Abstract
The genus Tenacibaculum encompasses several species pathogenic for marine fish. Tenacibaculum dicentrarchi and “Tenacibaculum finnmarkense” (Quotation marks denote species that have not been validly named.) were retrieved from skin lesions of farmed fish such as European sea bass or Atlantic salmon. They cause a condition referred to as tenacibaculosis and severe outbreaks and important fish losses have been reported in Spanish, Norwegian, and Chilean marine farms. We report here the draft genomes of the T. dicentrarchi and “T. finnmarkense” type strains. These genomes were compared with draft genomes from field isolates retrieved from Chile and Norway and with previously published Tenacibaculum genomes. We used Average Nucleotide Identity and core genome-based phylogeny as a proxy index for species boundary delineation. This work highlights evolution of closely related fish-pathogenic species and suggests that homologous recombination likely contributes to genome evolution. It also corrects the species affiliation of strain AYD7486TD claimed by Grothusen et al. (2016).
Keywords: Tenacibaculum, tenacibaculosis, genomes, fish pathogens, virulence, evolution
Introduction
The rapid development of intensive aquaculture has been associated with a dramatic increase in outbreaks of infectious diseases (FAO 2016; Bayliss et al. 2017). The rapid international spread of pathogens through the trade of fish and eggs or as a response to environmental changes has been documented (Brynildsrud et al. 2014; Rahmati-Holasoo et al. 2016). In this context, the success and sustainability of aquaculture largely depend on the control of pathogens. Among those, several species of the genus Tenacibaculum are responsible for diseases collectively designated tenacibaculosis (Avendaño-Herrera et al. 2006; Suzuki 2015). Tenacibaculum dicentrarchi and “Tenacibaculum finnmarkense” are two among those fish-associated, recently described species. The former was first isolated from European sea bass (Dicentrarchus labrax) in Spain (Piñeiro-Vidal et al. 2012) and recently also identified from Atlantic salmon (Salmo salar) in Chile (Avendaño-Herrera et al. 2016) and Norway (Olsen et al. 2017) and from red conger eel (Genypterus chilensis) in Chile (Irgang et al. 2017). “Tenacibaculum finnmarkense” was isolated from Atlantic salmon with ulcerative disease in Norway (Småge et al. 2016).
Identification of the causative agent of tenacibaculosis was first based on the isolation of bacteria from tissues of diseased fish and their characterization by phenotypic, biochemical, and serological methods (Piñeiro-Vidal, Carballas et al. 2008; Piñeiro-Vidal, Riaza et al. 2008). The use of 16 S rDNA sequencing improved the identification reliability (Cepeda et al. 2003; Fringuelli et al. 2012). However, these methods usually cannot differentiate closely related bacterial species. MLST was developed (Habib et al. 2014) and used to demonstrate the presence of T. dicentrarchi in Chile (Avendaño-Herrera et al. 2016) and to reveal the variety of Tenacibaculum spp. in a number of sea-farmed fish species in Norway (Olsen et al. 2017).
In this study, we present seven draft genomes of Tenacibaculum strains, including the T. dicentrarchi (USC 3509T) and “T. finnmarkense” (HFJT) type strains, as well as five field isolates from Chile and Norway selected on the basis of available MLST data (Olsen et al. 2017). Comparison has been performed with available Tenacibaculum genomes from Genbank, including strain AYD7486TD originally described as T. dicentrarchi (Grothusen et al. 2016). We used Average Nucleotide Identity (ANI) to delineate species boundaries and core genome analysis to infer phylogenetic relationships between these strains. We also correct the species affiliation of strain AYD7486TD.
