Abstract
Rare diseases are often misdiagnosed or receive a delayed diagnosis; thus, unfortunately, affected individuals may not receive optimal medical management. Here, we report a case of two siblings with a severe phenotype of progressive pseudorheumatoid dysplasia (PPD). Their onset of symptoms began at the age of 3 yr. Both were neglected in the past, and the patients presented with a very severe phenotype and unmitigated natural history. PPD is a rare autosomal recessive skeletal dysplasia characterized by progressive joint stiffness, swelling, and pain. Because of observed muscle wasting, weakness, and the lack of laboratory testing, the case had been initially misdiagnosed by the local physicians. We aimed to provide diagnostic support by a targeted next-generation sequencing gene panel (Illumina TruSight One) for Mendelian diseases (Mendeliome), and we identified a homozygous frameshift mutation in the gene WISP3 (c.868_869delAG, p.Ser290Leufs*12). Thus, early diagnosis and intervention may have decreased the severity and complication of the disease.
Keywords: joint swelling, progressive joint destruction
INTRODUCTION
Rare genetic disorders, particularly in developing countries, are frequently misdiagnosed because of the lack of adequate medical facilities and genetic diagnosis. Consequently, many of these cases present with fatal and progressed phenotypes, showing an unbiased natural history without intervention unlike those cases diagnosed in more developed countries (Nampoothiri et al. 2014; Rai et al. 2016).
Progressive pseudorheumatoid dysplasia (PPD; MIM 208230) primarily affects the articular cartilage and follows an autosomal recessive mode of inheritance. WNT1 inducible signaling pathway protein 3 (WISP3), the gene mutated in PPD, is located on Chromosome 6q22. It has five coding exons that encode a 354-amino acid protein. The protein is essential for skeletal growth and cartilage homeostasis, as it contributes to the regulation of type II collagen and aggrecan in chondrocytes (Sen et al. 2004).
The current family was recruited from a remote area in Yemen during an outreach genetic clinic. Although the index cases had their first symptoms 20 years ago, they did not receive the right diagnosis or treatment. Here, we show that PPD can evolve as a severe destructive and disabling disease unless proper treatment and rehabilitation are received.
RESULTS
Clinical Presentation and Family History
Our male case P1 is a 24-yr-old male born to consanguineous Yemeni parents. He was healthy at birth, and as a toddler, he achieved developmental milestones at normal age. By the age of 3, he started to have difficulties during walking and developed progressive knee deformities. Over several years, he developed progressive symmetric joint pain, stiffness, and swelling. The first joints involved were the knees, followed by hips, elbows, and hand joints. The pain involved almost all joints but was most severe in the hips and lower back.
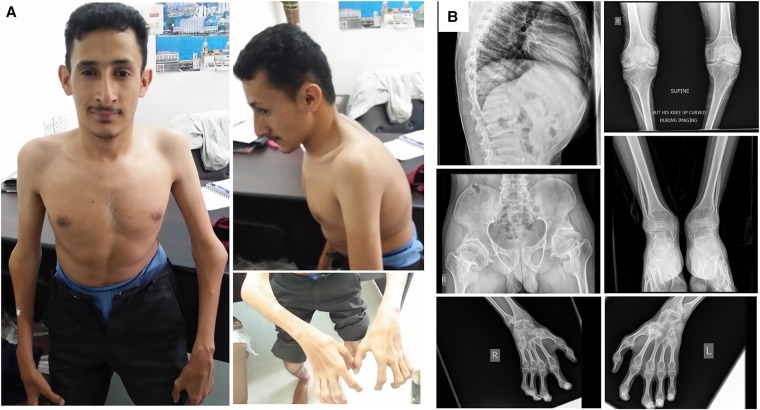
The patient took nonsteroidal anti-inflammatory drugs (NSAIDs) whenever needed for severe pain but has never been on steroid therapy. There were no symptoms of numbness or tingling in the extremities and there were no problems in passing urine. At our exam, he exhibited a flexed posture in the trunk and extremities and abnormal gait (Fig. 1A). We found enlargement of joints, which were more prominent in the interphalangeal, elbow, and knee joints (Fig. 1A,B), but there were no signs of inflammation such as tenderness or redness. The range of movement of all joints was extremely restricted, including neck, spine, shoulder, elbow, wrist, knee, ankle, and interphalangeal joints of hands and feet (see Supplemental Video 1). His mental status, vision, hearing, and speech were normal.
Figure 1.
