological environments, such as high biodiversity of predators, competitors, and overgrowths, as well as natural sponge-microbial associations in tropical sea water [49]. In addition to the postulation of the biogenetic relationships for new compounds in Scheme 1, complex hetercycles of pyrrole-imidazole alkaloids, such as aldizine-type and phakellin-type alkaloids, were supposed to be generated biogenetically from oroidin as a key intermediate through different manners of rearrangement and cyclization [44,50]. Although sponges have large symbiotic bacterial populations, oroidin and its analogues have been proved to be derived from sponge origin producing in its spherulous cells [51]. The structure variation is suggested to be an active enzymatic process involving the induction by symbiotic microorganisms. The prominent alkaloids, including oroidin (10), hymenidin (11), 10Z or 10E hymenialdisine (18 or 19), and debromohymenialdisine (24) from the sponge distributed at various biogeographic locations across Indo-Pacific reefs, suggesting that these typical alkaloids could be employed as chemical marks for taxonomy.
In the naturally occurring pyrrole-imidazole alkaloids, as derived from marine sponges, very few have been shown to modulate biofilm formation without killing the bacteria or disrupting their growth [8]. Oroidin and bromoageliferin were able to interfere with the bacterial attachment of Rhodospirillum salexigens and Vibrio vulnificus [52,53], where R. salexigens is a marine bacterium that is known to form biofilms. Oroidin exhibited modest anti-biofilm activity against two strains (PAO1 and PA14) of Pseudomonas aeruginosa, with IC50 values of 190 and 166 μM, whereas its analogue, dihydrosventrin, showed stronger inhibition against PAO1 and PA14 with IC50 values of 52 and 111 μM, respectively [54]. In our work, oroidin was inactive toward the anti-biofilm formation of E. coli, but dispacamide 1 (12) showed weak activity, with 10.4% inhibition rate at 100 μg/mL without microbicidal effect. These findings suggested that different pyrrole-imidazole alkaloids could play separate roles in ecological defense. Phakellin-based alkaloids dibromoisophakellin and dibromophakellin exerted significant reduction against the biofilm formation of E. coli with weak inhibition against the growth inhibition of planktonic bacteria, indicating the inhibitory effects via non-microbicidal mechanism. The molecular mechanism of the active compounds to eliminate biofilm formation remains to be assessed.
4. Materials and Methods
4.1. General Experimental Procedures
Optical rotations were measured on a Rudolph IV Autopol automatic polarimeter. IR spectra were determined on a Thermo Nicolet Nexus 470 FT-IR spectrometer. 1D and 2D NMR spectra were recorded on a Bruker Avance 400 NMR spectrometer (400 MHz for 1H and 100 MHz for 13C, respectively). Chemical shifts were referenced to the solvent peaks at δH 2.50 and δC 39.8 for dimethyl sulfoxide (DMSO-d6), respectively, and coupling constants were in Hz. ESIMS and HRESIMS spectra were obtained from a Thermo Scientific LTQ Orbitrap XL instrument. Column chromatography was carried out with silica gel (160–200 mesh, 200–300 mesh), and HF254 silica gel for TLC was obtained from Qingdao Marine Chemistry Co. Ltd. (Qingdao, China), ODS gel (50 μm), and Sephadex LH-20 (18–110 μm) were obtained from YMC (Kyoto, Japan) and Amersham Pharmacia Biotech AB, Uppsala, Sweden. HPLC chromatography was performed on an Alltech instrument (426-HPLC pump), equipped with an Alltech UVIS-200 detector and semipreparative reversed-phase columns (Grace Prevail C18, 5 μm, 250 mm × 10 mm). Analytical HPLC chromatography was performed on Prominence LC-20AD (Shimadzu, Kyoto, Japan) equipped with pump LC-20AD, detector Prominence SPD-M20A PDA and Shimadzu VP-ODS column (150 mm × 4.6 mm), Thermo BDS Hypersil C18 column (150 mm × 4.6 mm, 5 μm).
4.2. Animal Material
Sponge Stylissa massa was collected from the inner coral reef at a depth of around 8 m in Hainan Island of China, in May 2013, and the samples were frozen immediately after collection. The specimen was identified by Dr. Nicole J. de Voogd (Naturalis Biodiversity Center (Leiden, The Netherlands)). The voucher specimens (XSA-24) are deposited at the State Key Laboratory of Natural and Biomimetic Drugs, Peking University, China.