Materials and Methods
Bacterial Strains
The T. dicentrarchi type strain USC 3509T (Piñeiro-Vidal et al. 2012) was obtained from Dr Y. Santos (University of Santiago de Compostela, Spain) and the “T. finnmarkense” strain HFJT (Småge et al. 2016) was obtained from Dr H. Duesund (Cermaq Group AS, Bergen, Norway). Strains TNO006, TNO010, and TNO020 were isolated from skin ulcers of Atlantic salmon in Norway whereas strain TNO021 was isolated from mouth ulcer of a corkwing-wrasse (Symphodus melops) also in Norway (Habib et al. 2014; Olsen et al. 2017). Strain TdChD05 was retrieved from external lesion of an Atlantic salmon in Chile (Avendaño-Herrera et al. 2016). All strains were routinely grown on marine agar 2216 (Difco) and in the corresponding broth at 170 rpm and 15 °C (TNO006) or 22 °C (all other strains).
Genome Sequencing and Annotation
Genomic DNA was extracted with the Wizard Genomic DNA Purification Kit (PROMEGA). All strains were sequenced with Illumina (HiSeq 2x100 pair-end reads with 300 bp insert size). Sequencing reads were assembled using Velvet (Zerbino and Birney 2008) or SPAdes (Bankevich et al. 2012). Genome annotation, including manual curation, and genome comparison including core genome computation were performed using the MicroScope platform (Médigue et al. 2017 and references therein).
ANI and Phylogenetic Reconstruction
ANIs analyses were performed using the ANIm method described by Richter and Rosselló-Móra (2009) and implemented in the Python module Pyani (https://github.com/widdowquinn/pyani). Digital DNA–DNA hybridization was performed using the GGDC website (http://ggdc.dsmz.de/) and Formula 2 (Auch et al. 2010). Logistic regression was used for reporting the probabilities that DDH is ≥70% and thus accounting for bacteria belonging to the same species. Pairwise alignments were computed using the MUMer software (Kurtz et al. 2004). For phylogenetic reconstruction, comparison of the gene content between strains was done by pairwise proteome similarity search using BlastP Bidirectional Best Hit and the MicroScope default parameters (i.e., >80% protein identity, >80% coverage). A set of 895 groups of orthologous proteins was retained and multiple alignments on individual orthologous proteins were performed using MUSCLE (Edgar 2004) implemented in the msa R package (Bioconductor). The resulting alignments were manually checked and concatenated for tree reconstruction using UGENE and PhyML with default parameters (Okonechnikov et al. 2012). Tree rendering was achieved using the Figtree software (http://tree.bio.ed.ac.uk/software/figtree/). The neighbor-net analysis in the Splits Tree 4 software (http://splitstree.org/; Huson and Bryant 2006) provided a phylogenetic network representing possible evolutionary relationships between the concatenated sequences of core genome genes. Minimal recombination breakpoints were identified using the four-gamete test (Hudson and Kaplan 1985). Putative recombination events were indicated as pairwise homoplasy index (PHI; Bruen et al. 2006) calculated by Splits Tree 4.
Results and Discussion
General Genome Features
A summary of the genomes analyzed in this study is presented in supplementary table 1, Supplementary Material online. Strikingly, the sizes of the genomes reported here are the smallest among those available to date for the genus Tenacibaculum (e.g., T. maritimum: 3.4 Mb, T. soleae: 3.0 Mb, T. ovolyticum: 4.1 Mb). The average genome size is 2.7 Mb and 2.9 Mb for T. dicentrarchi and “T. finnmarkense,” respectively; at 2.4 Mb, strain TNO020 has the smallest genome. All strains studied are devoid of plasmid.
