Abstract
Pancreatic neuroendocrine tumors (PNETs) are a genomically and clinically heterogeneous group of pancreatic neoplasms often diagnosed with distant metastases. Recurrent somatic mutations, chromosomal aberrations, and gene expression signatures in PNETs have been described, but the clinical significance of these molecular changes is still poorly understood, and the clinical outcomes of PNET patients remain highly variable. To help identify the molecular factors that contribute to PNET progression and metastasis, and as part of an ongoing clinical trial at the BC Cancer Agency (clinicaltrials.gov ID: NCT02155621), the genomic and transcriptomic profiles of liver metastases from five patients (four PNETs and one neuroendocrine carcinoma) were analyzed. In four of the five cases, we identified biallelic loss of MEN1 and DAXX as well as recurrent regions with loss of heterozygosity. Several novel findings were observed, including focal amplification of MYCN concomitant with loss of APC and TP53 in one sample with wild-type MEN1 and DAXX. Transcriptome analyses revealed up-regulation of MYCN target genes in this sample, confirming a MYCN-driven gene expression signature. We also identified a germline NTHL1 fusion event in one sample that resulted in a striking C>T mutation signature profile not previously reported in PNETs. These varying molecular alterations suggest different cellular pathways may contribute to PNET progression, consistent with the heterogeneous clinical nature of this disease. Furthermore, genomic profiles appeared to correlate well with treatment response, lending support to the role of prospective genotyping efforts to guide therapy in PNETs.
Keywords: neoplasm of the gastrointestinal tract, neuroendocrine neoplasm
INTRODUCTION
Pancreatic neuroendocrine tumors (PNETs) are rare tumors of the endocrine pancreas, accounting for <5% of all pancreatic malignancies (Halfdanarson et al. 2008). The classification of PNETs is based on TNM stage and immunohistological examination, taking into account morphological tumor differentiation and grade (Klimstra et al. 2010). Poorly differentiated tumors are characterized by a high proliferation index (Ki-67 > 20%) and mitotic count (>20 per 10 high power field [HPF]), and are classified as neuroendocrine carcinomas (NECs). Until recently, well-differentiated PNETs were classified as either low-grade (G1) or intermediate-grade (G2) based on proliferation index (Ki-67 staining < 3% vs. 3%–20% of tumor cells). However, it is now recognized that there is a small subset of well-differentiated tumors with a discordantly high proliferation index that are biologically distinct from poorly differentiated NECs and are classified as high-grade (G3) PNETs (Coriat et al. 2016; Tang et al. 2016).
The molecular landscape of PNETs was initially described in a seminal paper by Jiao et al. (2011), in which whole-exome sequencing identified recurring mutations in MEN1, the chromatin remodeling genes DAXX and ATRX, and mechanistic target of rapamycin (mTOR) pathway genes, most commonly PTEN, TSC2, and PIK3CA. Mutations in these and other genes may occur in familial syndromes, including multiple endocrine neoplasia 1 (MEN1), tuberous sclerosis complex (TSC1/2), neurofibromatosis type 1 (NF1), and von Hippel–Lindau (VHL) disease, that increase susceptibility to PNET development (Jensen et al. 2008). More recently, whole-genome sequencing of 102 primary PNETs identified recurrent mutations in genes involved in chromatin remodeling, DNA damage repair, mTOR pathway activation, and telomere maintenance. Unexpectedly, 17% of the patients harbored germline mutations not only in previously described MEN1 and VHL but also in tumor suppressor gene CHEK2, DNA repair gene MUTYH, and cell cycle regulator CDKN1B (Scarpa et al. 2017).
The molecular characteristics of PNET metastases have not been reported in detail, but they have important implications for prognosis and treatment. Here, we describe genomic and transcriptomic profiles for five patients (four with metastatic G1–G2 PNETs and one G3 pancreatic NEC) who underwent sequencing analyses as part of an ongoing clinical trial at the BC Cancer Agency (clinicaltrials.gov ID: NCT02155621). The objectives of this report are to characterize the molecular landscapes of these metastatic samples, to identify known and novel molecular aberrations and pathway alterations, and to associate molecular data with clinical presentation and outcomes.
RESULTS
Clinical Presentation and Treatment Outcomes
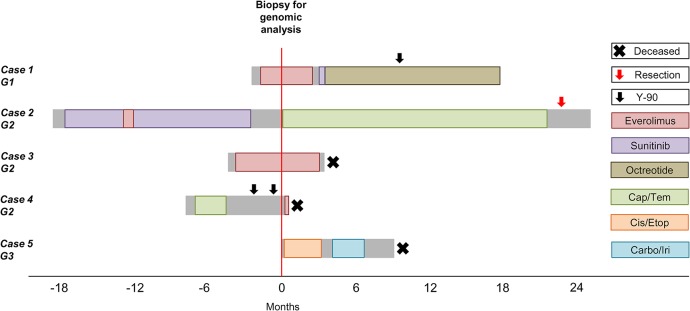
Clinical vignettes are described for each case below and summarized in Table 1 and Figure 1. Radiological responses are defined per RECIST version 1.1 (Response Evaluation Criteria in Solid Tumors) (Eisenhauer et al. 2009).
Table 1.
Baseline characteristics of five patients with metastatic pancreatic neuroendocrine tumors and carcinomas enrolled in the Personalized OncoGenomics program
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | |
|---|---|---|---|---|---|
| Age (yr), gender | 69, male | 52, female | 46, female | 36, male | 67, male |
| Metastatic sites | Liver | Liver | Liver | Liver, lymph nodes | Liver |
| Functional? | Yes | No | No | No | No |
| Grade | Low (G1) | Intermediate (G2) | Intermediate (G2) | Intermediate (G2) | High (G3) |
| Ki-67 index (%) | <2 | 10–15 | 15 | 20 | >70 |
| Baseline chromogranin A | 110 µg/l | 92 U/l (ULN <40) | 5200 µg/l | 250 µg/l | 4920 µg/l |
| 111In-labeled octreotide scan avidity? | Yes | Yes | Not done | Yes | Not done |
Figure 1.
Clinical evolution and treatment of five patients with metastatic pancreatic neuroendocrine tumors and carcinomas enrolled in the Personalized Oncogenomics program. Cap/Tem, capecitabine with temozolomide; Cis/Etop, cisplatin with etoposide; Carbo/Iri, carboplatin with irinotecan.
