Abstract
Our laboratory has studied Ras and Ras-like proteins since the discovery of the Ras oncogene 35 years ago. In this review, I will give an account of what we have done in these 35 years and indicate the main papers that have guided our research. Our efforts started with the early analysis of mutant Ras in human tumors followed by deciphering of the role of Ras in signal transduction pathways. In an attempt to interfere in Ras signaling we turned to Rap proteins. These proteins are the closest relatives of Ras and were initially identified as Ras antagonists. However, our studies revealed that the Rap signaling network primarily is involved in spatiotemporal control of cell adhesion, in part through regulation of the actin cytoskeleton. More recently we returned to Ras, trying to interfere in Ras signaling by combinatorial drug testing using the organoid technology.
THE BEGINNING
In 1982, the stunning result that a single-point mutation converted a normal H-Ras gene into an oncogene was published by the Weinberg laboratory (Parada et al. 1982). The experimental setup was based on the assumption that cancer is caused by mutations in genes and that these “oncogenes,” when transfected into normal cells, should be able to convert these cells in tumor cells. For this assay, Weinberg and colleagues transferred genomic DNA from a human kidney tumor cell line into NIH-3T3 mouse fibroblasts. This resulted in morphological transformation of NIH-3T3 cells, suggesting the presence of an oncogene in the genomic DNA of these tumor cells. However, the identification of such an oncogene was a painstaking process that requested multiple rounds of selection, to reduce the amount of human DNA present in the transformed mouse cell. The next step was the separation of the human DNA from the mouse DNA based on human-specific repetitive sequences, followed by cloning and sequence analysis. Many groups followed a similar path, resulting in the discovery of the H-ras-related genes K-ras and N-ras (Hall et al. 1983; Shimizu et al. 1983). Many tumor cell lines and tumor tissues were subsequently screened for oncogenes. But the assay remained extremely labor-intensive and the success rate was low. At that time, I was working in the laboratory of Alex van der Eb on cell transformation by the adenovirus E1 region. To switch to human oncogenes would have been an obvious choice, but jumping on a very labor-intensive bandwagon was not very attractive. I was, however, struck by a paper of Bruce Wallace showing a method to identify specific mutations in sickle cell anemia using synthetic oligonucleotides (Conner et al. 1983). The basic principle is that the thermal stability of a 20-mer oligonucleotide annealed to genomic DNA is reduced by a couple of degrees when one base pair is mismatched. As mutations were only found in codon 12 and 61 in H-, K-, and N-Ras, we needed a limited number of oligonucleotides representing all possible single base pair changes. At that time oligonucleotide synthesis was possible in only a few laboratories, one of which was the laboratory of Jacques van Boom next door. With some help from my thesis supervisor Piet Borst, I convinced him to make the required 72 different oligonucleotides, covering all possible codon 12 and 61 mutations. Detecting mutant Ras with this approach turned out to work quite well for cell lines and for really pure tumor samples, but it also had its limits of detection (Bos et al. 1984). The discovery of the polymerase chain reaction (PCR) method (Saiki et al. 1985) resolved this limitation and we were able to identify Ras mutations in a large variety of tumors by simple dot-blot assays (Verlaan-de Vries et al. 1986). As a PCR machine, we used a robot arm moving between water baths of different temperatures as our first PCR machine, with homemade Klenow polymerase to be added after each cycle. One of the highlights was the colon study with Bert Vogelstein showing K-ras mutations to occur in 50% of the colon tumors (Bos et al. 1987), an observation independently made by Manual Perucho and coworkers (Forrester et al. 1987). Interestingly, Ras mutations were also present in 50% of benign adenomas. Together with other genetic alterations detected by Vogelstein, this led to the postulation of the sequential activation of oncogenes in colorectal cancer (Vogelstein et al. 1988). Another highlight was the study of lung adenocarcinoma together with Sjoerd Rodenhuis, revealing 30% K-ras mutations (Rodenhuis et al. 1987). However, the clinical relevance/diagnostic value of Ras mutations became only apparent with the development of epidermal growth factor receptor (EGFR) inhibitors like cetuximab and erlotinib. The presence of a Ras mutation is currently an exclusion criterion for these drugs (Eberhard et al. 2005; Karapetis et al. 2008). Ras mutations were most frequently found in pancreatic cancer, as first described by Perucho and coworkers (Almoguera et al. 1988).
In these early studies, we already observed genetic heterogeneity in tumors. For instance, in melanoma we detected two different Ras mutations in one primary tumor that separated in the metastatic lesions (van ’t Veer et al. 1989). In seminomas, a morphologically very homogeneous tumor, Ras mutations were found in some parts of the primary tumor, but not in other parts (Mulder et al. 1989). In acute myeloid leukemias (AMLs), 30% of which carry N-Ras mutations, we observed that in some cases the Ras mutation remained detectable after complete remission. Interestingly, in these cases, Ras mutations were also present in the apparently unaffected lymphoid lineage, suggesting that the Ras mutation is present in an early hematopoietic stem cell not affected by the treatment. In other cases of AML, the Ras mutation disappeared after treatment, and did not recur after relapse. We concluded from these studies, first, that Ras mutations can contribute to the oncogenic process at different stages, second, that mutant Ras apparently has no effect on the lymphoid lineage, and third, that treatment kills all the aggressive blast cells of the leukemia, but not the underlying premalignant cell clone (Yunis et al. 1989). These and other early studies have been summarized in Bos (1989).
