Abstract
Plasma lipid levels are risk factors for cardiovascular disease, a leading cause of death worldwide. While many studies have been conducted on lipid genetics, they mainly focus on Europeans and thus their transferability to diverse populations is unclear. We performed SNP- and gene-level genome-wide association studies (GWAS) of four lipid traits in cohorts from Nigeria and the Philippines and compared them to the results of larger, predominantly European meta-analyses. Two previously implicated loci met genome-wide significance in our SNP-level GWAS in the Nigerian cohort, rs34065661 in CETP associated with HDL cholesterol (P = 9.0 × 10−10) and rs1065853 upstream of APOE associated with LDL cholesterol (P = 6.6 × 10−9). The top SNP in the Filipino cohort associated with triglyceride levels (rs662799; P = 2.7 × 10−16) and has been previously implicated in other East Asian studies. While this SNP is located directly upstream of well known APOA5, we show it may also be involved in the regulation of BACE1 and SIDT2. Our gene-based association analysis, PrediXcan, revealed decreased expression of BACE1 and decreased expression of SIDT2 in several tissues, all driven by rs662799, significantly associate with increased triglyceride levels in Filipinos (FDR <0.1). In addition, our PrediXcan analysis implicated gene regulation as the mechanism underlying the associations of many other previously discovered lipid loci. Our novel BACE1 and SIDT2 findings were confirmed using summary statistics from the Global Lipids Genetic Consortium (GLGC) meta-GWAS.
Keywords: GWAS, PrediXcan, Lipids, Population genetics, Gene expression
Introduction
Though 99.9% of the genome between humans is identical, millions of variant sites exist in different frequencies between populations, which leads to differences in gene expression and other complex traits (Brown et al., 2016). Since most GWAS have been conducted in European cohorts and most databases are built upon European data, the results may not accurately extrapolate to other global populations, which could lead to disparity within medicine (Bustamante, De La Vega & Burchard, 2011). This discrepancy is alarming considering urgent health issues worldwide, such as obesity and cardiovascular disease. Lipid levels as a complex trait are increasingly concerning due to the growing global rate of obesity caused by the increasing availability of high-fat foods and rapid urbanization (Ellulu et al., 2014). Decreased high density lipoprotein (HDL) cholesterol levels and increased low density lipoprotein (LDL) cholesterol and triglyceride (TRIG) levels are associated with cardiovascular disease, the leading cause of death in the United States (Go et al., 2013). Studies such as the Global Lipids Genetic Consortium (GLGC) acquire information predominantly from Europeans, but lack information from other populations (Coram et al., 2015; Willer et al., 2013). We aim to help remedy this issue by studying lipid traits in diverse populations. We chose two populations with lipid phenotypes available to study from the database of Genotypes and Phenotypes: Yoruba in Ibadan, Nigeria, (Yoruba) and Filipino in Cebu, Philippines (Cebu) (Hall et al., 2006; Adair et al., 2011; Wu et al., 2013).
At the time of our study, one of the largest available cholesterol SNP meta-analyses is the GLGC (Willer et al., 2013). The cohort in that study consists of 188,577 European-ancestry individuals and 7,898 non-European-ancestry individuals. One hundred fifty-seven loci were found to be significantly associated with total cholesterol (CHOL), HDL, TRIG, or LDL levels, and they conducted further gene set enrichment analysis with MAGENTA (Ayellet et al., 2010). However, gene-level association studies that integrate transcriptome data, like PrediXcan and TWAS, were not performed (Gamazon et al., 2015; Gusev et al., 2016). Summary statistics from GLGC were used as a replication and base set in our analyses of the Yoruba and Cebu cohorts.
Both the Cebu and Yoruba cohorts have been used in genetic studies of lipids previously (Hall et al., 2006; Wu et al., 2013). These studies, both using the same data we study here, focused on APOE, a well-known gene that is associated with lipid levels and Alzheimer’s disease (Middelberg et al., 2011). Previously, SNP-level GWAS in the Cebu and other East Asian cohorts attributed rs662799 as affecting function of APOA5, which is located 571 bases upstream (Wu et al., 2013; Lu et al., 2016; Spracklen et al., 2017). Beyond the APOE candidate gene study, no full GWAS has been conducted in the Yoruba cohort.
In this study, we performed a genome wide association study (GWAS) in each population using linear mixed modeling (Zhou & Stephens, 2012) and a conditional and joint analysis (Yang et al., 2012). Subsequently, we calculated the genetic correlation for each lipid trait between the populations at the SNP-level using bivariate REML analysis (Yang et al., 2011). We also used cross-population empirical Bayes (XPEB) modeling to improve power to detect SNPs with similar effects as previously found in larger European meta-analyses (Coram et al., 2015). Finally, we used the transcriptome-informed method PrediXcan to implicate genes in CHOL, HDL, LDL, and TRIG (Gamazon et al., 2015). Using our data and those from previous European studies, we confirm previously known loci and implicate new loci through the mechanism of gene expression regulation in Filipinos (Willer et al., 2013). Our gene-based association study for triglyceride levels in the Cebu cohort suggests that rs662799 may affect the expression of BACE1 and SIDT2 rather than that of APOA5.
Materials and Methods
Cohorts
We obtained data from both cohorts through the database of Genotypes and Phenotypes with Institutional Review Board approval (Mailman et al., 2007) (Table 1). Yoruba consists of 1,251 adults of Yoruba ethnicity age 73 to 103 years old, living in Ibadan, Nigeria in 2001, who were originally studied in the Indianapolis Ibadan Epidemiological Study of Dementia (Ogunniyi et al., 1997). The Cebu population consists of 1,799 Filipino mothers, who gave birth between May 1, 1983 and April 30, 1984 in the metropolitan area of Cebu, Philippines. This cohort was originally studied in the Cebu Longitudinal Health and Nutrition Survey (Adair et al., 2011), and at the time of data collection in 2005, the mothers were age 34 to 70. The Yoruba cohort was genotyped with the Illumina HumanOmni2.5 array and the Cebu cohort was genotyped with the Affymetrix Genomewide Human SNP Array 5.0. Both cohorts had CHOL, HDL, LDL, and TRIG levels measured after fasting (mg/dL) and we subsequently rank normalized each trait (Aulchenko et al., 2007) (Table S2). See https://github.com/aandaleon/px_chol for all scripts used in our analyses.
Table 1. Data analyzed.
| Yoruba | Cebu | |
|---|---|---|
| Accession number | phs000378.v1.p1 | phs000523.v1.p1 |
| Type of genotyping | Whole genome genotyping | Whole genome genotyping |
| Source platform | Illumina, HumanOmni2.5 | Affymetrix, Genomewide Human SNP Array 5.0 |
| Pre-QC SNPs | 2,443,179 | 440,792 |
| Pre-QC individuals | 1,251 | 1,799 |
| Post-QC SNPs | 1,522,836 | 369,185 |
| Post-QC individuals | 1,017 | 1,765 |
| Post-imputation GWAS SNPs | 12,553,142 | 4,496,603 |
Summary statistics from the Global Lipids Genetic Consortium meta-analysis (Willer et al., 2013) were downloaded from http://csg.sph.umich.edu/abecasis/public/lipids2013/. Though the offspring of the Cebu cohort are included in the GLGC cohort, they form a small portion of the dataset (1,771/188,577) and thus do not drive the signal for the entire dataset.
Quality control
We performed quality control on the genotypes in these cohorts with PLINK following a standard quality control pipeline (Turner et al., 2001; Purcell et al., 2007). Starting with the dbGaP PLINK binary files, we removed SNPs with call rates < 99% in the individual populations. Subsequently, SMARTPCA within Eigensoft was used to map individuals on their first 10 principal components, and individuals with excess (±5 standard deviations) from the population mean on the first two components were removed (Patterson, Price & Reich, 2006) (Figs. S1 and S2). This was followed by removing individuals with excess heterozygosity (±3 standard deviations), leaving the Yoruba cohort with 1,017 individuals with genotype and lipid phenotype data, including 1,522,836 SNPs. The Cebu cohort retained 1,757 individuals with genotype and lipid phenotype data, including 369,185 SNPs. Both cohorts were mapped to hg19, which included performing a liftover in the Cebu cohort from hg18 to hg19. We used an imputation preparation tool on the Cebu cohort available at http://www.well.ox.ac.uk/ wrayner/tools/ that adjusted the data by matching the strand, alleles, position, ref/alt assignments, and frequency differences to the 1000G reference panel.
Imputation
Yoruba SNPs were imputed on the Sanger Imputation Server with EAGLE2 and PBWT using the African Genomes Reference Panel to improve genome coverage (Delaneau, Marchini & Zagury, 2012; Durbin, 2014; McCarthy et al., 2016). We imputed the Cebu SNPs using the Michigan Imputation Server with the 1000 Genomes phase 3 reference panel and EAGLE2 (Auton et al., 2015; Das et al., 2016; Loh et al., 2016). The output from the imputation was filtered to remove SNPs with R2 < 0.8 and minor allele frequency <0.01, leaving 12,553,142 SNPs for analysis in Yoruba and 4,496,603 SNPs for analysis in Cebu.
SNP-level genome-wide association study
The imputed genotype dosages were used in a genome-wide association study performed with Genome-Wide Efficient Mixed Model Analysis (GEMMA) software using a univariate linear mixed model for each of the four phenotypes (Zhou & Stephens, 2012). SNPs with P < 5 × 10−8 using the Wald test were considered genome-wide significant. The top SNPs from the GEMMA analysis were plotted using LocusZoom to depict their proximity to various genes (Pruim et al., 2011). For each phenotype tested, we also used GEMMA to obtain the percent variance explained (PVE) by all the SNPs, i.e., the “chip heritability” (Zhou & Stephens, 2012). Conditional and joint analyses were then performed using GCTA-COJO (Yang et al., 2011; Yang et al., 2012) to identify the lead SNP or SNPs at each locus.