Case 1
A 69-yr-old man presented with hypercalcemia and was found on computed tomography (CT) imaging to have a pancreatic mass and liver lesions. Biopsy of the pancreatic mass obtained under endoscopic ultrasound guidance confirmed a G1 NET with Ki-67 < 2%. Serum chromogranin A (CgA) was slightly elevated at 110 µg/l (range of <94) and the liver and pancreatic lesions were intensely avid on octreotide scan. He commenced treatment with everolimus and underwent core biopsy of a liver metastasis for molecular analyses after 7 wk of therapy. He had radiologically stable disease, but treatment was discontinued after 4 mo because of grade 2 pneumonitis. Second-line systemic therapy with sunitinib was initiated but also discontinued early because of congestive heart failure. He eventually received yttrium 90 (Y-90) radioembolization therapy to his liver metastases and had a partial radiological response. His hypercalcemia, which was initially refractory to bisphosphonates and required several hospital admissions for management, normalized and remained stable 5 mo after Y-90 therapy. At the time of writing (20 mo after diagnosis), he remains well on maintenance monthly long-acting octreotide.
Case 2
A 52-yr-old female presented with several years of abdominal pain, which eventually led to abdominal ultrasound and CT that showed a pancreatic tail mass and multiple liver lesions. Liver core biopsy showed a G2 metastatic PNET with Ki-67 staining 10%–15% of tumor cell nuclei. Serum CgA was slightly elevated at 92 U/l (range of <40) and imaging with indium-111-labeled octreotide showed uptake in her known disease sites. She commenced treatment with sunitinib but required several dose reductions for grade 2 hand–foot syndrome. She had a brief trial of everolimus but switched back to sunitinib after 3 wk because of mucositis and pneumonitis. She had a symptomatic response and radiologically stable disease that was sustained for 15 mo before developing progressive liver metastases, which did not accumulate 18F-DOPA. She underwent a liver biopsy for genomic analysis prior to starting second-line systemic therapy with capecitabine and temozolomide. She achieved a partial radiographic response and, after 20 mo of chemotherapy, proceeded to resection of her pancreatic primary along with radiofrequency ablation of the liver metastases. Pathologic review of the resected pancreatic primary showed a well-differentiated G2 NET with <2 mitoses/10 HPF and proliferation index of 8.5%. Three of nine lymph nodes were involved. At the time of writing (43 mo after diagnosis), she remains well with disease stability off chemotherapy.
Case 3
A 46-yr-old female with a prior history of tuberous sclerosis complex, resected renal angiomyolipoma, and childhood epilepsy was noted to have liver masses on routine surveillance imaging. She was initially reluctant to have these investigated but agreed to a liver biopsy 9 mo after initial presentation due to progressive fatigue, leg edema, and abdominal discomfort related to massive hepatomegaly. Core needle biopsy was consistent with a G2 NET with a Ki-67 index of 15%. Importantly, immunostains did not support metastatic angiomyolipoma. In addition, PAX8 immunohistochemical stain was positive, favoring a pancreatic origin (Sangoi et al. 2011). Serum CgA was markedly elevated at 5200 µg/l (range of <94). She commenced treatment with everolimus and underwent biopsy of a liver metastasis for molecular analyses after 16 wk of therapy. She had an early symptomatic and marker response, where serum CgA decreased to 3780 µg/l from a peak of 8200 µg/l. Interestingly, her tuberous sclerosis skin lesions (presumed facial angiofibromas), also improved on mTOR inhibitor therapy. Radiological response was unable to be accurately assessed, as pretreatment imaging was performed 8 mo prior to the start of therapy. Her treatment course was complicated by recurrent anemia, diarrhea, and renal impairment, and after almost 7 mo of therapy, she developed clinical progression and died 8 mo after initial pathologic diagnosis. Referral to the Hereditary Cancer Program for clinical genetic testing had been discussed, but she had no significant family history, so this was not pursued.
Case 4
A 36-yr-old man presented with a short history of right shoulder and chest pain and was found on CT to have multiple liver lesions and a pancreatic tail mass. He underwent core liver biopsy, which was consistent with a well-differentiated NET both morphologically and on immunoprofile, with positive staining for synaptophysin and cytokeratin AE1/AE3. Although the tumor had <2 mitoses/10 HPF, the Ki-67 index was up to 20% in some areas; hence this was classified as a G2 NET. Octreotide scan identified additional disease within the retroperitoneal and supraclavicular lymph nodes, and serum CgA was modestly elevated at 250 µg/l (range of <94). He commenced first-line systemic therapy with capecitabine and temozolomide but progressed after three cycles. He then received Y-90 radioembolization to the liver but progressed within 3 mo. At this time, he underwent liver biopsy for genomic analysis prior to starting second-line systemic therapy with everolimus. He was referred for peptide receptor radionuclide therapy (PRRT), but his clinical status declined rapidly, and he died 10 d after commencing everolimus, 8.4 mo after initial diagnosis.
Case 5
A 67-yr-old male presented with a short history of diarrhea and constitutional symptoms and was found on CT to have multiple liver masses and a pancreatic tail solid lesion that appeared suspicious for pancreatic ductal adenocarcinoma. He consented for genomic analysis at the time of his diagnostic liver biopsy, which showed a G3 NEC, large cell type with positive staining for synaptophysin, chromogranin, and cytokeratin 19. Ki-67 stained >70% of tumor cell nuclei. Serum CgA was markedly elevated at 4920 µg/l (range of <94). He started treatment with cisplatin and etoposide chemotherapy and received four cycles before developing disease progression. He then received four cycles of carboplatin and irinotecan but also had upfront disease progression. He was referred to the phase I clinical trial program but had ongoing clinical deterioration and died 9 mo after initial diagnosis.
Genomic Analyses
Whole-genome sequencing of liver metastases and blood from the five patients was performed to identify chromosomal aberrations (Fig. 2) and sequence variants (Fig. 3A; Supplemental Data 1). Germline alterations in 98 cancer susceptibility genes were evaluated, as approved by the research ethics board. RNA sequencing was performed to identify altered gene expression and molecular pathways (Supplemental Data 2). Expression levels of select genes were converted into percentile ranks against a collection of tumor transcriptomes from The Cancer Genome Atlas project (https://portal.gdc.cancer.gov) (Fig. 3A). To identify potential upstream regulators (Fig. 3B) and pathways affected (Supplemental Data 2), normalized gene expressions (Supplemental Data 3) were compared to a compendium of 16 normal tissue transcriptomes from the Illumina Human BodyMap 2.0 project (https://www.ebi.ac.uk/gxa/experiments/E-MTAB-513/Results). Hereafter, description of individual gene expression level specifically refers to comparison with the TCGA tumor compendium unless otherwise noted.