Ras REGULATION
After identification of mutated Ras as oncogene, much effort was put in understanding the function of Ras, a small GTPase that normally cycles between an active GTP-bound and an inactive GDP-bound state, but when oncogenically mutated, is stalled in the active GTP-bound state. The obvious rationale being that understanding the normal function of Ras would provide insight into how to treat tumors harboring oncogenic Ras. The kinetics of Ras cycling between a GTP- and a GDP-bound state in vitro indicated the existence of accessory proteins to regulate Ras loading. The first regulatory protein identified was the CDC25 protein, a guanine nucleotide exchange protein for Ras in yeast (Broek et al. 1987; Robinson et al. 1987). Soon after, Frank McCormick discovered a p120 GTPase-activating protein (p120GAP) guided by the observation that the in vivo GTPase activity of Ras was much higher than the in vitro activity (Trahey and McCormick 1987; Wong et al. 1992).
At that time, GDP/GTP cycling of Ras was studied in vitro and a method measuring Ras activation in vivo was initially lacking. Julian Downward developed a method based on labeling of cells with radioactive orthophosphate followed by immunoprecipitation of Ras proteins. GDP and GTP were eluted and separated by thin layer chromatography. Using this procedure, it was first found that T-cell receptor engagement was a strong inducer of Ras activity (Downward et al. 1990). We followed by showing that insulin could activate Ras, connecting Ras to receptor tyrosine kinases (Burgering et al. 1991). Interestingly, many other extracellular stimuli were shown to activate Ras, including stimuli that use G-protein-coupled receptors (van Corven et al. 1993). This showed that Ras activation was not part of a signaling pathway induced by one specific ligand, but contributes to signaling induced by many factors controlling cellular responses. Subsequent genetic studies by others revealed several mammalian counterparts to the yeast CDC25 protein that mediate Ras activation, such as the Sos proteins, all having a characteristic CDC25 homology domain as catalytic domain. The various Ras guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) and their mode of action have been reviewed extensively (Bos et al. 2007).
Ras EFFECTORS
In the early 1990s, we learned from Chris Marshall that extracellular regulated kinase (ERK), also known as microtubule-associated protein kinase (MAPK), was activated in cells transfected with mutant Ras (Leevers and Marshall 1992). At that time, we were developing a system pioneered by Marino Zerial to use vaccinia virus to introduce dominant negative Ras (RasN17) into cells. Indeed, in cells expressing RasN17 insulin-induced ERK activation was completely blocked: This made ERK the first “proven” downstream biochemical effect of Ras in mammalian cells (de Vries-Smits et al. 1992). However, Ras and ERK did not directly interact and subsequently Raf was found to be the effector of Ras that, through MAPK/ERK kinase (MEK), activates ERK (Moodie et al. 1993).
Soon hereafter, multiple Ras effectors were identified, including phosphatidylinositol 3-kinase (PI3K) (Kodaki et al. 1994) and Ral guanine nucleotide dissociation stimulator (RalGDS) (Hofer et al. 1994). The discovery by Boudewijn Burgering and Paul Coffer in our laboratory that the product of PI3K, phosphatidylinositol 3,4,5, triphosphate, activates protein kinase B (Burgering and Coffer 1995) opened a completely new line of research, resulting in the identification of FoxO transcription factors as main targets for protein kinase B–mediated phosphorylation (Kops et al. 1999).
The importance of multiple effectors for Ras function was beautifully shown by Michael White using effector domain mutants that selectively bind to the various effectors: when transfected separately they had hardly any transforming activity, but in combination they did (White et al. 1995).
In the years thereafter, many aspects of the signaling networks in which Ras plays a central role and the role of Ras in cancer were elucidated (Cox et al. 2014; Papke and Der 2017). The most updated version of the Ras signaling network can be found at the National Cancer Institute (www.cancer.gov/research/key-initiatives/ras/ras-central/blog/2017/mccormick-ras-pathway-v3).
HOW TO INTERFERE IN MUTANT Ras SIGNALING
Knowledge on the regulation of Ras rapidly accumulated but did not yet result in treatment options for cancer patients. Initially, it was thought that Ras action could be inhibited by displacing the GTP moiety by a small molecule, but this idea was rapidly abandoned when it became clear that the affinity of GTP (and GDP) for Ras is within the picomolar range. Instead, most attention was focused on interfering in the lipid modification of Ras, particularly the carboxy-terminal farnesylation and later geranyl-geranylation. When it became clear that Ras belongs to a superfamily of small GTPases with over a hundred members almost all similarly modified, the enthusiasm for this approach diminished. These studies have recently been discussed by Cox and coworkers (Cox et al. 2014). Nowadays most attention is focused on interfering in signaling downstream of Ras (see below).
Rap PROTEINS
Rap proteins were first identified by Veronique Pizon and Pierre Chardin and, based on their similarity to Ras, the authors suggested that they may interact with the same effectors as Ras (Pizon et al. 1988). Subsequently, Noda and coworkers identified K-ras revertant 1 (K-rev1), which was identical to Rap1, in a screen for genes that can revert morphological transformation of mutant K-ras cells (Kitayama et al. 1989). This led to the hypothesis that Rap1 might be a decoy that traps Ras effectors in an inactive complex and could be a possible way to interfere in mutant Ras tumors? We became interested in Rap after our finding that in certain cell types cyclic adenosine monophosphate (cAMP) can inhibit growth-factor-induced (and Ras-mediated) activation of ERK (Burgering et al. 1991). Combined with the observation of Daniel Altschuler that cAMP could activate Rap1 (Altschuler et al. 1995), this suggested a role for Rap1 in inhibiting Ras following cAMP stimulation of cells. However, despite the fact that in vitro Rap1 can bind to the Ras-binding domain of Raf1, we were unable to show a direct involvement of Ras effector signaling (Zwartkruis et al. 1998).
