Abstract
Crustins form a large family of antimicrobial peptides (AMPs) in crustaceans composed of four sub-groups (Types I-IV). Type II crustins (Type IIa or “Crustins” and Type IIb or “Crustin-like”) possess a typical hydrophobic N-terminal region and are by far the most representative sub-group found in penaeid shrimp. To gain insight into the molecular diversity of Type II crustins in penaeids, we identified and characterized a Type IIb crustin in Litopenaeus vannamei (Crustin-like Lv) and compared Type II crustins at both molecular and transcriptional levels. Although L. vannamei Type II crustins (Crustin Lv and Crustin-like Lv) are encoded by separate genes, they showed a similar tissue distribution (hemocytes and gills) and transcriptional response to the shrimp pathogens Vibrio harveyi and White spot syndrome virus (WSSV). As Crustin Lv, Crustin-like Lv transcripts were found to be present early in development, suggesting a maternal contribution to shrimp progeny. Altogether, our in silico and transcriptional data allowed to conclude that (1) each sub-type displays a specific amino acid signature at the C-terminal end holding both the cysteine-rich region and the whey acidic protein (WAP) domain, and that (2) shrimp Type II crustins evolved from a common ancestral gene that conserved a similar pattern of transcriptional regulation.
Keywords: invertebrate immunity, host defense peptide, crustacean, WAP domain, molecular diversity, host-pathogen interaction
1. Introduction
Crustins are cysteine-rich antimicrobial peptides (AMPs) holding a typical whey acidic protein (WAP) domain [1]. The WAP domain is a conserved motif containing eight cysteine residues, forming a characteristic four-disulfide core (4DSC) arrangement, that is also found in many proteins exhibiting protease inhibitory properties or regulatory functions in growth and tissue differentiation [2]. These bioactive molecules were originally isolated from the hemolymph of the shore crab Carcinus maenas as a cationic 11.5-kDa peptide with specific activity against marine Gram-positive bacteria [3]. Homologues of the C. maenas 11.5-kDa peptide were identified some years later in two shrimp species (Litopenaeus vannamei and Litopenaeus setiferus) and tentatively named as “Crustins” [4], a nomenclature that was subsequently accepted and adopted for this AMP family [1,5,6,7]. Indeed, crustins comprise a large and diverse family of gene-encoded AMPs in decapod crustaceans [8], but they are also present in non-decapod species, such as amphipods, copepods and isopods, and even in some hymenopteran insects [1].
Based on the presence or absence of structural domains lying at the N-terminal region, Smith et al. [5] classified crustins into three sub-groups, designated as Types I to III. Type I crustins are most related to the C. maenas 11.5-kDa peptide (later termed as “carcinin” [9]) and are characterized by the presence of four conserved cysteine residues, the cysteine-rich domain, upstream to the C-terminal WAP domain. They occur mainly in Pleocyemata (crabs, lobsters and crayfish), but some members were also reported in penaeid shrimp [10,11]. On the other hand, Type II crustins are mainly present in penaeids (Dendrobranchiata) and harbor a hydrophobic region rich in glycine residues, the glycine-rich domain, positioned at the N-terminal end of the cysteine-rich domain that is also found in Type I crustins. By contrast, Type III crustins (also known as single WAP domain-containing proteins or SWD) are only composed of a single WAP domain, and display both antimicrobial and antiprotease activities [12]. Proteins containing two WAP domains (double WAP domain-containing proteins or DWD) and crustin homologues from hymenopteran insects have been recently classified as Type IV and Type V, respectively [13].
Due to the economic importance of penaeid shrimp worldwide, Type II is the most well characterized sub-group of crustins. They are usually active against Gram-positive bacteria, but in vivo studies using gene silencing revealed that Type II crustins play a key role in shrimp defense against pathogenic Gram-negative bacteria [14,15,16]. Interestingly, the knockdown of Type II crustins in the Pacific white shrimp L. vannamei causes an increase in mortality after infections with the bacterial pathogen Vibrio penaeicida, but not in response to the fungal pathogen Fusarium oxysporum [14]. The antibacterial activity of Type II crustins appears to be related to the WAP domain. Indeed, it has been shown that Type II crustins with an incomplete WAP domain have impaired antimicrobial activities [17]. This large crustin sub-group showed to be diverse in terms of both structure and function. For instance, in the black tiger shrimp Penaeus monodon, at least ten different Type II crustins (crustinPm1-10) have been identified, displaying diverse molecular structures and biological activities [7]. In the literature, this heterogeneous and complex crustin Type has been also subdivided into two sub-types, namely Type IIa or “Crustins”, most related to the first glycine-rich peptides identified in Litopenaeus species (Crustin Lv and Crustin Ls) [4], and Type IIb or “Crustin-like”, homologues of the CruFc peptide from Fenneropenaeus chinensis [17] and the Crus-likePm (or crustinPm7) peptide from P. monodon [18]. Interestingly, while Type IIa crustins have been identified in different penaeids [4,19,20,21], Type IIb crustins were only reported in Oriental (Asian) species [16,17,18,22].
Although a distinction between Type IIa and Type IIb crustins has been previously established (based on differences in the amino acid length between the cysteine-rich and WAP domains) [7], the current classification of these sub-types in penaeid shrimp remains confused and controversial, leading to misleading categorizations. In order to explore and refine this issue, we have firstly identified and characterized at both molecular and transcriptional levels a Type IIb crustin from the most commonly cultivated penaeid shrimp (L. vannamei), and then provided amino acid signatures specific to Type IIa and Type IIb crustins in penaeid shrimp. By taking advantage of publicly accessible databases, we have identified seven nucleotide sequences in L. vannamei transcriptomes showing high similarities to Type IIb crustins that were opportunely named Crustin-like Lv. L. vannamei Type IIa and Type IIb crustins are encoded by distinct genomic sequences and differ not only in the length of their structural domains, but with each sub-type possessing a specific amino acid signature at the C-terminal region containing the 12 conserved cysteine residues. A comparative gene expression analysis showed that L. vannamei Type IIa (Crustin Lv) and Type IIb (Crustin-like Lv) crustins are mainly expressed in circulating hemocytes and gills of juveniles and displayed a similar transcriptional pattern in response to two unrelated shrimp pathogens, the Gram-negative Vibrio harveyi and the White spot syndrome virus (WSSV). Finally, as observed for Crustin Lv, Crustin-like Lv transcripts were detected in all stages of L. vannamei development, from fertilized eggs to larval and postlarval stages. We provided here the first molecular characterization of a Type IIb crustin in an Occidental (non-Asian) species and the identification of molecular amino acid signatures specific to Type II crustins in penaeid shrimp.
2. Results and Discussion
2.1. Identification and Molecular Characterization of Type IIb Crustins in L. vannamei
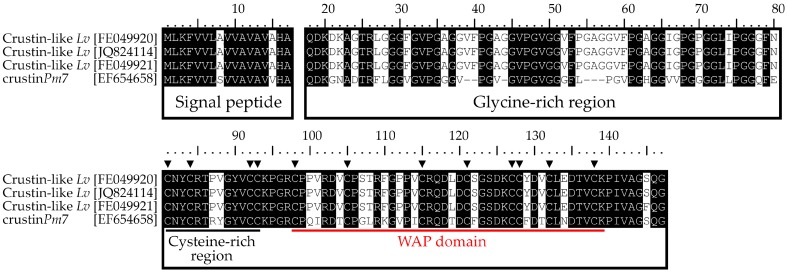
By taking advantage of publicly accessible databases, we have characterized for the first time Type IIb crustins in the most commonly cultivated penaeid shrimp, L. vannamei. In silico analysis led to the identification of three complete (one from the GenBank Nucleotide database and two from Expressed Sequence Tag libraries) and four incomplete/partial (Transcriptome Shotgun Assembly database) nucleotide sequences. All sequences corresponded to Type IIb crustins that were tentatively named Crustin-like Lv. L. vannamei Type IIa crustins were previously termed as Crustin Lv [4]. The complete full-length cDNA sequences (GenBank: JQ824114, FE049920 and FE049921) encode 147 amino acid precursors, starting with a predicted 17-residue signal peptide followed by a putative cationic (pI ~8) mature peptide of 130 amino acid residues (Figure 1). The presence of this signal peptide is necessary to direct the precursors to the endoplasmic reticulum and the Golgi apparatus. Then, the mature peptides are targeted to intracellular granules where they are stored [23]. All mature peptides possess a glycine-rich region containing three repeats of the pentapeptide Val-Phe-Pro-Gly-Ala (VFPGA) at the N-terminal end, followed by a C-terminal region containing twelve conserved cysteine residues, eight of them comprising a single WAP domain (Figure 1). For the incomplete Crustin-like Lv sequences (GenBank: GETD01016911, HAAW01014776, GETZ01011995 and GDUV01030786), only the C-terminal region, holding both the cysteine-rich region and the WAP domain, was identified. These incomplete sequences lack the signal peptide and part of the glycine-rich region (Figure S1).