4.3. Isolation and Purification
Sponge tissue (3.5 kg, wet weight) was extracted with 95% EtOH for three times, and the crude extract was partitioned between MeOH and n-hexane to remove lipids. The MeOH layer was further partitioned between H2O and n-BuOH to afford n-BuOH soluble extract (11.4 g). The DAD-HPLC-ESIMS and the NMR data revealed that n-BuOH fraction contains a profile of brominated alkaloids. The silica gel chromatography of n-BuOH-fraction (4.9 g) was eluted with CH2Cl2-MeOH (a gradient from 6:1 to 2:1) to afford five fractions (F1 to F5). F1 (336 mg) was chromatographed on a silica gel column eluting with petroleum ether (PE)-acetone (from 3:1 to 1:1, gradient), to yield 7 (6.0 mg), 17 (11.3 mg), 13 (8.6 mg), and 16 (29.3 mg). Part of F2 (140 mg) was subjected to a silica gel column using CH2Cl2-MeOH (13:1) as a mobile phase followed by HPLC purification using a mobile phase of CH3CN-H2O (28:72, 1‰ CF3COOH) to afford 25 (11.7 mg) and 26 (117.5 mg). F3 (795 mg) was separated on ODS column eluting with MeOH-H2O (1:5) to afford six subfractions (SF31–SF36). SF31 (146 mg) was purified by a semi-preparative HPLC eluting with CH3CN-H2O (14:86, 3‰ CF3COOH) to afford 1 (21.1 mg) and 27 (30.8 mg). Following the same separation protocol by the semi-preparative HPLC eluting with MeOH-H2O (3:1), 28 (1.2 mg) was purified from SF32 (82.8 mg). SF33 (248 mg) was separated on Sephadex LH-20 column eluting with MeOH to afford 11 (26 mg). SF34 (28 mg) was purified by the semi-preparative HPLC eluting with CH3CN-H2O (1:4, 1‰ CF3COOH) to afford 18 (3.3 mg), 12 (7.2 mg), and 10 (99.2 mg). F4 (515 mg) was separated on ODS column and eluted with a gradient of MeOH-H2O (25–50%) to afford five subfractions (SF41–SF45). Following the semi-preparative HPLC separation, 19 (6.7 mg) was obtained from SF42 (177 mg) (CH3CN-H2O 12:88, 1‰ CF3COOH), 15 (17.5 mg), 20 (13.5 mg), 8 (3.1 mg), and 21 (7.6 mg) were obtained from SF43 (120 mg). SF44 (337 mg) was separated by semi-preparative HPLC eluting with 32% CH3CN-H2O (1‰ CF3COOH) to yield 22 (2.9 mg), 23 (17.5 mg), and 32 (3.6 mg). F5 (2.9 g) was subsequently subjected to MPLC, and then purified by semi-preparative HPLC eluting with 36% MeOH-H2O to afford 24 (9.9 mg), 31 (2.0 mg), 30 (39.7 mg), 29 (11.9 mg), 9 (39.1 mg), 15 (9.6 mg), 2/3 mixture (5.2 mg), 4/5 (4.1 mg) mixture, and 6 (4.7 mg). The separation of 2/3 (a ratio of 1:1) and 4/5 (a ratio of 5:4) was achieved by the chiral HPLC column (Daciel CHIRALPAK IC column) eluting with n-hexane-isopropanol (78:22) to obtain 2 (2.4 mg), 3 (2.5 mg), 4 (2.0 mg), and 5 (1.8 mg).
Stylisine A (1): light yellow solid, [α] − 9.4 (c 0.14, MeOH), IR (KBr film) νmax 3322, 3149, 3079, 2925, 1679, 1657, 1585, 1205, 1136 cm−1, 1H and 13C NMR (DMSO-d6), see Table 1, ESIMS m/z 310.03 [M + H]+ and HRTOFMS m/z 310.0311 [M + H]+ (calcd. for C11H13N5OBr, 310.0303).
Stylisine B (2): light yellow amorphous, [α] + 30.8 (c 0.05, MeOH), IR (KBr film) νmax 3424, 1677, 1205, 1138 cm−1; 1H and 13C NMR (DMSO-d6), see Table 1, ESIMS m/z 401.92 [M + H]+ and HRTOFMS m/z 401.9199 [M + H]+ (calcd. for C11H10N5O2Br2, 401.9201).
Stylisine C (3): light yellow amorphous, [α] − 31.2 (c 0.05, MeOH), IR (KBr film) νmax3424, 2853, 1677, 1439, 1206, 1138 cm−1, 1H and 13C NMR (DMSO-d6), see Table 1, ESIMS m/z 401.92 [M + H]+ and HRTOFMS m/z 401.9199 [M + H]+ (calcd. for C11H10N5O2Br2, 401.9201).