ANI Delineates T. dicentrarchi and “T. finnmarkense” and Allocates Strain AY7486TD to “T. finnmarkense”
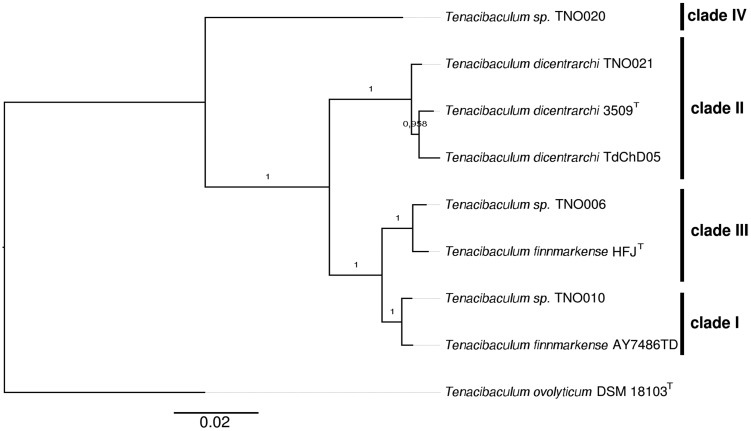
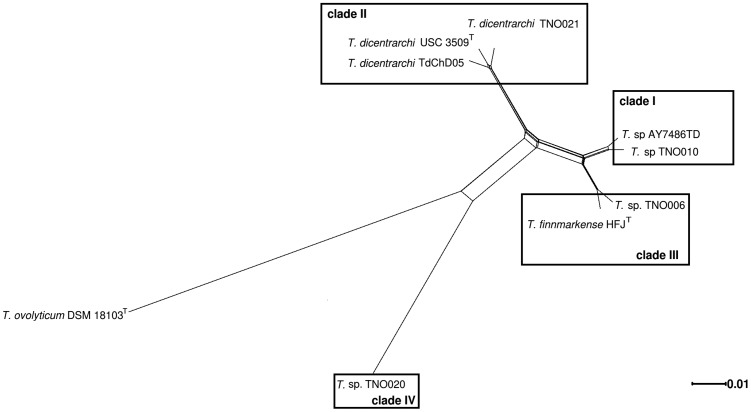
ANI was reported to be an accurate and practical method for species delineation and a 95–96% identity was proposed as the threshold (Rodriguez-R and Konstantinidis 2014). ANI comparisons were computed using T. ovolyticum (Suzuki et al. 2001) as an outgroup and the results were plotted as a heatmap (supplementary fig. 1A and 1B, Supplementary Material online). Strains TdChD05 from Chile and TNO021 from Norway, both previously allocated to the species T. dicentrarchi (Piñeiro-Vidal et al. 2012; Olsen et al. 2017; Avendaño-Herrera et al. 2016), indeed form a highly cohesive group with T. dicentrarchi USC 3509 T (98% ANI and ≥88% alignment coverage). In contrast, strain AY7486TD, also originally described as T. dicentrarchi (Grothusen et al. 2016), displays an ANI value of only 93% with the type strain of this species and consequently does not fall within the T. dicentrarchi cluster. Instead, strain AYD7486TD forms a cluster with strains TNO006, TNO010 and “T. finnmarkense” HFJT (≥96% ANI and ≥85% alignment coverage, above the species delineation threshold). ANI values delineate two subclusters, one grouping strains TNO010 and AYD7486TD (99% ANI) and the other grouping strain TNO006 and “T. finnmarkense” HFJT (98% ANI). Supplementary figure 1, Supplementary Material online, also shows that although T. dicentrarchi and “T. finnmarkense” are distinct species they obviously display significant proximity in terms of sequences identity (93–94% ANI) and fraction of shared genomes (77–88%). Strain TNO020 does not belong to any of the previously defined clusters (88–89% ANI; 55–68% alignment coverage) and therefore likely belongs to a yet undescribed Tenacibaculum species as previously suggested (Habib et al. 2014; Olsen et al. 2017). The same conclusions (supplementary fig. 1C, Supplementary Material online) were drawn using genome-to-genome distance calculator (Auch et al. 2010). As expected, T. ovolyticum DSM 18103T behaves as an outgroup displaying lower sequence identity (85%) and poor alignment coverage (17–34%) with all other strains studied. Using 895 core genome-encoding protein sequences, we constructed a phylogenetic tree from the concatenation of each individual alignment (fig. 1). Bootstrap values strongly support the division between T. dicentrarchi and “T. finnmarkense” and the core genome-based phylogenetic tree perfectly matches the ANI dendogram. Furthermore, a correlation between the MLST clades defined by Olsen et al. (2017) and the clusters observed in figure 1 is obvious: “T. finnmarkense” HFJT and strain TNO006 belong to clade III, strains TNO010 and AYD7486TD to clade I, all three T. dicentrarchi strains to clade II and strain TNO020 to clade IV. Using Splits Tree analysis, a reticulated network structure between the four clades, indicative of within and between species recombination events (fig. 2), is observed. The dense network joining clade I and clade III is in good agreement with the grouping of “T. finnmarkense” strains in two connected subclusters as previously observed in the MLST data set of Olsen et al. (2017). Whatever the group of strains considered (i.e., clades I and III strains, clades I, II, and III strains as well as clades I, II, III, and IV strains), the PHI test P-value is 0.0, indicating significant evidence of recombination. In addition, the high number of recombination breakpoints found in T. dicentrarchi and “T. finnmarkense” genomes (10 per kilobase in average) is another clue suggesting recombination events in these species.
