Abstract
Background
Skeletal muscle in livestock develops into meat, an important source of protein and other nutrients for human consumption. The muscle is largely composed of a fixed number of multinucleated myofibers determined during late gestation and remains constant postnatally. A population of postnatal muscle stem cells, called satellite cells, gives rise to myoblast cells that can fuse with the existing myofibers, thus increasing their size. This requires a delicate balance of transcription and growth factors and specific microRNA (miRNA) expressed by satellite cells and their supporting cells from the muscle stem cell niche. The role of transcription and growth factors in bovine myogenesis is well-characterized; however, very little is known about the miRNA activity during this process. We have hypothesized that the expression of miRNA can vary between primary cultures of skeletal muscle cells isolated from the semitendinosus muscles of different cattle breeds and subjected to myogenic differentiation.
Results
After a 6-day myogenic differentiation of cells isolated from the muscles of the examined cattle breeds, we found statistically significant differences in the number of myotubes between Hereford (HER)/Limousine (LIM) beef breeds and the Holstein-Friesian (HF) dairy breed (p ≤ 0.001). The microarray analysis revealed differences in the expression of 23 miRNA among the aforementioned primary cultures. On the basis of a functional analysis, we assigned 9 miRNA as molecules responsible for differentiation progression (miR-1, -128a, -133a, -133b, -139, -206, -222, -486, and -503). The target gene prediction and functional analysis revealed 59 miRNA-related genes belonging to the muscle organ development process.
Conclusion
The number of myotubes and the miRNA expression in the primary cultures of skeletal muscle cells derived from the semitendinosus muscles of the HER/LIM beef cattle breeds and the HF dairy breed vary when cells are subjected to myogenic differentiation. The net effect of the identified miRNA and their target gene action should be considered the result of the breed-dependent activity of satellite cells and muscle stem cell niche cells and their mutual interactions, which putatively can be engaged in the formation of a larger number of myotubes in beef cattle-related cells (HER/LIM) during in vitro myogenesis.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4492-5) contains supplementary material, which is available to authorized users.
Keywords: microRNA, Myogenesis, Bovine satellite cells, Myoblast differentiation, Cattle
Background
Skeletal muscle in livestock occupies approximately 40% of the animal body weight and develops into an important source of protein and other nutrients for human consumption [1, 2]. Skeletal muscle is largely composed of multinucleated muscle cells called myofibers. In cattle, the number of muscle fibers, a key determinant of the postnatal growth rate, is fixed late during the gestation period and remains constant postnatally, but each myofiber grows in size by the fusion of satellite cells [3, 4]. Activation of satellite cells (a population of postnatal muscle stem cells) give rise to myoblast cells that undergo multiple divisions before terminal differentiation. This can lead to fusion with the existing myofibers [5], which require a delicate balance between myogenic differentiation, myoblast proliferation, and the activity of transcription/growth factors and specific microRNA (miRNA). Signals from the surrounding cells, such as macrophages, fibroblasts, and muscle-resident stem cells support satellite cells in their action [6]. The effects of the involvement of transcription factors, such as MRF (MYF5, MYOD, myogenin, MRF4), and MEF2 families (reviewed in [7]), and the influence of the muscle stem cell niche on satellite cell behavior during skeletal muscle development is well-documented (reviewed in [8]), but little is known about the miRNA involvement in the regulation of this process, particularly in cattle.
Since the discovery of the first miRNA in 1993, the knowledge about miRNA’s impact on the development of different tissues has grown exponentially, including that in the skeletal muscle field [9, 10]. MicroRNA represents a class of ~ 22 nucleotide endogenous non-coding RNA molecules. These molecules modulate gene expression by base pairing to the 3’UTR regions of the target mRNA, leading to translational repression and/or mRNA cleavage [10]. Most of the miRNA regulating myogenic differentiation are found to be upregulated during this process [11]. Among them, the well-known muscle-specific miRNAs miR-1, -133a, -133b, and -206; also called myomiRs [11]. They play a fundamental role during muscle proliferation and differentiation by influencing a number of transcription factors and signaling molecules required for normal myogenesis progression.
Despite the increasing number of articles describing miRNA engagement in satellite cell activation, proliferation, and differentiation, the knowledge about their action, particularly in the development of bovine skeletal muscle, is still obscure. In this study, we focused on the differences in miRNA expression in a primary culture of skeletal muscle cells originating from the semitendinosus muscle of bulls of varying breeds and performance and subjected to myogenic differentiation.
Methods
Animals and tissue sampling
The experiment was conducted on primary cultures of skeletal muscle cells isolated from the semitendinosus muscle of 15-month-old bulls. The experimental groups were composed of Hereford (HER; high meat high-fat, maturating early, n = 4) and Limousin (LIM; high-meat low-fat, maturing late, n = 4) beef cattle bulls. Holstein-Friesian dairy cattle bulls (HF; typical dairy breed, maturing early with relatively poor carcass quality and a lower dressing percentage than beef bulls, n = 4) were used as the reference group. LIM and HER bulls were born in beef herds, while HF bulls in dairy herds. At the age of 2–3 months, the animals were transferred to the farm of Institute of Genetics and Animal Breeding Polish Academy of Sciences where they were housed in a loose barn until slaughter. The bulls were fed a total mixed ration (TMR) consisting of corn silage (75%), concentrates (20%), and hay (5%) and had access to water ad libitum [12, 13]. At the age of 15 months, all bulls were slaughtered after 24-h fasting. Samples of m. semitendinosus were immediately collected accordingly to the procedure described by Szcześniak et al. [14] and stored in liquid nitrogen until use.
Skeletal muscle cell isolation, proliferation, and differentiation
The collected skeletal muscle tissue samples were thawed, washed in phosphate buffered saline (PBS; Sigma-Aldrich, USA) and suspended in the incubation medium: Dulbecco’s Modified Eagle Medium (DMEM; Life Technologies, USA), Pronase from Streptomyces griseus (Sigma-Aldrich, USA) at 0.5 mg/ml, and penicillinum crystallisatum TZF (Polfa Tarchomin, Poland) at 10000 IU/100 mL, and pH 7.3. The samples were incubated for 1.5 h in 37 °C, subjected to repeated pipetting every 15 min. Then, the suspension was filtered through a 70-μm cell strainer (Becton Dickinson, USA) to separate the tissue debris. The filtrate was centrifuged thrice (20 min, 350 g), and the pellet was resuspended each time in a growth medium (GM: 10% fetal bovine serum [FBS, Life Technologies, USA]/DMEM/1% penicillin-streptomycin and 0.5% amphotericin B). As muscle tissue is mainly composed of myofibers, we decided to use 1-h preplating to increase the number of myoblasts in the culture. The preplating method is based on the fact that myoblasts adhere to a polystyrene culture dish slower than other cells such as fibroblasts [15]. The first preplating was executed after the abovementioned final centrifugation. After 1-h of preplating, the supernatant was centrifuged and the pellet resuspended in GM, transferred into 25-cm3 Primaria tissue culture flasks (Becton Dickinson, USA), and cultured at 37 °C in 5% CO2 and 95% humidity. The growth medium was changed every 48 h. During cell proliferation, on day 6, 8, and 10, the cells were trypsinized, centrifuged, resuspended in GM, subjected to 1-h preplating, and sieved into the Primaria tissue culture flasks. The four-time preplating allowed for 60%–70% myoblast purity, measured as a percentage of the M-cadherin positive cells (data not shown) [16]. In brief, the cells were fixed with 4% parafomaldehyde (Sigma-Aldrich, USA) for 30 min and washed with PBS. They were incubated with monoclonal anti-M-cadherin (611,100, BD Biosciences, USA) at 1:100 dilution with PBS-1% BSA (Sigma-Aldrich, USA) for 1 h, and then, exposed to the AlexaFluor488-conjugated goat anti-mouse antibody (Invitrogen, USA) diluted 1:500 with PBS. Finally, the cells were incubated in a 7-AAD (Sigma-Aldrich, USA) solution for nuclei staining and observed with a FV-500 laser scanning confocal microscope (Olympus, USA). The number of M-cadherin positive cells was determined with respect to the total number of nuclei in the primary cell culture.
