Abstract
Aquaporin-4, encoded by AQP4, is the major water channel in the central nervous system and plays an important role in the brain's water balance, including edema formation and clearance. Using genomic copy-number analysis and trio-exome sequencing, we investigated a male patient with intellectual disability, hearing loss, and progressive gait dysfunction and found a de novo missense change Ser111Thr in AQP4 as the only suspicious finding. Perinatally, signs of brain ischemia were detected in relation to acute collapse 2 h after birth that resolved a few days later. At the age of 3 mo, cardiac hypertrophy was detected that persisted through childhood but was completely resolved by age 16. In theory, this neurodevelopmental disorder with transient cardiomyopathy could be caused by a disturbance of cellular water balance. Ser111 is an extremely conserved residue in the short cytoplasmic loop between AQP4 transmembrane helix 2 and 3, present across all AQP isoforms from plants to mammals, and it does not appear to be a phosphorylation site. We found that the Ser111Thr change does not affect water permeability or protein stability, suggesting another and possibly regulatory role. Although causality remains unproven, this case study draws attention to AQP4 as a candidate gene for a unique developmental disorder and to a specific serine as a residue of possibly great functional importance in many AQPs.
Keywords: brain atrophy; cerebral ischemia; concentric hypertrophic cardiomyopathy; congenital sensorineural hearing impairment; gait imbalance; intellectual disability, mild; macrocephaly at birth; relative macrocephaly; spastic gait
INTRODUCTION
Aquaporins (AQPs) are water channels that enhance membrane water permeability by forming pores. They are found in bacteria, plants, and mammals and constitute a family of transmembrane proteins with a water channel in the middle, just permeable to a single file (string) of water molecules (Agre and Kozono 2003; Verkman 2011). In the plasma membrane, AQPs form tetramers. For AQP4, these tetramers may assemble into supramolecular square arrays, also called orthogonal arrays of particles (Furman et al. 2003; Crane et al. 2010). The rationale for assembling water channels into such large arrays is not clear (Crane et al. 2010).
There are several AQP genes of which AQP4 encodes the main AQP in the central nervous system. Expression is high in cells supporting neurons—for example, astroglia and supportive cells of sensory neurons in the eye and ear (Verkman 2012). AQP4 is also expressed in cardiomyocytes (Rutkovskiy et al. 2013). AQP4 accumulation can be seen in the perivascular endfeet of glial cells (i.e., in the blood–brain barrier) as well as in the ependymal–CSF barrier, which borders the ventricular and subarachnoid spaces (Verkman 2012). Although AQP4 KO mice appeared largely normal, conditions of stress revealed more subtle abnormalities. The KO mice displayed delayed cytotoxic edema formation, while vasogenic edema formation or conditions with hydrocephalus worsened (Papadopoulos and Verkman 2007; Rutkovskiy et al. 2014). In addition, KO mice had memory impairment (Skucas et al. 2011; Szu and Binder 2016).
There are two known human aquaporinopathies: a rare form of diabetes insipidus due to AQP2 mutations causing impaired water resorption in the renal collecting tubules (OMIM #125800), and neuromyelitis optica, an autoimmune disease with simultaneous inflammation and demyelination of the optic nerve and spinal cord and with the presence of autoantibodies against AQP4 (Verkman 2012). Here, we describe a possible third aquaporinopathy in a patient with a de novo missense variant Ser111Thr in AQP4 together with intellectual disability (ID) and a remarkable phenotype comprising both transient (brain ischemia and cardiac hypertrophy) and progressive features (gait disturbances).
