Abstract
The ability to predict transcription factors based on sequence information in regulatory elements is a key step in systems-level investigation of transcriptional regulation. Here, we have developed a novel tool, IMAGE, for precise prediction of causal transcription factors based on transcriptome profiling and genome-wide maps of enhancer activity. High precision is obtained by combining a near-complete database of position weight matrices (PWMs), generated by compiling public databases and systematic prediction of PWMs for uncharacterized transcription factors, with a state-of-the-art method for PWM scoring and a novel machine learning strategy, based on both enhancers and promoters, to predict the contribution of motifs to transcriptional activity. We applied IMAGE to published data obtained during 3T3-L1 adipocyte differentiation and showed that IMAGE predicts causal transcriptional regulators of this process with higher confidence than existing methods. Furthermore, we generated genome-wide maps of enhancer activity and transcripts during human mesenchymal stem cell commitment and adipocyte differentiation and used IMAGE to identify positive and negative transcriptional regulators of this process. Collectively, our results demonstrate that IMAGE is a powerful and precise method for prediction of regulators of gene expression.
Genome-wide mapping of transcriptional enhancers and assessment of their activity under different conditions constitutes a powerful first step in the analysis of transcriptional networks. The sequence information in the enhancers that change activity can subsequently be used to predict transcriptional regulators involved in mediating the transcriptional response. In this context, the sequence specificity, or motif, of a transcription factor is typically represented by a position weight matrix (PWM), which is a mathematical model that describes the log-likelihood of a transcription factor to bind to any DNA sequence.
The first generation of methods to predict transcription factors involved in a particular transcriptional response utilizes these PWMs to predict binding sites in enhancers in the vicinity of regulated genes. These methods range from simple PWM matching (Kel et al. 2003; Heinz et al. 2010; Grant et al. 2011; Gama-Castro et al. 2016; Tan and Lenhard 2016) to much more complex modeling-based approaches (Pique-Regi et al. 2011; Zhong et al. 2013; Sherwood et al. 2014; Kähärä and Lähdesmäki 2015; Jankowski et al. 2016; Chen et al. 2017). However, even with the best prediction of binding sites, the identification of causal transcription factors based only on motif enrichment in regulated enhancers is generally associated with a high false-positive rate, thereby leaving experimentalists with too many candidate factors to investigate. The false-positive rate can be significantly reduced by integrating gene expression data, such that only factors with a change in mRNA expression corresponding to the change in enhancer activity at predicted binding sites pass through the filter (Schmidt et al. 2016). However, this clearly biases the analysis toward transcription factors whose activity is regulated primarily by the expression level.
Recently, a new method, ‘Motif Activity Response Analysis’ (MARA), for prediction of transcription factors involved in gene regulation has been developed. The core function of MARA is to model the contribution of a specific motif to gene expression, the so-called ‘motif activity,’ based on motif occurrence and gene expression data using a machine learning approach (The FANTOM Consortium & Riken Omics Science Center 2009; Balwierz et al. 2014). This represents a significant improvement over traditional methods, as the identification of causal transcription factors in MARA is not based on motif enrichment analysis but a direct prediction of the effect of a motif. However, there are still several limitations of this method. First, MARA does not allow for integration of gene expression and enhancer activity (determined by, e.g., MED1 occupancy) or accessibility data (determined by, e.g., DNase I sensitivity). Instead, it uses motif occurrence within promoter regions to model the contribution of motifs to gene expression or, alternatively, motif occurrence within enhancer regions to model the contribution of motifs to enhancer activity. Thus, it cannot be used to predict transcription factors causal for gene expression based on sequence information in enhancers. Second, the identification of motifs is based on PWM matching using a log-likelihood scoring scheme and a limited database of PWMs. This approach may be biased because the precision of the log-likelihood scoring scheme is biased by the length and the complexity of the PWMs (Dabrowski et al. 2015), which means that the method is not equally sensitive toward all PWMs.
Here, we present a novel tool termed ‘Integrated analysis of Motif Activity and Gene Expression changes of transcription factors’ (IMAGE), which overcomes many of the current limitations associated with PWM-based prediction of causal transcription factors. To build IMAGE, we collated a large number of publicly available PWMs, implemented solutions to predict binding specificities of transcription factors with no known PWM, and evaluated different methods for PWM scoring. Based on this compendium of PWMs and enhancer and gene expression data, IMAGE uses machine learning to predict causal transcription factors, as well as their binding sites and target genes, with high confidence. Thus, IMAGE represents a powerful, precise, and novel method for unbiased prediction of key transcription factors driving specific gene programs.
Results
Creation of an extended PWM database
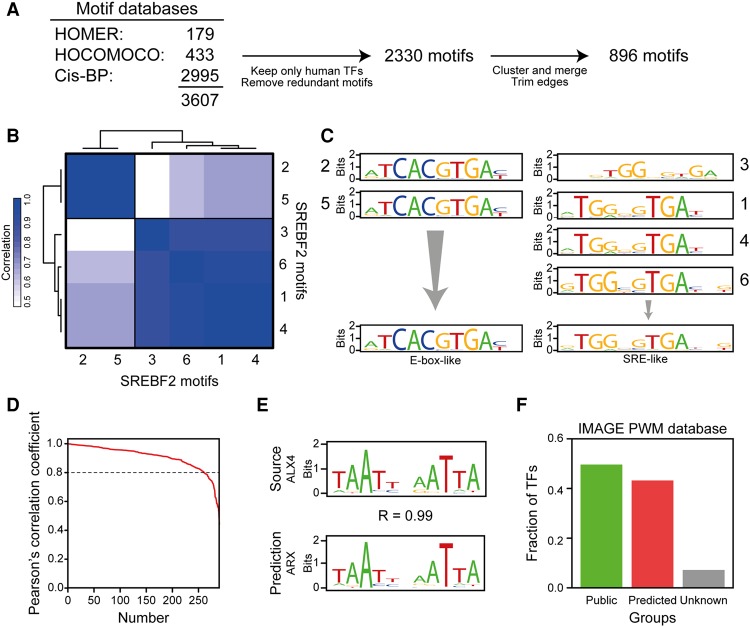
In the current consensus annotation, the human genome is predicted to contain approximately 1447 transcription factors (Wingender et al. 2013; Zhang et al. 2015). A comprehensive search through the published motif databases revealed a total of 3607 experimentally determined position weight matrices (Heinz et al. 2010; Weirauch et al. 2014; Kulakovskiy et al. 2016). These PWMs were collapsed to 2330 by removal of redundant PWMs, as well as removal of PWMs of transcription factors with no human paralogs (Fig. 1A). Since there are more nonredundant PWMs than there are transcription factors, the binding specificity of some transcription factors must be described by several PWMs. Manual inspection revealed that, although they are not redundant, many PWMs for a specific transcription factor are very similar, and therefore, they can be collapsed without significant loss of information. To distinguish between PWMs for a given factor that can be collapsed and those that represent distinct binding modes, we submitted the PWMs for each transcription factor to hierarchical clustering. This approach assumes that small differences in PWMs arise from technical variation, whereas large differences represent different binding modes. Consistent with this assumption, binding sites predicted only by a constituent motif, but not the collapsed motif itself, are not more associated with transcription factor binding than expected by random (Supplemental Fig. S1A). This suggests that genuine sequence specificity is not lost during motif collapsing. For example, the PWMs describing the binding specificity of sterol regulatory element binding transcription factor 2 (SREBF2) partition into two clusters (Fig. 1B). One of the clusters of PWMs is similar to an E-box-like motif, and the other is similar to the sterol-response element (SRE) (Fig. 1C). This is consistent with experimental data showing that SREBF2 binds to both E-box-like sequences and SREs (Amemiya-Kudo et al. 2002; Zeng et al. 2004). Clustering and merging of all PWMs yielded a total of 896 PWMs (Fig. 1A). Thus, the clustering approach reduces the complexity of the PWM library considerably (61.5%) but retains biologically meaningful differences in binding specificities.