Figure 1.
Litopenaeus vannamei Type IIb crustins. Amino acid sequences alignment of Type IIb crustins from L. vannamei (Crustin-like Lv) and the crustinPm7 from Penaeus monodon. Identical residues are highlighted in black. Triangles (▼) indicate the 12 conserved cysteine residues found in crustins. The whey acidic protein (WAP) domain is underlined by a solid red line.
Crustin-like Lv deduced amino acid sequences were compared with Type II crustins from penaeid shrimp and with crustins from other crustacean species (Figure S1). Mature Crustin-like Lv sequences shared a high amino acid sequence identity with other Type IIb crustins, such as Crustin-like from Marsupenaeus japonicus (75%; [16]), the Fi-crustin from Fenneropenaeus indicus (73%; [22]), crustinPm7 (Crus-likePm) from P. monodon (70%; [18]) and CruFc from F. chinensis (69%; [17]). Within Type IIa crustins, Crustin-like Lv was 44% identical to Crustin Lv sequences [4] and 40% to 54% identical to other Type IIa crustins from penaeids [10,19,20,21]. Regarding Type II crustins from non-penaeid species (Pleocyemata), Crustin-like Lv showed 43% to 51% amino acid identity with the crustins (PJC1 to PJC4) from the Japanese spiny lobster Panulirus japonicus [24]. Finally, L. vannamei Type IIb sequences showed less identity with the other crustins sub-groups found in crustaceans, namely Type I (30–43%), Type III (20–25%), and Type IV (25–27%).
2.2. L. vannamei Type II Crustins Are Encoded by Distinct Genes
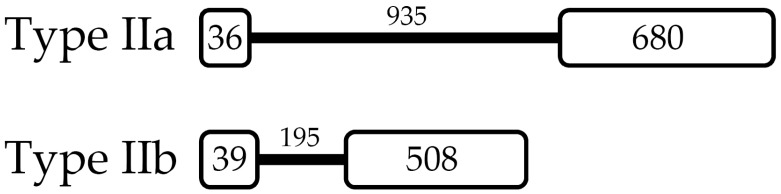
The L. vannamei Type IIa gene that encodes Crustin Lv is 1651 bp in size and consists of two exons (36 bp and 680 bp, respectively) interrupted by a single intron (935 bp) (Figure S2). The first exon covers the 5′ untranslated region (5′-UTR) and only two codons of the signal peptide (ATGAAG), while the second exon encodes the remainder of the signal peptide sequence, and the complete mature peptide and the 3′ untranslated region (3′-UTR) (Figure 2 and Figure S2). Likewise, the Type IIb gene (742 bp) (Figure S2), that encodes Crustin-like Lv, is also composed of two exons (39 bp and 508 bp, respectively) separated by one intron (195 bp). However, in the Type IIb gene, whereas the first exon covers only the 5′-UTR, the second exon covers the signal peptide, the complete mature peptide and the 3′-UTR (Figure 2 and Figure S2). All splice sites in both Type II crustin genes followed the canonical GT/AG splicing recognition rule. Interestingly, L. vannamei Type IIb genomic organization was quite similar to that found in the crustinPm7, a Type IIb crustin gene from P. monodon [18]. By contrast, the P. monodon Type IIa crustinPm5 gene [25] showed an exon-intron organization different from that found in the Type IIa gene from L. vannamei (Figure 2). Altogether, these results suggest that penaeid Type II crustins are encoded by distinct genomic sequences, and that the molecular diversity found in this crustin sub-group was driven by gene duplication and mutations [26].
Figure 2.
Litopenaeus vannamei Type II crustins are encoded by distinct genomic sequences. A not-to-scale representation of L. vannamei Type II crustins, Type IIa (Crustin Lv) and Type IIb (Crustin-like Lv). White boxes indicate the exons and the black line indicates the intron. The numbers show the size (in base pairs) of the exons and introns.
2.3. Gene Expression Distribution of Type II Crustins in Shrimp Tissues
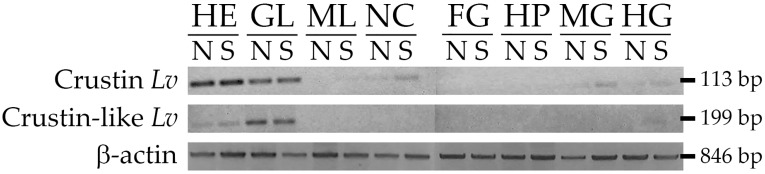
In our transcriptional analyses, we compared the L. vannamei Type II crustins in terms of (1) tissue distribution, (2) transcriptional response to two unrelated pathogens, and (3) expression profile during shrimp development. Firstly, the gene expression distribution of Crustin Lv (Type IIa) and Crustin-like Lv (Type IIb) were evaluated in eight different tissues (hemocytes, gills, muscle, nerve cord, foregut, hepatopancreas, midgut and hindgut) from naïve (unchallenged) animals and shrimp at 48 h post-stimulation with heat-killed bacteria (V. harveyi). Interestingly, both L. vannamei Type II crustin sub-types showed a similar gene expression distribution in shrimp tissues. Transcripts of Crustin Lv and Crustin-like Lv were detected in circulating hemocytes and gills of both unchallenged and Vibrio-stimulated animals (Figure 3). Remarkably, whereas the highest expression levels of the Crustin Lv gene (Type IIa) were found in circulating hemocytes, the Crustin-like Lv gene (Type IIb) showed to be predominantly expressed in gills (Figure 3). In Vibrio-stimulated animals, the expression of the Crustin Lv gene was also detected in the nerve cord and in the two gut portions (midgut and hindgut) (Figure 3). For both Type II crustins, no signals were observed in muscle, foregut, and hepatopancreas (Figure 3). Comparatively, Type II crustins from P. monodon were heterogeneously distributed among shrimp tissues. For instance, while crustinPm5 appeared to be constitutively transcribed in the epipodite and eyestalks [25], the other P. monodon Type II crustins (crustinPm1, crustinPm4 and crustinPm7) were mainly expressed in hemocytes [10,18,27].
Figure 3.
Gene expression distribution of Type II crustins in shrimp tissues. Semiquantitative reverse transcriptase polymerase chain reaction (RT-PCR) analysis of Crustin Lv (Type IIa) and Crustin-like Lv (Type IIb) transcript levels in different tissues from naïve (N) and Vibrio-stimulated (S) shrimp. The expression of the β-actin gene was used as an endogenous control. HE: hemocytes, GL: gills, ML: muscle, NC: nerve cord, FG: foregut, HP: hepatopancreas, MG: midgut, HG: hindgut.