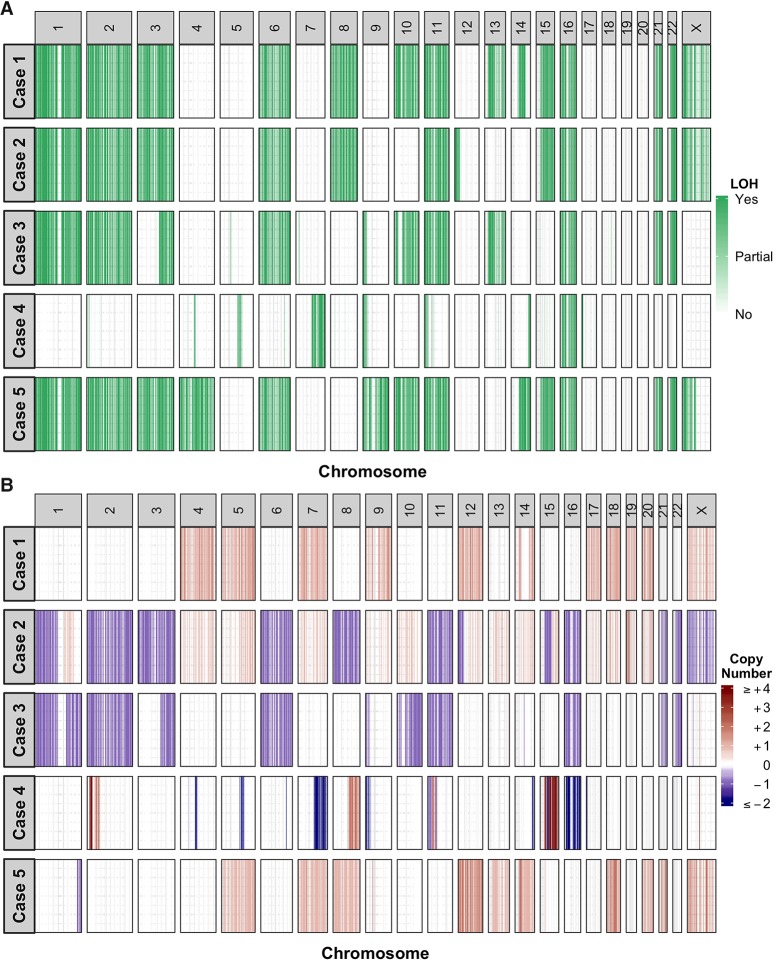
Figure 2.
Genome-wide copy-number architectures across the five cases. (A) Chromosomal regions with loss of heterozygosity events are depicted in green. The zygosity states at these regions are not discriminated. (B) Ploidy-corrected copy-number changes in protein-coding regions are depicted in red (copy gain) or blue (copy loss). The magnitude of copy-number gains is capped at +4.
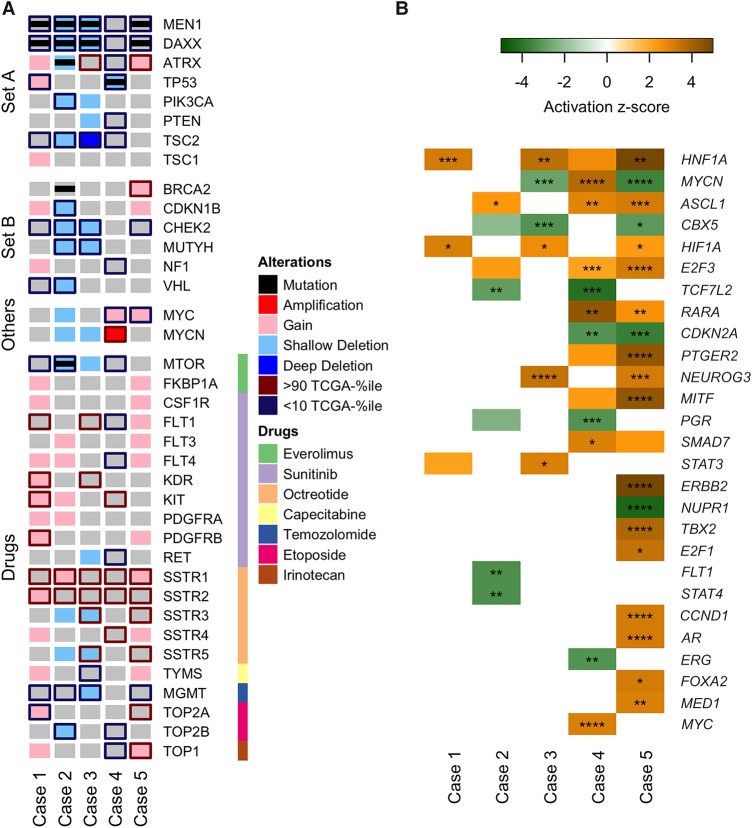
Figure 3.
Key molecular alterations and predicted upstream regulators across the five cases. (A) An OncoPrint depicting alterations in genes that have either been implicated in PNETs or are targets of conventional therapeutic agents used to treat PNETs. All alterations described here are somatic, aside from Case 3, which had germline one copy loss of TSC2. “Set A” contains genes recurrently mutated in sporadic PNETs (Jiao et al. 2011). “Set B” contains genes in which germline mutations were recently reported in PNET patients (Scarpa et al. 2017). “Others” contains genes of interest in this study. “Drugs” contains genes with protein products that are the molecular targets (color-coded) of the indicated therapeutic agents used to treat PNETs. Nonsynonymous mutations (black bar), copy-number aberrations (red/blue shade), and up- or down-regulated expressions (red/blue edge) of these genes are shown. All genes have gray background by default to facilitate visualization. Shallow and deep deletions refer to one- or two-copy losses, respectively. Up-regulated genes are defined as those expressed at levels >90% of TCGA tumor compendium, and down-regulated genes as those expressed at levels <10% of TCGA tumor compendium. (B) Using the Upstream Regulator Analysis tool from Ingenuity Pathway Analysis software, the activation states of prospective upstream regulators in each sample were predicted based on differentially regulated genes. Only upstream regulators with activation score of >3 or <−3 were selected. Color and intensity indicate the predicted activation state and effect size, respectively. The overlap p-value of each prediction is also indicated. *, p < 0.05; **, p < 0.01; ***, p< 0.001; ****, p < 0.0001.