After the final passage, cells were transferred (100,000 per well) on to a 6-well Collagen I coated plate (Becton Dickinson, USA) and cultured in GM in 5% CO2 at 37 °C and 95% humidity. After reaching 80% cell confluence, GM was replaced by a differentiation medium (DM: 2%HS/DMEM/1% penicillin-streptomycin and 0.5% amphotericin B) in which cells were incubated for the next 6 days. DM was replaced every 48 h. On day 6, differentiated cells with visible myotubes were washed with PBS and stored at − 80 °C until the analysis.
Myoblast fusion
To assess the number of myotubes on day 6 of the differentiation, the DM medium was discarded, cells were washed twice in ice-cold PBS, fixed in 75% methanol (15 min), stained with Giemsa dye (0.04% w/v in methanol; 5 min), and rinsed in distilled water (Sigma-Aldrich, USA). Nuclei were counted using a phase-contrast microscope (CK40, Olympus) randomly in five fields of view for each cell culture. Structures with at least three nuclei were considered myotubes (n = 4; see statistical analysis).
RNA isolation and validation
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol and validated using a Nanodrop spectrophotometer (Nanodrop Technologies, USA); its integrity was checked using a Bioanalyzer 2100 (Agilent Technologies, USA). Samples with RIN ≥ 9.5 were subjected to further analysis.
miRNA microarray analysis
In the present study, custom Bovine miRNA microarrays (8 × 60 K) (Agilent Technologies, USA) containing probes for 763 Bos taurus miRNAs were used (National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO): GPL19028). For the miRNA profiling, 100 ng of the total RNA from each sample (n = 4, see statistical analysis) was labeled and hybridized using miRNA Complete Labeling and Hyb Kit (Agilent Technologies, USA), according to the manufacturer’s protocol. RNA Spike-In Kit (Agilent Technologies, USA) was used as the internal control. Slides were scanned using Agilent Microarray Scanner (G2505C), and features were extracted using the Agilent Feature Extraction image analysis tool version 10.7.3.1 with default settings and default normalization scheme for Agilent one-color data (75th percentile scaling). The microarray data were statistically analyzed using Gene Spring 13 (Agilent Technologies, USA) and the default protocol for miRNA experiments, where one-way analysis of variance (ANOVA) with Benjamini-Hochberg multiple testing correction adjustment (FDR) was applied. MiRNAs with FDR ≤ 0.05 were selected as significantly differentially expressed. MiRNA with fold change of FC ≥ 2.0 were chosen as common for both HER/LIM primary cultures. The microarray experiment was performed according to the MIAME guidelines [17]. The data obtained in the microarray experiment were deposited in the NCBI GEO database and numbered GSE73778.
qPCR validation
To validate the microarray results, selected miRNAs were examined using a real-time polymerase chain reaction technique (qPCR). First strand cDNA was synthesized using 10 ng of the total RNA (n = 4) and the miRCURY LNA™ Universal RT cDNA Synthesis Kit II (Exiqon, Denmark) with UniSp6 Spike-in used as the internal control. qPCR analyses were performed using a SYBR® Green master mix, Universal RT (Exiqon, Denmark), as follows: polymerase activation at 95 °C for 10 min; amplification (40 cycles) including denaturation at 95 °C for 10 s, annealing at 60 °C for 1 min; melting curve included: denaturation at 95 °C for 0 s, annealing at 65 °C for 15 s, continuous melting at 95 °C for 0 s; and cooling at 40 °C for 30 s. Primers for the selected miRNA and reference U6 snRNA were provided by Exiqon (Denmark). Their sequences and miRBase accession numbers are listed in Table 1. Each sample was tested twice with a Stratagene Mx3005P thermal cycler (Agilent Technologies, USA).
Table 1.
miRNA primers used for validation of microarray results (Exiqon, Denmark)
| miRNA Primer | Target sequence | Accession (Exiqon) |
|---|---|---|
| hsa-miR-1 LNA™ PCR primer set, UniRT | UGGAAUGUAAAGAAGUAUGUAU | MIMAT0000416 |
| bta-miR-9-5p LNA™ PCR primer set, UniRT | UCUUUGGUUAUCUAGCUGUAUG | MIMAT0009389 |
| hsa-miR-128-3p LNA™ PCR primer set, UniRT | UCACAGUGAACCGGUCUCUUU | MIMAT0000424 |
| hsa-miR-133a-3p LNA™ PCR primer set, UniRT | UUUGGUCCCCUUCAACCAGCUG | MIMAT0000427 |
| hsa-miR-139-5p LNA™ PCR primer set, UniRT | UCUACAGUGCACGUGUCUCCAGU | MIMAT0000250 |
| hsa-miR-145-5p LNA™ PCR primer set, UniRT | GUCCAGUUUUCCCAGGAAUCCCU | MIMAT0000437 |
| hsa-miR-206 LNA™ PCR primer set, UniRT | UGGAAUGUAAGGAAGUGUGUGG | MIMAT0000462 |
| hsa-miR-486-5p LNA™ PCR primer set, UniRT | UCCUGUACUGAGCUGCCCCGAG | MIMAT0002177 |
| cfa-miR-503 LNA™ PCR primer set, UniRT | UAGCAGCGGGAACAGUACUG | MIMAT0006746 |
| bta-miR-660 LNA™ PCR primer set, UniRT | UACCCAUUGCAUAUCGGAGCUG | MIMAT0004344 |
|
U6 snRNA LNA™ PCR primer set, UniRT
(reference RNA) |
GUGCUCGCUUCGGCAGCACAUAU ACUAAAAUUGGAACGAUACAGAG AAGAUUAGCAUGGCCCCUGCGCA AGGAUGACACGCAAAUUCGUGAA GCGUUCCAUAUUUUU |
without MIMAT # |
A qPCR analysis for selected myogenesis-related genes that could interact with the identified miRNAs, such as myogenin (Myog, accession no. NM_001111325), Myod (accession no. NM_001111325), Myf5 (accession no. NM_001111325), and Mstn (accession no. NM_001111325), was also performed. For this purpose, primers were designed using the Primer3 software and Bos taurus mRNA sequences obtained from the NCBI Nucleotide database. Specificity of primers and secondary structure formation were checked as described previously [18]. First strand cDNA synthesis was performed using 1 μg of the total RNA and the Transcriptor First Strand cDNA Synthesis Kit (Roche, USA) with a mixture of oligo(dT)18 and random hexamer primers, according to the manufacturer’s protocol. qPCR primers sequences and conditions are listed in Additional file 1. The analysis was performed according to the methodology described earlier with glyceraldehyde-3-phosphate dehydrogenase (Gapdh, accession no. NM_001034034) used for normalization as the non-regulated reference gene [19]. Each sample was tested twice in a Stratagene Mx3005P thermal cycler (Agilent Technologies, USA). The amplification efficiency (E = 10(− 1/slope) - 1) was determined by plotting a comparative quantitation standard curve and was ≥0.9 for each gene. The qPCR analysis was conducted according to a standardized approach [20].
Target gene prediction
Target genes for differentially expressed miRNA were identified using TargetScan release 7.1 (http://www.targetscan.org/vert_71/). The TargetScan database was chosen because of the availability of bovine species for prediction. Scanning was performed for the target score (total context score) of ≤ − 0.3 and conserved/non-conserved miRNA families and target sites. Moreover, the Pathway Studio Web (Elsevier, Netherlands) with Enrichment Analysis of Selected Entities tool was also used for target gene searching. In the Pathway Studio Web analysis, the literature data were considered, proving real miRNA targets, or if not available, prediction was done on the basis of the PicTar and miRanda databases. Only genes found in both analyses were considered targets for the identified miRNAs.
Functional analysis
An ontological analysis of miRNAs and their direct/indirect target genes was done using The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 tool (http://david.ncifcrf.gov/) where Gene Ontology Enrichment was calculated using the EASE Score corrected for multiple hypotheses testing using the Benjamini and Hochberg false discovery rate (FDR ≤ 0.05). Further, a similar analysis was performed using the PANTHER database (http://pantherdb.org/) and the functional classification tool. The relevance networks of the identified miRNAs and the selected target genes were prepared in Pathway Studio Web by using the Pathway Builder tool.