Clinical photos and radiologic images of the patient. (A) Enlargement of elbows, widening of the thoracic wall, and severe contractures of the fingers are visible. Note that the posture of the patient is altered because of severe joint involvement. (B) X-ray analysis of the whole body. Increased anteroposterior diameter of thoracic wall, platyspondyly, and irregularity of end plates. An anteroposterior radiograph of the hips showed narrowing of joint spaces and mild sclerotic and cystic changes on the acetabulum and the head of the femur. A hand radiograph demonstrated severe enlargement and cystic changes on proximal interphalangeal (PIP), distal interphalangeal (DIP), and metacarpophalangeal (MCP) joints and carpal bones and metaphyseal cupping of the radius and ulna. Foot and ankle radiographs also showed severe enlargement and cystic changes of tarsal and metatarsal joints. A bilateral knee radiograph revealed narrowing of joint spaces and secondary osteoporosis.
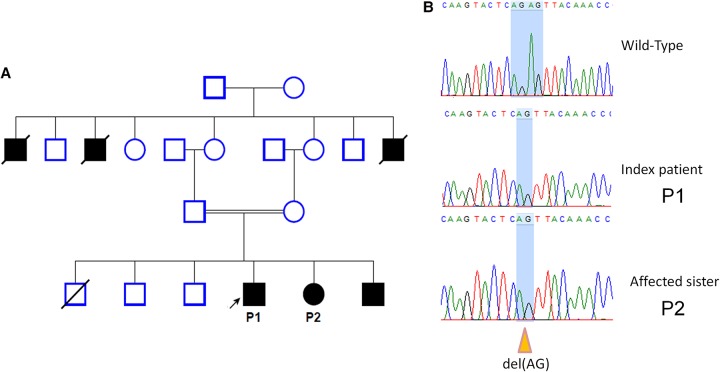
From the family history, two siblings (sister P2 and brother; 22 and 20 yr old, respectively) were affected with a similar manifestation, and three granduncles were reported, who died between the ages of 60 and 70 with a history of the same complaints (Fig. 2A). Although the disease is severe and disabling, it is mainly limited to the joints and does not affect other vital body organs, such as heart, liver, or brain and, consequently, life expectancy seems not to be reduced. In our studied family, the grandparents died between the ages of 60 and 70 yr and this is close to the average life expectancy in Yemen (estimated 65.5 yr according to WHO 2015; http://www.who.int/countries/yem/en/).
Figure 2.
Family pedigree and Sanger sequencing chromatograph. (A) The pedigree of a Yemeni family with PPD showing the consanguinity; the arrow indicates the index case P1. Case P2 underwent exome sequencing. (B) Sanger sequencing chromatograph confirming the mutation detected by Mendeliome (c: 868_869delAG, p.S290Lfs*12). This mutation has been reported in compound heterozygosity with another missense mutation in one Italian family (Garcia Segarra et al. 2012).
Radiologic Analysis
X-ray analysis was performed to thoracolumbar spine, hips, ankles, knees, wrists, and fingers. A lateral radiograph of the thoracolumbar spine showed an increase in anteroposterior diameter of the thoracic wall, platyspondyly, and irregularity of end plates. An anteroposterior radiograph of the hips showed narrowing of joint spaces and mild sclerotic and cystic changes on the acetabulum and the head of the femur. A hand radiograph revealed severe enlargement and cystic changes on proximal interphalangeal (PIP), distal interphalangeal (DIP), metacarpophalangeal (MCP) joints and carpal bones and metaphyseal cupping of the radius and ulna. Foot and ankle radiographs also showed severe enlargement and cystic changes in tarsal and metatarsal joints. A radiograph of the bilateral knees revealed narrowing of joint spaces and secondary osteoporosis (Fig. 1B).
Genomic Analyses
To uncover the underlying genetic defect, we have performed targeted next-generation sequencing using a comprehensive gene panel for Mendelian diseases. This panel is offered by Illumina under the trade name TruSight One and has been referred to as Mendeliome by several groups throughout the world (Saudi Mendeliome Group 2015; Fazeli et al. 2016; Moosa et al. 2016; Haliloglu et al. 2017; Weissbach et al. 2017).
We filtered the variants for homozygous recessive inheritance as predicted by the pedigree (Fig. 2A). The homozygous filtering showed a hit with a homozygous frameshift mutation in WISP3 (WNT1 inducible signaling pathway protein 3; NM_003880.3) in exon 5; c.868_869delAG, p.Ser290Leufs*12 (Table 1). Sanger sequencing was performed to confirm the mutation as well as the segregation of the mutation within the family (Fig. 2B).