Fig. 1.
—Core genome phylogeny. Phylogenetic tree inferred by PhyML with boostraping (100 replicates) using the concatenation of the 895 aligned orthologous genes. The MLST clades I–IV defined in Olsen et al. (2017) are reported.
Fig. 2.
—Detection of recombination. Reticulate evolutionary relationship between concatenated sequences of the 895 core genome genes visualized by the Splits Tree neighbor-net analysis. The clades defined in Olsen et al. (2017) are reported.
Comparative Genomics Highlights Intricate Relationships between T. dicentrarchi and “T. finnmarkense”
The average number of predicted CDS for T. dicentrarchi and “T. finnmarkense” strains is 2,381 and 2,536, respectively, which is in good agreement with the observed genome sizes. The core genome is composed of 2,013 and 1,947 CDS for T. dicentrarchi and “T. finnmarkense,” respectively (supplementary fig. 2A and B, Supplementary Material online). These small gene sets, about half those of Tenacibaculum agarivorans HZ1T (Xu et al. 2017) and Tenacibaculum jejuense KCTC 22618T (Ficko-Blean 2017), seem related to a deficient biopolymer-degrading ability of T. dicentrarchi and “T. finnmarkense.” Indeed, these genomes lack the pathways encoding for the degradation of marine carbohydrates (e.g., sulfatase, glycoside hydrolase, polysaccharide lyase) identified in the environmental species T. agarivorans and T. jejuense, in line with a restricted ecological niche (i.e., fish tissues) and an exclusive protein-based predicted regimen for these pathogenic species. Moreover, the presence of insertion sequences or their scars as well as genes remnants in T. dicentrarchi, “T. finnmarkense” and strain TNO020 argues for genome reduction trends in contrast to the horizontal transfer genes in T. jejuense and T. agarivorans. These findings support the expected small genome size of bacterial pathogens compared with their nonpathogenic relatives (Weinert and Welch 2017). The core genome of the seven strains belonging to both T. dicentrarchi and “T. finnmarkense” is composed of 1,818 CDS (supplementary fig. 2C, Supplementary Material online), a value close to those computed for each species. Each strain has ∼180 specific genes essentially composed of prophages remnants, restriction/modification systems, toxin/antitoxin systems and transposases encoding genes or their scars as well as genes required for the biosynthesis of exopolysaccharides that likely account for the minor intraspecies genome size differences previously mentioned. These strain-specific genes, representing the accessory genome, do not seem linked to bacterial pathogenicity as no bona fide toxin or virulence factor-encoding genes have been identified in this gene pool. Therefore, virulence-encoding genes likely belong to the core genome common to both T. dicentrarchi and “T. finnmarkense.” Among those, peptidases containing a carboxy-terminal protein domain (TIGR04183), predicted to be required for T9SS-mediated secretion and cell surface exposure (Veith et al. 2013), were identified. These peptidases (i.e., TFINN_2500013, TFINN_140038, and TFINN_60057 from “T. finnmarkense” HFJT and their orthologs) are likely involved in the breakdown of proteinaceous compounds and the destruction of host tissues. The presence of a M9 family protease-encoding gene (TFINN_140038 and orthologs), similar to the 120 kDa collagenase of Clostridium perfringens (Matsushita et al. 1994) but different from the M43 family collagenase (encoded by MARIT_1085) identified in T. maritimum (Pérez-Pascual et al. 2017), suggests convergent evolution for some virulence-linked functions in fish-pathogenic Tenacibaculum species.