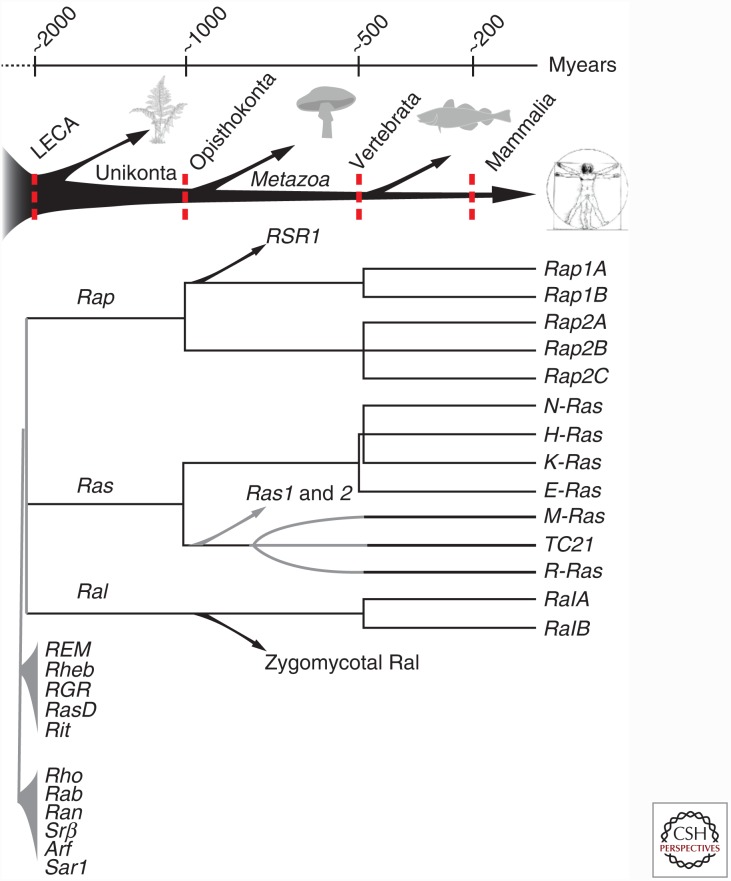
A function of Rap proteins distinct from Ras is further supported by the notion that Ras and Rap were already present as separate proteins in the hypothetical last eukaryotic common ancestor (van Dam et al. 2011). Later in evolution at the opisthokonta stage the Rap branch separated in Rap1 and Rap2 and at the vertebrata stage in Rap1A and B and Rap2A, B, and C (Fig. 1). Research on Rap orthologs in many species has provided pivotal knowledge about human Rap proteins (Frische and Zwartkruis 2010), but especially the analysis of the single Rap protein in budding yeast, Rsr1, has led to a framework for the regulation and function of these small GTPases. Budding always occurs adjacent to a scar of a previous bud. This bud site selection is however abolished in cells mutated for Rsr1 (Bud1), for a GEF (Bud5) or for a GAP (Bud2) for Rsr1. It was found that these proteins form a complex that recognizes special landmark proteins in the rim of the bud scar. The signal is further propagated to the CDC42 complex, which facilitates the local recruitment of the actin cytoskeleton (Bi and Park 2012). In addition, in Drosophila, Rap proteins play a role in cell migration and the proper localization of adherence junctions (Asha et al. 1999; Knox and Brown 2002). This led to the hypothesis that also mammalian Rap proteins are involved in spatial control of cell adhesion, most likely through actin-driven processes.
Figure 1.
Evolutionary timeline of the Ras family, Ras, Rap, and Ral proteins. The last eukaryotic common ancestor (LECA) likely contained already representatives of these proteins. Later duplications are responsible for the different members in mammalian cells. (From van Dam et al. 2011; adapted, with permission, from the authors.)
REGULATION OF Rap
Our initial studies were devoted to find stimuli that can activate Rap1 and we used platelets as they abundantly express Rap1. To measure Rap activity, we developed a nonradioactive pull-down assay, in which active GTP-bound Rap1 is precipitated with a GST-coupled Ras-binding domain (Franke et al. 1997). Such pull-down assays have subsequently been developed for Ras and many other GTPases and are now the most widely used assay to measure activity of a variety of small GTPases, including those from the Rho family. We found that all stimuli that resulted in a rapid activation of platelets also activated Rap1. As one of the hallmarks of platelet activation is increased integrin-mediated adhesion and spreading, we postulated that Rap1 may control integrin activation (Franke et al. 1997). Subsequent studies revealed that a large variety of stimuli are able to activate Rap proteins in all cell types tested.
The observation that, similar to Ras, Rap1 is activated by many stimuli opened the quest for guanine nucleotide exchange factors (GEFs) and GAPs that could regulate this activity. The first Rap-specific GEF, C3G, was identified by Michiyuki Matsuda and coworkers (Gotoh et al. 1995). Among others, it mediates signals from receptor tyrosine kinases through the adaptor protein Crk. Currently at least nine Rap-selective GEFs have been identified (Fig. 2), which have, similar to Ras GEFs, a CDC25 homology domain. However, both in vivo and in vitro there is a strong specificity for Rap, with CalDAGGEF3 (RasGrp3) as an exception as it acts both on Ras and Rap. In addition, almost all these proteins have protein–protein or protein–lipid interaction domains, suggesting multiple interaction partners. RapGAPs have a different structure and a different mechanism of catalysis compared to RasGAPs; whereas RasGAPs provide a critical arginine (arginine finger) for hydrolysis of GTP, RapGAPs provide an aspartate (aspartate thumb). Again, despite similarities between Ras and Rap, the GAPs are specific for their cognate GTPase (Bos et al. 2007). Exceptions are the GAP1(IP4BP) family members a Rap-like GAP, which upon membrane binding adopts activity toward Ras (Sot et al. 2013).