Table 1.
WISP3 variant
| Chr:position GRCh37(hg19) | NCBI reference sequence | HGVS cDNA | HGVS protein | Variant type | Genotype |
|---|---|---|---|---|---|
| 6:112390624-112390625 | NM_003880.3 | c.868_869delAG | p.Ser290Leufs*12 | Frameshift | Homozygous |
HGVS, Human Genome Variation Society.
The frameshift mutation is located in the carboxy-terminal cysteine knot (CTCK; amino acids 262–354) domain of the WISP3 protein. This domain is highly conserved in vertebrates and involved in the regulation of bone morphogenetic proteins and/or Wnt signaling, and more than one-third of pathogenic mutations of the WISP3 gene have been reported to occur in this domain (Dalal et al. 2012; Rai et al. 2016).
We also performed copy-number variant (CNV) calling; however, detected CNVs have not been considered as disease causing for this phenotype (Supplemental Table 1).
DISCUSSION
In developing countries, the focus has been on infectious disease and less attention has been given to rare genetic diseases. In our case, the initial diagnosis was made by the local physicians, classifying the phenotype as a neuromuscular disease because of severe contractures, muscle wasting, and the absence of further biochemical and radiological workup. In contrast, early diagnoses of many PPD cases in the literature were mainly rheumatoid arthritis (Spranger et al. 1983), because they were detected at a less severe stage of the disease progression. We believe that a reduction of life expectancy is not to be expected, even though the disabling course of the disease is dramatic. Notably, the other affected family members of our patient died at ages between 60 and 70 yr.
So far, around 70 families worldwide were confirmed to have PPD by mutation analysis of WISP3, which is the only gene known to be associated with PPD (Dalal et al. 2012). Several of these cases were reported in Arab families from Jordan, Saudi Arabia (Hurvitz et al. 1999), Syria, Lebanon, and Palestine (Delague et al. 2005), but this is the first PPD case report from Yemen.
The mutation that we found in WISP3 (c.868_869delAG; p.Ser290Leufs*12) is a frameshift mutation that leads to a premature termination codon in exon 5 located in the CTCK domain. It had already been reported before in a compound heterozygous status together with a missense mutation in an Italian family (Garcia Segarra et al. 2012). Unfortunately, no detailed clinical information was provided on this case in the report; thus, we were not able to compare the clinical phenotypes. The onset of symptoms usually starts between 3 and 6 yr of age and the most common presenting symptoms are progressive multiple joint stiffness, pain, contractures, and eventually gait disturbance. The frequency of PPD is estimated to be about 1 per million in the United Kingdom, but it is likely to be higher in some Arab countries and Turkey (Wynne-Davies et al. 1982; Garcia Segarra et al. 2012).
No clear genotype–phenotype correlation could be established for WISP3 mutations until now, and there is a rather small variation in age of onset, severity, and progression. However, it has been shown that patients with nonsense or frameshift mutations do not have a more severe phenotype than those with missense mutations (Garcia Segarra et al. 2012; review of 63 cases). Yet, it is challenging to draw a conclusion based on retrospective studies from different countries because of the large differences in the standard of medical care, such as early intervention and rehabilitation that may alter the progression of the disease.
It is very likely that negligence and lack of proper management, such as physical therapy and treatment with NSAIDs, led to the severe outcome of the cases reported here. This study also illustrates that comprehensive gene panel sequencing, such as the Mendeliome analysis, may provide an effective diagnostic tool for deciphering the cause of an unclear presentation of a disease with a strong genetic background and allow timely treatment for the well-being of patients.
METHODS
DNA was extracted from blood with standard protocols. About 50 ng of patient 2 (Fig. 2) DNA was enriched in a pool of three samples using the TruSight One (Illumina Inc.) according to manufacturer's instructions and a 2× 150-bp read length, followed by next-generation sequencing on a MiSeq sequencer. This resulted in 95.4% of uniquely mapped reads with a mean coverage of 82× and 20× coverage for 94.2% of target sequences (see Table 2 and Supplemental Material).
Table 2.