Comparison of populations
Both populations’ GEMMA results and summary statistics from GLGC (Willer et al., 2013) were used in a cross-population empirical Bayes model (XPEB) to compute false discovery rates for each SNP, with significance declared at FDR < 0.05. This model improves efficiency in GWAS by incorporating relevant results from larger (ex. GLGC) GWAS only when there are similar effect sizes between populations (Coram et al., 2015). Sample sizes in the GLGC results ranged from 50,000 to 187,365 depending on the SNP. Because XPEB assumes similar sample size in the base population across SNPs, we ran XPEB using only the SNPs with sample size between 80,000 and 95,000 in GLGC, which left us with 4,454,201 markers for each phenotype. SNP-level comparisons between populations were performed using Genome-wide Complex Trait Analysis (GCTA) software (Yang et al., 2011). We performed a bivariate restricted maximum likelihood (REML) analysis to estimate the genetic correlation between the Cebu and Yoruba cohorts for each phenotype (Lee et al., 2012).
Gene-based association study
PrediXcan, the gene-level association study, was performed using models built with cis-expression quantitative trait loci results from the Genotype-Tissue Expression Project (GTEx) (Ardlie et al., 2015; Wheeler et al., 2016; Barbeira et al., 2017). GTEx models (GTEx-V6p-HapMap-2016-09-08.tar.gz) were downloaded from PredictDB at http://predictdb.hakyimlab.org/. Significance for each tissue was determined as FDR <0.1 across all testable genes in all tissues (N = 198,970). In total, 44 GTEx models were tested in both cohorts (Table 2). Predicted expression levels were obtained and tested for association with the lipid phenotypes using PrediXcan software (Gamazon et al., 2015). Significant genes were further plotted using ggplot to depict the predicted gene expression against the observed phenotype (Wickham, Winston & RStudio, 2016). For the replication cohort, GLGC, Summary-PrediXcan (Barbeira et al., 2017) was used because only summary statistics were available.
Table 2. The number of genes tested in PrediXcan using expression prediction models built in GTEx Project tissues.
Genes tested had a cross-validated prediction performance R2 > 0.01.
| Tissue model | Tissue abbreviation | Genes tested | Tissue model | Tissue abbreviation | Genes tested |
|---|---|---|---|---|---|
| Adipose—Subcutaneous | ADPSBQ | 7,254 | Esophagus—Mucosa | ESPMCS | 7,710 |
| Adipose—Visceral (Omentum) | ADPVSC | 4,447 | Esophagus—Muscularis | ESPMSL | 6,338 |
| Adrenal Gland | ADRNLG | 3,785 | Heart—Atrial Appendage | HRTAA | 4,450 |
| Artery—Aorta | ARTAORT | 5,943 | Heart—Left Ventricle | HRTLV | 4,718 |
| Artery—Coronary | ARTCRN | 3,141 | Liver | LIVER | 2,502 |
| Artery—Tibial | ARTTBL | 7,074 | Lung | LUNG | 6,448 |
| Brain—Anterior cingulate cortex (BA24) | BRNACC | 2,430 | Muscle—Skeletal | MSCLSK | 6,520 |
| Brain—Caudate (basal ganglia) | BRNCDT | 3,325 | Nerve—Tibial | NERVET | 8,016 |
| Brain—Cerebellar hemisphere | BRNCHB | 4,077 | Ovary | OVARY | 2,673 |
| Brain—Cerebellum | BRNCHA | 5,066 | Pancreas | PNCREAS | 4,603 |
| Brain—Cortex | BRNCTXA | 3,334 | Pituitary | PTTARY | 3,094 |
| Brain—Frontal Cortex (BA9) | BRNCTXB | 3,138 | Prostate | PRSTTE | 2,491 |
| Brain—Hippocampus | BRNHPP | 2,508 | Skin—Not Sun Exposed (Suprapubic) | SKINNS | 5,471 |
| Brain—Hypothalamus | BRNHPT | 2,290 | Skin—Sun Exposed (Lower leg) | SKINS | 7,665 |
| Brain—Nucleus accumbens (basal ganglia) | BRNNCC | 2,984 | Small Intestine—Terminal Ileum | SNTTRM | 2,515 |
| Brain—Putamen (basal ganglia) | BRNPTM | 2,621 | Spleen | SPLEEN | 3,602 |
| Breast—Mammary Tissue | BREAST | 4,473 | Stomach | STMACH | 4,035 |
| Cells—EBV-transformed lymphocytes | LCL | 3,441 | Testis | TESTIS | 7,002 |
| Cells—Transformed fibroblasts | FIBRBLS | 7,543 | Thyroid | THYROID | 7,853 |
| Colon—Sigmoid | CLNSGM | 3,619 | Uterus | UTERUS | 2,058 |
| Colon—Transverse | CLNTRN | 4,729 | Vagina | VAGINA | 1,939 |
| Esophagus—Gastroesophageal Junction | ESPGEJ | 3,457 | Whole Blood | WHLBLD | 6,588 |
Backward elimination modeling
Because our PrediXcan analysis showed multiple genes associated with TRIG at the 11q23.3 locus in Cebu, we conducted a backward elimination analysis to determine the lead gene or genes. We used the R lm function to build all multiple linear regression models. The starting model included predicted expression terms for all genes with rs662799 or a linked SNP (r2 > 0.6) in its predictive model and the absolute value of the marginal t-statistic greater than 3. The term with the highest P-value was eliminated and the model rerun until only terms with P < 0.05 remained in the model.
Results
Yoruba SNP-level GWAS
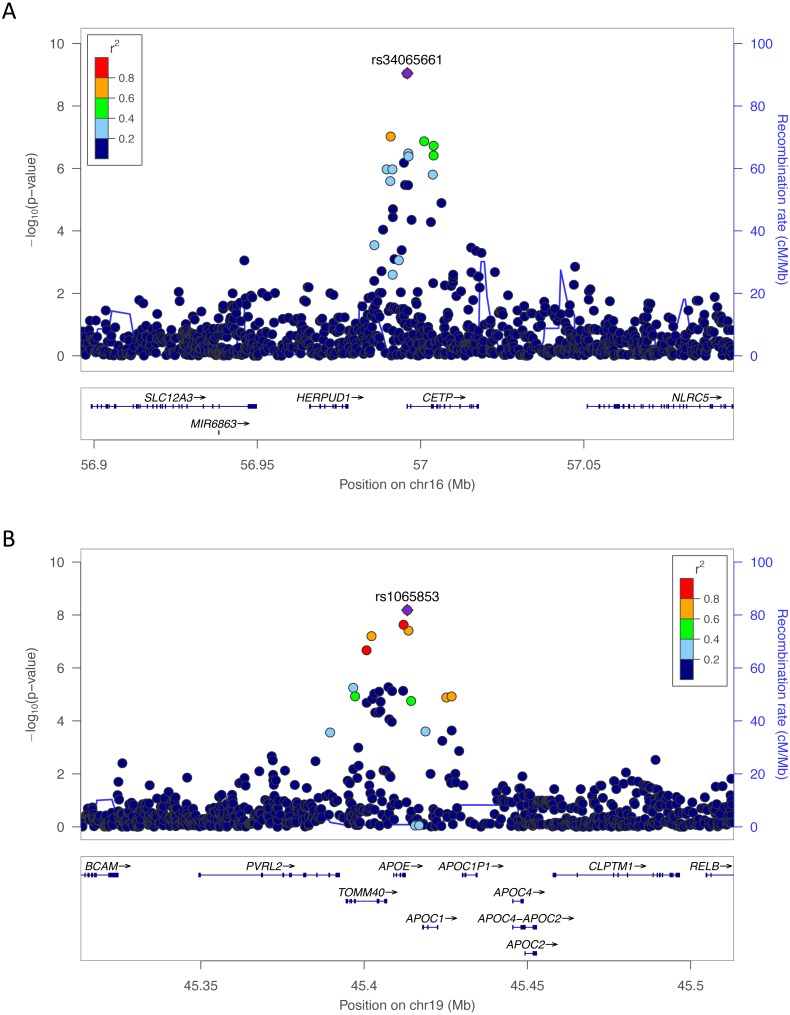
We sought to better understand the genetic architecture of lipid traits within and across populations. In the Yoruba cohort from Ibadan, Nigeria, which included 1,017 individuals and 12,553,142 SNPs, we conducted SNP-level GWAS for four lipid traits CHOL, HDL, LDL, and TRIG. For each lipid trait, we used a univariate linear mixed model, which accounts for relatedness within the populations (Zhou & Stephens, 2012). This was especially important because one-third of the Yoruba cohort is related to at least one other member (proportion identity by descent > 0.125). Across the four phenotypes, five SNPs surpassed the genome-wide significance threshold of P < 5 × 10−8 at two loci (Fig. 1). Conditional and joint analysis (Yang et al., 2011; Yang et al., 2012) did not reveal additional associated SNPs at these two loci and the top hits are shown in Table 3. rs34065661 is on chromosome 16q13 within an intron of CETP and rs1065853 is on chromosome 19q13.32 near APOE (Fig. 1). Both CETP and APOE are well-known and well-studied lipid genes (Buyske et al., 2012; Rasmussen-Torvik et al., 2012).
Figure 1. LocusZoom plots of the most significant SNPs in (A) HDL (rs34065661) and (B) LDL (rs1065853) in Yoruba.
The color of each dot represents the SNP’s linkage disequilibrium r2 with the labeled SNP in the 1000 Genomes African populations.
Table 3. SNP-level GWAS results in the Yoruba (Y) and Cebu (C) populations.