In L. vannamei, the expression of Type IIa crustins was restricted to both circulating and tissue-infiltrating hemocytes [28]. More specifically, the expression of this gene was apparently higher in semi-granular hemocytes than in the granular cells [29]. Thus, the detection of Crustin Lv transcripts in shrimp tissues was undoubtedly the result of infiltrating hemocytes. By contrast, in this study, Crustin-like Lv transcripts were mainly detected in gills. This could be the result of a distinct migratory behavior displayed by circulating hemocytes in shrimp tissues, or the presence of specific tissue-resident hemocyte populations. We cannot rule out the hypothesis of the existence of crustin-expressing hemocytes in shrimp gills distinct from those found circulating in hemolymph. Actually, the presence of tissue-specific subsets of a particular immune cell type has been reported in mammals [30]. Thus, the next import steps will be (1) to colocalize Type II crustins in shrimp hemocytes to verify whether they are expressed in the same hemocyte populations, and (2) to define the precise site of expression of the Crustin-like Lv gene by using physical mapping techniques (in situ hybridization and immunohistochemistry) as previously performed for other crustin genes [23,27,28].
2.4. Expression Profile of Type II Crustins in Response to Bacterial and Viral Infections
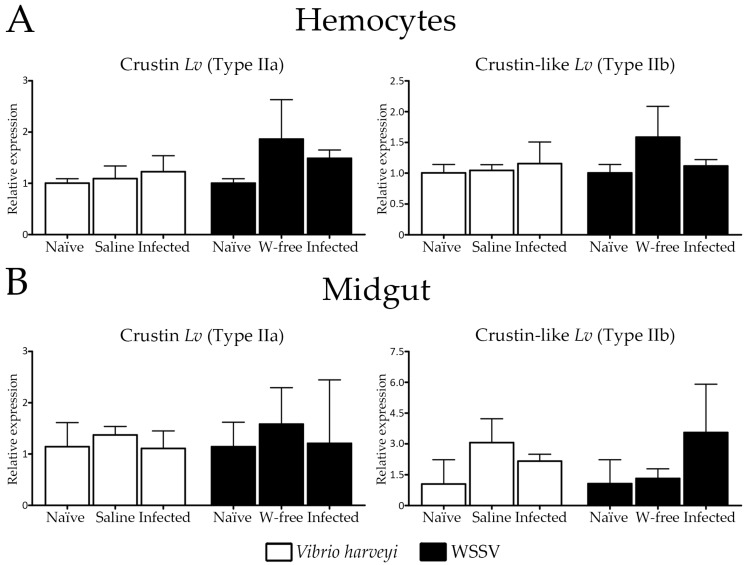
We have further investigated the transcriptional response of Type II crustins in two important immune tissues after experimental infections with the Gram-negative V. harveyi and the WSSV. Their expression levels were quantified at 48 h after infections by fluorescence-based reverse transcriptase quantitative PCR (RT-qPCR) in circulating hemocytes and midgut. In a previous study from our group, the expression of gene-encoded AMPs (penaeidins, crustins, anti-lipopolysaccharide factors and stylicins) showed to be modulated in L. vannamei shrimp at 48 h post-infections [31]. Importantly, whereas hemocytes are the main site for the expression of AMPs in shrimp [6], the midgut represents an important route of pathogen entry in invertebrates [32]. The transcriptional levels of both Type II crustins were not affected by the bacterial or by the viral infection in circulating hemocytes or in the midgut (Figure 4). Furthermore, no obvious differences in gene expression were observed between unchallenged shrimp and the controls (shrimp injected with SSW or the WSSV-free inoculum) (Figure 4). At this same time point, the expression of the Crustin Lv gene (Type IIa) showed to be drastically reduced in circulating hemocytes of shrimp succumbing to a lethal infection by the opportunistic filamentous fungus Fusarium solani but not by the WSSV [33]. In our study, the lack of induction of L. vannamei Type II crustins could be partly attributed to the time course response of Type II crustins in the analyzed tissues but also to the route of infection. Indeed, we have evaluated the transcriptional response of L. vannamei Type II crustins at 48 h post-infections and it is probable that the modulation of these genes has occurred earlier. For instance, in many shrimp species, the expression of crustins showed to be modulated in the first hours post-infections [10,11,12,18]. Moreover, our experimental procedure consisted of injection of the pathogens directly into the shrimp hemocel in order to standardize a same inoculum load per animal [33]. By using a natural route of infection (immersion method), Soonthornchai et al. [34] could observe an increase in crustin expression in the midgut of P. monodon shrimp experimentally infected with V. harveyi.
Figure 4.
Relative expression profile of Crustin Lv (Type IIa) and Crustin-like Lv (Type IIb) genes in (A) circulating hemocytes and (B) midgut of shrimp at 48 h after experimental infections with the Gram-negative Vibrio harveyi ATCC 14126 (white bars) or the White spot syndrome virus (black bars). Results are presented as mean ± standard deviation of relative expressions (three biological replicates). WSSV: White spot syndrome virus. W-free: tissue homogenate inoculum prepared from WSSV-free shrimp.
Despite the lack of regulation at the transcriptional level, both Type IIa and Type IIb crustins showed to be directly involved in shrimp antimicrobial defense against pathogenic Vibrio infections [14,15,16]. Besides, the regulation of these molecules may occur at the post-translational level (maturation and/or trafficking), rather than at the transcriptional level, as observed for penaeidins. Indeed, penaeidins are shrimp gene-encoded AMPs that are constitutively expressed and stored in circulating hemocytes, then released to hemolymph in response to microbial challenges [35]. As with penaeidins, crustins are also produced and stored in hemocytes [23,27], so it is plausible to suppose that they are also released towards the plasma by a regulated secretion pathway triggered by infectious agents.
2.5. Expression of Type IIb Crustins during Shrimp Ontogenesis
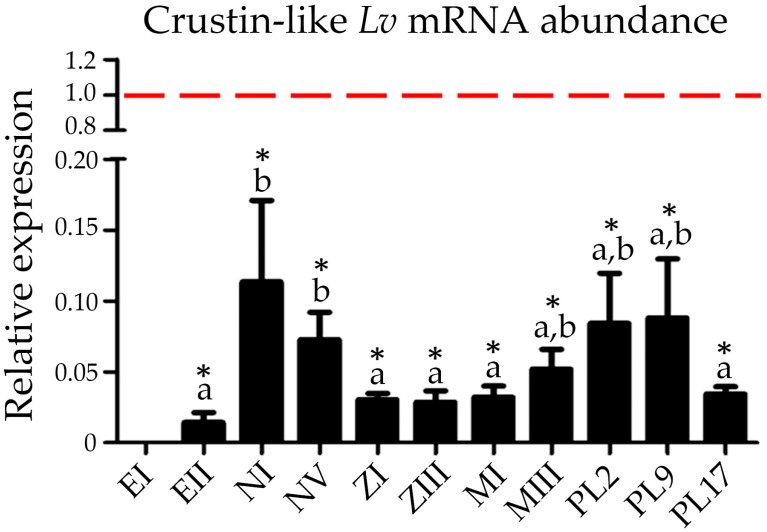
Another important result from this study was the first characterization of the transcriptional profile of a Type IIb crustin during shrimp development. The transcript abundance of the Crustin-like Lv gene (Type IIb) was quantified by RT-qPCR in the 12 developmental stages of L. vannamei, from fertilized eggs to larval and postlarval stages, and also in circulating hemocytes from juveniles. Crustin-like Lv expression was detected in all shrimp developmental stages, but only quantified from fertilized eggs at 7–11 h post-spawning (Figure 5). Interestingly, Crustin-like Lv gene expression profile during shrimp ontogenesis was quite similar to that which was previously observed for the Crustin Lv gene [36]. Both Type II crustins were transcribed at variable levels during shrimp development. However, the highest mRNA levels were observed in hemocytes from juveniles (Figure 5), which is in accordance with our results of tissue expression distribution (Figure 3). Moreover, transcripts for both genes were found in fertilized eggs, revealing a maternal contribution of these AMPs to the shrimp offspring [36]. Interestingly, Suleiman et al. [23] detected the presence of the carcinin antimicrobial peptide (Type I crustin) at protein levels in the ovaries and oocytes of the shore crab C. maenas. The expression of crustins in early larval stages of L. vannamei suggests the participation of gene-encoded AMPs in a critical phase where the immune system has not been fully developed. Therefore, the specific role of Type II crustins during shrimp development claims for further investigations.
Figure 5.