Sequencing coverage
| Enrichment kit | Illumina TruSight One |
|---|---|
| Read length | 151 |
| Mean Cov | 82 |
| Cov 2× | 98.8 |
| Cov 10× | 97.7 |
| Cov 20× | 94.2 |
| Cov 30× | 87 |
| Total reads | 16,198,128 |
| Unique reads (UR) | 13,594,734 |
| Unique mapped reads (UMR) | 12,966,845 |
| %UR | 83.9 |
| %UMR | 95.4 |
The data were analyzed using a data analysis pipeline developed at the Cologne Center for Genomics (CCG) (Kawalia et al. 2015). At first, raw sequencing reads were mapped to human genome reference (GRC37) using bwa-aln (version 0.6.2) (Li and Durbin 2009) followed by duplication marking using Picard (version 1.64) (http://broadinstitute.github.io/picard/). Thereafter, GATK's practice guidelines (Van der Auwera et al. 2013) were followed, in which base quality score recalibration and local indel realignment were performed using GATK (version 1.6.11) (McKenna et al. 2010). After the postalignment improvements, variant calling was performed using GATK's unified-genotyper (version 1.6.11) and SAMtools’ mpileup (version 0.1.18) (Li et al. 2009). The variant lists from both callers were merged and annotated using various public databases: dbSNP (Sherry et al. 2001), 1000 Genomes Project (The 1000 Genomes Project Consortium et al. 2010), Exome Variant Server (EVS http://evs.gs.washington.edu/EVS/), HGMD professional database (Stenson et al. 2014), dbVAR and DGVa (Lappalainen et al. 2013), GERP (Davydov et al. 2010), and Ensembl (Flicek et al. 2008). The functional effect of variants were predicted using PolyPhen (Adzhubei et al. 2013), SIFT (Kumar et al. 2009), and RVIS (Petrovski et al. 2013). All of this downstream analysis has been assembled together with a few scripts developed in-house. Moreover, splice site analysis based on Yeo et al. (2004) was performed. At last, the annotated variant list is uploaded to CCG's web interface and database varbank (https://varbank.ccg.uni-koeln.de).
Variant prioritization and data filtering were conducted using the varbank database and analysis pipeline. We filtered out common variants with allele frequencies of >1% in the following databases: 1000 Genomes (http://browser.1000genomes.org), Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS/), and Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/).
Additionally, three widely used CNV callers, ExomeDepth (Plagnol et al. 2012), XHMM (Fromer et al. 2012), and CoNIFER (Krumm et al. 2012), were used to obtain highly confident calls based on the consensus between the callers (see Supplemental Material; Supplemental File 1 for called CNVs and details about callers).
ADDITIONAL INFORMATION
Data Deposition and Access
The pathogenic WISP3 mutation has been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number SCV000612146.
Ethics Statement
Informed consent was obtained from the patient for genetic investigations and recording and publishing of the disease-related information. The study was approved by the institutional review board of the Ethics Committee of the University Hospital of Cologne.
Acknowledgments
This work is funded by the Deutsche Forschungsgemeinschaft (DFG) Emmy Noether Grant (CI218/1-1) to S.C. We thank all the staff of the Cologne Center for Genomics for their assistance in next-generation sequencing. The sequencing data were analyzed using the DFG-funded CHEOPS high performance compute cluster of the Regional Computing Center of the University of Cologne.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Peter N. Robinson
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Adzhubei I, Jordan DM, Sunyaev SR. 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter 7: Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal A, Bhavani G SL, Togarrati PP, Bierhals T, Nandineni MR, Danda S, Danda D, Shah H, Vijayan S, Gowrishankar K, et al. 2012. Analysis of the WISP3 gene in Indian families with progressive pseudorheumatoid dysplasia. Am J Med Genet A 158A: 2820–2828. [DOI] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delague V, Chouery E, Corbani S, Ghanem I, Aamar S, Fischer J, Levy-Lahad E, Urtizberea JA, Mégarbané A. 2005. Molecular study of WISP3 in nine families originating from the Middle-East and presenting with progressive pseudorheumatoid dysplasia: identification of two novel mutations, and description of a founder effect. Am J Med Genet A 138A: 118–126. [DOI] [PubMed] [Google Scholar]