Statistical analysis
A statistical analysis of the differences in the number of myotubes was performed using one-way ANOVA with Tukey post-hoc testing and a p value of ≤0.05. The microarray data analysis was performed using Gene Spring 13 (Agilent Technologies, USA) as described above. The miRNA and mRNA qPCR results were analyzed by the GenEx 6.0 (MultiD Analyses AB, Sweden) and Prism 5.0 (GraphPad Software, USA) software using the comparative Ct method [21]. Results with p ≤ 0.05 were considered to be statistically significant. Hierarchical clustering (Ward’s algorithm with Euclidean distance measure) and Spearman’s correlation coefficients were calculated using GenEx 6.0 (MultiD Analyses AB, Sweden). For each animal in the experimental groups (n = 4), two independent cell isolations were made (one from each half-carcass). Results from both half-carcasses were averaged for the animal, and then, the interbreed comparison and the statistical analysis were performed. In case of Gene Spring analysis, because of software limitations, it was not possible to average microarray results from both half-carcasses (two separate arrays) before the data processing. Gene Spring analysis was performed using 8 samples for each breed (4 animals × 2 half-carcasses).
Results
Differences in myotube formation in vitro
The myotube formation assessment on day 6 of the skeletal muscle cell differentiation revealed statistically significant differences in the number of myotubes among all examined breeds. The difference between cells originating from HER/LIM and HF bulls’ muscle was statistically significant (p ≤ 0.001) with a considerable increase in the number of myotubes in beef cattle-related cell cultures. A difference between both beef breeds was also noticed (p ≤ 0.05) (Fig. 1).
Fig. 1.
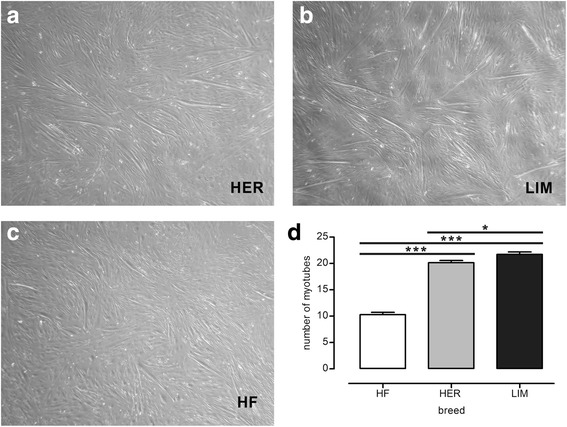
Myotube formation on day 6 of the differentiation of the primary cultures of skeletal muscle cells originating from the semitendinosus muscle of HER (a), LIM (b), and HF (c) breeds. Bar graph (d) presenting the difference in the number of myotubes between the examined cell cultures; results are shown as mean ± SEM with * and *** asterisks for p ≤ 0.05 and p ≤ 0.001, respectively; n = 4
Microarray analysis
The microarray analysis of the total RNA isolated from the aforementioned cell cultures revealed differences in the expression of 94 miRNAs (FDR ≤ 0.05) in the primary cultures originating from both beef breeds HER/LIM, when compared with the HF dairy breed-derived cells (Additional file 2). The 23 molecules whose expression was changed at least twice in both comparisons (HER vs. HF and LIM vs. HF; FDR ≤ 0.05, FC ≥ 2.0; both up- or downregulated) were classified as the miRNAs characteristic for beef cattle-originating primary cultures of skeletal muscle cells and as candidate molecules responsible for the significant increase in the number of myotubes observed in the HER/LIM cultures (Fig. 2). Among them, 17 miRNA molecules had an increased level of expression and 6 had a lower miRNA expression in the HER/LIM cells. The microarray results showed that the most upregulated expression changes were noticed for miR-139, miR-2469, and miR-486, while the highest downregulation was observed for miR-9, miR-29b, and miR-31 (Table 2). Further, as the number of myotubes varied between both beef breeds, a statistical analysis of the microarray results was performed for this comparison as well and revealed no differences in the miRNA expression between the HER and the LIM-derived cell cultures (FDR ≤ 0.05, t-test).
Fig. 2.
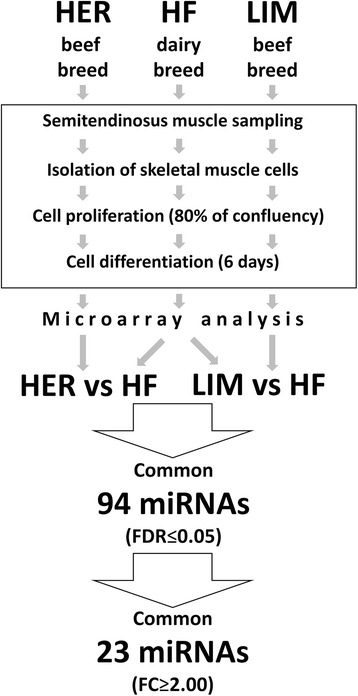
Experiment design and microarray results. HF - Holstein-Friesian; HER - Hereford; LIM - Limousine; n = 4
Table 2.
MiRNAs varying in expression in primary cultures of skeletal muscle cells isolated from semitendinosus muscle of both beef breed bulls (HER and LIM), when compared with the dairy breed (HF; the reference)
| No. | Systematic name | p (Corr) | FC (HER vs. HF) | Log FC (HER vs. HF) | Regulation (HER&LIM vs. HF) | FC (LIM vs. HF) | Log FC (LIM vs. HF) | Active Sequence | miRBase Accession No. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | bta-miR-139 | 3.07E-13 | 123.08 | 6.94 | up | 126.11 | 6.98 | ACTGGAGACACGTGC | MIMAT0003788 |
| 2 | bta-miR-2469 | 1.07E-03 | 83.81 | 6.39 | up | 11.25 | 3.49 | AAGCCGCAGGCCC | MIMAT0012059 |
| 3 | bta-miR-486 | 2.06E-03 | 51.74 | 5.69 | up | 35.07 | 5.13 | CTCGGGGCAGCTCA | MIMAT0009329 |
| 4 | bta-miR-2439-3p | 6.58E-03 | 31.43 | 4.97 | up | 7.98 | 3.00 | TCTGCCTACCTGTCTTC | MIMAT0012014 |
| 5 | bta-miR-449a | 4.82E-02 | 21.21 | 4.41 | up | 10.53 | 3.40 | ACCAGCTAACAATACACTGC | MIMAT0009320 |
| 6 | bta-miR-503-5p | 1.26E-02 | 15.69 | 3.97 | up | 15.71 | 3.97 | CAGTACTGTTCCCGC | MIMAT0025557 |
| 7 | bta-miR-1 | 1.87E-06 | 12.44 | 3.64 | up | 14.67 | 3.87 | ATACATACTTCTTTACATTCC | MIMAT0009214 |
| 8 | bta-miR-133a | 1.68E-06 | 11.63 | 3.54 | up | 12.57 | 3.65 | CAGCTGGTTGAAGGGGAC | MIMAT0009225 |
| 9 | bta-miR-206 | 1.07E-05 | 11.60 | 3.54 | up | 13.36 | 3.74 | CCACACACTTCCTTAC | MIMAT0009260 |
| 10 | bta-miR-133b | 5.70E-06 | 11.53 | 3.53 | up | 12.12 | 3.60 | TAGCTGGTTGAAGGGGACC | MIMAT0009226 |
| 11 | bta-miR-542-5p | 2.59E-02 | 8.77 | 3.13 | up | 25.04 | 4.65 | CTCGTGACATGATGATC | MIMAT0013594 |
| 12 | bta-miR-128 | 2.59E-02 | 4.51 | 2.17 | up | 4.78 | 2.26 | AAAGAGACCGGTTCACTGT | MIMAT0003541 |
| 13 | bta-miR-660 | 5.18E-04 | 2.49 | 1.32 | up | 2.26 | 1.18 | CAGCTCCGATATGCAA | MIMAT0004344 |
| 14 | bta-miR-378b | 2.00E-03 | 2.42 | 1.28 | up | 2.38 | 1.25 | GCCTTCTGACTCCAAG | MIMAT0025535 |
| 15 | bta-miR-378c | 4.33E-03 | 2.25 | 1.17 | up | 2.18 | 1.13 | ACTTCTGACTCCAAGTC | MIMAT0025551 |
| 16 | bta-miR-30a-5p | 6.14E-03 | 2.14 | 1.10 | up | 2.51 | 1.33 | AGCTTCCAGTCGAGG | MIMAT0003841 |
| 17 | bta-miR-30f | 1.45E-02 | 2.14 | 1.10 | up | 2.86 | 1.52 | AGCTGAGAGTGTAGGGT | MIMAT0009282 |
| 18 | bta-miR-9 | 2.77E-02 | −40.65 | −5.35 | down | −16.45 | −4.04 | TCATACAGCTAGATAACCA | MIMAT0009389 |
| 19 | bta-miR-29b | 2.08E-03 | −18.13 | −4.18 | down | −4.24 | −2.08 | AAACACTGATTTCAAATGGT | MIMAT0003828 |