Figure 1.
Construction of an extended PWM database. (A) Several motif databases were collated in IMAGE. The databases (listed in the figure) were combined, and only nonredundant motifs assigned to human transcription factors were kept. All motifs for a given transcription factor were compared by correlation using HOMER (Heinz et al. 2010). The correlation matrices were clustered by hierarchical clustering, and clustered motifs were merged using MATLIGN (Kankainen and Löytynoja 2007). The edges of the merged motifs were trimmed using MotIV (Mercier et al. 2011), and only motifs with a length ≥4 bp after trimming were included. (B) SREBF2 motifs fall into two distinct clusters. The heat map shows the clustering of the motifs (indicated by numbers 1 through 6) mapped to SREBF2. (C) SREBF2 motifs that cluster together are very similar, and each cluster represents a unique binding specificity. Each SREBF2 motif was visualized using seqLogo (http://bioconductor.org/packages/release/bioc/html/seqLogo.html). Each group of motifs corresponds to a cluster. (D) Pearson's correlation coefficient for each transcription factor-motif pair, ordered by the correlation coefficient, between the known motifs of the transcription factors and the motif predicted by DNA-binding domain alignment. Motifs were compared using HOMER (Heinz et al. 2010). (E) Comparison of the experimentally determined motif for ALX homeobox 4 (ALX4) with the motif predicted based on DBD similarity, the aristaless related homeobox (ARX) motif. (F) IMAGE contains at least one motif for the vast majority of transcription factors. The bars show the fraction of human transcription factors in the IMAGE database that has a publicly available motif, a motif predicted by IMAGE, or no motif information.
Extension of the PWM library by prediction
Of the 1447 putative transcription factors in the human genome, only 718 could be assigned to a PWM in the library of 896 collapsed PWMs. The majority (76.4%) of the 729 transcription factors with no PWM belong to the C2H2 class of transcription factors. The C2H2 class of transcription factors are particularly difficult to study with respect to DNA binding affinity, since these transcription factors, on average, contain around 10 DNA-binding domains (DBDs) that do not all contact DNA simultaneously (Najafabadi et al. 2015), and since the transcription factors belonging to this class are very diverse (Emerson and Thomas 2009; Stubbs et al. 2011). Recently, a powerful tool, ZifRC, that can predict the sequence specificity of these C2H2 transcription factors was developed (Najafabadi et al. 2015). We systematically analyzed all C2H2 transcription factors with no known motifs using this tool and added the resulting PWMs to our library. This means that IMAGE, in contrast to most other tools, is able to predict C2H2-class transcription factor involvement in controlling gene expression.
For the remaining transcription factors that could not be assigned to a PWM, we took advantage of the fact that transcription factors with closely related primary sequence of their DBDs tend to have similar sequence specificities (Berger et al. 2008; Noyes et al. 2008; Alleyne et al. 2009; Bernard et al. 2012; Weirauch et al. 2014; Zamanighomi et al. 2017). Therefore, PWMs of uncharacterized transcription factors can be predicted from the sequence specificity of transcription factors with similar DBDs. To do this, we first constructed a database of the primary amino acid sequences of the DBDs of all transcription factors with a known motif. Second, we matched the amino acid sequence of the DBD of all non-C2H2 transcription factors with no known motifs against this database using blast (Camacho et al. 2009) and inferred PWMs from the sequence specificity of the transcription factor with the highest DBD sequence similarity. Cross-validation of this approach for transcription factors with known PWMs demonstrated that the predicted PWM is highly similar (R ≥ 0.8) to the known PWM for more than 70% of the factors. Furthermore, all predictions have at least moderate correlation (R ≥ 0.5), and for the ones with only moderate correlation, the predicted PWM often displays a partial match to the true PWM (Fig. 1D,E). Importantly, by employing both PWM prediction and the ZifRC tool, 92.8% of all putative transcription factors in the human genome can be assigned to one or more PWMs in our extended library (Fig. 1F). To our knowledge, this constitutes the, to date, most complete PWM database.
Determination of uniform P-value-based thresholds
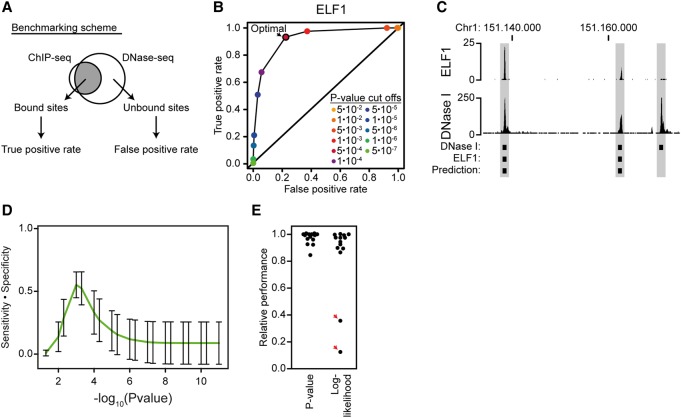
One of the caveats of PWM matching is that the commonly used log-likelihood scoring scheme is biased by the length and complexity of PWMs (Dabrowski et al. 2015). In principle, this means that a single log-likelihood threshold, where all PWMs predict binding sites with maximum sensitivity and specificity, does not exist. Alternative methods for scoring of PWM matches have been developed by others (Kel et al. 2003; Hertzberg et al. 2005; Touzet and Varré 2007; Pan 2008). Recently, P-value-based approaches have been adopted by large consortia, such as ENCODE (Kheradpour and Kellis 2014) and HOCOMOCO (Kulakovskiy et al. 2016). However, these approaches have, to our knowledge, not yet been formally benchmarked and are not incorporated in the commonly used tools for motif analysis. Here, we have applied and benchmarked a P-value-based approach that uses the score distribution to estimate thresholds for positive identification of individual PWMs (Touzet and Varré 2007). To test the validity of P-value-based scoring, we applied it to ENCODE data (The ENCODE Project Consortium 2012). Briefly, bound sites of a transcription factor were defined as the top 5000 peaks in a ChIP-seq experiment, and unbound background sites were defined as 20,000 random DNase I hypersensitive sites that do not overlap any sites with ChIP-seq signal (Fig. 2A). The false-negative and false-positive rates were estimated for several different P-value thresholds for PWMs matching for several transcription factor-motif combinations. As an example, prediction of ELF1 binding sites has optimal sensitivity (high true-positive rate) and specificity (high true-negative rate) at a P-value of 5 × 10−4 (Fig. 2B,C). Interestingly, this P-value threshold represents a local maximum, where all tested transcription factor-motif combinations, on average, have the maximum predictive performance (Fig. 2D). Similarly, we can also find a local maximum, where the transcription factor-motif combinations, on average, have the highest predictive performance using log-likelihood scoring (Supplemental Fig. S1B). However, the predictive power at the local maximum for the log-likelihood-based approach is lower than for the P-value-based approach, and the standard deviation is larger, indicating that the P-value-based approach, on average, performs better than the log-likelihood-based approach. Importantly, we find that at the local maxima of prediction power, all motifs perform close to their individual maximal power in the P-value-based approach, while there are significant outliers in the log-likelihood-based approach (Fig. 2E). This shows that the P-value-based method implemented in IMAGE has a single optimal threshold across many transcription factor-motif pairs and that, in contrast to the log-likelihood-based approach used by other methods, the P-value-based approach robustly scores PWMs with different lengths and complexity.