The approved therapeutic drugs for the treatment of PNETs in Canada include the mTOR inhibitor everolimus, the receptor tyrosine kinase (RTK) inhibitor sunitinib, the somatostatin analogs octreotide and lanreotide, and chemotherapy with capecitabine/temozolomide, whereas platinum-based chemotherapy is typically reserved for high-grade disease. To retrospectively study patient response to drug treatment based on the status of their respective molecular targets, we retrieved the list of proteins targeted by the aforementioned approved therapeutic agents from Santos et al. (2017) and examined the status of their gene and gene expression (Fig. 3A).
Shared Molecular Alterations
Whole-genome sequencing of tumor and blood identified several genomic alterations shared within our cohort. Loss of heterozygosity (LOH) events were frequently observed in cases 1, 2, 3, and 5, with Chromosomes 1, 2, 3, 6, 11, 16, 21, and 22 largely affected. These include regions encoding for MEN1 (11q13.1), DAXX (6p21.32), and TSC2 (16p13.3) (Fig. 2A). Frequent copy-number gains were observed in cases 1 and 5, whereas copy-number losses were more frequent in cases 2 and 3. Gene amplification events, defined as the ploidy-corrected copy-number gain being greater than the ploidy, were absent in cases 1, 2, and 3 but were observed in case 5 and more frequently in case 4 (Fig. 2B). Consistent with the reported low mutation burden in low- and intermediate-grade PNETs (Jiao et al. 2011), <50 nonsynonymous mutations in protein-coding genes were identified in three of the five cases (median = 39; range = 21–170; Supplemental Data 1). We identified somatic mutations in MEN1 and DAXX in cases 1, 2, 3, and 5. These comprised nonsense, frameshift, and splice site mutations as well as in-frame deletion or missense mutations predicted to be deleterious (Table 2). In conjunction with LOH events or copy-number losses, these mutations resulted in biallelic loss of MEN1 and DAXX in the four cases. The expression levels of both MEN1 and DAXX were low across all five cases, whereas TSC2 expression was low in cases 1, 2, 3, and 4. Tumor suppressor gene CHEK2 also had low expression in cases 1, 2, 3, and 5 (Fig. 3A).
Table 2.
Sequence variants identified from the sample cohort that have previously been associated with PNETs
| Case ID | Gene | Chr. | HGVS DNA | HGVS protein | Variant type | Predicted effect | dbSNP ID | Genotype |
|---|---|---|---|---|---|---|---|---|
| Case 1 | MEN1 | 11 | c.1579C>T | p.Arg527* | snv | stop_gained | rs104894261 | Homo |
| Case 1 | DAXX | 6 | c.1178delA | p.Lys393fs | del | frameshift_variant | Homo | |
| Case 2 | MEN1 | 11 | c.245_259delACCTGTCTATCATCG | p.Asp82_Ile86del | del | disruptive_inframe_deletion | ||
| Case 2 | DAXX | 6 | c.329_330delCT | p.Ser110fs | del | frameshift_variant | ||
| Case 2 | ATRX | X | c.1558G>T | p.Val520Phe | snv | missense_variant | Homo | |
| Case 2 | MTOR | 1 | c.6625C>G | p.Leu2209Val | snv | missense_variant | Homo | |
| Case 2 | BRCA2 | 13 | c.3504G>T | p.Met1168Ile | snv | missense_variant | Het | |
| Case 3 | MEN1 | 11 | c.798+1G>A | snv | splice_donor_variant+intron_variant | Homo | ||
| Case 3 | DAXX | 6 | c.850C>T | p.Pro284Ser | snv | missense_variant | Homo | |
| Case 4 | TP53 | 17 | c.818G>T | p.Arg273Leu | snv | missense_variant | rs28934576 | Homo |
| Case 5 | MEN1 | 11 | c.981_1006delCCACTGTCGCA ACCGCAATGTGCGGG | p.Tyr327fs | del | frameshift_variant | ||
| Case 5 | DAXX | 6 | c.801_824delTAACAGGCGCAT TGAGCGGCTCAT | p.Asn268_Ile275del | del | disruptive_inframe_deletion |
PNETs, pancreatic neuroendocrine tumors; del, deletion; snv, single-nucleotide variant; Homo, homozygous; Het, heterozygous.
The gene expression levels of the drug molecular targets varied across all five cases, but similar trends were observed in a few of the genes. All five samples had high expression of somatostatin receptors (SSTRs) 1 and 2, consistent with their neuroendocrine diagnosis and 111In-labeled octreotide scan avidity. The core kinase of the mTOR pathway MTOR was expressed at low levels in cases 1, 2, and 4. Low expression of O-6-methylguanine-DNA methyltransferase (MGMT), which confers sensitivity to temozolomide in glioblastoma (Hegi et al. 2005), was observed in cases 1, 2, 3, and 5 (Fig. 3A).
High expression of neuroendocrine markers and pancreas-specific transcription factors confirmed the pancreatic origin of the metastases (Supplemental Data 3). In all five cases, high expression levels of CHGA and SYP were observed, and NEUROD1 and PDX1 were expressed at high levels in four cases. In addition, each case exhibited increased expression of at least one of INS, GCG, GAS, SST, or VIP, favoring a pancreatic endocrine cell origin.
Case-Specific Molecular Alterations of Interest
Case 1
In addition to many of the shared molecular alterations, low expression levels of tumor suppressor genes TP53 and VHL were observed in this case. Several RTKs and targets of sunitinib, including FLT1 and KDR that encode for vascular endothelial growth factor receptors 1 and 2, respectively, were expressed at high levels (Fig. 3A). Moreover, several MAPK pathway genes including MAP3K10, MAP3K12, and MAP2K2 were highly expressed suggesting up-regulated activity of MAPK pathways (Supplemental Data 3). The use of sunitinib as second-line therapy was supported by the high expression of several RTKs; however, treatment response was not assessable because of early discontinuation of therapy.