Figure 2.
Rap guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs). Indicated are the domain structures of Rap-specific GEFs and GAPs. (From Bos et al. 2007; adapted, with permission, from the authors.)
EXCHANGE PROTEIN DIRECTLY ACTIVATED BY cAMP (Epac)
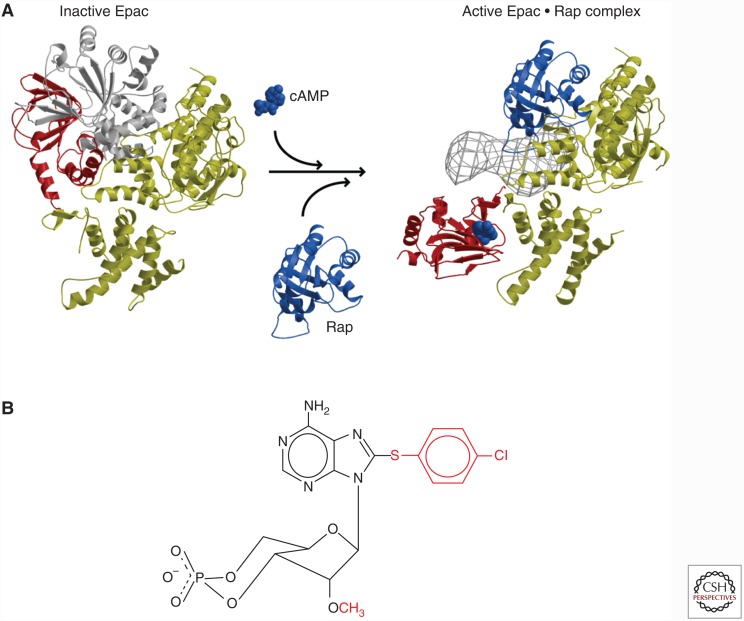
At one time, it was a dogma that all mammalian cAMP effects were mediated by protein kinase A. However, we noted that inhibition of protein kinase A did not affect cAMP-induced Rap1 activity. In a subsequent search in publicly available databases of the human genome project, we found cAMP-binding domains in close vicinity of CDC25 homology domains. Using purified protein encoded by the corresponding complementary DNA (cDNA), we could show a Rap-specific GEF with activity that was completely dependent on the presence of cAMP. We called this protein exchange protein directly activated by cAMP (Epac) (de Rooij et al. 1998). Two human Epac proteins were identified: one with a single cAMP-binding domain (Epac1) and one with two cAMP-binding domains (Epac2). Subsequent structural analysis, in collaboration with Fred Wittinghofer, revealed a detailed mechanism on how Epac is regulated (Fig. 3A). In the absence of cAMP, Epac is in a closed conformation with the cAMP-binding domain occluding the Rap-binding site. When cAMP binds, Epac adopts an open conformation, where Rap can bind and to become activated (Rehmann et al. 2006, 2008). In addition, our studies for the first time revealed how ingenuously cAMP acts: the cyclic phosphate group of cAMP releases a brake that allows a major conformational change that is further stabilized by interactions with the adenosine group (Rehmann et al. 2003). Simultaneously, cAMP also induces the translocation of Epac1 (but not Epac2) from the cytosol to the plasma membrane (Ponsioen et al. 2009). This is facilitated by a cAMP-induced conformational change in the Dishevelled, EGL-10, and pleckstrin (DEP) domain (Li et al. 2011), resulting in an increased affinity for membrane-bound phosphatidic acid (Consonni et al. 2012). Several additional membrane anchors have been identified for Epac1, including activated Ezrin, showing that, dependent on the conditions, Epac1 can localize differently (Gloerich et al. 2010).
Figure 3.
(A) Crystal structure of Epac 2 in the closed (left) and open (right) conformation. The gray area (right) represents the first cyclic adenosine monophosphate (cAMP) domain and Dishevelled, EGL-10, and pleckstrin (DEP) domain visualized by cryoelectron microscopy (Rehmann et al. 2006, 2008). (B) Structure of 8CPT-2′OMe-cAMP (007).
DEVELOPMENT OF 007
Further studies on the function of Epac1 were hampered by the fact that cAMP activates both protein kinase A (PKA) and Epac. However, Stein Døskeland pointed out to us a peculiarity in the cAMP-binding domain, where a highly conserved glutamate that forms hydrogen bounds with the 2′-hydroxyl of the ribose of cAMP was absent in the cAMP-binding domain of Epac1 and the high-affinity, second binding site of Epac2. This suggested that these hydrogen bonds, and thus the 2′OH of the ribose may be required specifically for cAMP to interact with PKA, but not with Epac. Together with Frank Schwede and Hans-Gottfried Genieser, we started to modify cAMP, resulting in a highly Epac-selective compound 8-CPT-2′OMe-cAMP (Fig. 3B), which we called 007 (Enserink et al. 2002). Using this compound, we demonstrated that cAMP through Epac is able to modify integrin-mediated cell-substratum adhesion and E-cadherin-mediated cell–cell adhesion (Rangarajan et al. 2003; Price et al. 2004). Subsequently, many studies have been published using this compound thereby showing the versatile role Epac proteins play in cAMP signaling, including insulin secretion, endothelial barrier function, memory, and heart rhythm (for further details, see Gloerich and Bos 2010).