Figure 2.
P-value-based threshold determination has a local maximum of sensitivity and specificity. (A) Scheme for benchmarking motif thresholds. Fourteen ENCODE ChIP-seq and DNase-seq data sets (The ENCODE Project Consortium 2012) from three different cell types were used to validate the P-value-based approach for cut-off determination by precision-recall statistics. Bound sites were defined as the top 5000 strongest sites in ChIP-seq. Unbound sites were defined as 20,000 random DNase I-sensitive sites that do not overlap a ChIP-seq peak. (B) Representative example of predictions across many cut-offs. The plot shows the true-positive rate and false-positive rate for motif-based prediction of ELF1 binding sites at 11 different P-value cut-offs. (C) Visualization of a representative region containing both positive and negative prediction. The screenshot was generated using the UCSC Genome Browser (Kent et al. 2002). It shows ELF1 binding sites (ChIP-seq), predicted ELF1 sites, and DNase-seq. (D) There is a local maximum in sensitivity multiplied by specificity using P-value-based PWM scoring. The line shows averaged prediction statistics across all 14 transcription factors (as indicated in A) at different P-value cut-offs. Error bars represent the standard deviation. (E) There are no outliers in relative performance for the P-value-based approach. The plot shows the predictive performance (sensitivity multiplied by specificity) of each of the 14 motifs that were calculated based on their P-value-based cut-offs and their log-likelihood-based cut-offs. The relative performance is the predictive performance at the local maximum for either P-value-based or log-likelihood-based cut-offs divided by the best predictive performance across all either P-value-based or log-likelihood-based cut-offs. The red arrows indicate factor predictions with low relative performance at the local maximum.
IMAGE—a novel tool for prediction of causal transcription factors
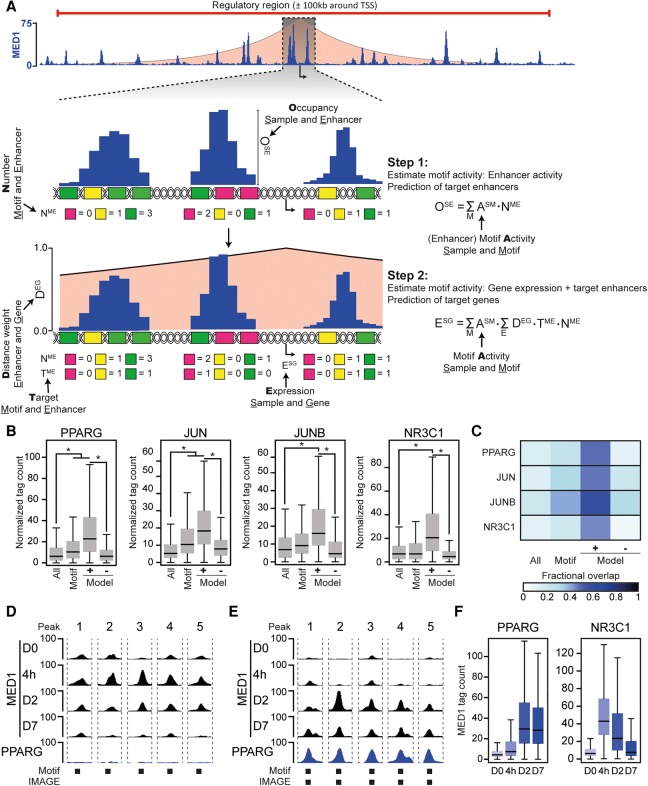
In order to fully utilize the power of the extended PWM database with a uniform P-value-based threshold, we designed a novel two-stage machine learning model that we termed IMAGE. To identify candidate transcription factors in IMAGE, the input sequences (i.e., regions that were predicted to be enhancers and promoters based on activating histone marks, chromatin accessibility, or cofactor binding) are scanned for motif occurrences using the extended PWM database and scored using the P-value-based approach. However, rather than looking for motif enrichment in a particular group of sites, the effect of a motif on gene expression, or the motif activity, is determined by modeling. This concept was recently pioneered in the Motif Activity Response Analysis tool, which predicts motif activity by modeling gene expression from promoter-based motifs (The FANTOM Consortium & Riken Omics Science Center 2009; Balwierz et al. 2014). In IMAGE, we have expanded this method to a two-stage modeling approach, that, in contrast to MARA, is not limited to promoters but models the effect of motifs on gene expression based on enhancer maps and integrates enhancer and gene expression data to arrive at highly precise predictions (Fig. 3A). In step 1, IMAGE identifies target enhancers of each motif. Here, IMAGE assumes that the enhancer activity, measured by occupancy, is defined by the sum of motif activities multiplied by the number of motif occurrences in each enhancer. In step 2, IMAGE leverages this information to model the effect of each motif on gene expression. Here, IMAGE assumes that the transcriptional output of a gene is defined by the sum of motif activities multiplied by the distance-weighted (weighted based on linear genomic distance between the enhancer and transcriptional start site such that distal enhancers are assigned a low weight and nearby enhancers a high weight [Wang et al. 2016]) sum of motif occurrences in all predicted target enhancers (from step 1) assigned to that gene. Thus, IMAGE allows for prediction of motif activity based on contribution to enhancer activity (step 1), as well as for prediction of motif activity based on contribution to gene expression (step 2).
Figure 3.
IMAGE predicts transcription factor binding sites with high confidence. The predictive power of IMAGE for identification of transcription factor binding sites was investigated using previously published data from 3T3-L1 preadipocyte differentiation (Siersbaek et al. 2011, 2017). (A) Schematic workflow of IMAGE. The shaded red area depicts the linear-distance association between enhancers and genes. The height illustrates the weight associated with each distance (y-axis on zoom-in). In step 1, the input sequences are scanned for motif occurrences using the P-value-based cut-off. Motifs that are only predicted to bind transcription factors that are expressed below one normalized tag per kilobase are filtered away. We define a variable M, which contains each remaining motif, a variable E which contains each enhancer, and a variable S which contains each sample. The motif activity of each remaining motif is determined using ridge regression to estimate ESM using the equation given in the figure. (OSE) Occupancy at enhancer E in sample S, (NME) number of motif M in enhancer E, (ESM) effect of motif M in sample S. Subsequently, target enhancers are predicted by leave-one-out analysis. In step 2, the effect of each motif on gene expression or motif activity is predicted using an additive model of transcriptional regulation and estimating ASM from the given equation. (DEG) Distance weight for enhancer E acting on gene G, (TME) target enhancer E of motif M, (NME) number of motif M at enhancer E, (ASM) motif activity of motif M in sample S. Subsequently, target genes are identified by leave-one-out analysis. (B,C) Validation of IMAGE binding site prediction during 3T3-L1 adipocyte differentiation using ChIP-seq. (B) Boxplots show the ChIP-seq occupancy of PPARG (6 d after induction of differentiation), JUN (4 h after induction of differentiation), JUNB (4 h after induction of differentiation), and NR3C1 (4 h after induction of differentiation) (Siersbaek et al. 2014) at either all putative enhancers defined by MED1 signal (all), all putative enhancers containing the respective transcription factor motif (motif), all enhancers predicted to be target enhancers of the transcription factor by IMAGE (Model+), or all enhancers predicted not to be target enhancers of the transcription factor by IMAGE (Model−). (*) denotes a significant (P ≤ 0.05) and nonnegligible effect (|Cohen's d| ≥ 0.2). (C) Heat map indicating the fraction of the MED1 binding sites defined in B that overlap with peak-detected sites based on ChIP-seq data. (D,E) Examples of MED1-bound enhancers with PPARG motifs that are unbound ([D] Peak 1: Chr 3: 151738674, Peak 2: Chr 8: 11273745, Peak 3: Chr 11: 51687839, Peak 4: Chr 2: 167420113, Peak 5: Chr 3: 41234389) or bound ([E] Peak 1: Chr 6: 144702629, Peak 2: Chr 6: 82598119, Peak 3: Chr 7: 29610142, Peak 4: Chr 11: 98447544, Peak 5: Chr 4: 108506609) by PPARG. Screenshots were generated using the USCS Genome Browser showing MED1 ChIP-seq data from day 0, 4 h, day 2, and day 7, and PPARG ChIP-seq data from day 6 following adipogenic stimulation of 3T3-L1 preadipocytes. Presence of PPARG motifs and IMAGE predicted binding sites is indicated below the screenshots. (F) Predicted enhancers of PPARG and NR3C1 have a temporal pattern of activation that is congruent with the activation of the transcription factors. The boxplots show the MED1 tag count at predicted target enhancers of PPARG and NR3C1 at the indicated time points during differentiation in 3T3-L1 cells.