Stylisine D (4): light yellow amorphous, [α] − 4.0 (c 0.05, MeOH), IR (KBr film) νmax 3452, 1735, 1719, 1674, 1653, 1381, 1367, 1205, 1139 cm−1; 1H and 13C NMR (DMSO-d6), see Table 1, ESIMS m/z 349.91 [M + H]+ and HRTOFMS m/z 349.9146 [M + H]+ (calcd. for C9H10N3O2Br2, 349.9140).
Stylisine E (5): light yellow amorphous, [α] + 4.0 (c 0.05, MeOH); IR (KBr film) νmax 3452, 1735, 1720, 1675, 1654, 1382, 1368, 1206, 1140 cm−1; 1H and 13C NMR (DMSO-d6), see Table 1, ESIMS m/z 349.91 [M + H]+ and HRTOFMS m/z 349.9146 [M + H]+ (calcd. for C9H10N3O2Br2, 349.9140).
Stylisine F (6): yellow solid, [α] − 19.0 (c 0.16, MeOH); IR (KBr film) νmax 3351, 2927, 1713, 1680, 1624, 1470, 1205, 1138 cm−1; 1H and 13C NMR (DMSO-d6), see Table 1, ESIMS m/z 221.09 [M + H]+ and HRTOFMS m/z 221.0931 [M + H]+ (calcd. for C11H13N2O3, 221.0926).
4.4. X-ray Crystallographic Analysis
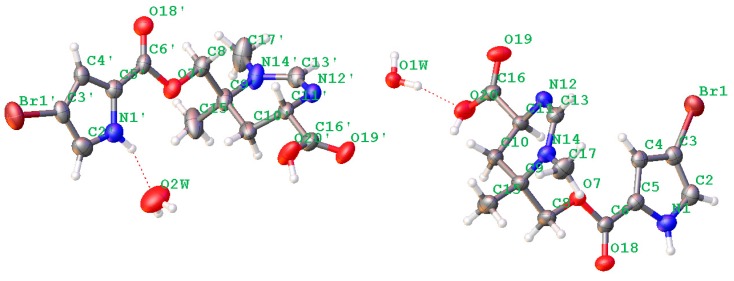
Crystal data were obtained on a RigakuMicroMax 002+ single-crystal X-ray diffractometer. Cell parameter measurements and data collection were performed with a Bruker APEX2 CCD diffractometer using the wavelength for Cu Kα (λ = 1.54184 A) radiation. The crystal structure of N-methylmanzacidin C was solved by direct methods (SHELXS-97), and subsequent Fourier difference techniques (SHELEX-97, version 6.10, Bruker AXS Inc. (Hong Kong, China)). The crystallographic data for the structures of N-methylmanzacidin C has been deposited in the Cambridge Crystallographic Data Center (CCDC number: 1543679).
Light yellow crystals of N-methylmanzacidin C were obtained from MeOH/H2O (1:1) using the vapor diffusion method. Crystal data for N-methylmanzacidin C, colorless needles, mp: 200.5–201.5 °C, C26H36N6O10Br2 (2 C13H16O4N3Br + 2 H2O), M = 752.43, orthorhombic, space group P212121, a = 7.17211(6) A, b = 18.32553(14) A, c = 25.1534(2) A, α = 90°, β = 90°, γ = 90°, V = 3305.98 (5) A3, Z = 4, T = 160.0 K, μ(Cu Kα) = 3.635 mm−1, Dcalc = 1.512 g/cm3, 28511 reflections measured (5.968 ≤ 2θ ≤ 137.558), 60,196 unique (Rint = 0.0334, Rsigma = 0.0202), which were used in all of the calculations. The final R1 was 0.0301 (I > 2σ (I)) and wR2 was 0.0745. Flack parameter = −0.019 (6).
4.5. ECD Calculation
ECD data were calculated in the B3LYP/6-31+G(d) level in MeOH. Conformational analyses were carried out via random searching in the Sybyl-X 2.0 version using the MMFF94S force field with an energy cutoff of 3.0 kcal/mol. The ECD spectra were simulated by the overlapping Gaussian function (half the bandwidth at 1/e peak height, 0.16–0.3 eV). The simulated spectra of the two lowest energy conformers for each structure were averaged according to the Boltzmann distribution theory and their relative Gibbs free energy (ΔG).