Case 2
Contrary to the other four cases where somatic mutations were only identified in MEN1 and DAXX, additional somatic mutations in genes associated with PNETs were identified in Case 2. These include a heterozygous missense mutation in BRCA2 and a homozygous missense mutation in ATRX, both of unknown significance (Table 2). Damaging effects of missense mutations prediction using PolyPhen-2 (Adzhubei et al. 2010) indicated that the BRCA2 variant was likely benign (score = 0.000), whereas the ATRX variant was likely damaging (score = 0.999). In addition, missense mutations resulting in amino acid substitutions adjacent to this variant (S519 and P521) have been reported and predicted to be pathogenic (http://cancer.sanger.ac.uk; COSM5878330, COSM5878331, COSM4993581, COSM4993582) (Forbes et al. 2015). Collectively, these predictions suggest that the identified mutation in ATRX likely resulted in loss of function. Mutations in DAXX and ATRX are typically mutually exclusive, and to our knowledge, this is the first report of concurrent homozygous DAXX and ATRX loss. Examination of allelic frequencies of DAXX and ATRX suggests that both variants were present in all tumor cells (Supplemental Data 1). The identified missense mutation in MTOR resulted in an amino acid substitution in the kinase domain of MTOR and had been previously characterized as an activating mutation that results in constitutively active mTOR signaling (Yamaguchi et al. 2015).
Case 3
In keeping with the clinical history of tuberous sclerosis complex, the TSC2 gene had a germline one-copy loss, and a somatic loss of the remaining copy, which caused complete loss of TSC2 with very low expression (0 percentile) in the tumor. The germline loss of TSC2 was related to structural variants in Chromosome 16, which included a large deletion spanning TSC2 and the base-excision repair gene NTHL1, and an inversion that caused NTHL1-TRAF7 fusion (Supplemental Fig. 1), predicted to be nonfunctional. Analysis of previously described mutational signatures (Alexandrov et al. 2013) showed an unusually high contribution of Signature 30, characterized by a high proportion of C>T transitions (Supplemental Fig. 2). Similar mutational profiles have been recently reported in NTHL1-mutated tumors related to polyposis syndromes (Rivera et al. 2015; Weren et al. 2015) and in NTHL1 knockout human colon organoids (Drost et al. 2017) but have not previously been described in PNETs. The tumor mutational burden was the highest among G1-2 PNETs in this cohort, with 7.47 somatic mutations per megabase versus 1.17–2.17 and C>T transition in 45% (77/170) of the coding variants.
Case 4
Unlike the other described PNET cases, LOH events and copy-number aberrations were infrequent in case 4 (Fig. 2). Moreover, no mutations were observed in MEN1, ATRX, and DAXX. Instead, the key genomic aberrations included a homozygous pathogenic TP53 mutation (COSM10779), copy-number loss, and a gene fusion event involving APC and MYCN amplification. Both TP53 and APC were expressed at low levels (Fig. 3A), consistent with the genomic aberrations. MYCN had a 38-copy gain and was overexpressed (99 percentile; 48-fold-change relative to the BodyMap compendium expression). Gene expression analyses predicted activation of MYCN (Fig. 3B) and identified enrichment of genes encoding ribosomal proteins (Supplemental Data 2), which are known to be regulated by MYCN (Boon et al. 2001).
In view of the unusual genomic findings, pathology review was undertaken and confirmed the diagnosis of G2 PNET in the liver biopsy samples taken at diagnosis and after progression on chemotherapy and Y-90. In particular, there was no evidence of transformation to a G3 NEC nor evidence of glandular or acinar components that might suggest an alternate diagnosis.
Case 5
Although case 5 was histologically diagnosed as a G3 NEC, its genomic landscape was similar to that of G1-2 PNETs in cases 1, 2, and 3. These include similar regions with LOH events (Fig. 2A) and copy-number aberrations (Fig. 2B), as well as biallelic loss of MEN1 and DAXX (Fig. 3A). Gene expression analyses suggested a number of transcription factors and receptors were uniquely affected in this case. The cell cycle regulator cyclin D1 was predicted to be activated (Fig. 3B), consistent with enrichment of genes involved in cell cycle pathway (Supplemental Data 2).
DISCUSSION
In this case series, we describe genomic and transcriptomic analyses of liver metastases from five metastatic PNET cases (1 G1 PNET, 3 G2 PNETs, and 1 G3 NEC). To our knowledge, this is the first study correlating response to systemic therapy with whole-genome and transcriptome analyses in PNET patients. Previous studies on resected primary PNETs have identified recurrent somatic mutations in MEN1, DAXX/ATRX, and mTOR pathway genes and germline mutations in genes involved in chromatin remodeling, DNA damage repair, mTOR signaling, and telomere maintenance (Jiao et al. 2011; Scarpa et al. 2017). Consistent with these reports, we identified biallelic loss of MEN1 and DAXX (due to deleterious mutations, copy-number losses, and/or LOH events) in four of our five cases and low mRNA expression of MEN1, DAXX, and TSC2 in all five cases. These likely contributed to tumor development and progression. In addition, one or more cases exhibited low expression levels of tumor suppressor genes CHEK2, CDKN1B, NF1, and VHL, in which germline mutations have been reported in PNETs (Scarpa et al. 2017). Although we only identified one germline deletion affecting TSC2 within our cohort (Case 3), the reduced expression of these genes likely also augmented disease development and progression.
Recent studies suggest that loss of nuclear DAXX/ATRX staining is associated with metastases, poor prognostic features, and shorter survival (Marinoni et al. 2014; Singhi et al. 2017). Although we are unable to make any definite conclusions about the prognostic impact of DAXX/ATRX in our cohort, it is clear that treatment and survival outcomes in PNET are highly variable, and other drivers are likely to contribute to disease progression and treatment response.
For example, we identified a patient (Case 4) with typical pathologic features of a G2 PNET, but unique genomic characteristics. The MYCN amplification and expression signature observed in this case have not previously been described in human PNETs, although ectopic targeted expression of MYCN in pancreatic islets or neural progenitor cells is sufficient to induce PNET development in zebrafish and mice, respectively (Yang et al. 2004; Fielitz et al. 2016). MYCN has been shown to promote cell proliferation, and MYCN amplification has been correlated with poor prognosis, particularly in neuroblastomas (Huang and Weiss 2013). Similarly, the clinical picture for this case was characterized by treatment resistance and poor survival that may have been compounded by TP53 loss, which may have negated the pro-apoptotic signals elicited by MYCN and augmented its oncogenic potential (Chen et al. 2010; Gamble et al. 2012). Of interest, MYC-targeting small interfering RNA is being investigated as a potential therapeutic strategy in PNET, where treatment responses observed in a phase I trial (Tolcher et al. 2015) have led to cohort expansion in this tumor type (NCT02110563). In this small cohort, MYC expression did not appear to correlate with treatment response; however, it would be important to explore any underlying molecular characteristics that may predict benefit to MYC-targeted therapy.