IMAGE accurately infers target sites of transcription factors
To determine the performance of the transcription factor binding site prediction during step 1 of IMAGE, we used IMAGE to predict transcription factor binding in enhancers during differentiation of 3T3-L1 preadipocytes based on our previously published data for gene expression (RNA-seq) and enhancer marks (MED1 ChIP-seq and DNase-seq) (Siersbaek et al. 2011, 2017). Potential binding sites for peroxisome proliferator-activated receptor gamma (PPARG), Jun proto-oncogene (JUN), JunB proto-oncogene (JUNB), and glucocorticoid receptor (NR3C1) were predicted using IMAGE, and the predictions were validated by comparison with our previously published ChIP-seq data for these factors (Siersbaek et al. 2014). For all four tested transcription factors, the binding sites predicted by IMAGE (Fig. 3B, Model+) are significantly more occupied by the transcription factor compared to both the occupancy at all enhancers and to the occupancy at enhancers with motifs for the given transcription factor (Fig. 3B, Motif). Furthermore, enhancers with motifs, but not predicted to be transcription factor binding sites by IMAGE (Fig. 3B, Model–), are less occupied by the transcription factor than all enhancers with motifs and the binding sites predicted by IMAGE (Fig. 3B, Model+). Consistently, the binding sites predicted by IMAGE have the highest overlap with peak-detected sites for each transcription factor (Fig. 3C, Model+), and visual inspection of the data shows that IMAGE is capable of differentiating between unbound (Fig. 3D) and bound (Fig. 3E) sites among enhancers of similar strength with motifs. Finally, consistent with the expected activation profile of PPARG and NR3C1, their IMAGE-predicted target sites displayed maximal MED1 occupancy late and early in adipogenesis, respectively (Fig. 3F). In order to assess the ability of IMAGE to leverage different sources of enhancer marks, we compared predictions based on MED1 ChIP-seq data to those based on DNase-seq data. We find that there is a large overlap, which is significantly higher than expected at random, between predicted target sites using either MED1 ChIP-seq or DNase-seq data (Supplemental Fig. S1C). Taken together, these analyses demonstrate that IMAGE identifies binding sites with high confidence from different types of enhancer signatures (cofactor ChIP-seq and DNase-seq) and that the prediction of binding sites by IMAGE is accurate.
IMAGE predicts causal regulators and their target genes with high confidence
To determine the ability of IMAGE to predict transcription factors that play a causal role in mediating a given transcriptional response, we applied the full IMAGE pipeline to our gene expression data (RNA-seq) and enhancer data (MED1 ChIP-seq) during 3T3-L1 differentiation, as described above. IMAGE step 1 predicts a total of 484 motifs that are bound by 530 transcription factors, which have significant changes in motif activity, i.e., are predicted to contribute to changes in enhancer activity, during adipogenesis.
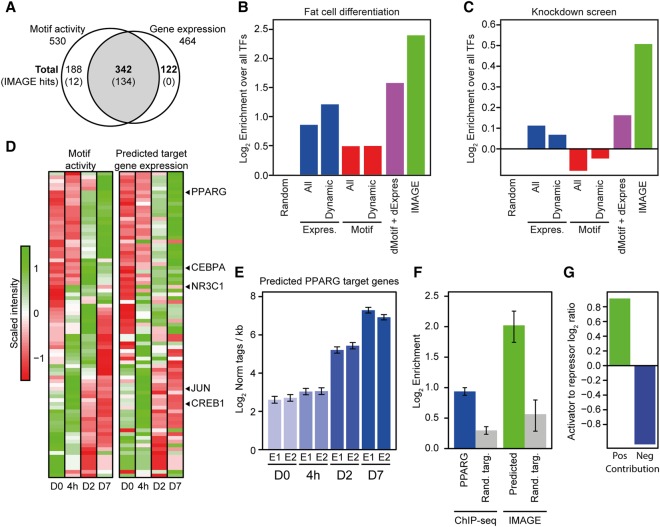
Out of these, 164 motifs (78 high confidence, 86 medium confidence) bound by 146 transcription factors (73 high confidence, 73 medium confidence) are identified by IMAGE step 2 as causal regulators of gene expression during 3T3-L1 differentiation. The majority of the transcription factors predicted to bind to these motifs (134/146, 91.8%) are both differentially expressed and have differential motif activities. The predicted transcription factors and their motifs constitute only 28.1% and 33.8% of all differentially expressed transcription factors and differentially regulated motif activities, respectively (Fig. 4A). This demonstrates that IMAGE predicts only a subset of the differentially regulated transcription factors to be causal for transcriptional regulation and that a significant proportion (8.2%) of the causal transcription factors are not differentially expressed during adipogenesis. In order to validate that the transcription factors identified using IMAGE play an important role in adipogenesis, we analyzed several sources of data, including GO terms, correlation between adipose tissue expression and BMI (Keller et al. 2011), a large-scale overexpression screen (Gubelmann et al. 2014), and a large-scale knockdown screen (Söhle et al. 2012). We find that the group of transcription factors predicted by IMAGE to be high confidence regulators of adipogenesis is highly enriched for transcription factors known to be involved in regulation of fat cell differentiation as defined by gene ontology (Fig. 4B). Furthermore, this group is highly enriched for transcription factors whose expression in adipose tissue correlates with BMI (Supplemental Fig. S1D) and which affect lipid accumulation upon both knockdown (Fig. 4C) and overexpression (Supplemental Fig. S1E). Notably, for the prediction of causal regulators of adipogenesis, IMAGE outperforms all other tested methods, including those focusing on differential expression of transcription factors combined with motif enrichment within dynamically regulated enhancers (Fig. 4B,C; Supplemental Fig. S1D,E). IMAGE achieves this high enrichment by having a higher precision compared to the other tested methods (Supplemental Fig. S1F). In a direct comparison between IMAGE and MARA, we find that both methods are useful for the identification of causal transcription factors but that IMAGE has higher precision than MARA, probably due to the more advanced model and data integration approach in IMAGE (Supplemental Fig. S1G). Overall, these results demonstrate that IMAGE offers more precise predictions of causal transcription factors than existing methods.
Figure 4.