Conclusion
Since the pioneering work of Wakabayashi and col. on T. maritimum in the eighties (Wakabayashi et al. 1986), many other Tenacibaculum species have been described. Some of them are important fish pathogens and an unexpected diversity at different levels (e.g., genetic, fish host, geographical) has been reported (Habib et al. 2014; Olsen et al. 2017). Tenacibaculum dicentrarchi strains were previously identified from several farmed fish species in Spain, Norway and Chile, whereas “T. finnmarkense” strains were exclusively isolated in Norway so far (Olsen et al. 2017). Thanks to genomes comparison, we were able to correct the affiliation of strain AYD7486TD which actually belongs to the species “T. finnmarkense” rather than to T. dicentrarchi as previously claimed (Grothusen et al. 2016). Importantly, this result demonstrates that “T. finnmarkense” is also present in Chilean fish farms. Our data set suggests that T. dicentrarchi strains form a cohesive group whereas “T. finnmarkense” strains are split into two subclusters. Similar subclusters, referred to as genomovars, were reported in Flavobacterium columnare, another fish pathogen of the family Flavobacteriaceae with a broad host range. Correlations between genomovars, fish hosts and virulence have been suggested (Evenhuis and LaFrentz 2016; Olivares-Fuster et al. 2007). Hence, the same type of genomic heterogeneity observed in “T. finnmarkense” may account for specific traits such as host specificity or level of virulence. Strikingly, the virulence factors described in the closely related species T. maritimum (i.e., a sphingomyelinase, a ceramidase, a chondroitin AC lyase, a sialidase and a M43 family collagenase; Pérez-Pascual et al. 2017) have not been identified in any of the genomes described in the present study. In full agreement with this observation, a parallel evolution of pathogenicity in the species encompassed in the genus Tenacibaculum has been proposed (Habib et al. 2014). However, the grouping of T. dicentrarchi and “T. finnmarkense” as well as T. soleae and T. ovolyticum (Habib et al. 2014; Olsen et al. 2017; Småge et al. 2016) in a single clade exclusively encompassing fish-associated bacteria suggests evolution of these four species from a pathogenic ancestor. In addition, our data support recombination as an important force shaping genome evolution of these fish pathogens as previously observed in other members of the family Flavobacteriaceae (Nicolas et al. 2008; Vos and Didelot 2009). The genome sequences reported here should therefore facilitate future epidemiological studies and provide new insights into pathogenicity and niche adaptation of emerging fish pathogens.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was funded by the EU EMIDA ERA-NET project “Control Flavobacteriaceae Infections in European Fish farms” and by the Agence Nationale pour la Recherche (contract ANR-14-CE19-0020). This work has benefited from the facilities and expertise of the high-throughput sequencing platform of I2BC (Centre de Recherche de Gif; http://www.i2bc.paris-saclay.fr/spip.php? article399&lang=en). We also wish to thank the following structures for providing computational resources: The INRA MIGALE bioinformatics platform (http://migale.jouy.inra.fr), the LABGeM, and the National Infrastructure “France Génomique” funded as part of the Investissement d’avenir program managed by Agence Nationale pour la Recherche (contract ANR-10-INBS-09). A.-H. acknowledges Grant FONDECYT 1150695 and the CONICYT/FONDAP/15110027 from the Comisión Nacional de Investigación Científica y Tecnológica (CONICYT, Chile). The authors are very grateful to Sophie Pasek and Mathilde Carpentier for fruitful discussion.
