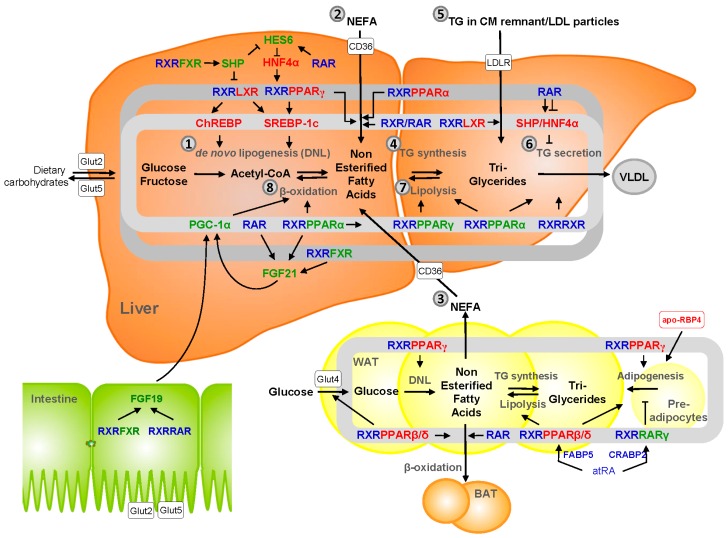
Figure 2.
Regulation of hepatic lipid metabolism by vitamin A metabolites. Triglyceride synthesis and breakdown is subdivided into eight steps: (1) de novo lipogenesis (DNL) in the liver, (2) influx of dietary lipids (delivered as non-esterified free fatty acids (NEFAs) or as triglycerides (TG) in chylomicrons), (3) influx of NEFAs produced by adipose tissue (primarily from white adipose tissue (WAT)), (4) esterification of lipids (mainly to TG) and packaging into lipid droplets, (5) influx of TG carried in CM remnants and low density lipoproteins (LDL), (6) efflux of TG carried in very low density lipoprotein (VLDL)-particles, (7) TG hydrolysis producing NEFAs, and (8) catabolism of NEFAs through mitochondrial and peroxisomal β-oxidation. Direct transcriptional regulation of lipogenic/lipolytic genes is shown in the inner (light gray) ring. Indirect transcriptional regulation is shown inside or outside the outer (dark gray) ring. Vitamin A-related factors are indicated in blue. Factors that promote lipogenesis are shown in red; factors promoting lipolysis in green. Relevant regulation and factors in adipose tissue and the intestine are also included (see main text for details about the specific genes that are regulated in each step). Additional abbreviations: FXR—Farnesoid X Receptor, SHP—Small Heterodimer Partner 1, HES6—Hes family BHLH transcription factor 6, HNF4α—Hepatocyte Nuclear Factor 4 alpha, LXR—Liver X Receptor, PPARγ—Peroxisome Proliferator-Activated Receptor gamma, chREBP—carbohydrate Response Element Binding Protein, SREBP-1c—Sterol Response Element Binding Protein-1c, PGC-1α—PPARγ-Coactivator 1-alpha, FGF—Fibroblast Growth Factor, BAT—Brown Adipose Tissue.