IMAGE identifies regulators of adipogenesis with high confidence. The predictive power of IMAGE for identification of important transcriptional regulators was investigated using previously published data from 3T3-L1 preadipocyte differentiation (Siersbaek et al. 2011, 2014, 2017). (A) IMAGE predicts causal regulators of 3T3-L1 adipocyte differentiation, including many that are not differentially expressed during differentiation. The Venn diagram shows the overlap between transcription factors with a significant change in IMAGE motif activity based on MED1 ChIP-seq (n = 2) and RNA-seq (n = 2) during 3T3-L1 adipogenesis, and transcription factors with a significant change in gene expression (RNA-seq, n = 2) at any point during differentiation. The numbers represent all transcription factors that have significant changes in motif activity and/or gene expression. Numbers in parentheses indicate the subset identified by IMAGE as high or medium confidence causal transcription factors. (B,C) Comparison of the predictive power of IMAGE and various other methods for predicting transcriptional regulators of adipogenesis previously defined as belonging to the GO term ‘fat cell differentiation’ (B), or previously identified in a knockdown screen to affect lipid accumulation during 3T3-L1 adipocyte differentiation (Söhle et al. 2012) (C). The bar plot shows the predictive power of the indicated methods as determined by the enrichment of the predicted factors over all transcription factors. The different methods compared with IMAGE are: (1) random selection (Random); (2) all expressed transcription factors (≥1 log2-transformed read per kilobase) (Expres., All); (3) all expressed and differentially expressed transcription factors (≥1 log2-transformed read per kilobase and Padj ≤ 0.05) (Expres., Dynamic); (4) all transcription factors whose motif is significantly enriched in MED1-bound enhancers compared to genomic background (Motif, All); (5) transcription factors whose motif is significantly enriched in differentially regulated enhancers (Padj ≤ 0.05) compared to genomic background (Motif, Dynamic); and (6) integration of the groups containing dynamically expressed transcription factors and transcription factors with motif enrichment in differentially regulated enhancers compared to genomic background (dMotif + dExpress). (D) Motif activities recapitulate the transcriptional waves during adipocyte differentiation in 3T3-L1 cells. Heat map of motif activity of all motifs with significant changes in motif activity during 3T3-L1 differentiation (left panel) and average expression of predicted target genes of these motifs (right panel) at the different time points following induction of differentiation, day 0, 4 h, and day 2 and day 7. (E) Example showing the average mRNA expression of genes predicted by IMAGE to be regulated by PPARG. Bar plot shows average gene expression for target genes of two replicates (E1 and E2) during 3T3-L1 differentiation. (F) Comparison of prediction of PPARG target genes based on IMAGE and based on PPARG ChIP-seq. The bar plot shows the enrichment of predicted PPARG target genes (predicted using either PPARG ChIP-seq data [Siersbaek et al. 2011] or IMAGE) among the group of PPARG-dependent genes (Schupp et al. 2009). PPARG-dependent genes are defined as genes significantly regulated during adipogenesis in 3T3-L1 control cells (Padj ≤ 0.01) but with at least twofold less repression or induction upon knockdown of PPARG in mature 3T3-L1 adipocytes. The enrichment of IMAGE-predicted or ChIP-seq-predicted (PPARG peak within ±25 kb of the transcription start site) target genes is calculated by comparing the fraction of predicted target genes that are experimentally defined as PPARG-dependent relative to a randomized control fraction using size-matched randomized groups of target genes and dependent genes. Background enrichment was estimated by 1000 permutations of randomizing the predicted target genes and calculating the same enrichment. The error bars indicate the standard deviation across 1000 permutations. (G) Motif activity can be used to distinguish between transcription activators and repressors. The bar plot shows the log2 ratio between the Jaccard similarity coefficient between motifs with significantly positive (P ≤ 0.05, motif activity ≥ 0.005) motif activity at day 7 of differentiation and transcription factors only marked as activators in the UniProt annotation, or motifs with significantly negative (P ≤ 0.05, motif activity ≤ −0.005) motif activity at day 7 of differentiation and transcription factors only marked as repressors in the UniProt annotation (Apweiler et al. 2004).
Importantly, we find that the temporal changes in motif activity throughout adipocyte differentiation of 3T3-L1 preadipocytes for many transcription factors are consistent with the reported changes in the activity of these factors during adipogenesis. For example, motif activities of JUN, NR3C1, and cAMP responsive element binding protein 1 (CREB1) peak during early adipogenesis, corresponding to the time at which these factors have been reported to be most active (Zhang et al. 2004; Steger et al. 2010; Siersbaek et al. 2011). In contrast, motif activity of peroxisome proliferator activated receptor gamma and CCAAT/enhancer binding protein alpha (CEBPA) peaks during late adipogenesis (Fig. 4D, left panel), when these factors are reported to be maximally active (Cao et al. 1991; Tontonoz et al. 1994; Yeh et al. 1995). This indicates that IMAGE accurately predicts motif activities and their changes. Furthermore, we find that there is generally a strong correlation between the temporal profile of the motif activity, as determined by genome-wide expression patterns, and the expression level of the subset of genes predicted to be target genes (Fig. 4D, left and right panel). This is exemplified by predicted PPARG target genes that, on average, are induced during late stages of adipogenesis (Fig. 4E) concurrently with the activation of PPARG itself. Importantly, the predicted target genes of PPARG are highly enriched for biological pathways such as the ‘PPAR signaling pathway’ and pathways related to lipid metabolism (Supplemental Fig. S1H), and they are also highly enriched among genes with significantly blunted expression upon knockdown of PPARG in 3T3-L1 adipocytes (Fig. 4F). In comparison to target genes predicted based on PPARG ChIP-seq data (i.e., by proximity between PPARG peaks and transcription start sites), we find that the IMAGE-predicted PPARG target genes are ∼2.5-fold more likely to be significantly blunted by knockdown (Fig. 4F). Taken together, this shows that IMAGE predicts target genes with very high precision and outperforms ChIP-seq in terms of accuracy. Interestingly, we find that transcription factors with a significant and strongly positive motif activity in 3T3-L1 adipocytes are more often transcriptional activators than repressors, whereas transcription factors with a significant and strongly negative motif activity are more often transcriptional repressors than activators (Fig. 4G). This suggests that IMAGE may also be able to help researchers determine the effect of transcription factors on their target genes through comparison of the temporal profiles of motif activity and target gene expression.