DOWNSTREAM TARGETS OF Rap1 PROTEINS
007 did not induce activation of ERK, indicating that B-raf is not a downstream target of Rap1 (Enserink et al. 2002). In this respect, a claim that B-raf is a downstream target of Rap1 is incorrect. The first genuine effector for a Rap protein identified was the Drosophila Canoe protein (Boettner et al. 2003). This protein is similar to the mammalian junctional protein AF6 with ubiquitin-like Ras association (RA) domains to bind active Rap1. Subsequently, AF6 was found to be one of the effectors of Rap1 that mediate some, but not all, responses of Rap1 activation. Indeed, of the many RA-domain-containing proteins multiple have been implicated to transduce Rap1 signaling. These effectors, together with the existence of multiple Rap1-GEFs and GAPs, prompted us to extend the hypothesis based on the yeast studies. We proposed that a cell can have multiple independent Rap modules consisting of a Rap protein, a GEF, and probably a GAP, each recognizing different landmarks (spatial cues) to regulate localized recruitment of the actin cytoskeleton in time and in space using a variety of effector systems. The presence of additional regulatory and interaction domains, particularly in the various GEFs, provide ample opportunities for specific landmark recognition and regulation. The consequence of this hypothesis was that we had to search for a specific Rap signaling module in a defined model system. Although we explored Rap1-induced integrin-mediated cell adhesion and Rap1-induced cell spreading in epithelial cells, we particularly focused our attention on endothelial barrier function. Endothelial cells are highly responsive to 007 as measured by, for instance, an increased electrical resistance of the monolayer. This increased barrier function is accompanied by a shift that is observed from radial stress fibers to circumferential actin, which could explain the increased tightness of endothelial cell–cell junctions. Using small-interfering RNA (siRNA) screens, we found that Rap1 interacted with a complex of two related RA-domain-containing proteins, Rasip1 and Radil. Interestingly, both Radil and Rasip1 are homologous to Canoe, the first effector found for Rap1 in Drosophila. Importantly, Radil directly interacts with ArhGAP29, a Rho-specific GAP, and we could show that ArhGAP29 mediates 007-induced decrease in radial stress fibers and increased barrier function (Fig. 4, left) (Post et al. 2013, 2015). Intriguingly, multiple Rap1 modules may control barrier function simultaneously. For instance, Mochizuki and coworkers discovered that circumferential actin formation is mediated by the CDC42-GEF FGD5 (Ando et al. 2013). How FGD5 is activated is still unclear, but, in overexpression studies, we observe that FGD5 also forms a complex with Radil, suggesting that the Radil/Rasip complex regulates both pathways. Also, the Rap1 effector AF6 is involved in this process (Birukova et al. 2013), but how these different effectors cooperate is still unclear. Surprisingly, Krit1, previously identified as a Rap1 effector in a similar assay in Huvec cells (Glading et al. 2007), did not affect barrier function in our experiments (Pannekoek et al. 2011).
Figure 4.
Graphical summary of (A) Epac1-mediated regulation of radial stress fibers through the Radil-Rasip1-ArgGAP29 complex, and of (B) Rap2A-mediated intestinal brush borders formation (for explanation see text).
Several other effectors for Rap1 have been identified, such as the RA-domain-containing protein Riam, which plays an important role in integrin-mediated cell adhesion (Lafuente et al. 2004).
Rap2
The early deviation of Rap1 and Rap2 in evolution suggests that they may have different functions and indeed, although Rap2 is regulated by the same GEFs and GAPs as Rap1, it is clearly different as Rap2(-GTP) binds to a different set of effector proteins lacking an RA domain: the Traf2 and Nck-interacting kinases (TNIKs) (Taira et al. 2004). We stumbled on Rap2A in the analysis of brush-border formation in W4 intestinal epithelial cells. In these cells, brush-border formation is induced by doxycycline-induced activation of the LKB–Strd1α complex (Baas et al. 2004). When we tested the involvement of Rap proteins in this process, we found that siRNA mediated depletion of Rap2A, but the other Rap proteins abolished this process. Subsequent studies revealed a pathway, which is induced by PIP2 enriched apical membrane formation. PIP2 serves as anchor for phospholipase D, which produces phosphatidic acid. Phosphatidic acid is then the anchor for the RapGEF PDZ-GEF. As a consequence, PDZ-GEF is recruited to the apical membrane, activates Rap2A, which in turn recruits and activates the serine-threonine kinase TNIK. TNIK activates the serine-threonine kinase Mst4, which induces the phosphorylation and activation of Ezrin. This activation of Ezrin is a critical step as active Ezrin can rescue Rap2 depletion. Ezrin recruits the actin cytoskeleton for brush-border formation (Fig. 4, right) (Gloerich et al. 2012). The similarity with the yeast model is evident: A landmark is recognized in two steps by a GEF for Rap followed by effector binding, which in several steps leads to the recruitment of actin.
ARE WE FINISHED WITH Ras?