| 20 | bta-miR-31 | 1.99E-03 | −16.57 | −4.05 | down | −213.81 | −7.74 | AGCTATGCCAGCATCTT | MIMAT0003548 |
| 21 | bta-miR-194 | 4.25E-02 | −14.83 | −3.89 | down | −2.55 | −1.35 | TCCACATGGAGTTGCT | MIMAT0009254 |
| 22 | bta-miR-145 | 3.22E-02 | −3.38 | −1.76 | down | −2.82 | −1.5 | AGGGATTCCTGGGAAAAC | MIMAT0003542 |
| 23 | bta-miR-222 | 4.74E-02 | −2.53 | −1.34 | down | −2.12 | −1.08 | ACCCAGTAGCCAG | MIMAT0003530 |
FC fold change, HF Holstein-Friesian, HER Hereford, LIM Limousine; FDR ≤ 0.05; FC ≥ 2.0; n = 4 (see statistical analysis)
Functional analysis of identified miRNAs
A functional analysis of 23 identified miRNA molecules using Pathway Studio Web (Elsevier, Netherlands) allowed the identification of muscle development-related biological processes in which HER/LIM cell-specific miRNAs could be involved. According to the aforementioned database, a majority of the analyzed molecules were engaged in the myogenesis process (miR-1, -29b, -128, -133a, -133b, -139, -206, -222, -449a, -486, and -503). Among them, 9 miRNA were classified as involved in myoblast differentiation (miR-1, -128, -133a, -133b, -139, -206, -222, -486, and -503). Moreover, some of identified molecules were also annotated as taking part in myoblast proliferation (miR-1, -128, -133a, -133b, -139, and -206); myocyte function (miR-31, -133a, -145, and -222); myoblast fusion (miR-206, -222, and -486); and satellite cell activation (miR-1 and -206) (Fig. 3).
Fig. 3.
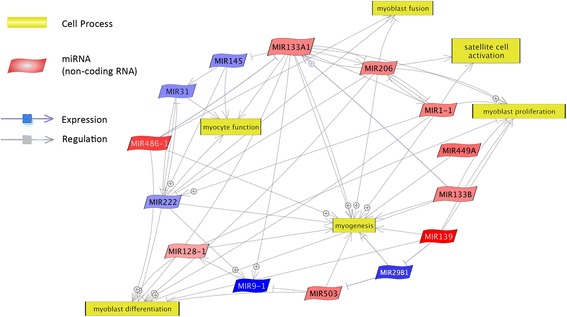
Relevance network of identified miRNAs and biological processes concerning skeletal muscle development. Upregulated and downregulated miRNAs were marked in red and blue, respectively (Pathway Studio Web; Elsevier, Netherlands)
Real-time qPCR validation
Based on the available literature and the Pathway Studio Web analysis, 10 out of the 23 miRNAs identified by the microarray technique were selected for the qPCR validation. Among them were muscle-specific myomiRs miR-1, -133a, -206, and miR-486 and non-myomiRs such as miR-9-5p, -128, -139, -145, -503, and -660. A statistical analysis of the qPCR results using the GenEx 6.0 software confirmed microarray data showing a significant upregulation of myomiRs in the HER/LIM cell cultures as compared to that in the HF cell cultures (Fig. 4A). For non-myomiRs, a statistically significant upregulation of miR-128 and -139 was confirmed. Statistically significant downregulation was confirmed for miR-9-5p and miR-145 only in the case of the HER cell cultures (for LIM, p ≤ 0.16 and p ≤ 0.053, respectively). The expression difference of miR-503 and miR-660 was not statistically significant in the qPCR analysis (Fig. 4B). All miRNAs validated by qPCR exhibited the same trend of expression change as in the microarray experiment.
Fig. 4.
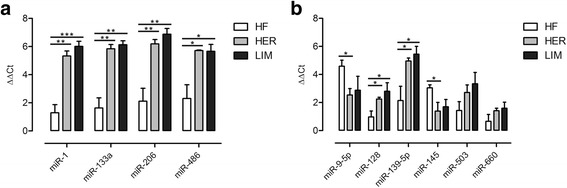
Real-time qPCR validation of microarray results for myomiRs (a) and non-myomiRs (b). Results are shown as mean ± SEM with *, **, and *** asterisks for p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively; n = 4
Expression of selected myogenesis-related genes
Based on previous publications, a few pivotal myogenesis-related genes (Myod, Myf5, myogenin and Mstn) described as influenced by some of the identified miRNAs were chosen. The qPCR analysis revealed an increased transcript level in both HER/LIM-derived cells only for myogenin mRNA (Fig. 5). The difference in the myogenin expression between HER and LIM was not statistically significant. Expression of the other examined genes was not statistically significant in both HER/LIM cell cultures as compared to that of the HF-derived cells.
Fig. 5.
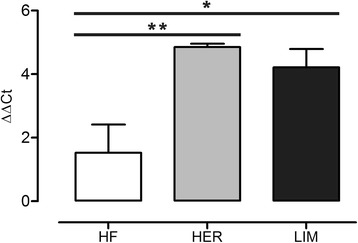
Expression of myogenin mRNA in primary cultures of skeletal muscle cells at 6th day of differentiation. Results are shown as mean ± SEM with * and ** asterisks for p ≤ 0.05 and p ≤ 0.01, respectively; n = 4
Hierarchical clustering and Spearman’s correlation coefficient
Hierarchical clustering for the validated miRNAs and myogenin was performed using GenEx 6.0 (MultiD Analyses AB, Sweden). A clear distinction was observed between the beef and the dairy cattle-derived cell cultures, showing a higher similarity between HER and LIM, than HF cells (Fig. 6). Further, Spearman’s correlation coefficients calculated for the validated miRNAs showed a strong or very strong uphill correlation among the myomiRs (miR-1, -133a, -206, and -486), miR-128, and miR-139. All the aforementioned correlations were significant with p ≤ 0.01 (Additional file 3). Moreover, strong uphill correlations were also shown for the aforementioned myomiRs and myogenin with p ≤ 0.05 (Additional file 3).
Fig. 6.
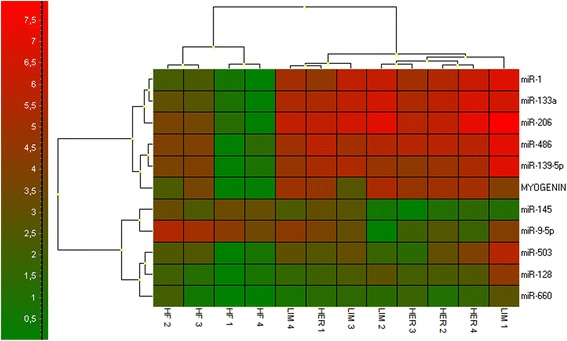
Hierarchical clustering of the validated miRNAs and myogenin (GenEx 6.0; MultiD Analyses AB, Sweden); n = 4
miRNA target gene prediction and functional analysis
The TargetScan database and the Pathway Studio algorithm (PicTar and miRanda) were used for the miRNA target gene prediction. TargetScan search resulted in 5142 records (total context score ≤ − 0.3) with 3918 unique targets (after the removal of duplicates). The Pathway Studio analysis revealed 3355 records and 2392 unique target genes for the set of the 23 identified miRNA molecules. Both sets of genes were compared, and the overlapping 1249 miRNA targets were considered significant (Additional file 4). The batch of all identified targets was loaded in the DAVID online software to find biological processes in which miRNA-related genes are involved. The functional analysis resulted in 192 processes (FDR ≤ 0.05) in which nearly 1250 genes were engaged (Additional file 5). Among them, 40 genes were involved in the muscle organ development process (Table 3). Moreover, the PANTHER online database was used for a functional analysis for the same group of targets, showing the involvement of 21 genes in the muscle organ development process (Table 3, Additional file 6). Both sets of targets belonging to the developmental process of a muscle organ were combined into one group of miRNA-related genes of interest. Surprisingly, only 5 target genes were similar for both analyses (DAVID/PANTHER) (Table 3).