IMAGE predicts novel transcriptional regulators with high accuracy
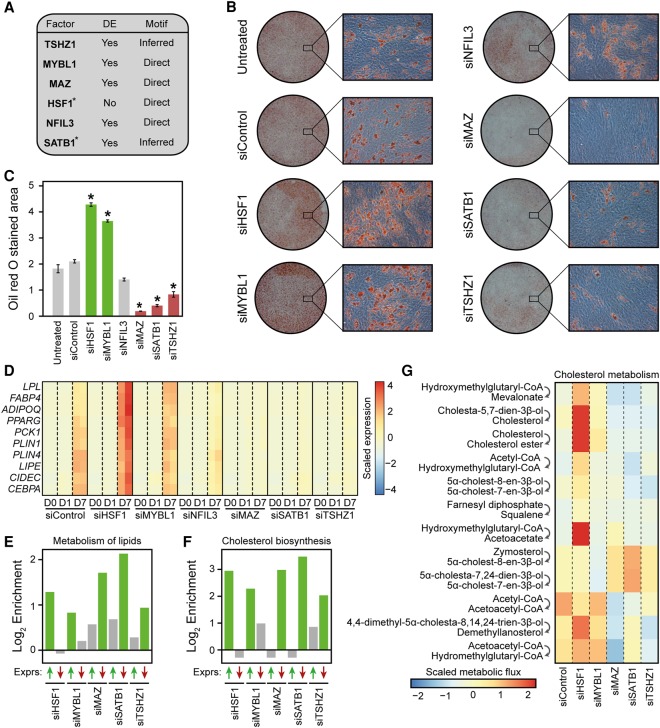
In order to further demonstrate the potential of IMAGE to predict causal transcription factors, we applied IMAGE to a much less characterized model system, the immortalized human mesenchymal stem cell (MSC) line, hMSC-TERT4 (Simonsen et al. 2002). The transcriptional network that drives the commitment and adipocyte differentiation of these multipotent stem cells is not well understood. Thus, we mapped enhancers and enhancer activity by MED1 ChIP-seq and transcriptional output by RNA-seq at different time points during commitment and differentiation of these cells, i.e., immediately prior to addition of differentiation cocktail (day 0), and at 4 h, 1 d, 3 d, 7 d, and 14 d after addition of the adipogenic cocktail. This data set provides insight into human adipocyte differentiation from MSC at hitherto unsurpassed temporal resolution. IMAGE identified 115 causal transcription factors (bound to 124 motifs) with high confidence and 122 additional transcription factors (bound to 125 motifs) with medium confidence. From this list of 237 transcription factors, we excluded all transcription factors which had previously been studied in the context of adipogenesis, and from the remaining transcription factors, we chose six transcription factors with different expression levels and patterns (Supplemental Fig. S2A) and different motif activity levels and patterns (Supplemental Fig. S2B). Importantly, three of the transcription factors predicted to bind to these selected motifs would most likely not have been included on the short list using other approaches, since one (heat shock transcription factor 1 [HSF1]) is not significantly regulated during adipogenesis, and two (teashirt zinc finger homeobox 1 [TSHZ1] and special AT-rich sequence-binding protein 1 [SATB1]) do not have public motifs, but were assigned to their motifs by IMAGE based on DBD similarity (Fig. 5A).
Figure 5.
IMAGE predicts transcription factors controlling human MSC commitment and differentiation with high confidence. (A) Table listing the six transcription factors chosen among the transcription factors predicted to be causally involved in adipocyte differentiation of hMSC-TERT4 cells based on IMAGE analyses of enhancer activity (MED1 ChIP-seq, n = 2) and gene expression (RNA-seq, n = 2). It is indicated which genes are differentially expressed (DE: Padj ≤ 0.05) during adipocyte differentiation and whether motifs were experimentally derived (Direct) or inferred based on similarity to other transcription factors. (*) indicates that the motif was identified with medium confidence. (B,C) The majority of the causal transcription factors predicted by IMAGE have an impact on lipid accumulation. (B) Representative images at low and high magnification of Oil Red O staining of hMSCs transfected with the indicated siRNAs and differentiated to adipocytes for 14 d. (C) Quantification of the Oil Red O stained area using ImageJ from a 3×3 grid (nine random locations, recorded at 10× magnification) for each replicate (n = 2). (D) The candidate factors regulate the expression of adipocyte marker genes. The heat map shows the expression of 10 well-known adipocyte marker genes (lipoprotein lipase [LPL], fatty acid binding protein 4 [FABP4], adiponectin [ADIPOQ], peroxisome proliferator activated receptor gamma [PPARG], phosphoenolpyruvate carboxykinase 1 [PCK1], perilipin 1 [PLIN1], perilipin 4 [PLIN4], hormone sensitive lipase [LIPE], cell death inducing DFFA like effector c [CIDEC], and CCAAT/Enhancer binding protein alpha [CEBPA]) as determined by RNA-seq in hMSC-TERT4 cells differentiated to adipocytes for the indicated time points with or without knockdown of each of the candidate factors. (E,F) The candidate factors regulate the expression of genes involved in both metabolism of lipids and cholesterol biosynthesis in a manner that correlates with their impact on lipid accumulation. Enrichment of genes belonging to the ‘Metabolism of lipids’ (E) or the ‘Cholesterol biosynthesis’ (F) pathways (Reactome database [Croft et al. 2014]) among genes that are up-regulated and down-regulated upon knockdown of the indicated transcription factors. The bar plot shows the log2 enrichment of genes that are expressed to a significantly (Padj ≤ 0.05) higher (green upward facing arrow) or lower (red downward facing arrow) level at day 7 of differentiation upon the indicated knockdown compared to control. The enrichment is calculated by comparing the fraction of genes within that pathway that are expressed to either a higher or a lower level upon knockdown compared to control relative to the fraction of all genes that are expressed to either a higher or a lower level upon knockdown compared to control. Significant enrichments (Padj ≤ 0.1) are denoted with green bars. (G) The change in metabolic flux through most reactions assigned to cholesterol metabolism or squalene and cholesterol synthesis correlate with the change in lipid accumulation upon knockdown of the candidate factors. The heat map shows the predicted scaled and centered metabolic flux of all metabolic reactions assigned to cholesterol metabolism or squalene and cholesterol synthesis that reached a flux of at least 0.1 µmol per g dw per h. Metabolic fluxes were predicted by the SPOT method (Kim et al. 2016) from transcriptome data at day 7 in the different knockdowns and in the control using a model of human metabolism (Recon 2) (Thiele et al. 2013). The top step converting HMG-CoA to mevalonate is the rate-limiting step.
We knocked down each of the six transcription factors (Supplemental Fig. S2C) and evaluated their impact on the ability of MSCs to undergo adipocyte differentiation as determined by lipid accumulation (Oil Red O) (Fig. 5B,C) and RNA-seq (Fig. 5D). Knockdown of five out of the six transcription factors significantly affects lipid accumulation during differentiation, two of the transcription factors (HSF1 and MYB proto-oncogene-like 1 [MYBL1]) lead to an increase in lipid accumulation compared to control cells, whereas knockdown of three of the transcription factors (MYC associated zinc finger protein [MAZ], SATB1, and TSHZ1) leads to a decrease in lipid accumulation (Fig. 5B,C). Knockdown of the last transcription factor (Nuclear Factor, Interleukin 3-regulated [NFIL3]) did not result in a significant change in the average Oil Red O stained area (Fig. 5B,C). Consistent with the effects on lipid accumulation, we find that the expression of known adipocyte marker genes as determined by RNA-seq is increased upon HSF1 knockdown and trends upward upon MYBL1 knockdown, whereas expression of these genes is decreased upon knockdown of MAZ, SATB1, or TSHZ1 (Fig. 5D). Importantly, we find that the predicted target genes of all of these five transcription factors are highly enriched for genes affected by their knockdown (Supplemental Fig. S2D). Furthermore, we find that binding sites of all five transcription factors (inferred using public ChIP-seq data) overlap significantly more than expected with MED1-bound enhancers in hMSC-TERT4 cells (Supplemental Fig. S2E). Collectively, this indicates that IMAGE accurately identifies novel transcriptional regulators associated with enhancers and predicts both their binding site specificity and gene regulatory events.