Shown are SNPs that reach genome-wide significance after conditional and joint analysis (Yang et al., 2011; Yang et al., 2012).
| Pop. | Pheno. | Chr. | Position | SNP ids | Nearest gene | Non- effect allele | Effect allele | EAF | Marginal Beta | Marginal P | Joint Beta | Joint P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y | HDL | 16 | 56995935 | rs34065661 | CETP | C | G | 0.093 | 0.468 | 9.0 × 10−10 | 0.468 | 8.7 × 10−10 |
| Y | LDL | 19 | 45413233 | rs1065853 | APOE | G | T | 0.113 | −0.405 | 6.6 × 10−9 | −0.405 | 6.4 × 10−9 |
| C | TRIG | 11 | 116663707 | rs662799 | APOA5 | A | G | 0.245 | 0.342 | 2.7 × 10−16 | 0.342 | 2.5 × 10−18 |
| C | TRIG | 2 | 27731212 | rs3817588 | GCKR | T | C | 0.309 | −0.202 | 2.6 × 10−7 | −0.202 | 2.1 × 10−8 |
Cebu SNP-level GWAS
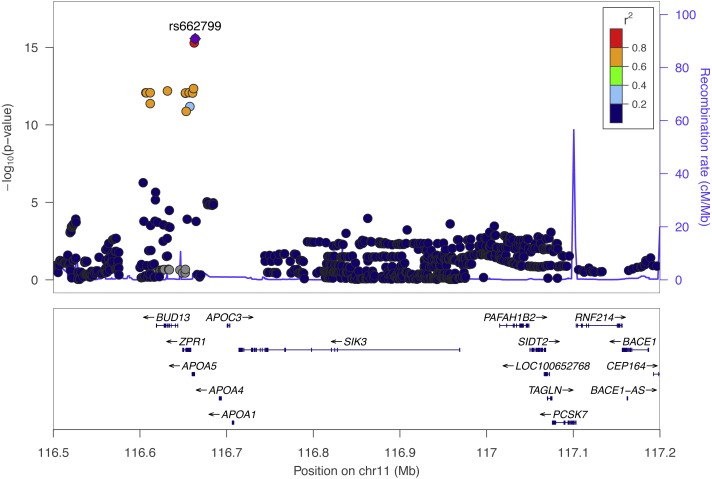
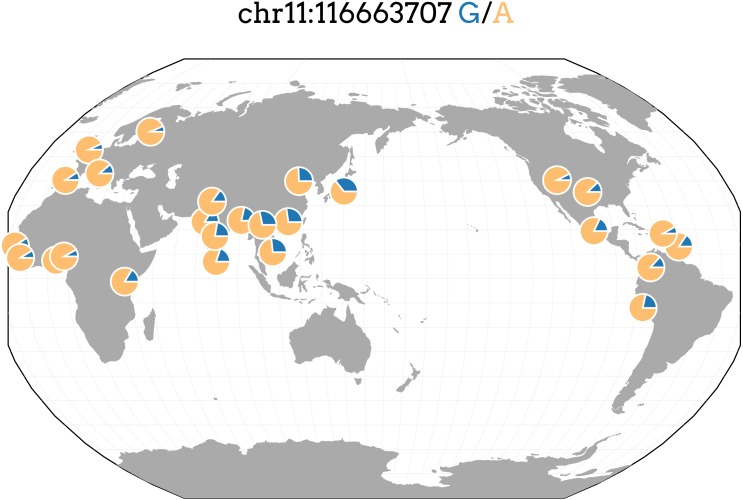
We performed SNP-level GWAS for the same four lipid phenotypes in 1,765 individuals from Cebu, Philippines, using 4.5 million imputed SNPs (see Methods). No SNPs met genome-wide significance for the CHOL, HDL, and LDL phenotypes. However, 44 SNPs were genome-wide significant (P < 5 × 10−8) for TRIG and all grouped on chromosome 11q23.3 (Fig. 2), a locus that includes various lipid genes such as APOA1 and APOA4. The most significant SNP in this group is rs662799, with a marginal P = 2.7 × 10−16. It is located 571 base pairs upstream of APOA5, and has been previously associated with cholesterol traits in Asian populations, possibly due to its high minor allele frequency within Asian populations, with MAF for minor allele G at 0.245 in 1000 Genomes EAS compared to 0.083 in EUR (Marcus & Novembre, 2016) (Fig. 3). Conditional and joint analysis (Yang et al., 2011; Yang et al., 2012) did not reveal additional associated SNPs at the 11q23.3 locus, but did reveal an additional genome-wide significant SNP on chromosome 2 in the GCKR gene (Table 3). As a positive control, we compared the results from the previous Cebu GWAS (Wu et al., 2013) and our GWAS and obtained largely the same significant results (Table S1).
Figure 2. The top Cebu GWAS signal, rs662799, which associated with TRIG levels is 571 bp upstream of APOA5.
The color of each dot represents the SNP’s linkage disequilibrium r2 with rs662799 in the 1000 Genomes East Asian populations.
Figure 3. Allele frequencies of TRIG associated SNP and driver of predicted expression models in multiple genes, rs662799, in 1000 Genome populations.
Figure generated with the Geography of Genetic Variants Browser (Marcus & Novembre, 2016).
Integrating larger European study results into Yoruba and Cebu SNP-level GWAS
The overlapping genetic architecture between populations for most traits is likely nonzero, but not 100% either due to differences in allele frequencies, effect sizes, and linkage disequilibrium patterns. Currently, European GWAS often have sample sizes 100 times larger than non-European studies. However, studies in diverse populations are growing (Spracklen et al., 2017; Wojcik et al., 2017). Traditional meta-analysis methods give the most weight to the GWAS with the largest sample size. Therefore, meta-analyses combining populations by traditional methods would be driven by the European results, drowning out any additional signal. The cross-population empirical Bayes (XPEB) method is designed to boost signal in a target (small, usually non-European) population whenever the base (large, usually European) population shares genetic architecture, but does not generate false positives when the signal is only present in the base population (Coram et al., 2015).
We used XPEB to improve power for mapping lipid traits in the Yoruba and Cebu cohorts by integrating results from the base population GLGC, a large lipid meta-analysis of European individuals (Willer et al., 2013). In XPEB, we input the SNPs and P-values from both our target GWAS (Yoruba or Cebu) and the base European GWAS. The output includes a population-wide estimate of the degree of genetic architecture overlap, κ1, and a new false discovery rate (FDR) for each individual SNP in common between the base and target GWAS.
In the Yoruba population, we found associated loci (FDR < 0.05) using XPEB for the CHOL, HDL, and LDL phenotypes (Table S3). These results reflect the estimated architecture overlap with GLGC, where κ1 was 0.65 for CHOL, 0.51 for HDL, 0.9 for LDL, and 0 for TRIG. The CETP locus, which was also significant in the Yoruba-only HDL GWAS was the most significant result in the XPEB analysis. In addition, several other previously implicated genes were significant in the XPEB analysis, including LDLR for CHOL and PCSK9, LPA, and SMARCA4 for LDL (Wu et al., 2013; Willer et al., 2013; Surakka et al., 2014).
In Cebu, CHOL, TRIG, and LDL each had κ1 = 0.90, while HDL had κ1 = 0.64. Tens or hundreds of SNPs had FDR < 0.05 for each phenotype, including those found in the Cebu-only GWAS (Table 3). Additional significant SNPs located within or near other well-known, previously studied lipid genes included CETP and LIPC in HDL; PLCG1 and TOP1 in LDL; and APOA5 and BUD13 in TRIG (Asselbergs et al., 2012; Spracklen et al., 2017; Wu et al., 2013; Zhou et al., 2013; Kim et al., 2011) (Table S4).
Heritability
As part of our GWAS study, we also estimated the percent variance explained (PVE) by all SNPs tested, i.e., “chip heritability”, using GEMMA and conducted further genetic correlation analysis using GCTA (Yang et al., 2011; Zhou & Stephens, 2012). By estimating heritability, we can help determine which portion of our phenotype is not explained through our analyses and is influenced by other factors, such as diet.
In the Yoruba cohort, no PVE estimate was significantly different than zero. All PVE estimates for Yoruba were low when compared to variance component studies in a Finnish cohort of 5,123 individuals (Sabatti et al., 2009; Zhou, 2017). Unlike in Yoruba, PVE estimates for two phenotypes, HDL and TRIG, within the Cebu were significantly different than zero and closer to the Finnish estimates (Table 4).
Table 4. Percent variance explained (PVE) and standard error as estimated in GEMMA for each trait compared.
| Yoruba (n = 1,017) | Cebu (n = 1,765) | European (n = 5,123)a | |
|---|---|---|---|
| CHOL | 0.040 ± 0.061 | 0.067 ± 0.093 | 0.29 ± 0.043 |
| HDL | 0.013 ± 0.037 | 0.217 ± 0.091 | 0.34 ± 0.043 |
| TRIG | 0.049 ± 0.110 | 0.140 ± 0.103 | 0.38 ± 0.041 |
| LDL | 0.029 ± 0.046 | 0 ± 0.096 | 0.19 ± 0.047 |
Notes.
We attempted to estimate the genetic correlation between all SNPs in the Cebu and Yoruba populations using bivariate REML analysis as implemented in GCTA (Lee et al., 2012). The only phenotype that converged was TRIG, with an estimated correlation of 0.644 ± 0.65, indicating there is shared architecture between the populations. However, the small sample sizes available in our study do not offer enough power to reliably estimate heritability and genetic correlation as indicated by the large standard errors.
Yoruba gene-based association study
While many GWAS have been performed on lipid traits, most of the significant SNPs found fall outside of protein coding regions and thus their mechanisms of action are not immediately apparent. PrediXcan is a gene-level association method that incorporates functional data on potential regulatory elements to provide mechanistic directionality for association of a gene with a phenotype (Gamazon et al., 2015). PrediXcan uses gene expression prediction models built from genome-transcriptome datasets such as the Genotype-Tissue Expression (GTEx) Project to predict gene expression from genotype and then tests the predicted expression levels for association with with trait of interest (Ardlie et al., 2015). We applied PrediXcan to our SNP-level GWAS results using models built in 44 GTEx tissues (Barbeira et al., 2017) (Table 2).
For each tissue, we declared associations significant if FDR <0.1, across all genes and tissues tested, to adjust for multiple testing. Of the four phenotypes for this cohort and 44 tissues with models available, one gene, PAX6, surpassed the significance threshold. PAX6 was not significant in Cebu or GLGC (Table 5). In humans, PAX6 has been associated with insulin production (Ahlqvist et al., 2012). Currently, there is little known about PAX6 and its potential association with lipid or cardiovascular phenotypes.