Table 3.
Muscle organ development process-related target genes identified in DAVID and PANTHER. Full sets of identified processes and the corresponding target genes are listed in Additional files 5 (DAVID) and 6 (PANTHER)
| Selected muscle-related biological processes (DAVID) | |||
| Term | Count | Genes | FDR |
| GO:0007517 muscle organ development | 40 | Sri, Cav2, Mef2a , Utrn, Eln, Cacnb2 , Pax3 , Tagln2, Itgb1, Pten, Tgfb2, Gphn, Atg5, Gata6, Hlx, Pax7 , Col6a3, Rhoa, Ppp3ca, Rarb, Nr2f2, Col11a1, Foxl2, Actc1, Met, Mstn, Igf1, Tbx1, Mbnl1 , Sirt1, Prox1, Foxp1, Foxp2, Fxr1, Mapk14, Six1, Pdgfrb, Hbegf, Foxc1, Serp1 | 1.11E-04 |
| Selected muscle-related biological process (PANTHER) | |||
| Term | Count | Genes | |
| muscle organ development (GO:0007517) | 21 | Hoxa9, Lef1, Otx2, Lpp, Pax7 , Hoxb4, Lhx5, Hoxa11, Dlx3, Myo1c, Pdlim2, Myo5a, Myo10, Cacnb2 , Mef2a , Hoxa13, Pax3, Lhx8, Cdh24, Hoxc13, Mbnl1 | |
Underlined are genes common for both the DAVID and the PANTHER analyses. DAVID The Database for Annotation, Visualization and Integrated Discovery v6.7, PANTHER Protein ANalysis THrough Evolutionary Relationships Classification System, FDR false discovery rate
Apart from their involvement in the muscle organ development process, the identified target genes were classified as participating in positive (49) and negative (45) regulation of cell differentiation. Target genes belonging to all the identified processes are listed in Additional file 5.
Using the DAVID algorithm, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was searched, showing the involvement of the predicted targets in 56 signaling pathways (Additional file 7). This included MAPK, insulin signaling, mTOR, ErbB, TGFβ, and Wnt pathways (Additional file 8), which are mentioned in discussion as possibly regulated by the identified miRNAs.
Discussion
MiRNAs and their role in skeletal muscle development have been studied in recent years showing that myogenesis is governed not only by myogenic regulatory factors, (such as MYF5, MYOD, myogenin, MRF4), growth factors and the related proteins, but also by small non-coding RNA. These small non-coding RNA fragments cooperate with the abovementioned regulatory factors in executing myogenic cell proliferation, differentiation, fusion, and myofiber maturation during skeletal muscle growth [8]. Albrecht et al. [22, 23] in their in vivo study reported a tendency showing a higher number of muscle fibers per muscle bundle in beef than in dairy cattle during both late gestation and postnatal muscle maturation. However, a statistically significant difference was observed only in the case of the Belgian Blue breed when compared to Holstein-Friesian. Rehfeldt et al. [24] suggested that animals with a larger number of muscle fibers of moderate size produce more meat of better quality. In a previous publication, we reported the major carcass traits of three cattle breeds, with the lowest dressing percentage for the Holstein-Friesian breed and significantly higher for the Hereford and Limousin breeds [12].
Because muscle mass is determined, at least in part, by the number of muscle fibers already formed during fetal life [4], we decided to examine the differences in the in vitro differentiation of skeletal muscle cells between beef and dairy cattle-derived primary cultures and to investigate the breed-specific miRNA pattern characteristic for HER/LIM cell cultures.
In the proposed experimental model, we did not separate all the non-myogenic cells from the culture to allow the influence of the muscle stem cell niche cells on the satellite cells present in the primary culture. As the cells were isolated from the same portion of the semitendinosus muscle, in two independent isolations from both half-carcasses of each animal, using the same amount of tissue and the same methodology, we considered such prepared primary cultures as characteristic for the specific breed. To increase the number of satellite cells that we wanted to subject to differentiation in the primary cultures, the preplating procedure was used four times. The preplating was done in the same manner for all the isolations from all the animals. This approach should provide a condition similar to this in muscle in vivo (except the number of myoblast cells), where proliferating, differentiating, and fusing myoblasts are under the influence of the factors and metabolites released by neighboring cells representing the skeletal muscle stem cell niche [8]. It must be kept in mind that the considered primary cultures of skeletal muscle cells were not pure satellite cell cultures but contained an enhanced percentage of myogenic cells and different breed-dependent amount of other cells present in the muscle tissue. This could be a main source of variation of the myotube number and miRNA expression in the examined primary cultures of skeletal muscle cells.
As a result of the myogenic differentiation of the isolated cells, enhanced myotube formation was observed in the HER/LIM-derived primary cultures as compared to the HF cells (Fig. 1). It was accompanied by a differential expression of the 23 miRNAs (FDR ≤ 0.05, FC ≥ 2.0; Fig. 2, Table 2). Real-time qPCR validation of the selected miRNA confirmed the trends observed in the microarray analysis; however, the expression differences of miR-503 and miR-660 were not statistically significant (Fig. 4). Moreover, a higher expression of myogenin mRNA was detected in both HER/LIM breed-related cells (Fig. 5). A functional analysis of the identified miRNAs confirmed their involvement in myogenesis, particularly in the differentiation processes (Fig. 3). Considering the above data, in the following discussion, it should be kept in mind that the obtained results relate to the interbreed differences in the miRNA expression in the primary cell cultures subjected to differentiation, and they are not a proliferation vs. differentiation comparison. It is also important to note that the isolated cells were not pure satellite cell cultures and the interbreed differences in myogenic/non-myogenic cell content in the primary culture could be the main source of the observed variation in the miRNA expression.
In the following discussion, symbols of genes and their protein products have been presented in their original form depending on the specificity of the cited paper, as lowercase italic with the first letters in uppercase or as uppercase letters when they refer to transcriptomic or proteomic research, respectively.
A few high-throughput studies have confirmed some of the identified miRNAs (miR-1, miR-128, miR-133a, miR-133b, miR-206, miR-222, and miR-503) as common for skeletal muscle development in mouse, human, pig, common carp [11], and cattle [25]. All the aforementioned miRNAs have manifested upregulation during myogenic differentiation, except the downregulated miR-222, both in the above-cited papers [11, 25] and in our study (Table 2).
MyomiRs - muscle-specific miRNAs
In our experiment, four myomiRs were identified as differentially expressed. Their expression in the HER/LIM-derived primary cultures was considerably higher than in HF cells (Table 2, Fig. 4). In general, the myomiRs expression is greatly enhanced during myogenesis and is required for its optimal progression [26]. Cell culture experiments have shown that miR-1 and miR-206 promote muscle cell differentiation, whereas miR-133 enhances cell proliferation. Dai et al. [27] confirmed the mechanism in which miR-1 and miR-206 positively regulate bovine skeletal muscle satellite cell myogenic differentiation via the downregulation of PAX7 and HDAC4. MiR-1 and miR-206 were also found to inhibit PAX3 [28] and NOTCH3 [29] allowing differentiation to proceed. Moreover, miR-206 directly targets cyclin D1 (CCND1) and DNA polymerase α (POLA1), reducing the proliferation rate of myogenic cells [30]. It might also indirectly downregulate DNA-binding protein inhibitors (IDs) and myogenic repressors (MYORs), the Myod inhibitors, and thus, regulate myoblast differentiation [10]. One study suggested that the rapid removal of SNAI1 and SNAI2 at the onset of differentiation is mediated by miR-30a and miR-206, respectively, resulting in the upregulation of myogenin and a dependent increase in the miR-30a and miR-206 expression [31]. Confirmed upregulation of myogenin mRNA in the HER/LIM cells could strengthen the possibility that similar mechanisms are executed in these cells (Fig. 5). It should be noted that miR-30a, which regulates the Snai1 expression, was also upregulated in our study (Table 2).