Relative abundance of Crustin-like Lv (Type IIb) transcripts during shrimp development. Results are present as mean ± standard deviation (three biological replicates). The red dotted line indicates the basal expression level of the Crustin-like Lv gene in hemocytes from juvenile shrimp. EI: fertilized eggs at 0–4 h post-spawning; EII: fertilized eggs at 7–11 h post-spawning; NI: nauplius I; NV: nauplius V; ZI: protozoea I; ZIII: protozoea III; MI: mysis I; MIII: mysis III; PL2: postlarva 2; PL9: postlarva 9; PL17: postlarva 17. Different letters indicate significant differences among the developmental stages from EII to PL17 (one-way ANOVA/Tukey, p < 0.05). Asterisks (*) shows significant differences between each developmental stage and hemocytes from juveniles (one-way ANOVA/Tukey, p < 0.05).
2.6. Molecular Signatures for Type II Crustins in Penaeid Shrimp
With the comparative transcriptional profiling of Crustin Lv and Crustin-like Lv genes in hand, we focused our attention on the molecular diversity of Type II crustins in penaeid shrimp (Decapoda: Penaeidae). Full coding sequences of Type II crustins were systematically collected from both annotated (GenBank Nucleotide database) and non-annotated (EST and TSA libraries) nucleotide databases and used for multiple-sequence alignments (Figure 6 and Figure S1) and phylogenetic reconstructions (Figure 7). An important contribution to this analysis was undoubtedly the identification of Crustin-like sequences in an Occidental penaeid species, L. vannamei, since Type IIb crustins have been only reported in the Asian shrimps P. monodon, F. chinensis, F. indicus and M. japonicus [16,17,18,22]. However, our in silico mapping method failed to recover Type IIb crustin sequences in other penaeid shrimp from the Western Hemisphere (as well as shrimp from other genera), due to the lack of genomic and transcriptomic data available for those species. Moreover, it is important to point out the confused nomenclature adopted in the literature concerning the use of the terms “crustin” and “crustin-like peptides” [15,37,38], evidencing that the terminology of this AMP family need to be fully revised.
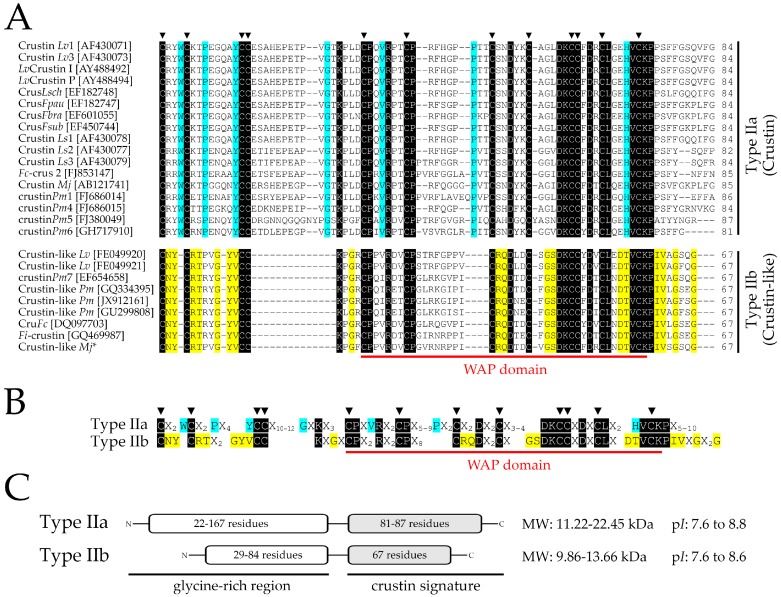
Figure 6.
The molecular signature of shrimp Type II crustins. (A) Amino acid sequence alignments of the C-terminal region holding the 12 conserved cysteine residues (the crustin signature [39]) of Type IIa and Type IIb crustins from penaeid shrimp: Litopenaeus vannamei (Crustin Lv1, Crustin Lv3, LvCrustin I, LvCrustin P, Crustin-like Lv), Litopenaeus setiferus (Crustin Ls1, Crustin Ls2, Crustin Ls3), Litopenaeus schmitti (CrusLsch), Farfantepenaeus paulensis (CrusFpau), Farfantepenaeus brasiliensis (CrusFbra), Farfantepenaeus subtilis (CrusFsub), Fenneropenaeus chinensis (Fc-crus 2, CruFc), Fenneropenaeus indicus (Fi-crustin), Penaeus monodon (crustinPm1, crustinPm4, crustinPm5, crustinPm6, crustinPm7, Crustin-like Pm), and Marsupenaeus japonicus (Crustin Mj, Crustin-like Mj). (B) Consensus amino acid sequence of shrimp Type IIa and Type IIb crustins. X indicates any amino acid. Identical residues are highlighted in black. Specific amino acid residues conserved in shrimp Type IIa and Type IIb crustins are highlighted in blue and yellow, respectively. Triangles (▼) indicate the 12 conserved cysteine residues found in crustins. The whey acidic protein (WAP) domain is underlined by a solid red line. (C) A not-to-scale representation of shrimp Type IIa and Type IIb crustins indicating the N-terminal glycine-rich region and the C-terminal crustin signature (cysteine-rich region + WAP domain). MW: molecular weight. pI: theoretical isoelectric point. * Sequence obtained from [16] (not deposited in any database).
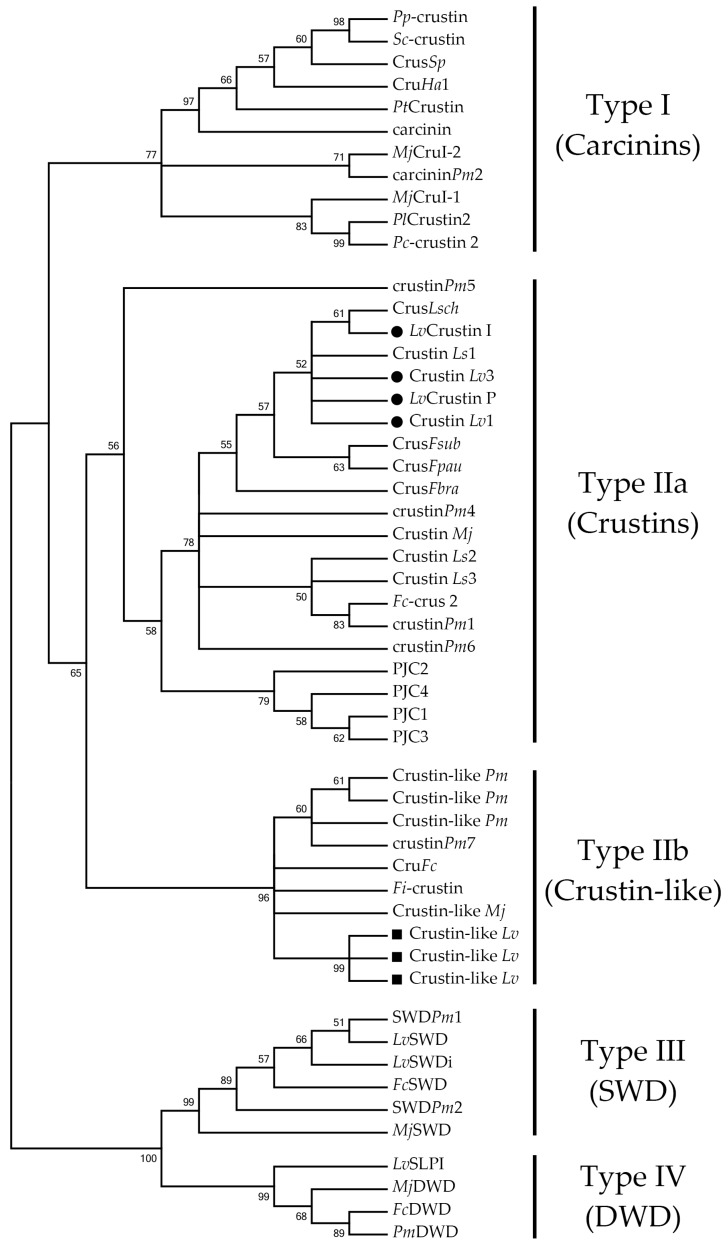
Figure 7.