The identification of five novel transcription factors (HSF1, MYBL1, MAZ, SATB1, and TSHZ1) affecting adipogenesis both positively and negatively prompted us to investigate whether we could identify common downstream regulatory pathways mediating the effects of these factors based on the RNA-seq data from knockdown experiments. We focused on metabolic pathways and correlated the effect of the transcription factors on gene expression with their effect on lipid accumulation. Intriguingly, we found only six metabolic pathways significantly affected by all five factors and correlating with lipid accumulation (Supplemental Table S1). As expected, the ‘metabolism of lipids’-pathway is significantly enriched for genes induced during differentiation (Supplemental Table S1). Furthermore, this pathway is also enriched for genes expressed at higher levels at day 7 of differentiation upon either HSF1 or MYBL1 knockdown compared to control and, for genes expressed at lower levels, upon MAZ, TSHZ1, or SATB1 knockdown compared to control (Fig. 5E), consistent with the effect of the knockdown of these factors on lipid accumulation. Interestingly, two of the six pathways, i.e., ‘cholesterol biosynthesis’ and ‘regulation of cholesterol biosynthesis by SREBP (SREBF),’ are not significantly enriched for genes regulated between day 0 and day 7 of differentiation (Supplemental Table S1), yet they are enriched for genes regulated in a manner that correlates with induction of differentiation markers and lipid accumulation upon knockdown of each of the five transcription factors (Fig. 5F). To further interrogate the regulation of cholesterol metabolism under knockdown conditions, we used the recently developed tool SPOT (Kim et al. 2016) to infer intracellular metabolic flux based on our transcriptomic data. As expected, the flux through the lipolysis and lipid synthesis pathways is increased at day 7 of differentiation compared to day 0 in control cells, and the flux positively correlates with induction of differentiation markers and lipid accumulation upon knockdown of the five transcription factors (Supplemental Fig. S2F). Consistent with the pathway analysis, we found that the predicted metabolic flux through many of the metabolic reactions involved in cholesterol synthesis, including the rate-limiting conversion of hydroxymethylglutaryl-CoA to mevalonate, is increased upon HSF1 or MYBL1 knockdown and decreased upon MAZ, SATB1, or TSHZ1 knockdown compared to control (Fig. 5G), indicating that these transcription factors affect cholesterol synthesis in a manner that correlates with their effect on adipogenesis. Collectively, these results demonstrate that IMAGE is a powerful tool for identification of novel causal transcription factors, several of which would have eluded detection by most other methods, and that the integrated analysis of these novel regulators can lead to prediction of novel regulatory pathways.
Discussion
In this study, we have developed a new and powerful computational tool, IMAGE, which, based on time-resolved genome-wide profiling of gene expression and enhancer activity, can predict transcription factors that are causally involved in transcriptional responses. IMAGE uses a novel machine learning model for transcriptional regulation that is based on the activity of both enhancers and promoters, and subsequently, integrates gene expression with motif activity analysis, thereby allowing for a prediction of causal transcription factors that is superior to existing methods with respect to precision of predictions.
We used public databases to collect the largest possible set of PWMs and collapsed the database by correlation analysis to reduce library complexity. Furthermore, the database was significantly expanded by including prediction of motifs for transcription factors with unknown binding preferences. The resulting database covers PWM for 92.8% of all human transcription factors and is to our knowledge currently the most comprehensive motif database. Often, positive identification of a motif depends on reaching a certain threshold on a log-likelihood scale. However, the precision of log-likelihood thresholds is dependent on the complexity and length of the PWM (Dabrowski et al. 2015), and it is therefore impossible to define a unified threshold across all PWMs. Here, we identify motif thresholds uniformly by applying a P-value-based approach. Importantly, benchmarking on ENCODE data demonstrates that this approach has a local maximum where all tested motifs perform close to their best performance. This indicates that the P-value-based approach is not affected by the length or complexity of the PWM. Collectively, the exhaustive prediction of PWMs, using established methods, forming an extended PWM library combined with uniform P-value-based scoring scheme represents a significant leap forward in the pursuit of unbiased identification of putative transcriptional regulators by motif analysis.
One important problem in traditional motif enrichment analysis is that each motif is analyzed independently, even though it is known that transcription factor binding to DNA is dependent on chromatin state and cross-talk with other transcription factors (Slattery et al. 2011; Arvey et al. 2012; Yáñez-Cuna et al. 2012). A previous method, MARA (Balwierz et al. 2014), has approached this problem by using modeling across all motifs to estimate motif activity, based on either motif occurrence in promoters and gene expression, or motif occurrence in enhancers and enhancer activity. Although this is a major step forward in the analysis of transcriptional regulation based on motifs, it is incomplete since it neglects the interplay between enhancers and promoters in gene expression analysis. Our novel tool, IMAGE, extends on MARA by utilizing a more complete combinatorial and additive model. This combinatorial approach is not restricted to modeling gene expression based only on promoters; instead, our model integrates the signal from all enhancers in the vicinity of each promoter using distance-weighting. Importantly, even with distance-weighting, enhancer-to-gene assignment is not supported by direct evidence. Thus, there is inevitably some noise derived from wrong assignment of enhancers to genes. However, in spite of that, IMAGE outperforms MARA for identification of causal transcription factors. This shows that the gain of precision by leveraging enhancers outweighs the addition of noise from wrong assignment of enhancers to promoters. In addition, IMAGE also integrates gene expression data with the enhancer maps to restrict the analysis to include only transcription factors that are expressed in the given cell type. Importantly, we designed IMAGE to be as user-friendly as possible while maintaining transparency. Thus, we wrote the IMAGE code in human-readable programming languages and provide user-friendly guidelines. Furthermore, we have designed IMAGE to process input files directly from easy-to-use upstream pipelines, such as iRNA-seq (Madsen et al. 2015) and the HOMER toolbox (Heinz et al. 2010). Notably, the use of input directly from iRNA-seq allows the user to use intron-based analysis of RNA-seq data for a more accurate prediction of transcriptional events.
IMAGE is widely applicable, as it can be used to predict causal transcription factors involved in the regulation of gene expression, e.g., in response to differentiation signals or metabolic signals, as well as multigroup comparison studies, such as cell type comparisons. We tested the performance of IMAGE extensively by analyzing the ability to predict transcriptional regulators of 3T3-L1 preadipocyte differentiation. This cell line constitutes an ideal model system for validation due to the many large data sets available for this cell system. Using this cell line, we confirm that the target sites predicted for each motif in step 1 of IMAGE on average display strong binding of the corresponding transcription factors and that the target enhancers are activated with a temporal pattern, which is consistent with the activation profiles of the respective transcription factors. Collectively, this indicates that IMAGE provides precise prediction of which motifs are bound in enhancers and which transcription factors bind to these. IMAGE identifies 81 motifs bound by 76 transcription factors as high confidence causal regulators of adipogenesis during 3T3-L1 differentiation. These 76 transcription factors are significantly enriched among transcription factors assigned to the ‘fat cell differentiation’ GO term, and importantly, the enrichment of IMAGE-predicted transcription factors is higher than enrichment of transcription factors predicted by other methods, including gene expression analysis, motif enrichment, simple integration of expression and motif enrichment, or MARA. Furthermore, transcription factors predicted by IMAGE also display a greater enrichment for factors that have a significant impact on lipid accumulation upon knockdown or overexpression compared with transcription factors predicted by other methods. These results demonstrate that IMAGE outperforms existing methods for prediction of key transcriptional regulators. Furthermore, we show that IMAGE is also capable of identifying target genes with greater accuracy than ChIP-seq-based prediction of target genes, as exemplified by the predicted target genes of PPARG. Collectively, this demonstrates that IMAGE predicts causal transcription factors as well as their binding sites and target genes with high precision.