RESULTS
Clinical Presentation and Diagnostic Investigations
A male patient with mild ID, sensorineural hearing loss, macrocephaly, and progressive gait problems underwent routine molecular diagnostic investigations for copy-number aberrations (on high-resolution Affymetrix SNP-array type 6.0) and de novo or recessive Mendelian causes of developmental delay. The latter was done by next-generation sequencing–based exome analysis comparing patient to parental DNA sequences. No copy-number aberrations or homozygosity regions of >5 Mb were found after analysis of the SNP-array data. His karyotype was also normal; 46,XY. On trio-exome sequencing, the only suspicious finding was heterozygosity of a de novo missense variant in the AQP4 gene: AQP4 (NM_001650.4) c.332G>C, p.(Ser111Thr). This de novo variant finding was verified by Sanger sequencing of patient and parents. The variant is not listed in the ExAC/gnomAD database of population variants (see http://gnomad.broadinstitute.org/gene/ENSG00000171885). AQP4 is a gene with high tolerance to both loss-of-function mutations and missense variation with a pLI of 0.02 and a Z-score of −0.25 in the ExAC database (see http://exac.broadinstitute.org/gene/ENSG00000171885 for further details) (Lek et al. 2016). The variant has been registered by us as open access in DECIPHER (patient #HUH323434), but so far no other patients with the same or overlapping AQP4 variants have been found in DECIPHER or through the MatchMaker Exchange hub of databases (Philippakis et al. 2015).
A more detailed disease history was thereafter obtained, and the patient was reexamined. He was born by cesarean section because of breech presentation 1 wk preterm after a normal pregnancy: birth weight 4370 g (P92/+1.4 SD), head circumference 38 cm (P98/+2.0 SD), and length 53 cm (P85/+1.1 SD). Normal Apgar scores, 8–9–10 after 1, 5, and 10 min, respectively. He was the fifth child of healthy unrelated parents and has four healthy siblings.
Two hours after birth he became cyanotic with marmorated skin, and examination showed marked hypotonia, a heart murmur, unstable blood pressure, and areflexia. He responded to sedation, vasopressors, and intubation and resolved completely within a few days. The cause of this acute event was never discovered: An echocardiogram was normal, and no signs of sepsis were found. Cerebral ultrasound indicated slightly enlarged and asymmetric ventricles with increased echogenicity of the left anterior horn, EEG was compatible with encephalopathy, possibly due to ischemic brain injury, and cerebral CT showed supratentorial changes with low attenuation, also compatible with ischemic brain injury. Follow-up EEG, including a recent sleep deprived study, was normal. Despite an increasing tendency to fall that has developed in the last years, he has never had any seizures. Hypertrophic obstructive cardiomyopathy was discovered at 3 mo of age. Later, his cardiomyopathy slowly resolved, but he was treated for cardiomyopathy until age 16. He had several attacks with normothermic tachypnoea and sweating as an infant. Cerebral MRIs showed nonprogressive findings: His ventricles were large and there were symmetrical atrophic changes in the lentiform nuclei (putamen and globus pallidus) and a thick cranium. The changes were interpreted as possibly related to perinatal asphyxia.
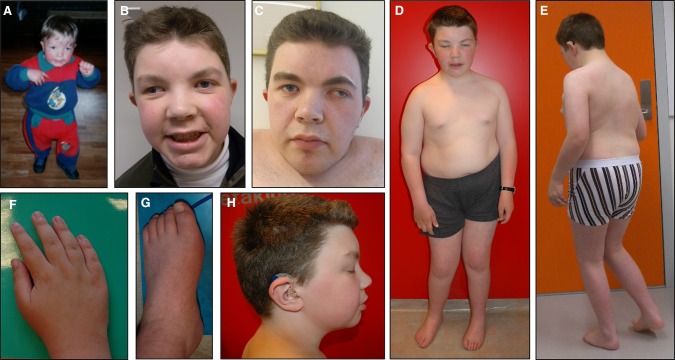
A metabolic screen and muscle biopsy were normal. He was hypotonic with developmental delay: walked at age 2.5 years and started talking at age 4.5 years. Vision has been normal but with alternating strabismus. He has a nonprogressive sensorineural hearing impairment accompanied by dysarthria. Cognitive skills were formally addressed to a level of mild ID at age 16, with normal behavior and social skills. On examination at age 24 years he is short, 8 cm below the 2.5th percentile (−3.7 SD), with a large head (61 cm, +3.1 SD) and mild facial dysmorphism (Fig. 1). He also has hypogonadotropic hypogonadism and tapering fingers. He has always been overweight, at age 15 his BMI was 24.6 (P92/+1.4 SD).