- Fazeli W, Karakaya M, Herkenrath P, Vierzig A, Dotsch J, von Kleist-Retzow JC, Sebahattin C. 2016. Mendeliome sequencing enables differential diagnosis and treatment of neonatal lactic acidosis. Mol Cell Ped 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T. 2008. Ensembl 2008. Nucleic Acids Res 36: D707–D714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O'Donovan MC, Owen MJ, et al. 2012. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 91: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Segarra N, Mittaz L, Campos-Xavier AB, Bartels CF, Tuysuz B, Alanay Y, Cimaz R, Cormier-Daire V, Di Rocco M, Duba HC, et al. 2012. The diagnostic challenge of progressive pseudorheumatoid dysplasia (PPRD): a review of clinical features, radiographic features, and WISP3 mutations in 63 affected individuals. Am J Med Genet C Semin Med Genet 160C: 217–229. [DOI] [PubMed] [Google Scholar]
- Haliloglu G, Becker K, Temucin C, Talim B, Küçükşahin N, Pergande M, Motameny S, Nürnberg P, Aydingoz U, Topaloglu H, et al. 2017. Recessive PIEZO2 stop mutation causes distal arthrogryposis with distal muscle weakness, scoliosis and proprioception defects. J Hum Genet 62: 497–501. [DOI] [PubMed] [Google Scholar]
- Hurvitz JR, Suwairi WM, Van Hul W, El-Shanti H, Superti-Furga A, Roudier J, Holderbaum D, Pauli RM, Herd JK, Van Hul EV, et al. 1999. Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat Genet 23: 94–98. [DOI] [PubMed] [Google Scholar]
- Kawalia A, Motameny S, Wonczak S, Thiele H, Nieroda L, Jabbari K, Borowski S, Sinha V, Gunia W, Lang U, et al. 2015. Leveraging the power of high performance computing for next generation sequencing data analysis: tricks and twists from a high throughput exome workflow. PLoS One 10: e0126321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, O'Roak BJ, Malig M, Coe BP, Quinlan AR, Nickerson DA, Eichler EE, Roak BJO, et al. 2012. Copy number variation detection and genotyping from exome sequence data. Genome Res 22: 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Lappalainen I, Lopez J, Skipper L, Hefferon T, Spalding JD, Garner J, Chen C, Maguire M, Corbett M, Zhou G, et al. 2013. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res 41(Database issue): D936–D941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short-read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1725–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa S, Obregon MG, Altmüller J, Thiele H, Nürnberg P, Fano V, Wollnik B. 2016. Novel IFT122 mutations in three Argentinian patients with cranioectodermal dysplasia: expanding the mutational spectrum. Am J Med Genet Part A Genet 170A: 1295–1301. [DOI] [PubMed] [Google Scholar]
- Nampoothiri S, Yesodharan D, Sainulabdin G, Narayanan D, Padmanabhan L, Girisha KM, Cathey SS, De Paepe A, Malfait F, Syx D, et al. 2014. Eight years’ experience from a skeletal dysplasia referral center in a tertiary hospital in Southern India: a model for the diagnosis and treatment of rare diseases in a developing country. Am J Med Genet A 164A: 2317–2323. [DOI] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. 2013. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 9: e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, Wood NW, Hambleton S, Burns SO, Thrasher AJ, et al. 2012. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 28: 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai E, Mahajan A, Kumar P, Angural A, Dhar MK, Razdan S, Thangaraj K, Wise CA, Ikegawa S, Pandita KK, et al. 2016. Whole exome screening identifies novel and recurrent WISP3 mutations causing progressive pseudorheumatoid dysplasia in Jammu and Kashmir-India. Sci Rep 6: 27684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudi Mendeliome Group. 2015. Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol 16: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen M, Cheng YH, Goldring MB, Lotz MK, Carson DA. 2004. WISP3-dependent regulation of type II collagen and aggrecan production in chondrocytes. Arthritis Rheum 50: 488–497. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger J, Albert C, Schilling F, Bartsocas C. 1983. Progressive pseudorheumatoid arthropathy of childhood (PPAC): a hereditary disorder simulating juvenile rheumatoid arthritis. Am J Med Genet 14: 399–401. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips AD, Cooper DN. 2014. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 133: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. 2012. A map of human genome variation from population-scale sequencing. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform 11: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach S, Reinert M-C, Altmüller J, Krätzner R, Thiele H, Rosenbaum T, Nürnberg P, Gärtner J. 2017. A new CUL4B variant associated with a mild phenotype and an exceptional pattern of leukoencephalopathy. Am J Med Genet Part A 173A: 2803–2807. [DOI] [PubMed] [Google Scholar]
- Wynne-Davies R, Hall C, Ansell BM. 1982. Spondylo-epiphysial dysplasia tarda with progressive arthropathy. A “new” disorder of autosomal recessive inheritance. J Bone Joint Surg Br 64: 442–445. [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB, Liebert MA, Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.