Although our analysis and those of many other research groups have provided much insight into the role Rap proteins play in cellular responses, the initial idea that Rap1 counteracts mutant Ras signaling and could be used as a route to interfere in mutant Ras signaling in cancers turned out to be too naïve. However, despite huge efforts by many groups and many pharmaceutical industries, the alternative approach to interfere in the Ras pathway by selective inhibitors has not worked either as we are still unable to treat Ras-mutant cancers with targeted drugs (Stephen et al. 2014). Ras itself is still largely “undruggable,” and the alternative to inhibit downstream targets of mutant Ras, most notably Raf, MEK, ERK, and PI3K, have thus far failed in the clinic, especially when single drugs are used. For such an approach to be successful, it is essential that mutant Ras-containing tumors cells do have a specific sensitivity for these pathway drugs. Indeed, genetic and drug screens in mutant cell lines did identify several possible targets for mutant Ras tumor cell lines, but these have not been translated into clinical targets (Downward 2015). Perhaps the commonly used mutant Ras-containing cell lines identify sensitivities that do not occur in tumors. We therefore shifted to the tumor organoid model system developed by Hans Clevers (Sato et al. 2009; van de Wetering et al. 2015). The advantage of this model system is that tumor organoids can be rapidly established from almost all individual (epithelial) tumors and thus can capture the genetic diversity of these tumors. Moreover, drug responses can be compared with organoids from normal tissue to determine the therapeutic window. Initial studies using a number of colon tumor organoids already indicated the power of this technology (van de Wetering et al. 2015). As a first proof-of-concept, we tested various targeted drugs that are currently being used in clinical trials for inhibiting mutant Ras-containing colorectal cancer, most notably pan-EGFR inhibitors in combination with MEK inhibitors (Sun et al. 2014). We found that these combinations gave a clear cell-cycle arrest of mutant K-ras-containing organoids, supporting the observation that this combination has an effect on mutant Ras tumor cells. However, organoids with nonmutated K-Ras remain far more sensitive for this drug combination. Furthermore, we noted that organoids from normal tissue were similarly sensitive for this combination than tumor organoids with nonmutated K-ras, and thus much more sensitive than mutant K-ras organoids, indicating that for this combination the therapeutic window is limited (Verissimo et al. 2016).
It is now more than 35 years ago that Ras was identified as an oncogene in human cancers and we still have not solved the problem of how to target mutant Ras in cancer. As a consequence, patients carrying mutant Ras in their tumors are excluded from therapies targeting the EGFR signaling cascade. Moreover, when patients carrying normal Ras in their tumors are treated, the appearance of mutant Ras is a frequent resistance mechanism. To make personalized medicine a success, we have to come up with a solution. New model systems like tumor organoids may be helpful in the identification of new drug combinations, but also in finding those patients that may uniquely respond to such combinations.
ACKNOWLEDGMENTS
I thank all members of my laboratory in past years for the tremendous contributions they have made and are still making to understand Ras-like small GTPases and hopefully to find a drug combination that works for mutant Ras-containing tumors. I also thank my colleagues Fried Zwartkruis, Willem Jan Pannekoek, and Boudewijn Burgering for critically reading this manuscript.
Footnotes
Editors: Linda VanAelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. 1988. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53: 549–554. [DOI] [PubMed] [Google Scholar]
- Altschuler DL, Peterson SN, Ostrowski MC, Lapetina EG. 1995. Cyclic AMP-dependent activation of Rap1b. J Biol Chem 270: 10373–10376. [DOI] [PubMed] [Google Scholar]
- Ando K, Fukuhara S, Moriya T, Obara Y, Nakahata N, Mochizuki N. 2013. Rap1 potentiates endothelial cell junctions by spatially controlling myosin II activity and actin organization. J Cell Biol 202: 901–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha H, de Ruiter ND, Wang MG, Hariharan IK. 1999. The Rap1 GTPase functions as a regulator of morphogenesis in vivo. EMBO J 18: 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, Clevers HC. 2004. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116: 457–466. [DOI] [PubMed] [Google Scholar]
- Bi E, Park HO. 2012. Cell polarization and cytokinesis in budding yeast. Genetics 191: 347–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birukova AA, Tian X, Tian Y, Higginbotham K, Birukov KG. 2013. Rap-afadin axis in control of ρ signaling and endothelial barrier recovery. Mol Biol Cell 24: 2678–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Harjes P, Ishimaru S, Heke M, Fan HQ, Qin Y, Van Aelst L, Gaul U. 2003. The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics 165: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. 1989. ras oncogenes in human cancer: A review. Cancer Res 49: 4682–4689. [PubMed] [Google Scholar]
- Bos JL, Verlaan-de Vries M, Jansen AM, Veeneman GH, van Boom JH, van der Eb AJ. 1984. Three different mutations in codon 61 of the human N-ras gene detected by synthetic oligonucleotide hybridization. Nucleic Acids Res 12: 9155–9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B. 1987. Prevalence of ras gene mutations in human colorectal cancers. Nature 327: 293–297. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129: 865–877. [DOI] [PubMed] [Google Scholar]
- Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powers S, Wigler M. 1987. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell 48: 789–799. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Coffer PJ. 1995. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376: 599–602. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Medema RH, Maassen JA, van de Wetering ML, van der Eb AJ, McCormick F, Bos JL. 1991. Insulin stimulation of gene expression mediated by p21ras activation. EMBO J 10: 1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner BJ, Reyes AA, Morin C, Itakura K, Teplitz RL, Wallace RB. 1983. Detection of sickle cell β S-globin allele by hybridization with synthetic oligonucleotides. Proc Natl Acad Sci 80: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni SV, Gloerich M, Spanjaard E, Bos JL. 2012. cAMP regulates DEP domain-mediated binding of the guanine nucleotide exchange factor Epac1 to phosphatidic acid at the plasma membrane. Proc Natl Acad Sci 109: 3814–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. 2014. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 13: 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J, Zwartkruis FJ, Verheijen MH, Cool RH, Nijman SM, Wittinghofer A, Bos JL. 1998. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396: 474–477. [DOI] [PubMed] [Google Scholar]
- de Vries-Smits AM, Burgering BM, Leevers SJ, Marshall CJ, Bos JL. 1992. Involvement of p21ras in activation of extracellular signal-regulated kinase 2. Nature 357: 602–604. [DOI] [PubMed] [Google Scholar]
- Downward J. 2015. RAS synthetic lethal screens revisited: Still seeking the elusive prize? Clin Cancer Res 21: 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J, Graves JD, Warne PH, Rayter S, Cantrell DA. 1990. Stimulation of p21ras upon T-cell activation. Nature 346: 719–723. [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH, et al. 2005. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 23: 5900–5909. [DOI] [PubMed] [Google Scholar]
- Enserink JM, Christensen AE, de Rooij J, van Triest M, Schwede F, Genieser HG, Doskeland SO, Blank JL, Bos JL. 2002. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat Cell Biol 4: 901–906. [DOI] [PubMed] [Google Scholar]
- Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. 1987. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature 327: 298–303. [DOI] [PubMed] [Google Scholar]
- Franke B, Akkerman JW, Bos JL. 1997. Rapid Ca2+-mediated activation of Rap1 in human platelets. EMBO J 16: 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frische EW, Zwartkruis FJ. 2010. Rap1, a mercenary among the Ras-like GTPases. Dev Biol 340: 1–9. [DOI] [PubMed] [Google Scholar]
- Glading A, Han J, Stockton RA, Ginsberg MH. 2007. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell–cell junctions. J Cell Biol 179: 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloerich M, Bos JL. 2010. Epac: Defining a new mechanism for cAMP action. Annu Rev Pharmacol Toxicol 50: 355–375. [DOI] [PubMed] [Google Scholar]
- Gloerich M, Ponsioen B, Vliem MJ, Zhang Z, Zhao J, Kooistra MR, Price LS, Ritsma L, Zwartkruis FJ, Rehmann H, et al. 2010. Spatial regulation of cyclic AMP-Epac1 signaling in cell adhesion by ERM proteins. Mol Cell Biol 30: 5421–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloerich M, ten Klooster JP, Vliem MJ, Koorman T, Zwartkruis FJ, Clevers H, Bos JL. 2012. Rap2A links intestinal cell polarity to brush border formation. Nat Cell Biol 14: 793–801. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Hattori S, Nakamura S, Kitayama H, Noda M, Takai Y, Kaibuchi K, Matsui H, Hatase O, Takahashi H, et al. 1995. Identification of Rap1 as a target for the Crk SH3 domain-binding guanine nucleotide-releasing factor C3G. Mol Cell Biol 15: 6746–6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Marshall CJ, Spurr NK, Weiss RA. 1983. Identification of transforming gene in two human sarcoma cell lines as a new member of the ras gene family located on chromosome 1. Nature 303: 396–400. [DOI] [PubMed] [Google Scholar]
- Hofer F, Fields S, Schneider C, Martin GS. 1994. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci 91: 11089–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. 2008. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765. [DOI] [PubMed] [Google Scholar]
- Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. 1989. A ras-related gene with transformation suppressor activity. Cell 56: 77–84. [DOI] [PubMed] [Google Scholar]
- Knox AL, Brown NH. 2002. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295: 1285–1288. [DOI] [PubMed] [Google Scholar]
- Kodaki T, Woscholski R, Hallberg B, Rodriguez-Viciana P, Downward J, Parker PJ. 1994. The activation of phosphatidylinositol 3-kinase by Ras. Curr Biol 4: 798–806. [DOI] [PubMed] [Google Scholar]
- Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398: 630–634. [DOI] [PubMed] [Google Scholar]
- Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. 2004. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell 7: 585–595. [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Marshall CJ. 1992. Activation of extracellular signal-regulated kinase, ERK2, by p21ras oncoprotein. EMBO J 11: 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tsalkova T, White MA, Mei FC, Liu T, Wang D, Woods VL Jr, Cheng X. 2011. Mechanism of intracellular cAMP sensor Epac2 activation: cAMP-induced conformational changes identified by amide hydrogen/deuterium exchange mass spectrometry (DXMS). J Biol Chem 286: 17889–17897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie SA, Willumsen BM, Weber MJ, Wolfman A. 1993. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science 260: 1658–1661. [DOI] [PubMed] [Google Scholar]
- Mulder MP, Keijzer W, Verkerk A, Boot AJ, Prins ME, Splinter TA, Bos JL. 1989. Activated ras genes in human seminoma: Evidence for tumor heterogeneity. Oncogene 4: 1345–1351. [PubMed] [Google Scholar]
- Pannekoek WJ, van Dijk JJ, Chan OY, Huveneers S, Linnemann JR, Spanjaard E, Brouwer PM, van der Meer AJ, Zwartkruis FJ, Rehmann H, et al. 2011. Epac1 and PDZ-GEF cooperate in Rap1 mediated endothelial junction control. Cell Signal 23: 2056–2064. [DOI] [PubMed] [Google Scholar]