Figure 1.
Patient pictures. Patient pictures from ages 3 (A), 16 (D–H), 18 (B), and 25 (C) years.
Neurological assessment at age 5 revealed finger tremor, delayed fine motor skills, slow gait, spasticity, and sensorineural hearing loss. Assessment at age 19 showed that his gait was abnormal with a tendency to scissoring and posturing of the left arm. Formal testing showed asymmetrical spasticity and hyperreflexia more prominent on the left, as well as bilateral Babinski signs. There was also slight rigidity in both arms. Repeat examination a few years later showed similar findings, albeit with increased gait difficulties.
Unlike the transient signs of brain ischemia and resolving cardiomyopathy, motor function has slowly deteriorated. As a teenager he played soccer as a 12th team member, but now he needs a wheelchair to get around, despite being treated with botulism injections in the Achilles tendons to avoid spasticity, and he has an increasing tendency to fall.
Functional AQP4 Studies
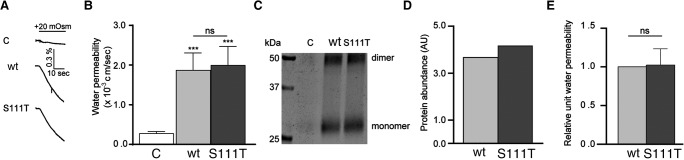
To determine if the Ser111Thr (S111T) change affected the water permeation pathway through AQP4, the human wild-type (wt) AQP4 and the AQP4-S111T were expressed in Xenopus laevis oocytes. Oocytes have a very low intrinsic water permeability, which increases 10–20-fold upon expression of AQP4 (Fenton et al. 2010). The water permeability of uninjected oocytes and oocytes expressing AQP4-wt or AQP4-S111T was determined by exposing the oocytes to a hyperosmotic challenge of +20 mOsm (obtained by addition of mannitol to the control solution) while observing their volume with a sensitive camera (see Fig. 2A for representative volume traces). For each of the three individual experiments, the osmotic water permeabilities were determined for the uninjected oocytes and oocytes expressing either AQP4-wt or AQP4-S111T. In the representative experiment illustrated in Figure 2, the water permeability of the AQP4-expressing oocytes was significantly higher than that of the uninjected oocytes: in ×10−3 cm/sec: 0.27 ± 0.05 for the uninjected, 1.87±0.43 for AQP4-wt, and 1.99±0.48 for AQP4-S111T (n = 4 oocytes in one representative experiment, P < 0.001). The water permeability of oocytes expressing AQP4-wt or AQP4-S111T was not significantly different. To determine the relative unit water permeability, semiquantification of the AQP4 plasma membrane protein abundance was obtained by surface biotinylation of the oocytes followed by western blotting and immunolabeling with an anti-AQP4 antibody (see Fig. 2C for a representative western blot). Densitometric analysis of the immunoreactive bands identified as AQP4 protein (monomers and dimers), after background correction, is shown for this representative experiment in Figure 2D.
Figure 2.
The relative unit water permeability of AQP4-wt and AQP4-S111T expressed in oocytes. (A) Representative volume traces from an uninjected oocyte (labeled C) and oocytes expressing AQP4-wt or AQP4-S111T challenged with a hyperosmotic gradient of +20 mOsm (indicated by a black bar). (B) The water permeabilities are given for uninjected control oocytes (labeled “C”), AQP4-wt-, and AQP4-S111T-expressing oocytes for one representative experiment (n = 4 oocytes, error bars given as ±SD, statistical significance above the histograms refers to comparison to uninjected oocytes). (C) A representative western blot of biotinylated oocyte plasma membranes from one experiment (n = 4 oocytes in each lane). (D) Quantification of the representative western blot in C. (E) The relative unit water permeability was obtained for each experiment by division of the AQP4-mediated water permeabilities from B with plasma membrane AQP4 abundance (D), normalized to AQP4-wt and summarized across individual experiments (n = 3, error bars given as ±SEM). Water permeabilities were compared using one-way ANOVA with Dunnett's multiple comparison post hoc test and the relative unit water permeabilities were compared using Student's paired t-test. ***, P < 0.001; ns, not significant.