Table 5. Top genes (FDR <0.1) in Yoruba and Cebu found using PrediXcan.
All results are for the TRIG phenotype. Results from both populations and the Global Lipids Genetic Consortium (GLGC) are shown. Six associations discovered in Cebu replicated in the GLGC.
| Chr. | Tissue | Gene name | Beta (Yoruba) | P (Yoruba) | FDR (Yoruba) | Beta (Cebu) | P (Cebu) | FDR (Cebu) | Beta (GLGC) | P (GLGC) |
|---|---|---|---|---|---|---|---|---|---|---|
| 2p23.3 | THYROID | FNDC4 | 0.05 | 0.50 | 0.99 | −0.24 | 1.0 × 10−6 | 0.037 | −0.11 | 7.7 × 10−83 |
| 11p13 | ARTAORT | PAX6 | −0.74 | 4.8 × 10−7 | 0.093 | 0.16 | 0.20 | 0.98 | 0 | 0.91 |
| 11q23.3 | ESPMCS | BACE1 | 0.22 | 0.33 | 0.99 | −3.30 | 1.7 × 10−15 | 3.0 × 10−10 | −0.17 | 7.3 × 10−19 |
| 11q23.3 | CLNSGM | APOA4 | 0.80 | 0.55 | 0.99 | −11.6 | 6.5 × 10−12 | 5.9 × 10−7 | −1.95 | 1.7 ×10−39 |
| 11q23.3 | BRNHPP | APOA1 | 0.015 | 0.88 | 0.99 | 0.50 | 3.2 × 10−7 | 0.019 | 0 | 0.31 |
| 11q23.3 | HRTLV | SIDT2 | 0.24 | 0.21 | 0.98 | 0.90 | 8.1 × 10−7 | 0.037 | 0.16 | 9.3 × 10−29 |
| 11q23.3 | THYROID | SIDT2 | 0.10 | 0.61 | 0.99 | −0.55 | 1.7 × 10−6 | 0.050 | −0.55 | 1.1 × 10−102 |
| 11q23.3 | WHLBLD | BACE1 | 0.00 | 0.99 | 0.99 | −0.79 | 3.4 × 10−6 | 0.087 | −0.28 | 4.0 × 10−28 |
Cebu gene-based association study
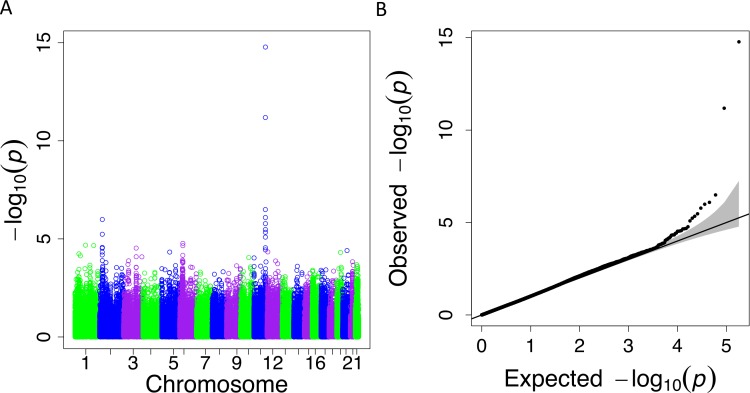
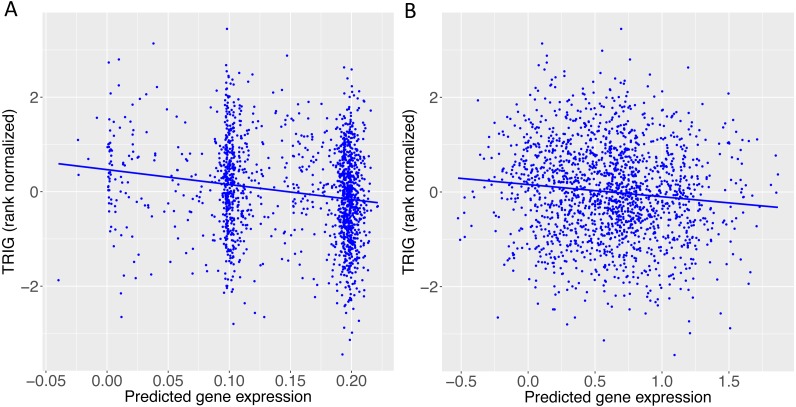
When we applied PrediXcan to the Cebu cohort, seven genes were found significant (Table 5). A few of these genes, such as FNDC4, APOA1 and APOA4, are well-documented in lipid traits, while genes such as SIDT2 have been previously implicated in Asians (Teslovich et al., 2010; Kim et al., 2011; Willer et al., 2013; Wu et al., 2013; Zhou et al., 2013; Gombojav et al., 2015; Lu et al., 2016; Spracklen et al., 2017). The association of BACE1 with TRIG was highly significant (FDR = 3.02 × 10−10, Fig. 4). The predicted increase in BACE1 expression and decrease in TRIG levels is an association not previously seen in humans, but has been observed in mice (Meakin et al., 2012; Baek et al., 2016) (Fig. 5). Additionally, SIDT2 has a similar effect across many tissue models, indicating its potential importance in regulating TRIG levels as well (Figs. 5 and 6). Both BACE1 and SIDT2 have increased gene expression associated with decreased TRIG levels in most tissues and share many SNPs in most of their prediction models.
Figure 4. PrediXcan results for the Cebu TRIG phenotype using gene expression models built in 44 GTEx tissues.
(A) Manhattan plot: −log10 P-values are plotted against the respective chromosomal position of each gene across all tissues. (B) QQ plot of observed versus expected −log10 P-values for each gene across all tissues.
Figure 5. TRIG levels vs. predicted expression of two genes in Cebu.
(A) BACE1 predicted expression using the GTEx ESPMCS prediction model. (B) SIDT2 predicted expression using the GTEx LCL prediction model.
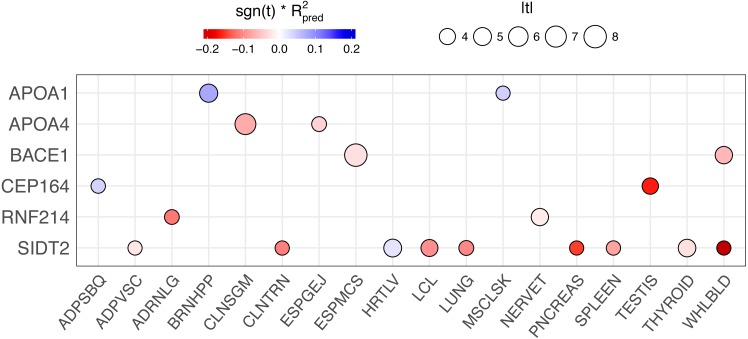
Figure 6. PrediXcan results of genes with rs662799 or linked SNPs (r2 > 0.6) in their predictive models across tissues.
The size of the circle is proportional to the absolute value of the t-statistic in the PrediXcan association test and the color indicates the direction of effect (sign of the t-statistic, −1 or 1) multiplied by the prediction performance of the model (R2) for each gene-tissue combination. Only genes are plotted for clarity.
We conducted a backward elimination analysis to determine the lead gene or genes at the 11q23.3 locus. The starting model include all gene-tissue combinations in Fig. 6, which includes all genes with rs662799 or a linked SNP (r2 > 0.6) in its predictive model and |t| > 3, where t is the association test statistic. The most significant gene in the final model was ESPMCS-BACE1 (P = 1 × 10−10), with residual effects present (P < 0.05) for LCL-SIDT2, MSCLSK-APOA1, TESTIS-CEP164, and WHLBLD-BACE1 (Table 6).
Table 6. Gene-tissue combinations from Fig. 6 with P < 0.05 in a backwards-elimination linear model including the TRIG phenotype and predicted gene expression terms.
| Tissue | Gene | Estimate | Std. Error | t value | P |
|---|---|---|---|---|---|
| ESPMCS | BACE1 | −3.7 | 0.57 | −6.4 | 1.7 × 10−10 |
| LCL | SIDT2 | −0.20 | 0.06 | −3.2 | 0.001 |
| MSCLSK | APOA1 | 1.5 | 0.52 | 2.9 | 0.003 |
| TESTIS | CEP164 | −0.17 | 0.08 | −2.2 | 0.025 |
| WHLBLD | BACE1 | 0.83 | 0.29 | 2.9 | 0.003 |
Comparing populations
In a direct comparison of the significant genes obtained through PrediXcan for the Cebu and Yoruba cohorts, there is no gene that shares significance (FDR <0.10) between populations (Table 5). There was no overlap in significant genes between Yoruba and GLGC, but there was overlap in significant genes between Cebu and GLGC in FNDC4, SIDT2, APOA4, and BACE1 with the same effect direction for all genes (Willer et al., 2013) (Table 5).
Discussion
Using genome-wide genotypes and lipid levels obtained in two diverse populations from Ibadan, Nigeria and Cebu, Philippines, we performed multiple genome-wide analyses with the goal of better understanding the underlying genetic architecture of cholesterol traits in both populations.
Top GWAS SNPs have been previously shown to associate with lipid traits
Within the Yoruba portion of our study, rs34065661 and rs17231520, SNPs close to or within CETP, were significant (Fig. 1A). CETP, cholesteryl ester transfer protein, is a well-known lipid gene previously implicated in lipid studies in African populations (Buyske et al., 2012; Elbers et al., 2012). CETP is involved in the transfer of cholesterol from HDL to other lipoproteins, and is a common genetic target for statins and other cholesterol-lowering drugs (Barter et al., 2003). It also has strong association with Alzheimer’s disease and other neurodegenerative diseases (Xiao et al., 2012). Additional significant SNPs in Yoruba, rs1065853, rs7412, and rs75627662, are located near or within APOE (Fig. 1B). APOE, apolipoprotein E, is another well-known cholesterol gene previously implicated in other lipid studies (Rasmussen-Torvik et al., 2012; Surakka et al., 2014; Mahley, 2016; Spracklen et al., 2017; Zhu et al., 2017). It acts as a lipid transport protein in high association with LDL receptors and is also strongly associated with neurodegenerative diseases (Moriarty et al., 2017).