Literature Cited
- Auch AF, Klenk H-P, Göker M.. 2010. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2(1):142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendaño-Herrera R, et al. 2016. Isolation, characterization and virulence potential of Tenacibaculum dicentrarchi in salmonid cultures in Chile. Transbound Emerg Dis. 63(2):121–126. [DOI] [PubMed] [Google Scholar]
- Avendaño-Herrera R, Toranzo AE, Magariños B.. 2006. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis Aquat Organ. 71(3):255–266. [DOI] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.http://dx.doi.org/10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss SC, et al. 2017. The promise of whole genome pathogen sequencing for the molecular epidemiology of emerging aquaculture pathogens. Front Microbiol. 8:121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D.. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172(4):2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsrud O, et al. 2014. Microevolution of Renibacterium salmoninarum: evidence for intercontinental dissemination associated with fish movements. ISME J. 8(4):746–756.http://dx.doi.org/10.1038/ismej.2013.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda C, García-Márquez S, Santos Y.. 2003. Detection of Flexibacter maritimus in fish tissue using nested PCR amplification. J Fish Dis. 26(2):65–70. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.http://dx.doi.org/10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenhuis JP, LaFrentz BR.. 2016. Virulence of Flavobacterium columnare genomovars in rainbow trout Oncorhynchus mykiss. Dis Aquat Organ. 120(3):217–224. [DOI] [PubMed] [Google Scholar]
- FAO. 2016. The State of World Fisheries and Aquaculture 2016. Available from: http://www.fao.org/3/a-i5555e.pdf. [Google Scholar]
- Ficko-Blean E, et al. 2017. Carrageenan catabolism is conferred by a complex regulon in marine heterotrophic bacteria. Nat Commun (forthcoming) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fringuelli E, et al. 2012. Development of a quantitative real-time PCR for the detection of Tenacibaculum maritimum and its application to field samples. J Fish Dis. 35(8):579–590.http://dx.doi.org/10.1111/j.1365-2761.2012.01377.x [DOI] [PubMed] [Google Scholar]
- Grothusen H, et al. 2016. First complete genome sequence of Tenacibaculum dicentrarchi, an emerging bacterial pathogen of salmonids. Genome Announc. 4(1):e01756–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib C, et al. 2014. Multilocus sequence analysis of the marine bacterial genus Tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Appl Environ Microbiol. 80(17):5503–5514.http://dx.doi.org/10.1128/AEM.01177-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 111(1):147–164. [DOI] [PMC free article] [PubMed]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23(2):254–267.http://dx.doi.org/10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Irgang R, et al. 2017. First identification and characterization of Tenacibaculum dicentrarchiisolated from Chilean red conger eel (Genypterus chilensis, Guichenot 1848). J Fish Dis. 40(12):1915–1920. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita O, Yoshihara K, Katayama S, Minami J, Okabe A.. 1994. Purification and characterization of Clostridium perfringens 120-kilodalton collagenase and nucleotide sequence of the corresponding gene. J Bacteriol. 176(1):149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C, et al. 2017. MicroScope-an integrated resource for community expertise of gene functions and comparative analysis of microbial genomic and metabolic data. Brief Bioinformatics bbx113, https://doi.org/10.1093/bib/bbx113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P, et al. 2008. Population structure of the fish-pathogenic bacterium Flavobacterium psychrophilum. Appl Environ Microbiol. 74(12):3702–3709. [DOI] [PMC free article] [PubMed]