The relative unit water permeability of AQP4-wt and AQP4-S111T was obtained by division of the water permeability (Fig. 2B) with the plasma membrane protein abundance (Fig. 2D) for each individual experiment, normalization to the AQP4-wt, and averaging across the three individual experiments. The unit water permeability for the AQP4-S111T was 102% ± 22% of the AQP4-wt (n = 3, P = 0.93), which illustrates that the Ser111Thr change does not affect the water permeation pathway through AQP4. This is also in line with Ser111 being located distant from the water channel in the three-dimensional protein structure (Supplemental Figs. S1, S2).
DISCUSSION
On average, one to two de novo variants are found in the exome per generation (Acuna-Hidalgo et al. 2016); thus, our finding of a single de novo variant in the exome of this patient is as expected. However, in light of this patient's phenotype, with transient cerebral and cardiac abnormalities, as well as mild ID with progressive gait disturbance, we considered the finding as a potential cause. The variant resides in the brain's major water channel gene, AQP4, a gene with particularly high expression in the cerebellum (Hubbard et al. 2015) but also in cardiomyocytes (Rutkovskiy et al. 2014). High expression is also found in the inner ear and brain pathways used for hearing (see the SHIELD database, https://shield.hms.harvard.edu; and Christensen et al. 2009). Thus, the expression pattern overlaps with affected organs (brain, ear, and heart).
Ser111 is an extremely conserved residue, not only in AQP4, but also in other aquaporins (AQP1, AQP2, AQP4, AQP5, AQP6, and AQP12; see Fig. 3). In Arabidopsis AQP2 the residue at this position is a threonine, but in a very different context than for other AQP2 genes (only a few of the surrounding amino acids are conserved; Fig. 3). Although it is unknown what functional role Thr111 may play in this plant AQP, one could imagine that loop interactions and potential interaction with cytosolic proteins may differ among different AQPs. The residue is not present in aquaglyceroporins, suggesting a specific role related to cellular or organ water balance. The question is what the role of this residue might be. One possibility is that Ser111 is a protein kinase A (PKA) phosphorylation site, somehow regulating AQP4 function (Song and Gunnarson 2012). However, although Ser111 sits in a motif (RKIS/T) with resemblance to a PKA phosphorylation site (RRXS/T), and the corresponding peptide can be phosphorylated by PKA in vitro, there is no evidence that the ability to phosphorylate Ser111 affects AQP4 function (Assentoft et al. 2013) or expression (Kitchen et al. 2015). Furthermore, missense changes to Arg108 (A108S) and Ile110 (I110F) have been found in the gnomAD database, also suggesting that conservation of a PKA phosphorylation site motif framing Ser111 is not critical. Finally, a phospho-specific antibody, exclusively recognizing AQP4 when phosphorylated on Ser111, failed to detect phosphorylation in cell lysates of rat brain stimulated by conditions proposed to induce phosphorylation of this residue (Assentoft et al. 2013). Putative phosphorylation sites are also present in the carboxy-terminal part of AQP4 (Ser276, Ser285, Ser316, Ser321, and Ser322). As for Ser111, mutating these phosphorylation sites did not affect water permeability nor membrane abundance of AQP4 (Assentoft et al. 2014), but Ser276 phosphorylation by PKA appears necessary for a normal AQP4 membrane translocation response (Kitchen et al. 2015). As for Ser111 (but unlike many other AQP4 serines), no variants to Ser276 have been registered in the gnomAD database.
Figure 3.

Conservation of Ser111 in AQP4 and between aquaporins.