The most significant SNP within the Cebu GWAS portion of our study is rs662799, at P = 2.7 ×10−16. It has been previously associated with cholesterol traits in Asian populations (Lu et al., 2016; Spracklen et al., 2017) (Fig. 2). APOA5 is a well-documented gene associated with triglyceride levels (Go et al., 2013). Other genes within this linkage group include ZPR1 and BUD13, which have both been previously implicated with triglyceride levels in East Asians as well, possibly due to its higher minor allele frequency in those populations (Kim et al., 2011; Lin et al., 2016) (Fig. 3).
The mechanism underlying the association of rs662799 with TRIG levels may include long distance regulation of BACE1 and SIDT2
In our PrediXcan analysis, BACE1 reached significance (FDR =3.0 × 10−10) in both the Cebu and GLGC replication cohorts, but it was not significant in Yoruba (Fig. 4). Currently, in humans, BACE1 is known to increase risk of Alzheimer’s disease with increased expression (Cole & Vassar, 2007). While not recognized as significant in the GLGC SNP meta-analysis (Willer et al., 2013), our application of PrediXcan to the GLGC GWAS results verified that the BACE1 association with TRIG is significant (P = 7.3 × 10−19) (Table 5).
This gene has been studied in terms of Alzheimer’s disease and weight gain in mice. In our results, we found increased predicted gene expression for BACE1 to be associated with lower TRIG levels (Table 5). For BACE1 knockout mice, there was no significant difference in triglyceride levels versus wild-type mice, but they did have lower average body weight (Meakin et al., 2012). Additionally, in mice, higher triglyceride levels were found to reduce BACE1 expression in a study concerning Alzheimer’s treatment (Baek et al., 2016). This latter result is consistent with our finding that increased BACE1 expression is associated with lower triglyceride levels (Fig. 5).
The SNP rs662799 is 571 bases upstream of the gene with which it is typically associated, APOA5 (Fig. 2). APOA5 was well-predicted in only one tissue, SNTTRM, and the prediction was not driven by SNPs linked to rs662799. Thus, APOA5 may not affect TRIG levels through the mechanism of variation in gene expression regulation. Even though rs662799 is located 493kb downstream of BACE1, it has the largest effect size, i.e., it is the driver SNP, in the predictive model for BACE1 in ESPMCS. SNPs closer to BACE1 are not linked to rs662799 (Fig. 2). The similar effects and significance of BACE1 in mouse studies, the Cebu cohort analysis, and GLGC PrediXcan results indicate the increased expression of BACE1 is associated with decreased TRIG levels and that variation in the regulation of BACE1 may contribute to differences in TRIG levels.
Another significant gene in our gene-based association study of TRIG in Cebu is SIDT2, which is 386 kb downstream of rs662799. Here, the effects of many SNPs, including some linked to rs662799, combine in the prediction model for SIDT2 gene expression, with no dominant driver SNP, as demonstrated by the lack of discernible clusters in the plot of TRIG levels versus predicted expression (Fig. 5). Additionally, SIDT2 exhibits more consistent effect sizes over more tissues than BACE1 (Fig. 6), in which predicted expression is associated with higher TRIG levels (Fig. 5).
SIDT2, along with others nearby on the same chromosome, has been previously implicated in East Asian GWAS (Gombojav et al., 2015). In our results, we found increased predicted gene expression for SIDT2 to be associated with lower TRIG levels in most models. SIDT2 has been associated with glucose and lipid metabolism in mice, as SIDT2 knockout mice have significantly higher serum levels of TRIG than wild-type mice (Gao et al., 2016). Since increased gene expression is associated with lower TRIG levels in our cohort for a majority of models, our models concur with the association present within mice. Significance of SIDT2 for TRIG in LCL was also replicated in GLGC at P = 1.1 ×10−102 (Table 5). The similar effects and significance of SIDT2 in knockout mice studies, the Cebu cohort analysis, and GLGC PrediXcan results indicate that increased expression of SIDT2 is associated with decreased TRIG levels. Therefore, variation in the regulation of SIDT2 may contribute to differences in TRIG levels.
In an attempt to disentangle the multiple genes associated with TRIG at the 11q23.3 locus, we performed backwards elimination modeling. This analysis showed that BACE1 has the strongest effect at the locus, with SIDT2, APOA1, and CEP164 contributing smaller effects.
rs662799 has a greater impact in East Asian populations
The significant SNPs in the GWAS portion of our study have been previously associated with lipid traits, but these prior studies did not conduct a robust gene-based association modeling (Hall et al., 2006; Buyske et al., 2012; Rasmussen-Torvik et al., 2012; Wu et al., 2013). In our PrediXcan analysis, we implicated a new gene (PAX6) in Yoruba and additional genes at the chromosome 11 locus in Cebu, including genes with predictive SNPs located hundreds of thousands of base pairs away from their transcription start sites.
rs662799 in particular is a significant SNP in cholesterol GWAS of East Asians (Teslovich et al., 2010; Wu et al., 2013; Lin et al., 2016; Lu et al., 2016; Spracklen et al., 2017), with its minor allele frequency as high as 0.37 in the 1000 Genomes Japanese (JPT) population, and it is also the top SNP, with a minor allele frequency of 0.245, in our own TRIG GWAS analysis for Cebu (Table 3). rs662799 has a low minor frequency in European and African populations, indicating that even if effects are similar, it has a lower allelic impact in European and African ancestry populations (Fig. 3). In Asian populations, therefore, rs662799 has a more significant genetic impact (Brown et al., 2016) due to its higher MAF, causing the difference in phenotypic effects. This demonstrates how prediction models may vary in utility across populations.
Conclusions
In this study, we use a transcriptome-informed approach to implicate new genes in lipid traits. Limitations arose from the small sample sizes in desired populations and the lack of population-specific transcriptome prediction models. Other lipid trait GWAS, such as the GLGC, included over 180,000 individuals of European ancestry (Willer et al., 2013), while each population in this study had less than 2,000 individuals. Current GTEx models are based on an 85% European-American and 15% African-American population, which cannot be fully extrapolated to diverse populations. While Yoruba in Ibadan, Nigeria is a HapMap population, and specific reference panels exist for African populations, there is a lack of publicly available data for southeast Asian and Pacific Islander populations, which is an issue due to the rarer variants in more isolated island populations (Loh et al., 2016). For example, the reference panel used for imputation, 1000 Genomes Phase 3, contains only Vietnamese, Chinese, and Japanese genotypes for East Asian populations (Auton et al., 2015). Without data collection and proper models for non-European populations, there is less potential for accurate implementation of precision medicine. To fully characterize the impact of genetic variation between populations, larger studies in non-European populations are needed.
Supplemental Information
Comparison of individual SNP results for the Cebu Longitudinal Health and Nutrition Survey in Wu et al. (2013) and our study.
Basic cohort data and raw summary statistics for non-transformed lipid phenotypes collected after fasting (mg/dL).
Results for XPEB performed on the Yoruba cohort using GLGC as a base population.
Results for XPEB performed on the Cebu cohort using GLGC as a base population.
The first two genotypic principal components for the Yoruba lipid cohort (GWAS) are plotted in comparison to HapMap populations YRI (Yoruba in Ibadan, Nigeria), CEU (European ancestry in Utah), and ASN (Chinese in Beijing and Japanese in Tokyo).
The first two genotypic principal components for the Cebu lipid cohort (GWAS) are plotted in comparison to HapMap populations YRI (Yoruba in Ibadan, Nigeria), CEU (European ancestry in Utah), and ASN (Chinese in Beijing and Japanese in Tokyo).
Acknowledgments
The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through accession numbers phs000378.v1.p1 and phs000523.v1.p1. Gene expression prediction models were obtained from PredictDB at http://predictdb.hakyimlab.org/.
Funding Statement
This work is supported by the National Institutes of Health National Human Genome Research Institute Academic Research Enhancement Award R15 HG009569 (PI: Heather E. Wheeler) and the Loyola University Chicago Carbon Undergraduate Research Fellowship (Angela Andaleon). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Angela Andaleon conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Lauren S. Mogil analyzed the data, contributed reagents/materials/analysis tools, reviewed drafts of the paper.
Heather E. Wheeler conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, reviewed drafts of the paper.
Data Availability
The following information was supplied regarding data availability:
The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through accession numbers phs000378.v1.p1 and phs000523.v1.p1. Gene expression prediction models were obtained from PredictDB at http://predictdb.hakyimlab.org/. See https://github.com/aandaleon/px_chol for all scripts used in our analyses.