Conversely, a second G2 PNET (Case 2) has had relatively good treatment outcomes, with prolonged responses to sunitinib (15 mo) and capecitabine with temozolomide (no progression after 20 mo). We observed low expression of MGMT in the tumor, which has previously been described and correlated with better responses to temozolomide in small PNET cohorts (Kulke et al. 2009; Cros et al. 2016). Three other patients in our cohort (Cases 1, 3, and 5) also had low MGMT expression but none were treated with temozolomide, whereas the one patient with normal MGMT expression (Case 4) did not respond to temozolomide.
Alterations and aberrant expression of mTOR pathway genes have been frequently reported in PNETs (Missiaglia et al. 2010; Jiao et al. 2011), and the mTOR inhibitor everolimus is well-established as effective therapy in G1-2 PNETs (Yao et al. 2011). However, no biomarkers that may predict for response to everolimus have yet been validated. Within our cohort, three patients received everolimus, and Case 3 had an early and sustained response to treatment, which would be supported by genomic findings of TSC2 inactivation. Decreased expression of TSC2 and/or PTEN was observed in the other two patients (Cases 1 and 4); however, no conclusions can be made regarding the impact of these findings on everolimus response.
Previous studies have reported genomically distinct findings that differentiate pancreatic NETs from poorly differentiated NECs, with the latter frequently associated with TP53 and RB1 alterations and rarely with DAXX and ATRX loss (Yachida et al. 2012). Conversely, in a study comparing genomic characteristics of NECs from different sites, the majority of patients with pancreatic NECs did not harbor alterations in TP53 or RB1 (Bergsland et al. 2016). The poorly differentiated G3 NEC in this study (Case 5) did not have TP53 and RB1 alterations but retained the pathognomonic mutations of well-differentiated PNETs, raising the hypothesis that it may have evolved from a PNET, and the patient would have been a candidate for therapy with targeted molecular therapy or temozolomide rather than platinum therapy, which did not induce a response. Other genomic features in this case included a higher mutation burden and gene expression profile indicative of cell cycle activation that was not seen in the other cases (Fig. 3B; Supplemental Data 2).
Of interest, the higher than expected mutation burden in a G2 tumor (Case 3), coupled with an unusual mutational signature, drew our attention to the germline fusion event causing NTHL1 loss. Germline mutations in NTHL1 have recently been described in the context of polyposis syndromes (Rivera et al. 2015; Weren et al. 2015), similar to those associated with MUTYH loss, another base-excision repair gene. Analogous to our case, MUTYH loss has also been associated with a novel mutational signature in PNET (Scarpa et al. 2017), highlighting the potential utility of examining mutational signatures to determine mechanisms of tumorigenesis in PNET. To our knowledge, this is the first reported PNET case associated with NTHL1 loss and a somatic mutation pattern that resembles signature 30.
The clinical significance of these molecular changes remains poorly understood, and to date, there are no known prognostic or predictive biomarkers that are applicable in clinical practice. Aside from one patient in whom high expression of several RTKs and components of the MAPK pathway supported the use of sunitinib, genomic findings generally did not inform therapy decisions in real time. This may partly have been due to biopsy timing and relatively short disease course in three of the patients, as retrospective review suggests that the genomic results could potentially have informed treatment decisions. For the two patients who remain on follow-up, high expression of SSTR2 would support the use of peptide receptor radionuclide therapy (PRRT) with radiolabeled somatostatin analogs (Hicks et al. 2017).
PNETs are associated with highly heterogeneous clinical outcomes and treatment sensitivities. This in-depth whole-genome and transcriptome analyses of five cases demonstrates a number of shared and also unique molecular aberrations that contribute to this observed heterogeneity. Further molecular analyses of metastatic PNET with associated treatment and outcome information will be critical to improve our understanding of the prognostic and predictive implications of these molecular features.
METHODS
Sample Collection and Processing
Following informed consent, patients underwent image-guided metastatic biopsies as part of the Personalized OncoGenomics program of British Columbia (NCT 02155621). As per protocol, biopsies could be undertaken at any time up to disease progression on first-line systemic therapy. Up to five biopsy cores were obtained using 18-22G biopsy needles and embedded in optimal cutting temperature (OCT) compound. Tumor sections were reviewed by a pathologist to confirm the diagnosis, evaluate tumor content and cellularity, and select areas most suitable for DNA and RNA extraction. Peripheral venous blood samples were obtained at the time of biopsy and leukocytes isolated for use as a germline reference.
Nucleic Acid Extraction and Library Construction
DNA and RNA were extracted for genomic and transcriptomic library construction, which has been previously described in detail (Sheffield et al. 2015).
Sequencing and Bioinformatics
Paired-end reads were generated on an Illumina HiSeq2500 sequencer and aligned to the human reference genome (GSCh37, available from http://www.bcgsc.ca/downloads/genomes/9606/hg19/1000genomes/bwa_ind/genome) by the BWA aligner (v0.5.7) (Li and Durbin 2010). Sequencing read lengths were 125 bp. Somatic single-nucleotide variants (SNVs) and small insertions/deletions were processed using SAMtools (Li et al. 2009) and Strelka (v0.4.6.2) (Saunders et al. 2012). Regions of copy-number aberration (CNA) were determined using CNASeq (v0.0.6) and LOH by APOLLOH (v0.1.1) (Ha et al. 2012). Tumor content and ploidy models were estimated from sequencing data through analysis of the CNA ratios and allelic frequencies of each chromosome. This was then compared to in-house theoretical models for different ploidy at various tumor contents. Tumor content and sequencing coverage for each case are provided in Table 3. Structural variation was detected by de novo assembly of tumor reads using ABySS and Trans-ABySS (Robertson et al. 2010), followed by variant discovery using DELLY (Rausch et al. 2012). RNA-seq reads were processed and gene expression analyzed as previously described in Sheffield et al. (2015).
Table 3.