MiR-133, another myomiR, has been confirmed to increase myoblast proliferation and regulate differentiation by targeting SRF, MAML1, nPTB, and UCP2 proteins [10, 32]. The combined action of miR-133 and myomiRs (miR-1 and -206) induces MYOD1, PAX7, and myogenin causing myoblast differentiation [33]. Upregulation of both miR-133 and myogenin under IGF-1 influence has also been described [34]. It is plausible that in HER/LIM cells, the differentiation progression is accelerated via similar mechanisms involving miR-1, miR-133, miR-206, and myogenin, resulting possibly in enhanced myotube formation observed in the primary cultures of the skeletal muscle with a HER/LIM origin (Fig. 1).
Finally, miR-486 was last of the myomiRs differing in expression between the HER/LIM and the HF cells (Table 2, Fig. 4). MiR-486 has been shown to influence myoblast differentiation by targeting Pax7 and promoting the IGF-1R/PI3K/AKT signaling pathway by repressing its negative regulators PTEN and FOXO1A [28, 35]. PTEN inhibition leads to the activation of mTOR and an increase in protein synthesis [36]. The downregulation of miR-486 in normal myoblasts results in an impaired migration and myoblast fusion [37].
Note that the feedback loop of transcription factors, including myogenin, has been confirmed to participate in myomiR regulation [38], which may indicate it to be a molecule specifically modulating the miRNA expression in HER/LIM-derived cells, which can in turn be reinforced by strong uphill correlations between myogenin and the myomiRs identified in this study (Fig. 6, Additional file 3).
Non-myomiRs engaged in myoblast differentiation
In addition to the aforementioned myomiRs, some of the identified miRNAs are also linked to myogenic differentiation. Among them is miR-139, which was expressed over 120 times more in the HER/LIM cells (Table 2, Fig. 4). Hasseine et al. [39] found that miR-139 directly targets Foxo1 mRNA and reduces the level of its protein. It is plausible that miR-139 supports the miR-486-dependent inhibition of Foxo1 translation, thereby increasing the mTOR-mediated protein synthesis. However, information about its role in skeletal muscle development is still scarce. Moreover, miR-139-5p was also shown to induce cell cycle arrest (prerequisite of differentiation) by targeting oncogenic nuclear receptor subfamily 5, group a, member 2 (Nr5a2) [40], which was confirmed to be developmentally regulated in bovine skeletal muscle [41].
Another molecule, miR-128a (Table 2, Fig. 4), known to be increased during myoblast differentiation, was upregulated in cells derived from beef cattle muscle. Overexpression of miR-128a in myoblasts impede cell proliferation by targeting IRS1 [42], while its inhibition promotes proliferation and myotube hypertrophy [43]. It has also been found to regulate the target genes involved in insulin signaling. MiR-128a is predicted to target the 3′-untranslated region of Foxo1 [43] and thus, regulate AKT signaling, which could potentiate the action of the earlier described miRNAs -139 and -486. Another miR-128 putative target engaged in muscle development, Nr5a2, was identified in chickens [44]. Further, we assume that, at least partially, the coordinated action of miR-128 and miR-139 could influence the HER/LIM cell differentiation in a similar manner, possibly via the inhibition of Foxo1 and Nr5a2. Both the aforementioned miRNAs have manifested strong and very strong uphill correlations between each other and myomiRs, respectively, and a moderate uphill correlation with myogenin (Fig. 6, Additional file 3).
The last miRNA in this group is miR-222. It has been downregulated in HER/LIM primary cultures. Cardinali et al. [45] reported miR-222 decrease in the differentiated quail myotubes. They confirmed the correlation of the miR-222 expression with the Ras/MAPK pathway and showed that its inhibition induced the expression of p27 (cell cycle inhibitor) and muscle-specific proteins, facilitating cell fusion and the assembly of contractile structures [45]. In recent studies, they also found that the overexpression of miR-222 and the consequent silencing of Rbm24 resulted in the inhibition of myoblast fusion [46]. Furthermore, it was found that miR-222 negatively contributes to myoblast differentiation, taking part in the regulatory axis that includes mTOR and IGF-II [47]. miR-221/222 overexpression could directly increase β-catenin and repress the Wnt pathway inhibitors, enhancing the activity of the classic Wnt/β-catenin signaling pathway, which was shown to induce satellite cell proliferation [48].
It seems that all the aforementioned miRNAs differentially expressed in the HER/LIM-originating primary cultures of skeletal muscle cells can direct myogenesis towards differentiation progression (by different but complementary mechanisms), which can result in the increased number of myotubes observed in the HER/LIM-related cell cultures (Fig. 1).
Other miRNAs
Besides the abovementioned miRNAs promoting myoblast differentiation, a few identified molecules such as miR-29b [49], miR-31 [50], miR-9 [51], miR-145 [52], miR-194 [53], miR-378 [54], miR-449 [55], miR-503 [11, 27], miR-542, [56], and miR-660 [11] were described in the literature as skeletal muscle-related. At this time, sufficient data have not been published concerning miR-2439-3p and miR-2469 engagement in the proliferation and differentiation processes of any cells. Their increased expression, particularly in HER-related cell cultures, suggests that they could play an important role in in vitro myogenesis in this breed.
Myogenesis-related miRNA target genes
A functional analysis of 1249 unique target genes predicted for 23 HER/LIM-specific miRNAs has shown nearly 200 processes in which they could be involved (DAVID, FDR ≤ 0.05). Among them is the muscle organ development process covering 40 target genes. Additionally, 21 genes were indicated in the PANTHER database to be participants of the same muscle organ development process (Table 3). Surprisingly, Mef2a, Pax3, Pax7, Mbnl1, and Cacnb2 were the only target genes common for both analyses. Based on aforementioned findings, a relevance network of the direct interactions of miRNAs and their targets was prepared in the Pathway Studio Web software (Elsevier, Netherlands). It shows a complex network of the miRNA regulation of their direct or indirect target genes, some of which are well-known as myogenesis process-associated genes (Fig. 7). We suppose that the modulation of the expression of these genes (56 in total) by the HER/LIM cell-specific miRNA could govern the differences in myotube formation seen in the HER/LIM primary cultures. However, because this is only a prediction, their effective involvement in this phenomenon needs to be checked.
Fig. 7.
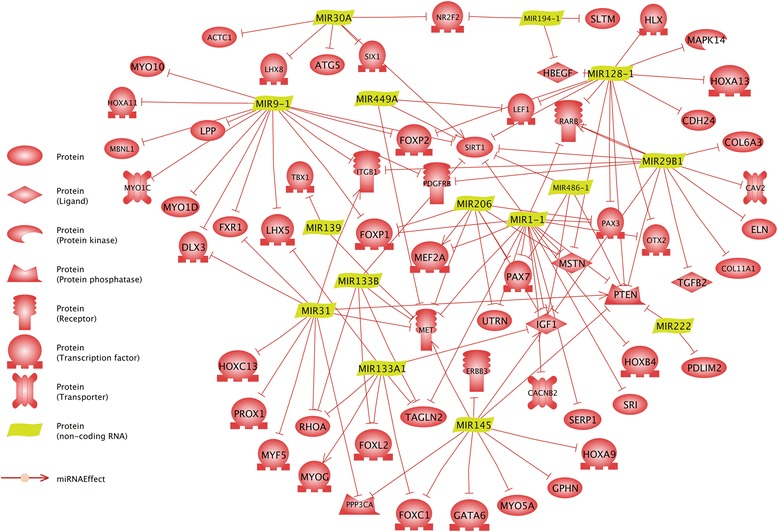
Relevance network of identified miRNAs and target genes belonging to the muscle organ development process (Pathway Studio Web, Elsevier, Netherlands)
In addition to the aforementioned muscle organ development process, general processes such as positive and negative regulation of differentiation were also identified (Additional file 5). This indicates a putative capability of the identified miRNAs in the regulation of dozens of genes undergoing expression at this stage of in vitro myogenesis and underlines the complexity of miRNA-dependent post-translational modifications in the primary cultures of muscle cells.