References
- Adair et al. (2011).Adair LS, Popkin BM, Akin JS, Guilkey DK, Gultiano S, Borja J, Perez L, Kuzawa CW, McDade T, Hindin MJ. Cohort profile: the Cebu longitudinal health and nutrition survey. International Journal of Epidemiology. 2011;40(3):619–625. doi: 10.1093/ije/dyq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlqvist et al. (2012).Ahlqvist E, Turrini F, Lang ST, Taneera J, Zhou Y, Almgren P, Hansson O, Isomaa B, Tuomi T, Eriksson K, Eriksson JG, Lyssenko V, Groop L. A common variant upstream of the PAX6 gene influences islet function in man. Diabetologia. 2012;55(1):94–104. doi: 10.1007/s00125-011-2300-8. [DOI] [PubMed] [Google Scholar]
- Ardlie et al. (2015).Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, Gelfand ET, Trowbridge CA, Maller JB, Tukiainen T, Lek M, Ward LD, Kheradpour P, Iriarte B, Meng Y, Palmer CD, Esko T, Winckler W, Hirschhorn JN, Kellis M, MacArthur DG, Getz G, Shabalin AA, Li G, Zhou Y-H, Nobel AB, Rusyn I, Wright FA, Lappalainen T, Ferreira PG, Ongen H, Rivas MA, Battle A, Mostafavi S, Monlong J, Sammeth M, Mele M, Reverter F, Goldmann JM, Koller D, Guigó R, McCarthy MI, Dermitzakis ET, Gamazon ER, Im HK, Konkashbaev A, Nicolae DL, Cox NJ, Flutre T, Wen X, Stephens M, Pritchard JK, Tu Z, Zhang B, Huang T, Long Q, Lin L, Yang J, Zhu J, Liu J, Brown A, Mestichelli B, Tidwell D, Lo E, Salvatore M, Shad S, Thomas JA, Lonsdale JT, Moser MT, Gillard BM, Karasik E, Ramsey K, Choi C, Foster BA, Syron J, Fleming J, Magazine H, Hasz R, Walters GD, Bridge JP, Miklos M, Sullivan S, Barker LK, Traino HM, Mosavel M, Siminoff LA, Valley DR, Rohrer DC, Jewell SD, Branton PA, Sobin LH, Barcus M, Qi L, McLean J, Hariharan P, Um KS, Wu S, Tabor D, Shive C, Smith AM, Buia SA, Undale AH, Robinson KL, Roche N, Valentino KM, Britton A, Burges R, Bradbury D, Hambright KW, Seleski J, Korzeniewski GE, Erickson K, Marcus Y, Tejada J, Taherian M, Lu C, Basile M, Mash DC, Volpi S, Struewing JP, Temple GF, Boyer J, Colantuoni D, Little R, Koester S, Carithers LJ, Moore HM, Guan P, Compton C, Sawyer SJ, Demchok JP, Vaught JB, Rabiner CA, Lockhart NC, Ardlie KG, Getz G, Wright FA, Kellis M, Volpi S, Dermitzakis ET. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergs et al. (2012).Asselbergs FW, Guo Y, Van Iperen EP, Sivapalaratnam S, Tragante V, Lanktree MB, Lange LA, Almoguera B, Appelman YE, Barnard J, Baumert J, Beitelshees AL, Bhangale TR, Chen YDI, Gaunt TR, Gong Y, Hopewell JC, Johnson T, Kleber ME, Langaee TY, Li M, Li YR, Liu K, McDonough CW, Meijs MF, Middelberg RP, Musunuru K, Nelson CP, O’Connell JR, Padmanabhan S, Pankow JS, Pankratz N, Rafelt S, Rajagopalan R, Romaine SP, Schork NJ, Shaffer J, Shen H, Smith EN, Tischfield SE, Van Der Most PJ, Van Vliet-Ostaptchouk JV, Verweij N, Volcik KA, Zhang L, Bailey KR, Bailey KM, Bauer F, Boer JM, Braund PS, Burt A, Burton PR, Buxbaum SG, Chen W, Cooper-Dehoff RM, Cupples LA, Dejong JS, Delles C, Duggan D, Fornage M, Furlong CE, Glazer N, Gums JG, Hastie C, Holmes MV, Illig T, Kirkland SA, Kivimaki M, Klein R, Klein BE, Kooperberg C, Kottke-Marchant K, Kumari M, Lacroix AZ, Mallela L, Murugesan G, Ordovas J, Ouwehand WH, Post WS, Saxena R, Scharnagl H, Schreiner PJ, Shah T, Shields DC, Shimbo D, Srinivasan SR, Stolk RP, Swerdlow DI, Taylor HA, Topol EJ, Toskala E, Van Pelt JL, Van Setten J, Yusuf S, Whittaker JC, Zwinderman AH, Anand SS, Balmforth AJ, Berenson GS, Bezzina CR, Boehm BO, Boerwinkle E, Casas JP, Caulfield MJ, Clarke R, Connell JM, Cruickshanks KJ, Davidson KW, Day IN, De Bakker PI, Doevendans PA, Dominiczak AF, Hall AS, Hartman CA, Hengstenberg C, Hillege HL, Hofker MH, Humphries SE, Jarvik GP, Johnson JA, Kaess BM, Kathiresan S, Koenig W, Lawlor DA, März W, Melander O, Mitchell BD, Montgomery GW, Munroe PB, Murray SS, Newhouse SJ, Onland-Moret NC, Poulter N, Psaty B, Redline S, Rich SS, Rotter JI, Schunkert H, Sever P, Shuldiner AR, Silverstein RL, Stanton A, Thorand B, Trip MD, Tsai MY, Van Der Harst P, Van Der Schoot E, Van Der Schouw YT, Verschuren WM, Watkins H, Wilde AA, Wolffenbuttel BH, Whitfield JB, Hovingh GK, Ballantyne CM, Wijmenga C, Reilly MP, Martin NG, Wilson JG, Rader DJ, Samani NJ, Reiner AP, Hegele RA, Kastelein JJ, Hingorani AD, Talmud PJ, Hakonarson H, Elbers CC, Keating BJ, Drenos F. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. American Journal of Human Genetics. 2012;91(5):823–838. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko et al. (2007).Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Auton et al. (2015).Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, Flicek P, Gabriel SB, Gibbs RA, Green ED, Hurles ME, Knoppers BM, Korbel JO, Lander ES, Lee C, Lehrach H, Mardis ER, Marth GT, McVean GA, Nickerson DA, Schmidt JP, Sherry ST, Wang J, Wilson RK, Gibbs RA, Boerwinkle E, Doddapaneni H, Han Y, Korchina V, Kovar C, Lee S, Muzny D, Reid JG, Zhu Y, Wang J, Chang Y, Feng Q, Fang X, Guo X, Jian M, Jiang H, Jin X, Lan T, Li G, Li J, Li Y, Liu S, Liu X, Lu Y, Ma X, Tang M, Wang B, Wang G, Wu H, Wu R, Xu X, Yin Y, Zhang D, Zhang W, Zhao J, Zhao M, Zheng X, Lander ES, Altshuler DM, Gabriel SB, Gupta N, Gharani N, Toji LH, Gerry NP, Resch AM, Flicek P, Barker J, Clarke L, Gil L, Hunt SE, Kelman G, Kulesha E, Leinonen R, McLaren WM, Radhakrishnan R, Roa A, Smirnov D, Smith RE, Streeter I, Thormann A, Toneva I, Vaughan B, Zheng-Bradley X, Bentley DR, Grocock R, Humphray S, James T, Kingsbury Z, Lehrach H, Sudbrak R, Albrecht MW, Amstislavskiy VS, Borodina TA, Lienhard M, Mertes F, Sultan M, Timmermann B, Yaspo M-L, Mardis ER, Wilson RK, Fulton L, Fulton R, Sherry ST, Ananiev V, Belaia Z, Beloslyudtsev D, Bouk N, Chen C, Church D, Cohen R, Cook C, Garner J, Hefferon T, Kimelman M, Liu C, Lopez J, Meric P, O’Sullivan C, Ostapchuk Y, Phan L, Ponomarov S, Schneider V, Shekhtman E, Sirotkin K, Slotta D, Zhang H, McVean GA, Durbin RM, Balasubramaniam S, Burton J, Danecek P, Keane TM, Kolb-Kokocinski A, McCarthy S, Stalker J, Quail M, Schmidt JP, Davies CJ, Gollub J, Webster T, Wong B, Zhan Y, Auton A, Campbell CL, Kong Y, Marcketta A, Gibbs RA, Yu F, Antunes L, Bainbridge M, Muzny D, Sabo A, Huang Z, Wang J, Coin LJM, Fang L, Guo X, Jin X, Li G, Li Q, Li Y, Li Z, Lin H, Liu B, Luo R, Shao H, Xie Y, Ye C, Yu C, Zhang F, Zheng H, Zhu H, Alkan C, Dal E, Kahveci F, Marth GT, Garrison EP, Kural D, Lee W-P, Fung Leong W, Stromberg M, Ward AN, Wu J, Zhang M, Daly MJ, DePristo MA, Handsaker RE, Altshuler DM, Banks E, Bhatia G, Del Angel G, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayellet et al. (2010).Ayellet VS, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLOS Genetics. 2010;6(8):e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek et al. (2016).Baek SH, Choi BY, Cho Y, Kim H, Jung GY, Park HJ, Han J, Bahn G, Jo D-G. The drug triglyceride reducing BACE1 expression level and preventing cognitive impairment in Alzheimer’s disease mice. Alzheimer’s & Dementia. 2016;12(7):P434–P435. doi: 10.1016/j.jalz.2016.06.834. [DOI] [Google Scholar]
- Barbeira et al. (2017).Barbeira A, Dickinson SP, Torres JM, Torstenson ES, Zheng J, Wheeler HE, Shah KP, Edwards T, Consortium G, Nicolae D, Cox NJ, Im HK. Integrating tissue specific mechanisms into GWAS summary results. BioRxiv. 2017 doi: 10.1101/045260. Article 045260. [DOI]
- Barter et al. (2003).Barter PJ, Brewer HB, Chapman MJ, Hennekens CH, Rader DJ, Tall AR. Cholesteryl ester transfer protein: a novel target for raising HDL and inhibiting atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2003;23(2):160–167. doi: 10.1161/01.ATV.0000054658.91146.64. [DOI] [PubMed] [Google Scholar]
- Brown et al. (2016).Brown BC, Ye CJ, Price AL, Zaitlen N. Transethnic genetic-correlation estimates from summary statistics. American Journal of Human Genetics. 2016;99(1):76–88. doi: 10.1016/j.ajhg.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, De La Vega & Burchard (2011).Bustamante CD, De La Vega FM, Burchard EG. Genomics for the world. Nature. 2011;475(7355):163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyske et al. (2012).Buyske S, Wu Y, Carty CL, Cheng I, Assimes TL, Dumitrescu L, Hindorff LA, Mitchell S, Ambite JL, Boerwinkle E, Buzkova P, Carlson CS, Cochran B, Duggan D, Eaton CB, Fesinmeyer MD, Franceschini N, Haessler J, Jenny N, Kang HM, Kooperberg C, Lin Y, Marchand L, Matise TC, Robinson JG, Rodriguez C, Schumacher FR, Voight BF, Young A, Manolio TA, Mohlke KL, Haiman CA, Peters U, Crawford DC, North KE. Evaluation of the metabochip genotyping array in African Americans and implications for fine mapping of gwas-identified loci: the PAGE study. PLOS ONE. 2012;7(4):32118–32119. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole & Vassar (2007).Cole SL, Vassar R. The Alzheimer’s disease Beta-secretase enzyme, BACE1. Molecular Neurodegeneration. 2007;2(1) doi: 10.1186/1750-1326-2-22. Article 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coram et al. (2015).Coram MA, Candille SI, Duan Q, Chan KHK, Li Y, Kooperberg C, Reiner AP, Tang H. Leveraging multi-ethnic evidence for mapping complex traits in minority populations: an empirical Bayes approach. American Journal of Human Genetics. 2015;96(5):740–752. doi: 10.1016/j.ajhg.