- Papke B, Der CJ. 2017. Drugging RAS: Know the enemy. Science 355: 1158–1163. [DOI] [PubMed] [Google Scholar]
- Parada LF, Tabin CJ, Shih C, Weinberg RA. 1982. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature 297: 474–478. [DOI] [PubMed] [Google Scholar]
- Pizon V, Lerosey I, Chardin P, Tavitian A. 1988. Nucleotide sequence of a human cDNA encoding a ras-related protein (rap1B). Nucleic Acids Res 16: 7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsioen B, Gloerich M, Ritsma L, Rehmann H, Bos JL, Jalink K. 2009. Direct spatial control of Epac1 by cyclic AMP. Mol Cell Biol 29: 2521–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post A, Pannekoek WJ, Ross SH, Verlaan I, Brouwer PM, Bos JL. 2013. Rasip1 mediates Rap1 regulation of Rhoρ in endothelial barrier function through ArhGAP29. Proc Natl Acad Sci 110: 11427–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post A, Pannekoek WJ, Ponsioen B, Vliem MJ, Bos JL. 2015. Rap1 spatially controls ArhGAP29 to inhibit Rho signaling during endothelial barrier regulation. Mol Cell Biol 35: 2495–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LS, Hajdo-Milasinovic A, Zhao J, Zwartkruis FJ, Collard JG, Bos JL. 2004. Rap1 regulates E-cadherin-mediated cell–cell adhesion. J Biol Chem 279: 35127–35132. [DOI] [PubMed] [Google Scholar]
- Rangarajan S, Enserink JM, Kuiperij HB, de Rooij J, Price LS, Schwede F, Bos JL. 2003. Cyclic AMP induces integrin-mediated cell adhesion through Epac and Rap1 upon stimulation of the β2-adrenergic receptor. J Cell Biol 160: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmann H, Prakash B, Wolf E, Rueppel A, de Rooij J, Bos JL, Wittinghofer A. 2003. Structure and regulation of the cAMP-binding domains of Epac2. Nat Struct Biol 10: 26–32. [DOI] [PubMed] [Google Scholar]
- Rehmann H, Das J, Knipscheer P, Wittinghofer A, Bos JL. 2006. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature 439: 625–628. [DOI] [PubMed] [Google Scholar]
- Rehmann H, Arias-Palomo E, Hadders MA, Schwede F, Llorca O, Bos JL. 2008. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature 455: 124–127. [DOI] [PubMed] [Google Scholar]
- Robinson LC, Gibbs JB, Marshall MS, Sigal IS, Tatchell K. 1987. CDC25: A component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science 235: 1218–1221. [DOI] [PubMed] [Google Scholar]
- Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. 1987. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med 317: 929–935. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. 1985. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230: 1350–1354. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Birnbaum D, Ruley MA, Fasano O, Suard Y, Edlund L, Taparowsky E, Goldfarb M, Wigler M. 1983. Structure of the Ki-ras gene of the human lung carcinoma cell line Calu-1. Nature 304: 497–500. [DOI] [PubMed] [Google Scholar]
- Sot B, Behrmann E, Raunser S, Wittinghofer A. 2013. Ras GTPase activating (RasGAP) activity of the dual specificity GAP protein Rasal requires colocalization and C2 domain binding to lipid membranes. Proc Natl Acad Sci 110: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, McCormick F. 2014. Dragging ras back in the ring. Cancer Cell 25: 272–281. [DOI] [PubMed] [Google Scholar]
- Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, et al. 2014. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep 7: 86–93. [DOI] [PubMed] [Google Scholar]
- Taira K, Umikawa M, Takei K, Myagmar BE, Shinzato M, Machida N, Uezato H, Nonaka S, Kariya K. 2004. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem 279: 49488–49496. [DOI] [PubMed] [Google Scholar]
- Trahey M, McCormick F. 1987. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238: 542–545. [DOI] [PubMed] [Google Scholar]
- van Corven EJ, Hordijk PL, Medema RH, Bos JL, Moolenaar WH. 1993. Pertussis toxin-sensitive activation of p21ras by G protein-coupled receptor agonists in fibroblasts. Proc Natl Acad Sci 90: 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJ, Bos JL, Snel B. 2011. Evolution of the Ras-like small GTPases and their regulators. Small GTPases 2: 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161: 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Veer LJ, Burgering BM, Versteeg R, Boot AJ, Ruiter DJ, Osanto S, Schrier PI, Bos JL. 1989. N-ras mutations in human cutaneous melanoma from sun-exposed body sites. Mol Cell Biol 9: 3114–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verissimo CS, Overmeer RM, Ponsioen B, Drost J, Mertens S, Verlaan-Klink I, Gerwen BV, van der Ven M, Wetering MV, Egan DA, et al. 2016. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife 5: e18489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan-de Vries M, Bogaard ME, van den Elst H, van Boom JH, van der Eb AJ, Bos JL. 1986. A dot-blot screening procedure for mutated ras oncogenes using synthetic oligodeoxynucleotides. Gene 50: 313–320. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. 1988. Genetic alterations during colorectal-tumor development. N Engl J Med 319: 525–532. [DOI] [PubMed] [Google Scholar]
- White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler MH. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80: 533–541. [DOI] [PubMed] [Google Scholar]
- Wong G, Muller O, Clark R, Conroy L, Moran MF, Polakis P, McCormick F. 1992. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell 69: 551–558. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Boot AJ, Mayer MG, Bos JL. 1989. Mechanisms of ras mutation in myelodysplastic syndrome. Oncogene 4: 609–614. [PubMed] [Google Scholar]
- Zwartkruis FJ, Wolthuis RM, Nabben NM, Franke B, Bos JL. 1998. Extracellular signal-regulated activation of Rap1 fails to interfere in Ras effector signalling. EMBO J 17: 5905–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]