Penaeid shrimp Type II crustins form two distinct phylogenetic clades. The phylogenetic tree was constructed using the Neighbor-Joining method in MEGA 6. Bootstrap sampling was reiterated 1000 times. Sequences included in analyses were the following: (i) Type I crustins (“Carcinins”): the Pp-crustin from Portunus pelagicus (GenBank: JQ965930), the Sc-crustin from Scylla serrata (GenBank: HQ638025), the CrusSp from Scylla paramamosain (GenBank: EU161287), the CruHa1 from Hyas araneus (GenBank: EU921641), PtCrustin from Portunus trituberculatus (GenBank: FJ612106), the carcinin (11.5-kDa peptide) from Carcinus maenas (GenBank: AJ427538), the PlCrustin2 from Pacifastacus leniusculus (GenBank: EF523613), the Pc-crustin 2 from Procambarus clarkii (GenBank: GQ301202), the MjCruI-1 and MjCruI-2 from Marsupenaeus japonicus [11], and the carcininPm2 from Penaeus monodon [10]; (ii) Type IIa crustins (“Crustins”): the LvCrustin I (GenBank: AY488492), LvCrustin P (GenBank: AY488494), Crustin Lv1 (GenBank: AF430071) and Crustin Lv3 (GenBank: AF430073) from Litopenaeus vannamei (indicated by black circles, ●), the Crustin Ls1 (GenBank: AF430078), Crustin Ls2 (GenBank: AF430077) and Crustin Ls3 (GenBank: AF430079) from Litopenaeus setiferus, the CrusLsch (GenBank: EF182748) from Litopenaeus schmitti, the CrusFpau (GenBank: EF182747) from Farfantepenaeus paulensis, the CrusFbra (GenBank: EF601055) from Farfantepenaeus brasiliensis, the CrusFsub (GenBank: EF450744) from Farfantepenaeus subtilis, the Crustin Mj (GenBank: AB121741) from M. japonicus, the crustinPm1 (GenBank: FJ686014), crustinPm4 (GenBank: FJ686015), crustinPm5 (GenBank: FJ380049) and crustinPm6 (GenBank: GH717910) from P. monodon, the Fc-crus 2 (GenBank: FJ853147) from Fenneropenaeus chinensis and the PJC1 (GenBank: FJ797417), PJC2 (GenBank: FJ797418), PJC3 (GenBank: FJ797419), and PJC4 (GenBank: FJ797420) from Panulirus japonicus; (iii) Type IIb crustins (“Crustin-like”): the Crustin-like Lv (GenBank: JQ824114, FE049920 and FE049921) from L. vannamei (indicated by black squares, ■), the Crustin-like Mj from M. japonicus [16], the CruFc (GenBank: DQ097703) from F. chinensis, the Fi-crustin (GenBank: GQ469987) from Fenneropenaeus indicus and the Crustin-like Pm (GenBank: GQ334395, JX912161 and GU299808) and crustinPm7 (GenBank: EF654658) from P. monodon; (iv) Type III crustins (“Single WAP domain-containing proteins or SWD”): the LvSWD (GenBank: AY464465) and LvSWDi [40] from L. vannamei, the SWDPm1 (GenBank: EU623979) and SWDPm2 (GenBank: EU623980) from P. monodon, the FcSWD (GenBank: EF216349) from F. chinensis and the MjSWD (GenBank: AU176270) from M. japonicus; (v) Type IV crustins (“Double WAP domain-containing proteins or DWD”): the LvSLPI (GenBank: EF467169) from L. vannamei, the PmDWD (GenBank: BI784457) from P. monodon, the FcDWD (GenBank: GQ303571) from F. chinensis, and the MjDWD (GenBank: EU095018) from M. japonicus.
The striking information given by the sequence analysis was the identification of amino acid signatures specific to Type IIa and Type IIb crustins in penaeid shrimp. We found a molecular pattern based on conserved amino acid residues that can successfully discriminate Type IIa and Type IIb crustins at the primary structure level (Figure 6). The classification of Type II crustins into two sub-groups (Type IIa and IIb) was initially proposed by Tassanakajon et al. [13], that is essentially based on differences in the amino acid length of the glycine-rich region and on the distance between the cysteine-rich region and the WAP domain. By contrast, multiple alignments of the amino acid sequences of all available shrimp Type II crustins revealed that the differences between Type IIa and Type IIb crustins lie in (1) the presence of specific amino acid residues positioned at precise locations of the C-terminal region holding the cysteine-rich region and the WAP domain, and (2) the arrangement (amino acid intervals) of the 12 conserved cysteine residues (Figure 6A,B). These 12 conserved cysteine residues (four from the cysteine-rich region and eight from the WAP domain) comprise a region previously termed as “crustin domain” (the crustin signature) [39] that is exclusively found in Type I (carcinins) and Type II (crustins and crustin-like peptides) crustins. On the other hand, Type III and Type IV crustins possess one and two WAP domains, respectively, but not the cysteine-rich region found in Type I and Type II crustins [5]. Thus, both Type III (SWD) and Type IV (DWD) crustins lack the crustin signature proposed by Zhao and Wang [39].
Type IIa crustins from penaeid shrimp contain three amino acid residues between the first two cysteine residues of the crustin signature (Cys1-Cys2) within a conserved tryptophan (Trp or W) preceding Cys2. Additionally, a sequence of 16–18 amino acids in length is found between Cys4-Cys5 and a sequence of 8–12 residues between Cys6-Cys7 (Figure 6A). Comparatively, Type IIb crustins contain an asparagine (Asn or N) and a tyrosin (Tyr or Y) between Cys1-Cys2 and only four residues between Cys4-Cys5 (Lys-Pro/Leu-Gly-Arg/Phe) (Figure 6A). Moreover, all Type IIb crustins contain a sequence of eight residues between Cys6-Cys7 and characteristic amino acid residues in the WAP domain that are not found in Type IIa crustins (Figure 6A). The specific amino acid signature of each sub-group of shrimp Type II crustins is presented in Figure 6B. Finally, whereas the N-terminal glycine-rich region of Type IIb crustins are composed of 29–84 amino acid residues, the glycine-rich region of Type IIa crustins can reach up to 167 residues in length (e.g., crustinPm4 from P. monodon [10]). On the other hand, whereas the amino acid length of the C-terminal region of Type IIa crustins showed to be variable (81–87 residues), the C-terminal region (holding the crustin signature) of all Type IIb crustins is composed of 67 amino acid residues (Figure 6C). Despite their differences in terms of size and molecular weight, shrimp Type II crustins are cationic peptides/polypeptides with theoretical isoelectric point (pI) ranging from 7.6 to 8.8.
Phylogenetic reconstructions clearly supported the distinction of shrimp Type II crustins into two separate groups. The obtained phylogenetic trees first showed that the four crustin Types found in crustaceans (Types I to IV) clustered into two distinct main clades: a first clade covering both Type I and Type II crustins and a second clade containing the sequences that lack the crustin signature (Type III and Type IV crustins) (Figure 7). Regarding the first clade, Type I and Type II crustins were split into distinct groups. Within the Type II group, shrimp Type IIa crustins (Dendrobranchiata) and crustins from the spiny lobster P. japonicus (Pleocyemata) formed a separate clade from Type IIb crustins (Figure 7). Indeed, as shrimp Type II crustins, P. japonicus crustins (PJC1 to PJC4) also harbor a portion rich in glycine residues at the N-terminal region [24]. From our analysis, P. japonicus crustins correspond to authentic Type II crustins since they hold the conserved amino acid residues found in the Sub-type IIa (Figure 6B). Moreover, while Type IIa crustins showed to be present in different decapod groups (Pleocyemata and Dendrobranchiata), Type IIb crustins appear to be exclusive of penaeid shrimp (Dendrobranchiata). Interestingly, the Type IIa crustinPm5 from P. monodon formed a distinct clade from the other Type IIa crustins from penaeid shrimp (Dendrobranchiata) and P. japonicus (Pleocyemata). Indeed, crustinPm5 is a unique crustin member within the Type IIa clade, apparently exclusive of the black tiger shrimp P. monodon, that is diverse not only in terms of sequence, but also in terms of genomic organization and tissue expression distribution [25]. Both sequence and phylogenetic data showed that Type IIa crustins display a high degree of diversity at both inter- and intraspecific levels when compared to Type IIb crustins. For instance, Type IIb crustins from distant shrimp species have been shown to be more similar to each other than Type IIa crustins from the same shrimp. This is the case of P. monodon Type IIa crustins that are diverse not only in terms of sequence, but also in genomic organization and gene expression regulation [10,27].