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das et al. (2016).Das S, Forer L, Schönherr S, Sidore C, Locke A, Kwong A, Vrieze S, Chew E, Levy S, McGue M, Schlessinger D, Stambolian D, Loh P, Iacono W, Swaroop A, Scott L, Cucca F, Kronenberg F, Boehnke M, Abecasis G, Fuchsberger C. Next-generation genotype imputation service and methods. Nature Genetics. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau, Marchini & Zagury (2012).Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nature Methods. 2012;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- Durbin (2014).Durbin R. Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT) Bioinformatics. 2014;30(9):1266–1272. doi: 10.1093/bioinformatics/btu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbers et al. (2012).Elbers CC, Guo Y, Tragante V, Van Iperen EPA, Lanktree MB, Castillo BA, Chen F, Yanek LR, Wojczynski MK, Li YR, Ferwerda B, Ballantyne CM, Buxbaum SG, Chen YDI, Chen WM, Cupples LA, Cushman M, Duan Y, Duggan D, Evans MK, Fernandes JK, Fornage M, Garcia M, Garvey WT, Glazer N, Gomez F, Harris TB, Halder I, Howard VJ, Keller MF, Kamboh MI, Kooperberg C, Kritchevsky SB, LaCroix A, Liu K, Liu Y, Musunuru K, Newman AB, Onland-Moret NC, Ordovas J, Peter I, Post W, Redline S, Reis SE, Saxena R, Schreiner PJ, Volcik KA, Wang X, Yusuf S, Zonderland AB, Anand SS, Becker DM, Psaty B, Rader DJ, Reiner AP, Rich SS, Rotter JI, Sale MM, Tsai MY, Borecki IB, Hegele RA, Kathiresan S, Nalls MA, Taylor HA, Hakonarson H, Sivapalaratnam S, Asselbergs FW, Drenos F, Wilson JG, Keating BJ. Gene-centric meta-analysis of lipid traits in African, East Asian and hispanic populations. PLOS ONE. 2012;7(12):1–14. doi: 10.1371/journal.pone.0050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellulu et al. (2014).Ellulu M, Abed Y, Rahmat A, Ranneh Y, Ali F. Epidemiology of obesity in developing countries: challenges and prevention. Global Epidemic Obesity. 2014;2(1):2. doi: 10.7243/2052-5966-2-2. [DOI] [Google Scholar]
- Gamazon et al. (2015).Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Nicolae DL, Cox NJ, Im HK. A gene-based association method for mapping traits using reference transcriptome data. Nature Genetics. 2015;47(9):1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao et al. (2016).Gao J, Zhang Y, Yu C, Tan F, Wang L. Spontaneous nonalcoholic fatty liver disease and ER stress in Sidt2 deficiency mice. Biochemical and Biophysical Research Communications. 2016;476(4):326–332. doi: 10.1016/j.bbrc.2016.05.122. [DOI] [PubMed] [Google Scholar]
- Go et al. (2013).Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics—2013 update: a report from the American Heart Association. Circulation. 2013;127(1):e6-e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombojav et al. (2015).Gombojav B, Lee SJ, Kho M, Song Y-M, Lee K, Sung J. Multiple susceptibility loci at chromosome 11q23.3 are associated with plasma triglyceride in East Asians. Journal of Lipid Research. 2015;57:318–324. doi: 10.1194/jlr.P063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev et al. (2016).Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, Jansen R, De Geus EJC, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, Alvarez M, Civelek M, Lusis AJ, Lehtimaki T, Raitoharju E, Kahonen M, Seppala I, Raitakari OT, Kuusisto J, Laakso M, Price AL, Pajukanta P, Pasaniuc B. Integrative approaches for large-scale transcriptome-wide association studies. Nature Genetics. 2016;48(3):245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall et al. (2006).Hall K, Murrell J, Ogunniyi A, Deeg M, Baiyewu O, Gao S, Gureje O, Dickens J, Evans R, Smith-Gamble V, Unverzagt FW, Shen J, Hendrie H. Cholesterol, APOE genotype, and Alzheimer disease: an epidemiologic study of Nigerian Yoruba. Neurology. 2006;66(2):223–227. doi: 10.1212/01.wnl.0000194507.39504.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim et al. (2011).Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, Hwang J-Y, Oh JH, Kim D-J, Kim NH, Kim S, Hong EJ, Kim J-H, Min H, Kim Y, Zhang R, Jia W, Okada Y, Takahashi A, Kubo M, Tanaka T, Kamatani N, Matsuda K, Park T, Oh B, Kimm K, Kang D, Shin C, Cho NH, Kim H-l, Han B-G, Lee J-Y, Cho YS. Large-scale genome-wide association studies in east Asians identify new genetic loci influencing metabolic traits. Nature Genetics. 2011;43(10):990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2012).Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin et al. (2016).Lin E, Kuo P-H, Liu Y-L, Yang AC, Kao C-F, Tsai S-J. Association and interaction of APOA5, BUD13, CETP, LIPA and health-related behavior with metabolic syndrome in a Taiwanese population. Nature Publishing Group. 2016;6(October) doi: 10.1038/srep36830. Article 36830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh et al. (2016).Loh P-R, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, Price AL. Reference-based phasing using the Haplotype Reference Consortium panel. Nature Genetics. 2016;48(11):1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2016).Lu X, Huang J, Mo Z, He J, Wang L, Yang X, Tan A, Chen S, Chen J, Charles Gu C, Chen J, Li Y, Zhao L, Li H, Hao Y, Li J, Hixson JE, Li Y, Cheng M, Liu X, Cao J, Liu F, Huang C, Shen C, Shen J, Yu L, Xu L, Mu J, Wu X, Ji X, Guo D, Zhou Z, Yang Z, Wang R, Yang J, Yan W, Peng X, Gu D. Genetic susceptibility to lipid levels and lipid change over time and risk of incident hyperlipidemia in chinese populations. Circulation: Cardiovascular Genetics. 2016;9(1):37–44. doi: 10.1161/CIRCGENETICS.115.001096. [DOI] [PubMed] [Google Scholar]
- Mahley (2016).Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. Journal of Molecular Medicine. 2016;94(7):739–746. doi: 10.1007/s00109-016-1427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman et al. (2007).Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nature Genetics. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus & Novembre (2016).Marcus JH, Novembre J. Visualizing the Geography of Genetic Variants. BioRxiv. 2016;(October) doi: 10.1101/068536. Article 068536. [DOI] [PMC free article] [PubMed]
- McCarthy et al. (2016).McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, Van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki A-E, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, De Bakker PIW, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R, Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nature Genetics. 2016;48(10):1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meakin et al. (2012).Meakin PJ, Harper AJ, Hamilton DL, Gallagher J, McNeilly AD, Burgess LA, Vaanholt LM, Bannon KA, Latcham J, Hussain I, Speakman JR, Howlett DR, Ashford MLJ. Reduction in BACE1 decreases body weight, protects against diet-induced obesity and enhances insulin sensitivity in mice. The Biochemical Journal. 2012;441(1):285–296. doi: 10.1042/BJ20110512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelberg et al. (2011).Middelberg RPS, Ferreira MAR, Henders AK, Heath AC, Madden PAF, Montgomery GW, Martin NG, Whitfield JB. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Medical Genetics. 2011;12 doi: 10.1186/1471-2350-12-123. Article 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty et al. (2017).Moriarty PM, Varvel SA, Gordts PLSM, McConnell JP, Tsimikas S. Lipoprotein(a) mass levels increase significantly according to APOE genotype: an analysis of 431,239 patients. Arteriosclerosis, Thrombosis, and Vascular Biology. 2017;37(3):580–588. doi: 10.1161/ATVBAHA.116.308704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunniyi et al. (1997).Ogunniyi A, Gureje O, Baiyewu O, Unverzagt F, Hall KS, Oluwole S, Osuntokun BO, Hendrie HC. Profile of dementia in a Nigerian community–types, pattern of impairment, and severity rating. Journal of the National Medical Association. 1997;89(6):392–396. [PMC free article] [PubMed] [Google Scholar]