Tumor content and sequencing coverage
| Case ID | Tumor genome sequencing coverage | Normal genome sequencing coverage | RNA sequencing coverage (no. of paired reads) | Tumor content (%) |
|---|---|---|---|---|
| Case 1 | 78× | 41× | 192M | 79 |
| Case 2 | 94× | 47× | 246M | 79 |
| Case 3 | 89× | 42× | 289M | 77 |
| Case 4 | 87× | 42× | 187M | 79 |
| Case 5 | 91× | 43× | 116M | 86 |
Gene Expression Analysis
In the absence of matched normal transcriptome data, the level of expression of each gene was determined as the number of reads per kilobase of transcript per million mapped reads (RPKM) and compared to a compendium of 16 normal transcriptomes from the Illumina BodyMap 2.0 project (https://www.ebi.ac.uk/gxa/experiments/E-MTAB-513/Results). Differential expression analysis between tumor and the normal compendium was performed as previously described (Sheffield et al. 2015). Up-regulated genes were defined as those with a fold change (FC) > 4 and a false-discovery rate-adjusted p-value < 0.05; down-regulated genes were defined as those with FC < −2 and a p-value < 0.1. Expression levels of select genes were converted into percentile ranks against a compendium of 5976 tumor transcriptomes across 25 cancer types from The Cancer Genome Atlas project. KEGG signaling pathway gene set enrichment analysis was performed using the Bioconductor (http://bioconductor.org) gage package (Luo et al. 2009). Upstream regulator analysis was performed using Ingenuity Pathway Analysis to predict the activation/inhibition states of G-protein-coupled receptors, ligand-dependent nuclear receptors, transcription regulators, kinases, and phosphatases from the list of differentially expressed genes (Krämer et al. 2014). To increase prediction confidence, only candidates with activation scores of >3 or <−3 were selected. Molecular targets of drugs mentioned in this study were retrieved from Santos et al. (2017).
Sequencing Result Visualization
LOH events and CNAs were illustrated using the Bioconductor package GenVisR (Skidmore et al. 2016). Heatmaps were generated using the Bioconductor package ComplexHeatmap (Gu et al. 2016) and R package gplots.
ADDITIONAL INFORMATION
Data Deposition and Access
DNA and RNA sequencing data have been deposited in the European Genome-phenome Archive (EGA) as part of the study EGAS00001001159, accession IDs EGAD00001003048, EGAD00001003089, EGAD00001002591, EGAD00001003069 and EGAD00001002607. The variants reported have been deposited in the ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) database with the following accession numbers: MEN1 c.1579C>T (SCV000611142), c.245_259delACCTGTCTATCATCG (SCV000611144), c.798+1G>A (SCV000611149), c.981_1006delCCACTGTCGCAACCGCAATGTGCGGG (SCV000611152); DAXX c.1178delA (SCV000611143), c.329_330delCT (SCV000611145), c.850C>T (SCV000611150), c.801_824delTAACAGGCGCATTGAGCGGCTCAT (SCV000611153); ATRX c.1558G>T (SCV000611146); MTOR c.6625C>G (SCV000611147); BRCA2 c.3504G>T (SCV000611148); TP53 c.818G>T (SCV000611151).
Ethics Statement
Patients provided written informed consent for metastatic biopsies, sequencing, and publication of results as part of the Personalized OncoGenomics program of British Columbia (NCT 02155621, University of British Columbia Clinical Research Ethics Board approval no. H12-00137).
Acknowledgments
We thank the patients and their families for participating in the Personalized OncoGenomics Program of BC.
Author Contributions
H.W., K.C.Y., Y.S., D.F.S., S.M.G., and D.J.R. contributed to study concept and design. H.W., K.C.Y., Y.S., E.Y.Z., J.M.L., H.F.K., S.E.K., J.M.K., H.J.L., A.J.M., X.F., J.M.D., K.S., C.Z., A.K., S.J.M.J., J.L., M.A.M., D.F.S., S.M.G., and D.J.R. contributed to acquisition, analysis, or interpretation of data. D.F.S. contributed to pathology oversight and interpretation. K.S., J.L., M.A.M., D.F.S., S.M.G., and D.J.R. contributed to study supervision. All authors have read and approved the final manuscript.
Funding
This work was supported by the British Columbia Cancer Foundation (BCCF) and the 2017 Neuroendocrine Tumor Research Foundation-AACR Grant, grant number 17-60-33-GORS.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. 2013. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 3: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland EK, Roy R, Stephens P, Ross JS, Bailey M, Olshen A. 2016. Genomic profiling to distinguish poorly differentiated neuroendocrine carcinomas arising in different sites. J Clin Oncol doi: 10.1200/JCO.2016.34.15_suppl.4020. [DOI] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus M-C, van Sluis P, Roobeek I, Weis I, Voûte PA, Schwab M, Versteeg R. 2001. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 20: 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Iraci N, Gherardi S, Gamble LD, Wood KM, Perini G, Lunec J, Tweddle DA. 2010. p53 is a direct transcriptional target of MYCN in neuroblastoma. Cancer Res 70: 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coriat R, Walter T, Terris B, Couvelard A, Ruszniewski P. 2016. Gastroenteropancreatic well-differentiated grade 3 neuroendocrine tumors: review and position statement. Oncologist 21: 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros J, Hentic O, Rebours V, Zappa M, Gille N, Theou-Anton N, Vernerey D, Maire F, Levy P, Bedossa P, et al. 2016. MGMT expression predicts response to temozolomide in pancreatic neuroendocrine tumors. Endocr Relat Cancer 23: 625–633. [DOI] [PubMed] [Google Scholar]
- Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, et al. 2017. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358: 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. 2009. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45: 228–247. [DOI] [PubMed] [Google Scholar]
- Fielitz K, Althoff K, De Preter K, Nonnekens J, Ohli J, Elges S, Hartmann W, Kloppel G, Knosel T, Schulte M, et al. 2016. Characterization of pancreatic glucagon-producing tumors and pituitary gland tumors in transgenic mice overexpressing MYCN in hGFAP-positive cells. Oncotarget 7: 74415–74426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. 2015. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 43: D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble LD, Kees UR, Tweddle DA, Lunec J. 2012. MYCN sensitizes neuroblastoma to the MDM2-p53 antagonists Nutlin-3 and MI-63. Oncogene 31: 752–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849. [DOI] [PubMed] [Google Scholar]