Why Type IIb crustins are quite similar among distant shrimp species is still an open question. On one hand, Type IIb crustins could be recently evolved from an ancestor shrimp Type II gene and, on the other hand, the interspecific conservation of Type IIb crustin sequences could be the result of selective pressures (environmental stressors, pathogens, etc.). In any cases, phylogenetic and maximum likelihood-based codon substitution analyses have shown that diversity in crustin family has been driven by successive gene duplications and positive Darwinian selection [26]. Our transcriptional data revealed that L. vannamei Type II crustins display a very similar pattern of gene expression in terms of tissue distribution, transcriptional response to pathogens, and mRNA abundance during shrimp development. Taken together, it is plausive to hypothesize that Type IIa and Type IIb crustins evolved by gene duplication from a common ancestral gene that conserved a similar pattern of transcriptional regulation. The apparent lack of Type IIb crustin sequences in available databases of other crustacean groups suggests that the gene duplication event relative to Type IIa–Type IIb divergence occurred exclusively in the Dendrobranchiata lineage.
3. Materials and Methods
3.1. Sequence Data Analysis and Phylogeny
Annotated Type IIb sequences (Crustin-like peptides) were systematically collected from publicly accessible databases and used for the search of homologous sequences in L. vannamei annotated (GenBank Nucleotide) and non-annotated EST (Expressed Sequence Tags) and TSA (Transcriptome Shotgun Assembly) databases. The complete nucleotide sequence of the Type II crustin genes was obtained by mining the genome of L. vannamei [41]. Homology searches were performed using BLAST at NCBI. All nucleotide sequences were manually inspected and translated using the ExPASy Translate Tool (http://web.expasy.org/translate/).
Prediction of signal peptide was performed with the SignalP 4.1 program (http://www.cbs.dtu.dk/services/SignalP/) and the theoretical isoelectric point (pI) and molecular weight (MW) of the mature peptides were predicted using the ExPASy ProtParam Tool (http://web.expasy.org/protparam/). Multiple alignments of the deduced amino acid sequences were generated using the BioEdit version 7.0 sequence alignment editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Phylogenetic analysis based on the amino acid sequences of crustins from different crustaceans were conducted in MEGA version 6.0 [41] using the Neighbor-Joining method (complete deletion option). Trees were resampled 1000 times.
3.2. Animals, Microbial Stimulation and Tissue Collection
Litopenaeus vannamei juveniles (10 ± 2 g) were obtained from the Laboratory of Marine Shrimps (Federal University of Santa Catarina, Brazil). Following acclimation (one week), five animals were injected with 5 × 107 colony-forming units (CFU)/animal of heat-killed (70 °C for 20 min) Vibrio harveyi ATCC 14126 under 100 µL sterile seawater (SSW). Unchallenged shrimp (n = 5) were used as control. At 48 h post-stimulation, circulating hemocytes were obtained as previously described [33] and shrimp organs (gills, muscle, nerve cord, foregut, hepatopancreas, midgut and hindgut) were harvested by dissection. Tissue samples were washed in a Tris-saline solution (10 mM Tris, 330 mM NaCl, pH 7.4), homogenized in TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and immediately processed for RNA isolation and tissue distribution analysis.
3.3. Experimental Infections
The experimental infections were performed with two shrimp pathogens, the Gram-negative Vibrio harveyi ATCC 14126 and the White spot syndrome virus (WSSV). For the bacterial infection, shrimp were injected with 6 × 107 CFU/animal of live V. harveyi ATCC 14126 (under 100 µL SSW) or with 100 µL SSW (aseptic injury control). For the viral infection, shrimp were injected with 100 µL of a WSSV inoculum containing 3 × 102 viral particles. The WSSV inoculum was prepared as previously described [33]. Animals injected with 100 µL of a tissue homogenate prepared from WSSV-free shrimp were used as injury control for the viral infection. At 48 h post-infections, circulating hemocytes and midguts were collected and separately pooled (three pools of five animals per condition) for total RNA isolation and fluorescence-based RT-qPCR analysis. Unchallenged animals (naïve shrimp at time 0 h) were used as control for all experimental conditions.
3.4. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis for Tissue Distribution of Gene Expression
Total RNA was purified using TRIzol reagent (Thermo Fisher Scientific) according to manufacturer’s specifications, treated with DNaseI (Thermo Fisher Scientific) for 15 min at 37 °C to remove remaining genomic DNA and precipitated with 0.3 M sodium acetate (pH 5.2) and isopropanol (1:1; v:v). First strand cDNA was synthesized from 1 µg of total RNA using RevertAid Reverse Transcriptase (Thermo Fisher Scientific) and oligo(dT)12–18 primers. PCR reactions were conducted in a 15-µL reaction volume using 1 µL of synthesized cDNA (diluted 1:10) as template. The primer sequences are listed in Table 1. The PCR conditions were as follows: 1 cycle of denaturation at 95 °C for 10 min followed by 40 cycles of 95 °C for 45 s, 60 °C for 45 s and 72 °C for 45 s, and a final extension step of 72 °C for 7 min. The PCR products were analyzed by electrophoresis in a 1.5% agarose and stained by ethidium bromide. The expression of the β-actin gene was used to normalize the RT-PCR data for comparison.
Table 1.
Nucleotide sequences of primers used in this study.
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Amplicon |
|---|---|---|---|
| Primers for Tissue Distribution Analysis (RT-PCR) | |||
| β-actin | TAATCCACATCTGCTGGAAGGTGG | TCACCAACTGGGATGACATGG | 846 bp |
| Crustin Lv | CGAACCAGAGACACCTGTTG | CAGCACACTTGTAGTCGTTG | 113 bp |
| Crustin-like Lv | GCAGGATAAAGACAAGGC | GTAATTGCAGTTGAATCCGCC | 199 bp |
| Primers for Quantitative Analysis of Gene Expression (RT-qPCR) | |||
| LvEF1α | TGGCTGTGAACAAGATGGACA | TTGTAGCCCACCTTCTTGACG | 103 bp |
| LvRpS6 | AGCAGATACCCTTGGTGAAG | GATGCAACCACGGACTGAC | 193 bp |
| LvL40 | GAGAATGTGAAGGCCAAGATC | TCAGAGAGAGTGCGACCATC | 104 bp |
| LvRpS3A | GGCTTGCTATGGTGTGCTCC | TCATGCTCTTGGCTCGCTG | 101 bp |
| β-actin | CCACGAGACCACCTACAAC | AGCGAGGGCAGTGATTTC | 142 bp |
| Crustin Lv | CGAACCAGAGACACCTGTTG | CAGCACACTTGTAGTCGTTG | 113 bp |
| Crustin-like Lv | GCAGGATAAAGACAAGGC | GTAATTGCAGTTGAATCCGCC | 199 bp |
3.5. Fluorescence-Based Reverse Transcription Real-Time Quantitative PCR (RT-qPCR)
RT-qPCR amplifications were performed using the StepOne Plus Real-time PCR System (Thermo Fisher Scientific) in a final volume of 15 µL containing 0.3 µM of each primer, 7.5 µL of reaction mix (Maxima SYBR Green/ROX qPCR Master Mix 2×; Thermo Fisher Scientific) and 1 µL of diluted cDNA (1:20). Primer sequences are listed in Table 1. The PCR conditions were as follow: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Melt curve analysis (60–95 °C at a temperature transition rate of 0.05 °C/s) for each primer pair was performed to ensure primer specificity with continuous fluorescence acquisition The efficiency of amplification was determined with a standard curve using a five-point dilution curve of a pool of cDNA samples. The eukaryotic translation elongation factor 1-alpha (LvEF1α) and the ribosomal proteins LvRpS6, LvL40 and LvRpS3A were used as reference genes for RT-qPCR data normalization using the 2−ΔΔCq method [42]. Statistical significance was considered at p < 0.05 by one-way ANOVA followed by Tukey’s multiple comparison test.