- Patterson, Price & Reich (2006).Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLOS Genetics. 2006;2(12):2074–2093. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim et al. (2011).Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ, Frishman D. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2011;27(13):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell et al. (2007).Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, De Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen-Torvik et al. (2012).Rasmussen-Torvik LJ, Pacheco JA, Wilke RA, Thompson WK, Ritchie MD, Kho AN, Muthalagu A, Hayes MG, Armstrong LL, Scheftner DA, Wilkins JT, Zuvich RL, Crosslin D, Roden DM, Denny JC, Jarvik GP, Carlson CS, Kullo IJ, Bielinski SJ, Mccarty CA, Li R, Manolio TA, Crawford DC, Chisholm RL. High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clinical and Translational Science. 2012;5(5):394–399. doi: 10.1111/j.1752-8062.2012.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti et al. (2009).Sabatti C, Service SK, Hartikainen A-L, Pouta A, Ripatti S, Brodsky J, Jones CG, Zaitlen NA, Varilo T, Kaakinen M, Sovio U, Ruokonen A, Laitinen J, Jakkula E, Coin L, Hoggart C, Collins A, Turunen H, Gabriel S, Elliot P, McCarthy MI, Daly MJ, Jarvelin M-R, Freimer NB, Peltonen L. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nature Genetics. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklen et al. (2017).Spracklen CN, Chen P, Kim YJ, Wang X, Cai H, Li S, Long J, Wu Y, Wang YX, Takeuchi F, Wu JY, Jung KJ, Hu C, Akiyama K, Zhang Y, Moon S, Johnson TA, Li H, Dorajoo R, He M, Cannon ME, Roman TS, Salfati E, Lin KH, Guo X, Sheu WH, Absher D, Adair LS, Assimes TL, Aung T, Cai Q, Chang LC, Chen CH, Chien LH, Chuang LM, Chuang SC, Du S, Fan Q, Fann CS, Feranil AB, Friedlander Y, Gordon-Larsen P, Gu D, Gui L, Guo Z, Heng CK, Hixson J, Hou X, Hsiung CA, Hu Y, Hwang MY, Hwu CM, Isono M, Jimmy Juang JM, Khor CC, Kim YK, Koh WP, Kubo M, Lee IT, Lee SJ, Lee WJ, Liang KW, Lim B, Lim SH, Liu J, Nabika T, Pan WH, Peng H, Quertermous T, Sabanayagam C, Sandow K, Shi J, Sun L, Tan PC, Tan SP, Taylor KD, Teo YY, Toh SA, Tsunoda T, Van Dam RM, Wang A, Wang F, Wang J, Wei WB, Xiang YB, Yao J, Yuan JM, Zhang R, Zhao W, Ida Chen YD, Rich SS, Rotter JI, Wang TD, Wu T, Lin X, Han BG, Tanaka T, Cho YS, Katsuya T, Jia W, Jee SH, Chen YT, Kato N, Jonas JB, Cheng CY, Shu XO, He J, Zheng W, Wong TY, Huang W, Kim BJ, Tai ES, Mohlke KL, Sim X. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Human Molecular Genetics. 2017;26(9):1770–1784. doi: 10.1093/hmg/ddx062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surakka et al. (2014).Surakka I, Horikoshi M, Mägi R, Sarin A-P. The impact of low-frequency and rare variants on lipid levels. Nature Genetics. 2014;47(6):589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich et al. (2010).Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee J-Y, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RYL, Wright AF, Witteman JCM, Wilson JF, Willemsen G, Wichmann H-E, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJG, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BWJH, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PKE, Lucas G, Luben R, Loos RJF, Lokki M-L, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw K-T, Kaprio J, Kaplan LM, Johansson A, Jarvelin M-R, Janssens ACJW, Ingelsson E, Igl W, Kees Hovingh G, Hottenga J-J, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, De Geus EJC, De Faire U, Crawford G, Collins FS, Chen Y-DI, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai E-S, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJP, Schadt EE, Rotter JI, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner et al. (2001).Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, De Andrade M, Doheny KF, Haines JL, Hayes G, Jarvik G, Jiang L, Kullo IJ, Li R, Ling H, Manolio TA, Matsumoto M, McCarty CA, McDavid AN, Mirel DB, Paschall JE, Pugh EW, Rasmussen LV, Wilke RA, Zuvich RL, Ritchie MD. Quality control procedures for genome-wide association studies. Current Protocols in Human Genetics. 2001;68:1–19. doi: 10.1002/0471142905.hg0119s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler et al. (2016).Wheeler HE, Shah KP, Brenner J, Garcia T, Aquino-Michaels K, Cox NJ, Nicolae DL, Im HK. Survey of the heritability and sparse architecture of gene expression traits across human tissues. PLOS Genetics. 2016;12(11):1–23. doi: 10.1371/journal.pgen.1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, Winston & RStudio (2016).Wickham H, Winston C, RStudio ggplot2: create elegant data visualisations using the grammar of graphics. Version 2.2.1https://cran.r-project.org/web/packages/ggplot2/index.html CRAN. 2016
- Willer et al. (2013).Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang H-Y, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikäinen L-P, Magnusson PKE, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CDCNA, Perola M, Petersen A-K, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney ASF, Döring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen A-L, Hayward C, Hernandez D, Hicks AA, Holm H, Hung Y-J, Illig T, Jones MR, Kaleebu P, Kastelein JJP, Khaw K-T, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimäki T, Lin S-Y, Lindström J, Loos RJF, Mach F, McArdle WL, Meisinger C, Mitchell BD, Müller G, Nagaraja R, Narisu N, Nieminen TVM, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancáková A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, Van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen Y-DI, Collins FS, Cooper RS, Danesh J, Dedoussis G, De Faire U, Feranil AB, Ferrières J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Järvelin M-R, Jula A, Kähönen M, Kaprio J, Kesäniem A, et al. Discovery and refinement of loci associated with lipid levels. Nature Genetics. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik et al. (2017).Wojcik G, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, Highland HM, Patel YM, Sorokin EP, Avery CL, Belbin GM, Bien SA, Cheng I, Hodonsky CJ, Huckins LM, Jeff J, Justice AE, Kocarnik JM, Lim U, Lin BM, Lu Y, Nelson SC, Park S-SL, Preuss MH, Richard MA, Schurmann C, Setiawan VW, Vahi K, Vishnu A, Verbanck M, Walker R, Young KL, Zubair N, Ambite JL, Boerwinkle E, Bottinger E, Bustamante CD, Caberto C, Conomos MP, Deelman E, Do R, Doheny K, Fernandez-Rhodes L, Fornage M, Heiss G, Hindorff LA, Jackson RD, James R, Laurie CA, Laurie CC, Li Y, Lin D-Y, Nadkarni G, Pooler LC, Reiner AP, Romm J, Sabati C, Sheng X, Stahl EA, Stram DO, Thornton TA, Wassel CL, Wilkens LR, Yoneyama S, Buyske S, Haiman C, Kooperberg C, Le Marchand L, Loos RJ, Matise TC, North KE, Peters U, Kenny EE, Carlson CS. Genetic diversity turns a new PAGE in our understanding of complex traits. BioRxiv. 2017 doi: 10.1101/188094. Article 188094. [DOI]
- Wu et al. (2013).Wu Y, Marvelle AF, Li J, Croteau-Chonka DC, Feranil AB, Kuzawa CW, Li Y, Adair LS, Mohlke KL. Genetic association with lipids in Filipinos: waist circumference modifies an APOA5 effect on triglyceride levels. Journal of Lipid Research. 2013;54(11):3198–3205. doi: 10.1194/jlr.P042077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao et al. (2012).Xiao Z, Wang J, Chen W, Wang P, Zeng H, Chen W. Association studies of several cholesterol-related genes (ABCA1, CETP and LIPC) with serum lipids and risk of Alzheimer’s disease. Lipids in Health and Disease. 2012;11(1) doi: 10.1186/1476-511X-11-163. Article 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2012).Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature Genetics. 2012;44(4):369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2011).Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2013).Zhou L, He M, Mo Z, Wu C, Yang H, Yu D, Yang X, Zhang X, Wang Y, Sun J, Gao Y, Tan A, He Y, Zhang H, Qin X, Zhu J, Li H, Lin X, Zhu J, Min X, Lang M, Li D, Zhai K, Chang J, Tan W, Yuan J, Chen W, Wang Y, Wei S, Miao X, Wang F, Fang W, Liang Y, Deng Q, Dai X, Lin D, Huang S, Guo H, Lilly Zheng S, Xu J, Lin D, Hu FB, Wu T. A genome wide association study identifies common variants associated with lipid levels in the Chinese population. PLOS ONE. 2013;8(12):e82420. doi: 10.1371/journal.pone.0082420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou (2017).Zhou X. A unified framework for variance component estimation with summary statistics in genome-wide association studies. BioRxiv. 2017 doi: 10.1101/042846. Article 042846. [DOI] [PMC free article] [PubMed]
- Zhou & Stephens (2012).Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nature Genetics. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu et al. (2017).Zhu Y, Zhang D, Zhou D, Li Z, Li Z, Fang L, Yang M, Shan Z, Li H, Chen J, Zhou X, Ye W, Yu S, Li H, Cai L, Liu C, Zhang J, Wang L, Lai Y, Ruan L, Sun Z, Zhang S, Wang H, Liu Y, Xu Y, Ling J, Xu C, Zhang Y, Lv D, Yuan Z, Zhang J, Zhang Y, Shi Y, Lai M. Susceptibility loci for metabolic syndrome and metabolic components identified in Han Chinese: a multi-stage genome-wide association study. Journal of Cellular and Molecular Medicine. 2017;21(6):1106–1116. doi: 10.1111/jcmm.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of individual SNP results for the Cebu Longitudinal Health and Nutrition Survey in Wu et al. (2013) and our study.
Basic cohort data and raw summary statistics for non-transformed lipid phenotypes collected after fasting (mg/dL).
Results for XPEB performed on the Yoruba cohort using GLGC as a base population.
Results for XPEB performed on the Cebu cohort using GLGC as a base population.
The first two genotypic principal components for the Yoruba lipid cohort (GWAS) are plotted in comparison to HapMap populations YRI (Yoruba in Ibadan, Nigeria), CEU (European ancestry in Utah), and ASN (Chinese in Beijing and Japanese in Tokyo).
The first two genotypic principal components for the Cebu lipid cohort (GWAS) are plotted in comparison to HapMap populations YRI (Yoruba in Ibadan, Nigeria), CEU (European ancestry in Utah), and ASN (Chinese in Beijing and Japanese in Tokyo).
Data Availability Statement
The following information was supplied regarding data availability:
The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through accession numbers phs000378.v1.p1 and phs000523.v1.p1. Gene expression prediction models were obtained from PredictDB at http://predictdb.hakyimlab.org/. See https://github.com/aandaleon/px_chol for all scripts used in our analyses.