4.6. Antibacterial Assay
The Gram positive bacteria involved Staphylococcus aureus ATCC 25923, Bacillus subtilis ATCC 11562, Staphylococcus haemolyticus ATCC 29970, while the Gram negative bacteria included Pseudomonas aeruginosa ATCC 9027, Escherichia coli ATCC 25922, Pseudomonas lachrymans ATCC 11921, Agrobacterium tumefaciens No. 8 ATCC 11158, Xanthomonas vesicatoria ATCC 11633, and Ralstonia solanacearum ATCC 11696. Each bacterium strain that was to be tested was streaked onto a lysogeny broth agar or Mueller-Hinton Broth agar plate, and was incubated at 37 °C for 24 h. One colony was then transferred to fresh lysogeny broth or Mueller-Hinton broth, and the cell density was adjusted to 104–105 cfu/mL. Compounds were dissolved in DMSO with 12.8 mg/mL, and then were serially diluted with freshly prepared microbial broth to give gradient concentrations of 128, 64, 32, 16, 8, and 4 μg/mL. An aliquot (160 μL) of each dilution was transferred to a 96-well plate. Each sample was repeated for four times with a final DMSO concentration of 0.03% in each well. The plates were incubated at 37 °C for 24 h and shaken at 200 rpm, the optical density of each well was measured at 620 nm. Each compound was screened against the Gram positive and negative bacteria, while chloramphenicol was used as positive control and 0.03% DMSO, 1% DMSO were used as negative control.
4.7. Inhibition of Biofilm Formation Assay
Inhibition assays were performed by taking an overnight culture of bacterial strain (Escherichia coli with Mueller-Hinton Broth, Pseudomonas aeruginosa with lysogeny broth) to an OD620 of 0.8–1.0 (1 × 108 CFU/mL), the bacterial suspension was diluted by the broth to the cell density with an OD620 of 0.02–0.05. Compounds were dissolved in DMSO with 10 mg/mL, while the solution was then serially diluted with freshly prepared microbial broth to give final concentrations. Each solution was transferred (140 μL) into the wells of the 96-well plate by an incubation, under stationary conditions for 24 h at 30 °C. After incubation, the medium was discarded from the wells and the plates were washed thoroughly with water. Plates were then stained with 160 μL of 0.1% solution of crystal violet (CV), and then incubated at ambient temperature for 30 min. Plates were washed with water again and the remaining stain was solubilized with 200 μL of 95% ethanol. Biofilm inhibition was quantitated by measuring the OD570 of each well and calculated. Gentamicin was used as positive control against Pseudomonas aeruginosa, and chloramphenicol was used as positive control against Escherichia coli.
Dispersion assays were performed by taking an overnight culture of bacterial strain (E. coli) to an OD620 of 0.8–1.0 (1 × 108 CFU/mL), the bacterial suspension was diluted by broth to the cell density, with an OD620 of 0.02–0.05. The bacterial suspension was transferred (130 μL) into the wells of a 96-well plate followed by an incubation under stationary conditions at 30 °C to establish the biofilms. After 24 h, the medium was discarded from the wells and the plates were washed thoroughly with water. Compounds were dissolved in DMSO with 10 mg/mL, while the solution was then serially diluted with freshly prepared microbial broth to give final concentrations. The solutions were aliquoted (140 μL) into the wells of the 96 well plate with the established biofilms. Medium alone was added to a subset of the wells to serve as a control. Plates were then incubated for 24 h at 30 °C. After incubation, the medium was discarded from the wells and the plates were washed thoroughly with water. Plates were then stained with 160 μL of 0.1% solution of crystal violet (CV), and then incubated at ambient temperature for 30 min. Plates were washed with water and the remaining stain was solubilized with 200 μL of 95% ethanol. Biofilm inhibition was quantitated by measuring the OD570 of each well and was calculated as a percentage of the blank control without the formation of biofilm.
4.8. Confocal Laser Scanning Microscope (CLSM) Assay
Inhibition assays were performed by taking an overnight culture of bacterial strain (Escherichia coli with Mueller-Hinton Broth) to an OD620nm of 0.8–1.0 (1 × 108 CFU/mL), the bacterial suspension was diluted by the broth to the cell density with an OD620 of 0.1. Compounds were dissolved in DMSO with 10 mg/mL. The compound solution (dibromophakellin and dibromoisophakellin) was then diluted with freshly prepared microbial broth to give a concentration of 200 μg/mL (positive control chloramphenicol of 40 μg/mL). Compound solution (1 mL) and bacterial solution (1 mL) were transferred into petri dishes to give final concentrations of 100 μg/mL (positive control chloramphenicol of 20 μg/mL, negative control of 100% DMSO). The dishes were incubated under stationary conditions for 24 h at 30 °C. After incubation, the medium was carefully discarded from the dishes, while the dishes were washed with 0.9% NaCl solution. Compound-treated biofilms were used for CLSM analysis. Biofilms were subsequently stained with the FilmtracerTM LIVE/DEAD® Biofilm Viability Kit for microscopy (invitrogen, molecular probes), according to the manufacturer’s instructions, and were observed by confocal laser scanning microscopy (Nikon A1plus) using 100× objective lens. The excitation/emission wavelength of SYTO9 stain was 488/525 nm and 561/595 nm for propidium iodide. The images were analyzed using NIS Elements software. This experiment was repeated for three times.