AQP4 has five exons and an open reading frame of 972 bp. There are two major isoforms, M1 and M23, named after the initiating methionine of the translated protein. M1 is translated from exon 1, whereas M23 is translated from exon 2. ENCODE data (https://www.encodeproject.org/) suggest that mRNA transcription can be initiated from promoters upstream of either exon 1 or exon 2. In addition, the longer transcript (M1) can also be a source of the M23 isoform by alternative initiation of translation (Pisani et al. 2011). AQP4 mRNA is 5.2 kb, mostly because of a large 3′ UTR. This suggests regulation of mRNA stability. M23 expression promotes square array assembly (e.g., large arrays of AQP4 tetramers) (Furman et al. 2003). Ser111 is a residue of both major isoforms (i.e., its presence is not restricted to AQPs capable of forming orthogonal arrays).
Our functional studies showed that the water permeability was not affected by the Ser111Thr amino acid replacement (Fig. 2). Previously, Ser111 has been mutated to alanine or aspartic acids (the latter to mimic phosphorylation), also without any effect on channel water permeability (Assentoft et al. 2013). Furthermore, structural modeling does not place Ser111 in contact with the AQP4 water channel (Supplemental Fig. 1). It is noteworthy that like Ser111, Ser180 and Ser188 have no missense variants in the gnomAD database, currently listing 246,000 alleles from individuals without severe pediatric disorders. These three serines mark positions of transfer from cytoplasmatic to intramembraneous parts of the AQP4 protein chain. Ser111 and Ser188 reside in cytoplasmatic corners of the AQP4 monomer, and Ser180 has contact with the water channel (Supplemental Fig. 2).
In summary, we present a single patient with a de novo AQP4 Ser111Thr variant and a phenotype that appears explainable by an AQP4 dysfunction. If there is a functional effect of replacing Ser111 in AQP4 with threonine, it is not on the level of protein stability or water permeability (Fig. 2). It is so far unknown why Ser111 is conserved not only in AQP4 but also among other AQP isoforms (Fig. 3). The biochemical change of replacing serine with threonine is mild, maybe because more radical missense changes would not be tolerated from a developmental point of view. Our case may be resolved when an answer to this question has been found, if other patients with the same mutation and phenotype are reported, or if, contrary to our expectations, the variant should be found in normal individuals.
METHODS
Trio-Exome Sequencing
Whole-exome sequencing was performed on trios on genomic DNA isolated from blood. DNA samples were prepared using the SeqCap EZ HGSC VCRome (Roche NimbleGen) and followed by paired-end 150-nt sequencing on the Illumina NextSeq500. Alignment and variant calling was performed as described in Bredrup et al. (2015). Median coverage of the target region was 95× with 96% of target region covered with at least 10 reads (Table 1). Data annotation and interpretation were performed using the Cartagenia Bench Lab, NGS module (Cartagenia). Filtering was set to find de novo, autosomal recessive, and X-linked causes of disease (the latter not excluding variants carried by the mother). Two de novo variants were found in the exome: AQP4(NM_001650.4) c.332G>C, p.(Ser111Thr) with 32/34 reads, and NPEPL1(NM_024663.3) c.1302+1G>T with 17/12 reads (wild-type read count listed first). The latter gene probably encodes an aminopeptidase that removes unsubstituted amino-terminal amino acids from various peptides, and the gene has high tolerance for LoF with a pLI of 0.00 (Lek et al. 2016). No homozygosity or compound heterozygosity for possibly pathogenic variants was detected, including when only rare missense variants were evaluated. The AQP4 variant was verified by Sanger sequencing. Despite discrepant scores by in silico variant assessment programs (MutationTaster score 1—disease causing, SIFT score 0.04—deleterious, PolyPhen score HumDiv 0.015—benign, PolyPhen score HumVar 0.040—benign) (Table 2), this de novo variant was considered interesting because the patient's phenotype was suggestive of a problem that in theory could be caused by water imbalance in the brain and heart.
Table 1.