Conclusions
Thus far, this is the first study showing the interbreed comparison of miRNA expression in a primary culture of skeletal muscle cells subjected to myogenic differentiation; these cells were isolated from semitendinosus muscle of bulls of different breeds and performance. We demonstrated an increase in the number of myotubes on day 6 of the differentiation of muscle-derived cells of the HER/LIM beef breed origin as compared to that in the case of the HF dairy breed-related cells. Further, differences in the miRNA expression in these cultures were demonstrated.
Because the primary cultures were not pure and contained breed-dependent number of myogenic/non-myogenic cells, the net effect of the identified miRNA action should be considered to be the resultant effect of the activity and mutual interactions of satellite cells and muscle stem cell niche cells, which putatively led to the formation of a relatively large number of myotubes in beef cattle-related cells (HER/LIM) during in vitro myogenesis.
As the number of specific cells was not equalized in the primary cultures among breeds (a proportional increase in the number of satellite cell numbers brought about by four iterations of preplating), it is possible that the variation in the number of myotubes and the identified miRNA expression differences between HER/LIM and HF were the effect of the breed-specific cell content in the primary cultures of the skeletal muscle-derived cells. A co-culture system in which myogenic cells and other residents of the stem cell niche will be cultured separately but with the possibility to interact with each other would be a great opportunity to validate the obtained results. It would allow the identification of the myogenic or non-myogenic sources of the identified miRNA, by checking the examined cells separately. It would also be interesting to confirm the aforementioned interbreed miRNA expression differences in an in vivo study of the different stages of bovine myogenesis.
Additional files
Real-time qPCR primers for selected genes. (XLS 38 kb)
Differences in miRNA expression in primary cultures of skeletal muscle cells originating from both HER/LIM breeds, when compared to the HF breed (FDR ≤ 0.05). (XLS 69 kb)
Spearman’s correlation coefficients (A) and their statistical importance (B) for qPCR-validated miRNAs (GenEx 6.0; MultiD Analyses AB, Sweden). (PDF 234 kb)
Target gene prediction for identified miRNAs (TargetScan; Pathway Studio Web, Elsevier, Netherlands). (XLS 2245 kb)
Functional analysis of miRNA target genes using the DAVID database (BP - biological processes). (XLS 145 kb)
miRNA target genes belonging to the muscle organ development process (PANTHER). (XLS 43 kb)
Functional analysis of miRNA target genes using KEGG (DAVID). (XLS 58 kb)
Selected pathways in which the identified target genes are involved (KEGG). (PDF 819 kb)
Acknowledgments
This work was supported by the National Science Centre (Poland), grant no. 2011/03/B/NZ9/03987. Publication of this manuscript was supported by KNOW (Leading National Research Centre) Scientific Consortium “Healthy Animal - Safe Food”, decision of Ministry of Science and Higher Education No. 05-1/KNOW2/2015.
We would like to thank Dr. Anita Dombkowski for help during manuscript preparation.
Availability of data and materials
The data obtained in the microarray experiment are deposited in the NCBI GEO database and numbered GSE73778. The bovine miRNA microarray platform design is deposited in the NCBI GEO and numbered GPL19028.
Abbreviations
- DAVID
The Database for Annotation, Visualization and Integrated Discovery
- FC
Fold Change
- FDR
False Discovery Rate
- HER
Hereford
- HF
Holstein-Friesian
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LIM
Limousin
- NCBI GEO
National Center for Biotechnology Information Gene Expression Omnibus database
- PANTHER
Protein ANalysis THrough Evolutionary Relationships Classification System
- RIN
RNA Integrity Number
- SEM
Standard Error of the Mean
Authors’ contributions
TS designed the study; supervised the project; performed muscle sampling, statistical analysis of microarray and RT-qPCR data, functional analysis, and interpretation of the obtained data; and wrote the manuscript. AC carried out bovine skeletal muscle cell isolation and culture, RNA isolation, and microarray and qPCR analyses. JO supervised the animal slaughter, and dissection. EB carried out RNA isolation and qPCR analyses. All authors read and approved the final manuscript.
Ethics approval
This study complies with national and institutional guidelines of the use of animals in research according to the Polish Legal Act of January 21, 2005. Since sample collection was performed during routine slaughter and no additional procedures that were harmful or painful for the animals were applied, this study did not require a formal ethics approval.
Consent for publication
Not Applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4492-5) contains supplementary material, which is available to authorized users.
Contributor Information
Tomasz Sadkowski, Email: tomasz_sadkowski@sggw.pl.
Anna Ciecierska, Email: anna_ciecierska@sggw.pl.
Jolanta Oprządek, Email: j.oprzadek@ighz.pl.
Edyta Balcerek, Email: edy.balcerek@gmail.com.
References
- 1.Gillis WA. Structural Variations in the muscle fiber in relation to tenderness of meat: a review. Proceedings of the Oklahoma Academy of Science. 1968;48:243–250. [Google Scholar]
- 2.Kotula AW, Lusby WR. Mineral composition of muscles of 1- to 6-year-old steers. J Anim Sci. 1982;54(3):544–548. doi: 10.2527/jas1982.543544x. [DOI] [PubMed] [Google Scholar]
- 3.Russell RG, Oteruelo FT. An Ultrastructural Study of the Differentiation of Skeletal Muscle in the Bovine Fetus. Anat Embryol. 198l;162:403–17. [DOI] [PubMed]
- 4.Robelin J, Lacourt A, Béchet D, Ferrara M, Briand Y, Geay Y. Muscle differentiation in the bovine fetus: a histological and histochemical approach. Growth Dev Aging. 1991;55:151–160. [PubMed] [Google Scholar]
- 5.Bischoff R. Regeneration of single skeletal muscle fibers in vitro. Anat Rec. 1975;182(2):215–235. doi: 10.1002/ar.1091820207. [DOI] [PubMed] [Google Scholar]
- 6.Kuang S, Gillespie MA, Rudnicki MA. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell. 2008;2(1):22–31. doi: 10.1016/j.stem.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93(1):23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee RC, Feinbaum RL, Ambros V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 10.Sharma M. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front Physiol. 2014;5:239. doi: 10.3389/fphys.2014.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dmitriev P, Barat A, Polesskaya A, O’Connell MJ, Robert T, Dessen P, et al. Simultaneous miRNA and mRNA transcriptome profiling of human myoblasts reveals a novel set of myogenic differentiation-associated miRNAs and their target genes. BMC Genomics. 2013;14(1):265. doi: 10.1186/1471-2164-14-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadkowski T, Jank M, Zwierzchowski L, Oprzadek J, Transcriptomic MT. Index of skeletal muscle of beef breeds bulls. J Physiol Pharmacol. 2009;60(1):15–28. [PubMed] [Google Scholar]
- 13.Sadkowski T, Jank M, Zwierzchowski L, Oprządek J, Motyl T. Comparison of skeletal muscle transcriptional profiles in dairy and beef breeds bulls. J Appl Genet. 2009;50(2):109–123. doi: 10.1007/BF03195662. [DOI] [PubMed] [Google Scholar]
- 14.Szcześniak KA, Ciecierska A, Ostaszewski P, Sadkowski T. Transcriptomic profile adaptations following exposure of equine satellite cells to nutriactive phytochemical gamma-oryzanol. Genes Nutr 2016;11(1):1–14. [DOI] [PMC free article] [PubMed]
- 15.Qu Z, Balkir L, van Deutekom JCT, Robbins PD, Pruchnic R, Huard J. Development of approaches to improve cell survival in myoblast transfer therapy. J Cell Biol. 1998;142(5):1257–1267. doi: 10.1083/jcb.142.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kook SH, Choi KC, Son YO, Lee KY, Hwang IH, Lee HJ, Chang JS, Choi IH, Lee JC. Satellite cells isolated from adult Hanwoo muscle can proliferate and differentiate into myoblasts and adipose-like cells. Mol Cells. 2006;22(2):239–245. [PubMed] [Google Scholar]