5. Conclusions
In summary, a comprehensively chromatographic separation of marine sponge Stylissa massa resulted in the isolation of 32 alkaloids with diverse scaffolds, including six new alkaloids. Bioassay results revealed that oroidin exhibited the inhibitory effects against a panel of bacteria. In addition, phakellin-based alkaloids dibromoisophakellin and dibromophakellin showed significant reduction against the biofilm formation of E. coli with weak inhibition toward planktonic bacteria, validating that the alkaloids were selectively killing biofilm bacteria to be acting via non-microbicidal mechanism. Present work firstly reported phakellin-based alkaloids to be new natural scaffold for biofilm inhibition.
Acknowledgments
This work was financially supported by the grants from 973 program (2015CB755906) and National Natural Science Foundation of China (81630089, 41376127, 21302005, 21672010, U1606403).
Supplementary Materials
The following are available online at www.mdpi.com/1660-3397/16/1/9/s1, HRESIMS, 1H, 13C NMR, HSQC, HMBC, ROESY, and IR spectra of the new alkaloids. Physical and spectroscopic data, HPLC chromatographs, 1H and 13C NMR, and 2D NMR spectra for known alkaloids.
Author Contributions
J.S. and J.W. isolated all compounds; B.A. performed the primary bioassay; N.J.d.V. helped for the taxonomic identification of sponge; W.C. assayed the anti-biofilm of compounds; W.L. elucidated the structures of new compounds and edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Vaccari L., Allan D.B., Sharifi-Mood N., Singh A.R., Leheny R.L., Stebe K.J. Films of bacteria at interfaces: Three stages of behavior. Soft Matter. 2015;11:6062–6074. doi: 10.1039/C5SM00696A. [DOI] [PubMed] [Google Scholar]
- 2.Kolter R. Biofilms in lab and nature: A molecular geneticist’s voyage to microbial ecology. Int. Microbiol. 2010;13:1–7. doi: 10.2436/20.1501.01.105. [DOI] [PubMed] [Google Scholar]
- 3.Koerdt A., Godeke J., Berger J., Thormann K.M., Albers S.J. Crenarchaeal biofilm formation under extreme conditions. PLoS ONE. 2010;5:e14104. doi: 10.1371/journal.pone.0014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barros J., Grenho L., Manuel C.M., Ferreira C., Melo L.F., Nunes O.C., Monteiro F.J., Ferraz M.P. A modular reactor to simulate biofilm development in orthopedic materials. Int. Microbiol. 2013;16:191–198. doi: 10.2436/20.1501.01.193. [DOI] [PubMed] [Google Scholar]
- 5.Stewart P.S., Franklin M.J., Williamson K.S., Folsom J.P., Boegli L., James G.A. Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2015;59:3838–3847. doi: 10.1128/AAC.00433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebeaux D., Chauhan A., Rendueles O., Beloin C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens. 2013;2:288–356. doi: 10.3390/pathogens2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart P.S. Antimicrobial tolerance in biofilms. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MB-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stowe S.D., Richards J.J., Tucker A.T., Thompson R., Melander C., Cavanagh J. Anti-biofilm compounds derived from marine sponges. Mar. Drugs. 2011;9:2010–2035. doi: 10.3390/md9102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balskus E.P. Sponge symbionts play defense. Nat. Chem. Biol. 2014;10:611–612. doi: 10.1038/nchembio.1588. [DOI] [PubMed] [Google Scholar]
- 10.Davies D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 11.Fusetani N. Biofouling and antifouling. Nat. Prod. Rep. 2004;21:94–104. doi: 10.1039/b302231p. [DOI] [PubMed] [Google Scholar]
- 12.Laport M.S., Santos O.C.S., Muricy G. Marine sponges: Potential sources of new antimicrobial drugs. Curr. Pharm. Biotechnol. 2009;10:86–105. doi: 10.2174/138920109787048625. [DOI] [PubMed] [Google Scholar]