Whole-exome sequencing coverage
| Total number of reads | Average read length (bp) | Aligned reads (%) | Average exome coverage | AQP4, average coverage | AQP4, fraction at 20× coverage (%) |
|---|---|---|---|---|---|
| 57,807,596 | 73.66 | 98.06 | 58.39× | 63.55× | 100 |
Table 2.
AQP4 variant information
| Gene | Chromosome | Nucleotide change | Amino acid change (predicted) | Variant type | Predicted effect |
dbSNP | Genotype | ClinVar ID | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | Align GVGD | MutationTaster | PolyPhen-2 (HumVar) | ||||||||
| AQP4 | 18 | c.332C>G | p.(Ser111Thr) | Substitution | Del | Class C55 | Disease-causing | Benign | – | Heterozygous (de novo) | SCV000611625.1 |
Del, deleterious.
Heterologous Expression of hAQP4 Isoforms in Xenopus laevis Oocytes
cDNA encoding the M23 isoform of human AQP4-wt and AQP4-S111T mutant subcloned into the oocyte expression vector pXOOM was purchased from GenScript, and 10 ng of AQP4 cRNA was microinjected into defolliculated Xenopus laevis oocytes. Xenopus laevis frogs were obtained from Nasco or EcoCyte Bioscience. The surgical protocol by which the oocytes were retrieved was approved by The Danish National Committee for Animal Studies and has previously been described in Fenton et al. (2010). Oocytes were kept at 19°C in Kulori medium (in mM: 90 NaCl, 1 KCl, 1 CaCl2, 1 MgCl2, 5 HEPES [Tris-buffered pH 7.4]) for 3–4 d before experiments.
Osmotic Water Permeability Measurements
The experimental setup for measuring water permeability of oocytes was previously described by Zeuthen et al. (2007). Briefly, the oocytes were placed in a small chamber with a glass bottom and perfused with a base solution (in mM: 100 NaCl, 2 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES, 20 mM mannitol [Tris-buffered pH 7.4, 220 mOsm]). Micrographs of the cross-sectional area of the oocytes were continuously captured at a rate of 25 images/sec by use of a long-distance objective viewing the oocytes from below. The oocytes were challenged with a hyperosmotic solution (addition of 20 mM mannitol to the base solution), and the osmotic water permeability was determined by Lp = (Jv)/(A × Δπ × Vw), in which Jv is the initial water flux during an osmotic challenge, A is the membrane surface areas (nine times the apparent area due to membrane folding) (Zampighi et al. 1995), Δπ is the osmotic challenge, and Vw is the partial molal volume of water (18 cm3/mol). Osmolarities of the solutions were verified with an accuracy of 1 mOsm with an osmometer Type 15 (Löser Messtechnik).
Cell Surface Biotinylation and Immunoblotting
Oocytes used for water permeability measurements were subsequently exposed to sulfo-NHS-SS-biotin for 45 min at 4°C and solubilized in lysis buffer (150 mM NaCl, 50 mM Tris/HCl, 1% (w/v) Triton X-100, 0.05% SDS, 5 mM EDTA, and protease inhibitors [0.4 mM pefablock, 8 µM leupeptin; Sigma-Aldrich]) for 30 min on ice. The resulting lysates were sonicated and centrifuged at 10,000g for 5 min at 4°C after which the supernatant was incubated with Neutravidin beads (VWR) for 60 min at room temperature (RT). After a brief centrifugation (1000g for 1 min), the supernatant was removed and the beads washed to remove unspecific proteins from the bead slurry. The biotinylated proteins were retrieved by incubation with SDS sample buffer (4% SDS, 0.006% bromophenol blue, 8.7 % glycerol, 0.25% Tris-base and 0.75% DTT [pH 6.8, adjusted with HCl]), electrophoresed on SDS/PAGE (12%), and transferred to PVDF membranes. Proteins were detected by use of a primary polyclonal anti-AQP4 antibody from Alomone labs (#AQP4-004) and a secondary antibody from Licor Biosciences (Licor 800; 1:10.000). The membranes were scanned (Licor) to allow for visualization of immunoreactive bands. Protein abundances were analyzed by densitometry using Image Studio 5.2 software (Licor).