- Okonechnikov K, Golosova O, Fursov M.. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8):1166–1167.http://dx.doi.org/10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- Olivares-Fuster O, et al. 2007. Host-specific association between Flavobacterium columnare genomovars and fish species. Syst Appl Microbiol. 30(8):624–633.http://dx.doi.org/10.1016/j.syapm.2007.07.003 [DOI] [PubMed] [Google Scholar]
- Olsen AB, et al. 2017. Multilocus sequence analysis reveals extensive genetic variety within Tenacibaculum spp. associated with ulcers in sea-farmed fish in Norway. Vet Microbiol. 205:39–45. [DOI] [PubMed] [Google Scholar]
- Olsen AB, et al. 2011. Tenacibaculum sp. associated with winter ulcers in sea-reared Atlantic salmon Salmo salar. Dis Aquat Organ. 94(3):189–199.http://dx.doi.org/10.3354/dao02324 [DOI] [PubMed] [Google Scholar]
- Pérez-Pascual D, et al. 2017. The complete genome sequence of the fish pathogen Tenacibaculum maritimum provides insights into virulence mechanisms. Front Microbiol. 8:1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro-Vidal M, Carballas CG, Gómez-Barreiro O, Riaza A, Santos Y.. 2008. Tenacibaculum soleae sp. nov., isolated from diseased sole (Solea senegalensis Kaup). Int J Syst Evol Microbiol. 58(Pt 4):881–885. [DOI] [PubMed] [Google Scholar]
- Piñeiro-Vidal M, Gijón D, Zarza C, Santos Y.. 2012. Tenacibaculum dicentrarchi sp. nov., a marine bacterium of the family Flavobacteriaceae isolated from European sea bass. Int J Syst Evol Microbiol. 62(Pt 2):425–429. [DOI] [PubMed] [Google Scholar]
- Piñeiro-Vidal M, Riaza A, Santos Y.. 2008. Tenacibaculum discolor sp. nov. and Tenacibaculum gallaicum sp. nov., isolated from sole (Solea senegalensis) and turbot (Psetta maxima) culture systems. Int J Syst Evol Microbiol. 58(Pt 1):21–25. [DOI] [PubMed] [Google Scholar]
- Rahmati-Holasoo H, et al. 2016. First detection of koi herpesvirus from koi, Cyprinus carpio L. experiencing mass mortalities in Iran: clinical, histopathological and molecular study. J Fish Dis. 39(10):1153–1163.http://dx.doi.org/10.1111/jfd.12448 [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R.. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 106(45):19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinidis KT.. 2014. Bypassing cultivation to identify bacterial species. Microbe 9(3):111–118. [Google Scholar]
- Småge SB, et al. 2016. Tenacibaculum finnmarkense sp. nov., a fish pathogenic bacterium of the family Flavobacteriaceae isolated from Atlantic salmon. Antonie Van Leeuwenhoek. 109(2):273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. 2015. Tenacibaculum. In: Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, Hedlund B, Dedysh S, editors. Bergey’s manual of systematics of archaea and bacteria. Chichester (United Kingdom: ): John Wiley & Sons, Ltd; p. 1–7. Available from: http://doi.wiley.com/10.1002/9781118960608.gbm00345 (accessed 28.08.17). [Google Scholar]
- Suzuki M, Nakagawa Y, Harayama S, Yamamoto S.. 2001. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int J Syst Evol Microbiol. 51(5):1639–1652.http://dx.doi.org/10.1099/00207713-51-5-1639 [DOI] [PubMed] [Google Scholar]
- Veith PD, et al. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res. 12(10):4449–4461.http://dx.doi.org/10.1021/pr400487b [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3(2):199–208. [DOI] [PubMed]
- Wakabayashi H, Hikida M, Masumura K.. 1986. Flexibacter maritimus sp. nov., a pathogen of marine fishes. Int J Syst Evol Microbiol. 36(3):396–398. [Google Scholar]
- Weinert LA, Welch JJ.. 2017. Why might bacterial pathogens have small genomes? Trends Ecol Evol. 32(12):936–947. [DOI] [PubMed] [Google Scholar]
- Xu Z-X, Yu P, Mu D-S, Liu Y, Du Z-J.. 2017. Tenacibaculum agarivorans sp. nov., an agar-degrading bacterium isolated from marine alga Porphyra yezoensis Ueda. Int J Syst Evol Microbiol. 67(12):5139–5143. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.http://dx.doi.org/10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.