- 13.Hertiani T., Edrada-Ebel R., Ortlepp S., van Soest R.W., de Voogd N.J., Wray V., Hentschel U., Kozytska S., Müller W.E.G., Proksch P. From anti-fouling to biofilm inhibition: New cytotoxic secondary metabolites from two Indonesian Agelas sponges. Bioorg. Med. Chem. 2010;18:1297–1311. doi: 10.1016/j.bmc.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 14.Tsukamoto S., Kato H., Hirota H., Fusetani N. Ceratinamides A and B: New antifouling dibromotyrosine derivatives from the marine sponge Pseudoceratina purpurea. Tetrahedron. 1996;52:8181–8186. doi: 10.1016/0040-4020(96)00387-0. [DOI] [PubMed] [Google Scholar]
- 15.Shalmali N., Ali M.R., Bawa S. Imidazole: An essential edifice for the identification of new lead compounds and drug development. Mini Rev. Med. Chem. 2017;17 doi: 10.2174/1389557517666170228113656. [DOI] [PubMed] [Google Scholar]
- 16.Lindel T. Chemistry and biology of the pyrrole-imidazole alkaloids. Alkaloids Chem. Biol. 2017;77:117–219. doi: 10.1016/bs.alkal.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Cychon C., Lichte E., Köck M. The marine sponge Agelas citrina as a source of the new pyrrole-imidazole alkaloids citrinamines A–D and N-methylagelongine. Beilstein J. Org. Chem. 2015;11:2029–2037. doi: 10.3762/bjoc.11.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Assmann M., Soest R.W.M., Kock M. New antifeedant bromopyrrole alkaloid from the Caribbean sponge Stylissa caribica. J. Nat. Prod. 2001;64:1345–1347. doi: 10.1021/np000482s. [DOI] [PubMed] [Google Scholar]
- 19.Melander R.J., Liu H., Stephens M.D., Bewley C.A., Melander C. Marine sponge alkaloids as a source of anti-bacterial adjuvants. Bioorg. Med. Chem. Lett. 2016;26:5863–5866. doi: 10.1016/j.bmcl.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodnik Z., Łos J.M., Zula A., Zidar N., Jakopin Z., Łos M., Dolenc M.S., Ilaš J., Wegrzyn G., Mašic L.P., et al. Inhibition of biofilm formation by conformationally constrained indole-based analogues of the marine alkaloid oroidin. Bioorg. Med. Chem. Lett. 2014;24:2530–2534. doi: 10.1016/j.bmcl.2014.03.094. [DOI] [PubMed] [Google Scholar]
- 21.Forte B., Malgesini B., Piutti C., Quartieri F., Scolaro A., Papeo G. A submarine journey: The pyrrole-imidazole alkaloids. Mar. Drugs. 2009;7:705–753. doi: 10.3390/md7040705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H., Khalil Z., Conte M.M., Plisson F., Capon R.J. A search for kinase inhibitors and antibacterial agents: Bromopyrrolo-2-aminoimidazoles from a deep-water Great Australian Bight sponge, Axinella sp. Tetrahedron Lett. 2012;53:3784–3787. doi: 10.1016/j.tetlet.2012.05.051. [DOI] [Google Scholar]
- 23.Autschbach J., Nitsch-Velasquez L., Rudolph M. Time-dependent density functional response theory for electronic chiroptical properties of chiral molecules. Top. Curr. Chem. 2011;298:1–98. doi: 10.1007/128_2010_72. [DOI] [PubMed] [Google Scholar]
- 24.Pescitelli G., Di Pietro S., Cardellicchio C., Capozzi M.A., Di Bari L. Systematic investigation of CD spectra of aryl benzyl sulfoxides interpreted by means of TDDFT calculations. J. Org. Chem. 2010;75:1143–1154. doi: 10.1021/jo902363c. [DOI] [PubMed] [Google Scholar]
- 25.Tsukamoto S., Kato H., Hirota H., Fusetani N. Mauritiamine, a new antifouling oroidin dimer from the marine sponge Agelas mauritiana. J. Nat. Prod. 1996;59:501–503. doi: 10.1021/np960113p. [DOI] [Google Scholar]
- 26.Mancini I., Guella G., Amade P., Roussakis C., Pietra F. Hanishin, a semiracemic, bioactive C9 alkaloid of the axinellid sponge Acanthella carteri from the Hanish Islands. A shunt metabolite? Tetrahedron Lett. 1997;38:6271–6274. doi: 10.1016/S0040-4039(97)01405-6. [DOI] [Google Scholar]