Data Presentation and Statistics
Data are presented as mean ± SD or SEM (as indicated in text and figure legend). Statistical significance was tested with one-way analysis of variance (ANOVA) with Dunnett's multiple comparison post hoc test or Student's paired t-test (as indicated in the Fig. 2 legend). P values < 0.05 were considered statistically significant. The number of experiments (n) corresponds to independent measurements from at least three different oocyte preparations.
Structural Modeling
The crystal structure was obtained from the RCSB PDB protein data bank (PDB ID: 3GD8; Ho et al. 2009) and subsequently analyzed by CAVER Analyst (Kozlikova et al. 2014). CAVER Analyst is a software tool for calculation, analysis and real-time visualization of access tunnels and channels in static and dynamic protein structures. First, a single channel was computed for a monomer using default values set in CAVER Analyst. Afterward, the symmetry defined in the PDF file was used to create the tetramer and its final visualization.
ADDITIONAL INFORMATION
Data Deposition and Access
The AQP4 variant has been registered in DECIPHER under accession number HUH323434 and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) under accession number SCV000611625.1.
Ethics Statement
Both the patient and his parents have consented to publication of this case history with patient pictures. Because the finding was a consequence of routine clinical evaluation and diagnostics, and further research did not require patient investigations that would not otherwise have been done, ethical review board evaluation is not required according to Norwegian rules, and IRB requests will in such cases be rejected (see Houge 2015, with response from the Norwegian health authorities).
Acknowledgments
We are grateful to the patient and his parents for their eagerness to collaborate.
Author Contributions
S.B. investigated the patient, wrote the clinical description, and was the family's contact person, T.L.T.-B. did and described AQP4 functional studies, I.A. found the mutation, J.B. and M.V. made structural AQP4 predictions, L.A.B. did primary and follow-up neurological investigations, N.M. was head of the AQP4 functional studies and contributed to write the manuscript, and G.H. put all the data together and was the principal author of the manuscript.
Funding
HelseVest (grant number 911967) supported a small part of this project.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Neal Sondheimer
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
References
- Acuna-Hidalgo R, Veltman JA, Hoischen A. 2016. New insights into the generation and role of de novo mutations in health and disease. Genome Biol 17: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agre P, Kozono D. 2003. Aquaporin water channels: molecular mechanisms for human diseases. FEBS Lett 555: 72–78. [DOI] [PubMed] [Google Scholar]
- Assentoft M, Kaptan S, Fenton RA, Hua SZ, de Groot BL, MacAulay N. 2013. Phosphorylation of rat aquaporin-4 at Ser(111) is not required for channel gating. Glia 61: 1101–1112. [DOI] [PubMed] [Google Scholar]
- Assentoft M, Larsen BR, Olesen ET, Fenton RA, MacAulay N. 2014. AQP4 plasma membrane trafficking or channel gating is not significantly modulated by phosphorylation at COOH-terminal serine residues. Am J Physiol Cell Physiol 307: C957–C965. [DOI] [PubMed] [Google Scholar]
- Bredrup C, Johansson S, Bindoff LA, Sztromwasser P, Kråkenes J, Mellgren AE, Brurås KR, Lind O, Boman H, Knappskog PM, et al. 2015. High myopia-excavated optic disc anomaly associated with a frameshift mutation in the MYC-binding protein 2 gene (MYCBP2). Am J Ophthalmol 159: 973–979 e972. [DOI] [PubMed] [Google Scholar]