- Ha G, Roth A, Lai D, Bashashati A, Ding J, Goya R, Giuliany R, Rosner J, Oloumi A, Shumansky K, et al. 2012. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res 22: 1995–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfdanarson TR, Rabe KG, Rubin J, Petersen GM. 2008. Pancreatic neuroendocrine tumors (PNETs): incidence, prognosis and recent trend toward improved survival. Ann Oncol 19: 1727–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. 2005. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352: 997–1003. [DOI] [PubMed] [Google Scholar]
- Hicks RJ, Kwekkeboom DJ, Krenning E, Bodei L, Grozinsky-Glasberg S, Arnold R, Borbath I, Cwikla J, Toumpanakis C, Kaltsas G, et al. 2017. ENETS consensus guidelines for the standards of care in neuroendocrine neoplasia: peptide receptor radionuclide therapy with radiolabelled somatostatin analogues. Neuroendocrinology 105: 295–309. [DOI] [PubMed] [Google Scholar]
- Huang M, Weiss WA. 2013. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med 3: a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RT, Berna MJ, Bingham DB, Norton JA. 2008. Inherited pancreatic endocrine tumor syndromes: advances in molecular pathogenesis, diagnosis, management, and controversies. Cancer 113: 1807–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331: 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra DS, Modlin IR, Coppola D, Lloyd RV, Suster S. 2010. The pathologic classification of neuroendocrine tumors: a review of nomenclature, grading, and staging systems. Pancreas 39: 707–712. [DOI] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J, Tugendreich S. 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulke MH, Hornick JL, Frauenhoffer C, Hooshmand S, Ryan DP, Enzinger PC, Meyerhardt JA, Clark JW, Stuart K, Fuchs CS, Redston MS. 2009. O6-methylguanine DNA methyltransferase deficiency and response to temozolomide-based therapy in patients with neuroendocrine tumors. Clin Cancer Res 15: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. 2009. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinoni I, Kurrer AS, Vassella E, Dettmer M, Rudolph T, Banz V, Hunger F, Pasquinelli S, Speel EJ, Perren A. 2014. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology 146: 453–460. [DOI] [PubMed] [Google Scholar]
- Missiaglia E, Dalai I, Barbi S, Beghelli S, Falconi M, della Peruta M, Piemonti L, Capurso G, Di Florio A, dell Fave G, et al. 2010. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. J Clin Oncol 28: 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera B, Castellsague E, Bah I, van Kempen LC, Foulkes WD. 2015. Biallelic NTHL1 mutations in a woman with multiple primary tumors. N Engl J Med 373: 1985–1986. [DOI] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. 2010. De novo assembly and analysis of RNA-seq data. Nat Methods 7: 909–912. [DOI] [PubMed] [Google Scholar]
- Sangoi AR, Ohgami RS, Pai RK, Beck AH, McKenney JK, Pai RK. 2011. PAX8 expression reliably distinguishes pancreatic well-differentiated neuroendocrine tumors from ileal and pulmonary well-differentiated neuroendocrine tumors and pancreatic acinar cell carcinoma. Mod Pathol 24: 412–424. [DOI] [PubMed] [Google Scholar]
- Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP. 2017. A comprehensive map of molecular drug targets. Nat Rev Drug Discov 16: 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. 2012. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28: 1811–1817. [DOI] [PubMed] [Google Scholar]
- Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, Lawlor RT, Johns AL, Miller DK, Mafficini A, et al. 2017. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 543: 65–71. [DOI] [PubMed] [Google Scholar]
- Sheffield BS, Tinker AV, Shen Y, Hwang H, Li-Chang HH, Pleasance E, Ch'ng C, Lum A, Lorette J, McConnell YJ, et al. 2015. Personalized oncogenomics: clinical experience with malignant peritoneal mesothelioma using whole genome sequencing. PLoS One 10: e0119689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhi AD, Liu T, Roncaioli JL, Cao D, Zeh HJ, Zureikat AH, Tsung A, Marsh JW, Lee KK, Hogg ME, et al. 2017. Alternative lengthening of telomeres and loss of DAXX/ATRX expression predicts metastatic disease and poor survival in patients with pancreatic neuroendocrine tumors. Clin Cancer Res 23: 600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skidmore ZL, Wagner AH, Lesurf R, Campbell KM, Kunisaki J, Griffith OL, Griffith M. 2016. GenVisR: Genomic Visualizations in R. Bioinformatics 32: 3012–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LH, Basturk O, Sue JJ, Klimstra DS. 2016. A practical approach to the classification of WHO grade 3 (G3) well-differentiated neuroendocrine tumor (WD-NET) and poorly differentiated neuroendocrine carcinoma (PD-NEC) of the pancreas. Am J Surg Pathol 40: 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolcher AW, Papadopoulos KP, Patnaik A, Rasco DW, Martinez D, Wood DL, Fielman B, Sharma M, Janisch LA, Brown BD, et al. 2015. Safety and activity of DCR-MYC, a first-in-class dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. J Clin Oncol doi: 10.1200/jco.2015.33.15_suppl.11006. [DOI] [Google Scholar]
- Weren RD, Ligtenberg MJ, Kets CM, de Voer RM, Verwiel ET, Spruijt L, van Zelst-Stams WA, Jongmans MC, Gilissen C, Hehir-Kwa JY, et al. 2015. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet 47: 668–671. [DOI] [PubMed] [Google Scholar]
- Yachida S, Vakiani E, White CM, Zhong Y, Saunders T, Morgan R, de Wilde RF, Maitra A, Hicks J, Demarzo AM, et al. 2012. Small cell and large cell neuroendocrine carcinomas of the pancreas are genetically similar and distinct from well-differentiated pancreatic neuroendocrine tumors. Am J Surg Pathol 36: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Kawazu M, Yasuda T, Soda M, Ueno T, Kojima S, Yashiro M, Yoshino I, Ishikawa Y, Sai E, Mano H. 2015. Transforming somatic mutations of mammalian target of rapamycin kinase in human cancer. Cancer Sci 106: 1687–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Kutok JL, Lee NH, Piao HY, Fletcher CDM, Kanki JP, Look AT. 2004. Targeted expression of human MYCN selectively causes pancreatic neuroendocrine tumors in transgenic zebrafish. Cancer Res 64: 7256–7262. [DOI] [PubMed] [Google Scholar]
- Yao JC, Shah MH, Ito T, Bohas CL, Wolin EM, Van Cutsem E, Hobday TJ, Okusaka T, Capdevila J, de Vries EG, et al. 2011. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med 364: 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.