- 27.Forenza S., Minale L., Riccio R., Fattorusso E. New bromopyrrole derivatives from the sponge Agela soroides. J. Chem. Soc. Chem. Commun. 1971;18:1129–1130. doi: 10.1039/c29710001129. [DOI] [Google Scholar]
- 28.Kobayashi J., Ohizumi Y., Nakamura H., Hirata Y. A novel antagonist of serotonergic receptors, hymenidin, isolated from the Okinawan marine sponge Hymeniacidon sp. Experientia. 1986;42:1176–1177. doi: 10.1007/BF01941300. [DOI] [PubMed] [Google Scholar]
- 29.Cafieri F., Fattorusso E., Mangoni A., Taglialatela-Scafati O. Dispacamides, anti-histamine alkaloids from caribbean Agelas sponges. Tetrahedron Lett. 1996;37:3587–3590. doi: 10.1016/0040-4039(96)00629-6. [DOI] [Google Scholar]
- 30.Nakamura H., Ohizumi Y., Kobayashi J., Hirata Y. Keramadine, a novel antagonist of serotonergic receptors isolated from the okinawan sea sponge Agelas sp. Tetrahedron Lett. 1984;25:2475–2478. doi: 10.1016/S0040-4039(01)81208-9. [DOI] [Google Scholar]
- 31.Williams D.E., Patrick B.O., Behrisch H.W., Soest R.V., Roberge M., Andersen R.J. Dominicin, a cyclic octapeptide, and laughine, a bromopyrrole alkaloid, isolated from the Caribbean marine sponge Eurypon laughlini. J. Nat. Prod. 2005;68:327–330. doi: 10.1021/np049711r. [DOI] [PubMed] [Google Scholar]
- 32.Fattorusso E., Taglialatela-Scafati O. Two novel pyrrole-imidazole alkaloids from the Mediterranean sponge Agela soroides. Tetrahedron Lett. 2000;41:9917–9922. doi: 10.1016/S0040-4039(00)01764-0. [DOI] [Google Scholar]
- 33.Fouad M.A., Debbab A., Wray V., Müller W.E.G., Proksch P. New bioactive alkaloids from the marine sponge Stylissa sp. Tetrahedron. 2012;68:10176–10179. doi: 10.1016/j.tet.2012.09.097. [DOI] [Google Scholar]
- 34.Tasdemir D., Mallon R., Greenstein M., Feldberg L.R., Kim S.C., Collins K., Wojciechowicz D., Mangalindan G.C., Concepción G.P., Harper M.K., et al. Aldisine alkaloids from the Philippine sponge Stylissa massa are potent inhibitors of mitogen-activated protein kinase kinase-1 (MEK-1) J. Med. Chem. 2002;45:529–532. doi: 10.1021/jm0102856. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi J., Ohizumi Y., Nakamura H., Hirata Y., Wakamatsu K., Miyazawa T. Hymenin, a novel α-adrenoceptor blocking agent from the Okinawan marine sponge Hymeniacidon sp. Experientia. 1986;42:1064–1065. doi: 10.1007/BF01940735. [DOI] [PubMed] [Google Scholar]
- 36.Albizatit K.F., Faulkner D.J. Stevensine, a novel alkaloid of an unidentified marine sponge. J. Org. Chem. 1985;50:4163–4164. doi: 10.1021/jo00221a041. [DOI] [Google Scholar]
- 37.Eder C., Proksch P., Wray V., Klaus Steube K., Bringmann G., Soest R.W.M., Ferdinandus E., Pattisina L.A., Wiryowidagdo S., Moka W. New alkaloids from the indopacific sponge Stylissa carteri. J. Nat. Prod. 1999;62:184–187. doi: 10.1021/np980315g. [DOI] [PubMed] [Google Scholar]
- 38.Meyer S.W., Köck M. NMR Studies of phakellins and isophakellins. J. Nat. Prod. 2008;71:1524–1529. doi: 10.1021/np8000706. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi J., Kanda F., Ishibashi M., Shigemori H. Manzacidins A–C, novel tetrahydropyrimidine alkaloids from the Okinawan marine sponge Hymeniacidon sp. J. Org. Chem. 1991;56:4574–4576. doi: 10.1021/jo00014a052. [DOI] [Google Scholar]
- 40.Tsukamoto S., Tane K., Ohta T., Matsunaga S., Fusetani N., Soest R.W.M. Four new bioactive pyrrole-derived alkaloids from the marine sponge Axinella brevistyla. J. Nat. Prod. 2001;64:1576–1578. doi: 10.1021/np010280b. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Y., Wang Y., Gu B., Yang F., Jiao W., Hu G., Yu H., Han B., Zhang W., Shen Y., et al. Antifungal bromopyrrole alkaloids from the South China Sea sponge Agelas sp. Tetrahedron. 2016;72:2964–2971. doi: 10.1016/j.tet.2016.04.020. [DOI] [Google Scholar]