- Christensen N, D'Souza M, Zhu X, Frisina RD. 2009. Age-related hearing loss: aquaporin 4 gene expression changes in the mouse cochlea and auditory midbrain. Brain Res 1253: 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JM, Tajima M, Verkman AS. 2010. Live-cell imaging of aquaporin-4 diffusion and interactions in orthogonal arrays of particles. Neuroscience 168: 892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton RA, Moeller HB, Zelenina M, Snaebjornsson MT, Holen T, MacAulay N. 2010. Differential water permeability and regulation of three aquaporin 4 isoforms. Cell Mol Life Sci 67: 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman CS, Gorelick-Feldman DA, Davidson KG, Yasumura T, Neely JD, Agre P, Rash JE. 2003. Aquaporin-4 square array assembly: opposing actions of M1 and M23 isoforms. Proc Natl Acad Sci 100: 13609–13614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JD, Yeh R, Sandstrom A, Chorny I, Harries WE, Robbins RA, Miercke LJ, Stroud RM. 2009. Crystal structure of human aquaporin 4 at 1.8 Å and its mechanism of conductance. Proc Natl Acad Sci 106: 7437–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houge G. 2015. [Where is the boundary between diagnostics and research?]. Tidsskr Nor Laegeforen 135: 1632. [DOI] [PubMed] [Google Scholar]
- Hubbard JA, Hsu MS, Seldin MM, Binder DK. 2015. Expression of the astrocyte water channel aquaporin-4 in the mouse brain. ASN Neuro 7: ii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen P, Day RE, Taylor LH, Salman MM, Bill RM, Conner MT, Conner AC. 2015. Identification and molecular mechanisms of the rapid tonicity-induced relocalization of the aquaporin 4 channel. J Biol Chem 290: 16873–16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlikova B, Sebestova E, Sustr V, Brezovsky J, Strnad O, Daniel L, Bednar D, Pavelka A, Manak M, Bezdeka M, et al. 2014. CAVER Analyst 1.0: graphic tool for interactive visualization and analysis of tunnels and channels in protein structures. Bioinformatics 30: 2684–2685. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos MC, Verkman AS. 2007. Aquaporin-4 and brain edema. Pediatr Nephrol 22: 778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C, et al. 2015. The matchmaker exchange: a platform for rare disease gene discovery. Hum Mutat 36: 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani F, Rossi A, Nicchia GP, Svelto M, Frigeri A. 2011. Translational regulation mechanisms of aquaporin-4 supramolecular organization in astrocytes. Glia 59: 1923–1932. [DOI] [PubMed] [Google Scholar]
- Rutkovskiy A, Valen G, Vaage J. 2013. Cardiac aquaporins. Basic Res Cardiol 108: 393. [DOI] [PubMed] [Google Scholar]
- Rutkovskiy A, Mariero LH, Vaage J. 2014. Deletion of the aquaporin-4 gene alters expression and phosphorylation of protective kinases in the mouse heart. Scand J Clin Lab Invest 74: 500–505. [DOI] [PubMed] [Google Scholar]
- Skucas VA, Mathews IB, Yang J, Cheng Q, Treister A, Duffy AM, Verkman AS, Hempstead BL, Wood MA, Binder DK, et al. 2011. Impairment of select forms of spatial memory and neurotrophin-dependent synaptic plasticity by deletion of glial aquaporin-4. J Neurosci 31: 6392–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Gunnarson E. 2012. Potassium dependent regulation of astrocyte water permeability is mediated by cAMP signaling. PLoS One 7: e34936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szu JI, Binder DK. 2016. The role of astrocytic aquaporin-4 in synaptic plasticity and learning and memory. Front Integr Neurosci 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkman AS. 2011. Aquaporins at a glance. J Cell Sci 124: 2107–2112. [DOI] [PubMed] [Google Scholar]
- Verkman AS. 2012. Aquaporins in clinical medicine. Annu Rev Med 63: 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampighi GA, Kreman M, Boorer KJ, Loo DD, Bezanilla F, Chandy G, Hall JE, Wright EM. 1995. A method for determining the unitary functional capacity of cloned channels and transporters expressed in Xenopus laevis oocytes. J Membr Biol 148: 65–78. [DOI] [PubMed] [Google Scholar]
- Zeuthen T, Zeuthen E, Macaulay N. 2007. Water transport by GLUT2 expressed in Xenopus laevis oocytes. J Physiol 579: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.