3.6. Quantitative Gene Expression Analysis during Shrimp Development
Three biological replicates of 12 development stages of L. vannamei were collected: fertilized eggs at 0–4 h (EI) and at 7–11 h post-spawning (EII), nauplius I and V (NI and NV), protozoea I and III (ZI and ZIII), mysis I and III (MI and MIII) and postlarvae aged of 2, 9 and 17 days (PL2, PL9 and PL17). Crustin-like Lv transcript levels during shrimp development were quantified by RT-qPCR and normalized with the gene expression of LvRpS6 and β-actin, as previously described [36]. Hemocyte samples from juvenile shrimp (three pools of five animals) were used as control for calibrating gene expression data. Statistical significance was considered at p < 0.05 by one-way ANOVA followed by Tukey’s multiple comparison test.
4. Conclusions
In conclusion, we have identified for the first time a Type IIb crustin (Crustin-like Lv) in an Occidental (non-Asian) penaeid, the Pacific white shrimp L. vannamei. The molecular characterization and comparison of Crustin-like Lv with other crustin sequences allowed to the identification of molecular amino acid signatures specific to Type II crustins in penaeid shrimp. Despite their similar patterns of gene expression, Type IIa and Type IIb form two distinct groups of crustins in penaeid shrimp with potential specific biological activities. Results from the in silico and phylogenetic analyses combined with our gene expression data suggested that shrimp Type II crustins evolved from a common ancestral gene that conserved a similar pattern of transcriptional regulation. The functional implication of the molecular diversity of Type II crustins in the shrimp immune response is an important task to be assessed by reverse genetic approaches. These bioactive molecules from marine invertebrates could find applications not only in aquaculture and shrimp farming, but also in both human and veterinary medicine.
Acknowledgments
We are grateful to the Laboratory of Marine Shrimp (Federal University of Santa Catarina—UFSC) for providing the shrimp used in this study. This work was funded by the Brazilian funding agencies CNPq (MEC/MCTI/CAPES/CNPq/FAPs PVE 401191/2014-1 and MCTI/CNPq Universal 406530/2016-5) and CAPES (CIMAR 1974/2014) and by the National High-Technology Research and Development Program of China (863 program; 2012AA10A404, 2012AA092205). C.B. and J.d.R.C. were supported by scholarships provided by CAPES.
Supplementary Materials
The following are available online at www.mdpi.com/1660-3397/16/1/31/s1, Figure S1: Amino acid sequence alignments of shrimp Type II crustins: Litopenaeus vannamei (LvCrustin P, LvCrustin I, Crustin Lv1, Crustin Lv3, Crustin-like Lv), Litopenaeus setiferus (Crustin Ls1, Crustin Ls2, Crustin Ls3), Farfantepenaeus paulensis (CrusFpau), Farfantepenaeus brasiliensis (CrusFbra), Farfantepenaeus subtilis (CrusFsub), Litopenaeus schmitti (CrusLsch), Marsupenaeus japonicus (Crustin Mj, Crustin-like Mj), Fenneropenaeus chinensis (Fc-crus 2, CruFc), Penaeus monodon (crustinPm1, crustinPm4, crustinPm5, crustinPm6, crustinPm7, Crustin-like Pm) and Fenneropenaeus indicus (Fi-crustin). The predicted signal peptides are in bold and underlined. Identical amino acid residues are shadowed with black backgrounds. Triangles (▼) indicate the 12 conserved cysteine residues found in crustins. * Sequence obtained from [16] (not deposited in any database). Figure S2: Genomic nucleotide and deduced amino acid sequences of Type IIa Crustin Lv (a) and Type IIb Crustin-like Lv (b). The predicted signal peptides are in bold and underlined. The exon and intron sequences are shown in black and grey, respectively. A dash (-) marks the stop codon.
Author Contributions
R.D.R. and L.M.P. conceived and designed the experiments; C.B. and J.d.R.C. performed the experiments; C.B., J.Y., J.X., L.M.P. and R.D.R. analyzed the data; C.B. and R.D.R. wrote the paper. All authors commented on the manuscript and discussed the data and implications.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Smith V.J., Dyrynda E.A. Antimicrobial proteins: From old proteins, new tricks. Mol. Immunol. 2015;68:383–398. doi: 10.1016/j.molimm.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Ranganathan S., Simpson K.J., Shaw D.C., Nicholas K.R. The whey acidic protein family: A new signature motif and three-dimensional structure by comparative modeling. J. Mol. Graph. Model. 1999;17:106–113. doi: 10.1016/S1093-3263(99)00023-6. [DOI] [PubMed] [Google Scholar]
- 3.Relf J.M., Chisholm J.R., Kemp G.D., Smith V.J. Purification and characterization of a cysteine-rich 11.5-kDa antibacterial protein from the granular haemocytes of the shore crab, Carcinus maenas. Eur. J. Biochem. 1999;264:350–357. doi: 10.1046/j.1432-1327.1999.00607.x. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett T.C., Cuthbertson B.J., Shepard E.F., Chapman R.W., Gross P.S., Warr G.W. Crustins, homologues of an 11.5-kDa antibacterial peptide, from two species of penaeid shrimp, Litopenaeus vannamei and Litopenaeus setiferus. Mar. Biotechnol. 2002;4:278–293. doi: 10.1007/s10126-002-0020-2. [DOI] [PubMed] [Google Scholar]
- 5.Smith V.J., Fernandes J.M.O., Kemp G.D., Hauton C. Crustins: Enigmatic WAP domain-containing antibacterial proteins from crustaceans. Dev. Comp. Immunol. 2008;32:758–772. doi: 10.1016/j.dci.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Rosa R., Barracco M. Antimicrobial peptides in crustaceans. Invertebr. Surviv. J. 2010;7:262–284. [Google Scholar]
- 7.Tassanakajon A., Rimphanitchayakit V., Visetnan S., Amparyup P., Somboonwiwat K., Charoensapsri W., Tang S. Shrimp humoral responses against pathogens: Antimicrobial peptides and melanization. Dev. Comp. Immunol. 2017 doi: 10.1016/j.dci.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Destoumieux-Garzón D., Rosa R.D., Schmitt P., Barreto C., Vidal-Dupiol J., Mitta G., Gueguen Y., Bachère E. Antimicrobial peptides in marine invertebrate health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016 doi: 10.1098/rstb.2015.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockton V., Hammond J., Smith V. Gene characterisation, isoforms and recombinant expression of carcinin, an antibacterial protein from the shore crab, Carcinus maenas. Mol. Immunol. 2007;44:943–949. doi: 10.1016/j.molimm.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Donpudsa S., Visetnan S., Supungul P., Tang S., Tassanakajon A., Rimphanitchayakit V. Type I and type II crustins from Penaeus monodon, genetic variation and antimicrobial activity of the most abundant crustinPm4. Dev. Comp. Immunol. 2014;47:95–103. doi: 10.1016/j.dci.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Jiang H.-S., Jia W.-M., Zhao X.-F., Wang J.-X. Four crustins involved in antibacterial responses in Marsupenaeus japonicus. Fish Shellfish Immunol. 2015;43:387–395. doi: 10.1016/j.fsi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Amparyup P., Donpudsa S., Tassanakajon A. Shrimp single WAP domain (SWD)-containing protein exhibits proteinase inhibitory and antimicrobial activities. Dev. Comp. Immunol. 2008;32:1497–1509. doi: 10.1016/j.dci.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Tassanakajon A., Somboonwiwat K., Amparyup P. Sequence diversity and evolution of antimicrobial peptides in invertebrates. Dev. Comp. Immunol. 2015;48:324–341. doi: 10.1016/j.dci.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Shockey J.E., O’Leary N.A., de la Vega E., Browdy C.L., Baatz J.E., Gross P.S. The role of crustins in Litopenaeus vannamei in response to infection with shrimp pathogens: An in vivo approach. Dev. Comp. Immunol. 