- 42.Andrade P., Willoughby R., Pomponi S.A., Kerr R.G. Biosynthetic studies of the alkaloid, stevensine, in a cell culture of the marine sponge Teichaxinella morchella. Tetrahedron Lett. 1999;40:4775–4778. doi: 10.1016/S0040-4039(99)00881-3. [DOI] [Google Scholar]
- 43.Stout E.P., Wang Y., Romo D., Molinski T.F. Pyrrole aminoimidazole alkaloid metabiosynthesis with marine sponges Agelas conifera and Stylissa caribica. Angew. Chem. Int. Ed. 2012;51:4877–4881. doi: 10.1002/anie.201108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genta-Jouve G., Cachet N., Holderith S., Oberhansli F., Teyssie J.L., Jeffree R., Mourabit A.A., Thomas O.P. New insight into marine alkaloid metabolic pathways: Revisiting oroidin biosynthesis. ChemBioChem. 2011;12:2298–2301. doi: 10.1002/cbic.201100449. [DOI] [PubMed] [Google Scholar]
- 45.Richards J.J., Reed C.S., Christian Melander C. Effects of N-pyrrole substitution on the anti-biofilm activities of oroidin derivatives against Acinetobacter baumannii. Bioorg. Med. Chem. Lett. 2008;18:4325–4327. doi: 10.1016/j.bmcl.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 46.Ballard T.E., Richards J.J., Aquino A., Reed C.S., Melander C. Antibiofilm activity of a diverse oroidin library generated through reductive acylation. J. Org. Chem. 2009;74:1755–1758. doi: 10.1021/jo802260t. [DOI] [PubMed] [Google Scholar]
- 47.Stiefel P., Rosenberg U., Schneider J., Mauerhofer S., Maniura-Weber K., Ren Q. Is biofilm removal properly assessed? Comparison of different quantification methods in a 96-well plate system. Appl. Microbiol. Biotechnol. 2016;100:4135–4145. doi: 10.1007/s00253-016-7396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Y., Stojicic S., Haapasalo M. Bacterial viability in starved and revitalized biofilms: Comparison of viability staining and direct culture. J. Endod. 2010;36:1820–1823. doi: 10.1016/j.joen.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 49.Sauleau P., Moriou C., Mourabit A.A. Metabolomics approach to chemical diversity of the Mediterranean marine sponge Agelas oroides. Nat. Prod. Res. 2017;31:1625–1632. doi: 10.1080/14786419.2017.1285298. [DOI] [PubMed] [Google Scholar]
- 50.Pina I.C., White K.N., Cabrera G., Rivero E., Crews P. Bromopyrrole carboxamide biosynthetic products from the Caribbean sponge Agelas dispar. J. Nat. Prod. 2007;70:613–617. doi: 10.1021/np0606042. [DOI] [PubMed] [Google Scholar]
- 51.Richelle-Maurer E., De Kluijver M.J., Feio S., Gaudencio S., Gaspar H., Gomez R., Tavares R., Van de Vyver G., Van Soest R.W.M. Localization and ecological significance of oroidin and sceptrin in the Caribbean sponge Agelas conifer. Biochem. Syst. Ecol. 2003;31:1073–1091. doi: 10.1016/S0305-1978(03)00072-3. [DOI] [Google Scholar]
- 52.Kelly S.R., Jensen P.R., Henkel T.P., Fenical W., Pawlik J.R. Effects of Caribbean sponge extracts on bacterial attachment. Aquat. Microb. Ecol. 2003;31:175–182. doi: 10.3354/ame031175. [DOI] [Google Scholar]
- 53.Yamada A., Kitamura H., Yamaguchi K., Fukuzawa S., Kamijima C., Yazawa K., Kuramoto M., Wang G.-Y.-S., Fujitani Y., Uemura D. Development of chemical substances regulating biofilm formation. Bull. Chem. Soc. Jpn. 1997;70:3061–3069. doi: 10.1246/bcsj.70.3061. [DOI] [Google Scholar]
- 54.Richards J.J.T., Ballard E.B., Huigens R.W., Melander C. Synthesis and screening of an oroidin library against Pseudomonas aeruginosa biofilms. ChemBioChem. 2008;9:1267–1279. doi: 10.1002/cbic.200700774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.