- 17.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 18.Szcześniak KA, Ciecierska A, Ostaszewski P, Sadkowski T. Characterisation of equine satellite cell transcriptomic profile response to β-hydroxy-β-methylbutyrate (HMB) Br J Nutr. 2016;116(8):1315–1325. doi: 10.1017/S000711451600324X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadkowski T, Ciecierska A, Majewska A, Oprządek J, Dasiewicz K, Ollik M, et al. Transcriptional background of beef marbling—novel genes implicated in intramuscular fat deposition. Meat Sci. 2014;97(1):32–41. doi: 10.1016/j.meatsci.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative realtime PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Albrecht E, Teuscher F, Ender K, Wegner J. Growth- and breed-related changes of muscle bundle structure in cattle. J Anim Sci. 2006;84(11):2959–2964. doi: 10.2527/jas.2006-345. [DOI] [PubMed] [Google Scholar]
- 23.Albrecht E, Lembcke C, Wegner J, Maak S. Prenatal muscle fiber development and bundle structure in beef and dairy cattle. J Anim Sci. 2013;91(8):3666–3673. doi: 10.2527/jas.2013-6258. [DOI] [PubMed] [Google Scholar]
- 24.Rehfeldt C, Fiedler I, Dietl G, Ender K. Myogenesis and postnatal skeletal muscle cell growth as influenced by selection. Livest Prod Sci. 2000;66(2):177–188. [Google Scholar]
- 25.Wang YM, Ding XB, Dai Y, Liu XF, Guo H, Zhang Y. Identification and bioinformatics analysis of miRNAs involved in bovine skeletal muscle satellite cell myogenic differentiation. Mol Cell Biochem. 2015;404(1–2):113–122. doi: 10.1007/s11010-015-2371-9. [DOI] [PubMed] [Google Scholar]
- 26.Hitachi K, Nakatani M, Tsuchida K. Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int J Biochem Cell Biol. 2014;47:93–103. doi: 10.1016/j.biocel.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Dai Y, Wang YM, Zhang WR, Liu XF, Li X, Ding XB, et al. The role of microRNA-1 and microRNA-206 in the proliferation and differentiation of bovine skeletal muscle satellite cells. In Vitro Cell Dev Biol Anim. 2016;52(1):27–34. doi: 10.1007/s11626-015-9953-4. [DOI] [PubMed] [Google Scholar]
- 28.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12(6):349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 29.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 MicroRNAs in differentiating myoblasts. J Biol Chem. 2012;287(48):40360–40370. doi: 10.1074/jbc.M112.378414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alteri A, De Vito F, Nessina G, Pompili M, Calconi A, Visca P, et al. Cyclin D1 is a major target of miR-206 in cell differentiation and transformation. Cell Cycle. 2013;12(24):3781–3790. doi: 10.4161/cc.26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CEM, van Ijcken WFJ. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in Myogenesis. Mol Cell. 2012;47(3):457–468. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar S, Dey BK, Dutta A. MiR-322/424 and-503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21(13):2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14(10):2495–2505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang M-B, Xu H, Xie S-J, Zhou H, Qu L-H. Insulin-like growth Factor-1 receptor is regulated by microRNA-133 during skeletal Myogenesis. PLoS One. 2011;6(12):e29173. doi: 10.1371/journal.pone.0029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31(1):203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weigl LG. Lost in translation: regulation of skeletal muscle protein synthesis. Curr Opin Pharmacol. 2012;12(3):377–382. doi: 10.1016/j.coph.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 37.Horak M, Novak J, Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev Biol. 2016;410(1):1–13. doi: 10.1016/j.ydbio.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Luo W, Nie Q, Zhang X. MicroRNAs involved in skeletal muscle differentiation. J Genet Genomics. 2013;40(3):107–116. doi: 10.1016/j.jgg.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Hasseine LK, Hinault C, Lebrun P, Gautier N, Paul-Bellon R, Van Obberghen E. miR-139 impacts FoxO1 action by decreasing FoxO1 protein in mouse hepatocytes. Biochem Biophys Res Commun. 2009;390(4):1278–1282. doi: 10.1016/j.bbrc.2009.10.135. [DOI] [PubMed] [Google Scholar]
- 40.Liu R, Yang M, Meng Y, Liao J, Sheng J, Pu Y, Yin L, Kim SJ. Tumor-suppressive function of miR-139-5p in esophageal squamous cell carcinoma. PLoS One. 2013;8(10):e77068. doi: 10.1371/journal.pone.0077068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson NJ, Lyons RE, Reverter A, Greenwood PL, Dalrymple BP. Inferring the in vivo cellular program of developing bovine skeletal muscle from expression data. Gene Expr Patterns. 2013;13(3–4):109–125. doi: 10.1016/j.gep.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Hitachi K, Tsuchida K. Role of microRNAs in skeletal muscle hypertrophy. Front Physiol. 2013;4:408. doi: 10.3389/fphys.2013.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci. 2013;126(12):2678–2691. doi: 10.1242/jcs.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li T, Wu R, Zhang Y, Zhu D. A systematic analysis of the skeletal muscle miRNA transcriptome of chicken varieties with divergent skeletal muscle growth identifies novel miRNAs and differentially expressed miRNAs. BMC Genomics. 2011;12(1):186. doi: 10.1186/1471-2164-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cardinali B, Castellani L, Fasanaro P, Basso A, Alemà S, Martelli F, et al. Microrna-221 and Microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4(10):e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cardinali B, Cappella M, Provenzano C, Garcia-Manteiga JM, Lazarevic D, Cittaro D, et al. MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 2016;7(2):e2086. doi: 10.1038/cddis.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erriquez D, Perini G, Ferlini A. Non-coding RNAs in muscle dystrophies. Int J Mol Sci. 2013;14(10):19681–19704. doi: 10.3390/ijms141019681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W-X, Hu Q, Qiu M-T, Zhong S-L, Xu J-J, Tang J-H, et al. miR-221/222: promising biomarkers for breast cancer. Tumor Biol. 2013;34(3):1361–1370. doi: 10.1007/s13277-013-0750-y. [DOI] [PubMed] [Google Scholar]
- 49.Chen J-F, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for Myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11(1):118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 51.Lu L, Zhou L, Chen EZ, Sun K, Jiang P, Wang L, et al. A novel YY1-miR-1 regulatory circuit in skeletal Myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS One. 2012;7(2):e27596. doi: 10.1371/journal.pone.0027596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Y, Chai J, Zhang H, Chu W, Liu L, Ma L, et al. miR-194 promotes burn-induced hyperglycemia via attenuating IGF-IR expression. Shock. 2014;42(6):578–584. doi: 10.1097/SHK.0000000000000258. [DOI] [PubMed] [Google Scholar]
- 54.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem. 2011;286(22):19431–19438. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pelosi L, Coggi A, Forcina L, Musarò A. MicroRNAs modulated by local mIGF-1 expression in mdx dystrophic mice. Front Aging Neurosci. 2015;7:69. doi: 10.3389/fnagi.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGregor RA, Poppitt SD, Cameron-Smith D. Role of microRNAs in the age-related changes in skeletal muscle and diet or exercise interventions to promote healthy aging in humans. Ageing Res Rev. 2014;17:25–33. doi: 10.1016/j.arr.2014.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time qPCR primers for selected genes. (XLS 38 kb)
Differences in miRNA expression in primary cultures of skeletal muscle cells originating from both HER/LIM breeds, when compared to the HF breed (FDR ≤ 0.05). (XLS 69 kb)
Spearman’s correlation coefficients (A) and their statistical importance (B) for qPCR-validated miRNAs (GenEx 6.0; MultiD Analyses AB, Sweden). (PDF 234 kb)
Target gene prediction for identified miRNAs (TargetScan; Pathway Studio Web, Elsevier, Netherlands). (XLS 2245 kb)
Functional analysis of miRNA target genes using the DAVID database (BP - biological processes). (XLS 145 kb)
miRNA target genes belonging to the muscle organ development process (PANTHER). (XLS 43 kb)
Functional analysis of miRNA target genes using KEGG (DAVID). (XLS 58 kb)
Selected pathways in which the identified target genes are involved (KEGG). (PDF 819 kb)
Data Availability Statement
The data obtained in the microarray experiment are deposited in the NCBI GEO database and numbered GSE73778. The bovine miRNA microarray platform design is deposited in the NCBI GEO and numbered GPL19028.