2009;33:668–673. doi: 10.1016/j.dci.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Hipolito S.G., Shitara A., Kondo H., Hirono I. Role of Marsupenaeus japonicus crustin-like peptide against Vibrio penaeicida and white spot syndrome virus infection. Dev. Comp. Immunol. 2014;46:461–469. doi: 10.1016/j.dci.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Sun B., Wang Z., Zhu F. The crustin-like peptide plays opposite role in shrimp immune response to Vibrio alginolyticus and white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol. 2017;66:487–496. doi: 10.1016/j.fsi.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J., Li F., Wang Z., Xiang J. Cloning and recombinant expression of a crustin-like gene from Chinese shrimp, Fenneropenaeus chinensis. J. Biotechnol. 2007;127:605–614. doi: 10.1016/j.jbiotec.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Amparyup P., Kondo H., Hirono I., Aoki T., Tassanakajon A. Molecular cloning, genomic organization and recombinant expression of a crustin-like antimicrobial peptide from black tiger shrimp Penaeus monodon. Mol. Immunol. 2008;45:1085–1093. doi: 10.1016/j.molimm.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Supungul P., Klinbunga S., Pichyangkura R., Jitrapakdee S., Hirono I., Aoki T., Tassanakajon A. Identification of immune-related genes in hemocytes of black tiger shrimp (Penaeus monodon) Mar. Biotechnol. 2002;4:487–494. doi: 10.1007/s10126-002-0043-8. [DOI] [PubMed] [Google Scholar]
- 20.Rosa R.D., Bandeira P.T., Barracco M.A. Molecular cloning of crustins from the hemocytes of Brazilian penaeid shrimps. FEMS Microbiol. Lett. 2007 doi: 10.1111/j.1574-6968.2007.00866.x. [DOI] [PubMed] [Google Scholar]
- 21.Sun C., Du X.-J., Xu W.-T., Zhang H.-W., Zhao X.-F., Wang J.-X. Molecular cloning and characterization of three crustins from the Chinese white shrimp, Fenneropenaeus chinensis. Fish Shellfish Immunol. 2010;28:517–524. doi: 10.1016/j.fsi.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Antony S.P., Bright Singh I.S., Philip R. Molecular characterization of a crustin-like, putative antimicrobial peptide, Fi-crustin, from the Indian white shrimp, Fenneropenaeus indicus. Fish Shellfish Immunol. 2010;28:216–220. doi: 10.1016/j.fsi.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Pisuttharachai D., Fagutao F.F., Yasuike M., Aono H., Yano Y., Murakami K., Kondo H., Aoki T., Hirono I. Characterization of crustin antimicrobial proteins from Japanese spiny lobster Panulirus japonicus. Dev. Comp. Immunol. 2009;33:1049–1054. doi: 10.1016/j.dci.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Vatanavicharn T., Supungul P., Puanglarp N., Yingvilasprasert W., Tassanakajon A. Genomic structure, expression pattern and functional characterization of crustinPm5, a unique isoform of crustin from Penaeus monodon. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2009;153:244–252. doi: 10.1016/j.cbpb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Padhi A. Adaptive evolution of crustin antimicrobial peptides in decapods. Genetica. 2012;140:197–203. doi: 10.1007/s10709-012-9671-8. [DOI] [PubMed] [Google Scholar]
- 26.Supungul P., Tang S., Maneeruttanarungroj C., Rimphanitchayakit V., Hirono I., Aoki T., Tassanakajon A. Cloning, expression and antimicrobial activity of crustinPm1, a major isoform of crustin, from the black tiger shrimp Penaeus monodon. Dev. Comp. Immunol. 2008;32:61–70. doi: 10.1016/j.dci.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y.-C., Chang P.-S., Chen H.-Y. Tissue expressions of nine genes important to immune defence of the Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2007;23:1161–1177. doi: 10.1016/j.fsi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Yang C.-C., Lu C.-L., Chen S., Liao W.-L., Chen S. Immune gene expression for diverse haemocytes derived from pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 2015;44:265–271. doi: 10.1016/j.fsi.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Davies L.C., Taylor P.R. Tissue-resident macrophages: Then and now. Immunology. 2015;144:541–548. doi: 10.1111/imm.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suleiman S., Smith V.J., Dyrynda E.A. Unusual tissue distribution of carcinin, an antibacterial crustin, in the crab, Carcinus maenas, reveals its multi-functionality. Dev. Comp. Immunol. 2017;76:274–284. doi: 10.1016/j.dci.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Saraiva R.G., Kang S., Simões M.L., Angleró-Rodríguez Y.I., Dimopoulos G. Mosquito gut antiparasitic and antiviral immunity. Dev. Comp. Immunol. 2016;64:53–64. doi: 10.1016/j.dci.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Goncalves P., Guertler C., Bachère E., de Souza C.R.B., Rosa R.D., Perazzolo L.M. Molecular signatures at imminent death: Hemocyte gene expression profiling of shrimp succumbing to viral and fungal infections. Dev. Comp. Immunol. 2014 doi: 10.1016/j.dci.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 33.Soonthornchai W., Rungrassamee W., Karoonuthaisiri N., Jarayabhand P., Klinbunga S., Söderhäll K., Jiravanichpaisal P. Expression of immune-related genes in the digestive organ of shrimp, Penaeus monodon, after an oral infection by Vibrio harveyi. Dev. Comp. Immunol. 2010;34:19–28. doi: 10.1016/j.dci.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Destoumieux D., Muñoz M., Cosseau C., Rodriguez J., Bulet P., Comps M., Bachère E. Penaeidins, antimicrobial peptides with chitin-binding activity, are produced and stored in shrimp granulocytes and released after microbial challenge. J. Cell Sci. 2000;113:461–469. doi: 10.1016/j.bbagen.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Quispe R.L., Justino E.B., Vieira F.N., Jaramillo M.L., Rosa R.D., Perazzolo L.M. Transcriptional profiling of immune-related genes in Pacific white shrimp (Litopenaeus vannamei) during ontogenesis. Fish Shellfish Immunol. 2016;58:103–107. doi: 10.1016/j.fsi.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Hauton C., Brockton V., Smith V.J. Cloning of a crustin-like, single whey-acidic-domain, antibacterial peptide from the haemocytes of the European lobster, Homarus gammarus, and its response to infection with bacteria. Mol. Immunol. 2006;43:1490–1496. doi: 10.1016/j.molimm.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 37.Christie A.E., Rus S., Goiney C.C., Smith C.M., Towle D.W., Dickinson P.S. Identification and characterization of a cDNA encoding a crustin-like, putative antibacterial protein from the American lobster Homarus americanus. Mol. Immunol. 2007;44:3333–3337. doi: 10.1016/j.molimm.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X., Wang J. The antimicrobial peptides of the immune response of shrimp. Invertebr. Surviv. J. 2008;5:162–179. [Google Scholar]
- 39.Du Z.-Q., Yuan J.-J., Ren D.-M. A novel single WAP domain-containing protein isoform with antibacterial relevance in Litopenaeus vannamei. Fish Shellfish Immmunol. 2015;44:478–484. doi: 10.1016/j.fsi.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Abdelrahman H., ElHady M., Alcivar-Warren A., Allen S., Al-Tobasei R., Bao L., Beck B., Blackburn H., Bosworth B., Buchanan J., et al. Aquaculture genomics, genetics and breeding in the United States: Current status, challenges, and priorities for future research. BMC Genom. 2017;18:191. doi: 10.1186/s12864-017-3557-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.