In order to further demonstrate the ability of IMAGE to predict novel key transcriptional regulators, we profiled transcriptional output and mapped enhancers and enhancer activity during commitment and adipocyte differentiation of human MSCs at hitherto unsurpassed temporal resolution. When IMAGE was applied to this data set, IMAGE predicted 237 transcription factors (115 with high confidence, 122 with medium confidence) to be involved in controlling gene expression during this process. Several of these have already established roles in adipogenesis; however, others have not previously been identified as being involved in MSC commitment or adipocyte differentiation. From these, we chose six different transcription factors with currently unknown roles. We purposely chose transcription factors that have inferred motifs, as well as nondynamic transcription factors, as these are not normally identified by alternative approaches. Knockdown of each of these factors showed that five out of six affect lipid accumulation as determined by lipid accumulation and RNA-seq, thereby demonstrating that IMAGE predicts causal transcription factors with very high precision. Interestingly, further interrogation of gene expression showed that all these transcription factors regulate pathways related to cholesterol biosynthesis in a manner that correlates positively with their effect on adipogenesis. This is particularly interesting since these cholesterol pathways are not normally regulated during adipocyte differentiation of these human MSCs. This suggests that these pathways are required for adipogenesis even though they are not significantly induced at the transcriptional level during adipogenesis. Consistent with this, it has been shown that treatment of preadipocytes with statins inhibits differentiation into mature adipocytes (Nakata et al. 2006; Nicholson et al. 2007).
In conclusion, we have developed a novel tool, IMAGE, for precise prediction of transcription factors and target enhancers that are causally involved in driving specific transcriptional responses in time series or multigroup comparison studies, such as comparisons of cell types. IMAGE offers several advantages over existing tools and strategies, including a more complete database of transcription factor motifs and a more advanced model of transcriptional regulation to identify causal regulators. Importantly, the tool is easy to use, transparent, and flexible in terms of input data.
Methods
Motif collection and cut-off determination
A list of all human transcription factors was generated by overlapping TFClass (Wingender et al. 2013) and AnimalTFDB 2.0 (Zhang et al. 2015). Motifs were collected from Cis-BP (Weirauch et al. 2014), HOMER (Heinz et al. 2010), and HOCOMOCO (Kulakovskiy et al. 2016). Only motifs that could be mapped to a human transcription factor were included, and redundant motifs were removed. For transcription factors with several motifs, a correlation score between the motifs was calculated using HOMER (Heinz et al. 2010) and clustered using hierarchical clustering. Clusters were defined using a tree height of 0.5, and all motifs within a cluster were aligned and merged using MATLIGN (Kankainen and Löytynoja 2007). The edges of all motifs were trimmed to the first position with information content of at least 0.3 using MotIV (Mercier et al. 2011). Only motifs of at least 4 bp in length after trimming were included. For each motif, cut-offs were calculated using TFMPvalue using a max granularity of 1 × 10−6 to allow efficient calculation of long motifs (Touzet and Varré 2007).
Motif prediction and cross-validation
The primary amino acid sequence of all human transcription factors was extracted from Cis-BP (Weirauch et al. 2014) and UniProt (Apweiler et al. 2004). For all transcription factors with a known motif, we constructed a database of the primary amino acid sequences of their DBDs. To predict the motif of a non-C2H2 zinc finger transcription factor, we extracted its DBD and searched for the best match in the DBD database using BLAST (Camacho et al. 2009). Predictions were only included if the sequence similarity had an E-score less than 1. For C2H2 zinc finger transcription factors, the protein sequence was submitted to ZifRC (Najafabadi et al. 2015) using a motif span of 0, and the longest motif was extracted. To cross-validate the DBD-based predictions, we predicted the motif of 290 transcription factors with an already known motif, excluding self-hits. The predicted motif was compared to the source motif by correlation using HOMER (Heinz et al. 2010).
Motif search, calculation of motif activities, and target prediction
IMAGE searches for motifs using our extended PWM database with P-value-based cut-offs using HOMER (Heinz et al. 2010). Subsequently, motifs without any hits in the supplied sequences and motifs mapping to transcription factors with low expression (default threshold: 1 normalized read per kilobase) are removed. To predict target enhancers, IMAGE performs ridge regression. The motif matrix is centered, and the user-supplied enhancer activity matrix of a normalized tag is centered and scaled. For each sample, ridge regression is performed using glmnet (Friedman et al. 2010) with 10-fold cross-validation. The model that is solved is:
OE,S is the sample- and enhancer-specific occupancy. AS,M is the sample- and motif-specific motif activity. NE,M is the enhancer- and motif-specific motif frequency. In other words, the enhancer activity at a specific position in a particular sample is given by the sum of all motif activities multiplied by their motif frequency at that site. Target enhancers are identified by leave-one-out-based analysis. We define target enhancers of each motif as sites where the motif is present and where the accuracy of the IMAGE model decreases upon leaving out that motif of the analysis. IMAGE uses the predicted sites to calculate motif activities for gene expression using an integrated model of enhancers using a similar ridge regression scheme to solve:
EG,S is the sample- and gene-specific mRNA expression. AS,M is the sample- and motif-specific motif activity. NE,M is the enhancer- and motif-specific motif frequency. TE,M is the enhancer- and motif-specific target prediction. DE,G is the enhancer- and gene-specific distance-weight calculated as in Wang et al. (2013) but subsequently scaled between 0 and 1. Target genes are identified by leave-one-out-based analysis. We calculate a P-value-like score based on the drop in prediction accuracy decreases upon leaving out that motif, as well as the predicted presence of binding sites near the gene. Genes with a score below 0.005 that are differentially regulated, as well as expressed above 1 normalized reads per kb, are defined as target genes.
Validation of IMAGE
For testing sets, we used our previously published MED1 ChIP-seq and RNA-seq (GSE95533) (Siersbaek et al. 2017) and DNase-seq (GSE27826) (Siersbaek et al. 2011) data from different time points during differentiation of 3T3-L1 preadipocytes. Transcription factor ChIP-seq data (GSE56872 and GSE27826) (Siersbaek et al. 2014) from the same time points were used for validation of predicted binding sites. RNA-seq from 3T3-L1 adipocytes treated with siPPARG or siControl (GSE14004) (Schupp et al. 2009) were processed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). Prediction of target genes was validated by comparison to large-scale screening by overexpression (Gubelmann et al. 2014) or knockdown (Söhle et al. 2012) (analyzed using redundant siRNA activity analysis [Konig et al. 2007]), as well as GO annotation (Blake et al. 2015) and genes whose expression in human adipose tissue correlate with BMI (Keller et al. 2011). Transcription factor annotations were extracted from the UniProt database (Apweiler et al. 2004). For overlap analysis between MED1-bound enhancers and the binding sites of candidate transcription, putative peaks (GSE105189, GSE91636, GSE66248, GSE91585, GSE44588) were downloaded. The two data sets from mouse were lifted to the human genome using liftOver, and all five were overlapped with MED1-bound enhancers using HOMER (Heinz et al. 2010).
Culture, differentiation, and transfection of human mesenchymal stem cells
Immortalized human mesenchymal stem cells, hMSC-TERT4, were cultured and differentiated essentially as previously described (Simonsen et al. 2002). Briefly, 2 d post-confluency, cells were induced to undergo adipogenesis by switching the cells to DMEM supplemented with 10% fetal bovine serum, 10 µg/mL insulin, 1 µM rosiglitazone, 100 mM dexamethasone, and 500 µM isobutylmethylxanthine. Differentiation media was replaced on day 2, 4, 7, 9, and 11. Knockdown of selected TFs was done 3 d prior to induction of the differentiation using reverse transfection with DharmaFECT (Thermo Fisher Scientific) and a pool of three different siRNA sequences (MISSION, Sigma).
Oil Red O staining
Following 14 d of differentiation, cells were fixed with 4% paraformaldehyde for 30 min, stained with Oil Red O (300 mg/mL Oil Red O in isopropanol diluted 60:40 v/v in water) for 30 min and washed in PBS. Whole-well images were captured with a Nikon D100 camera, and microscopic images were captured in color using a 10× phase contrast objective using a Leica DM IRB/E microscope. Image quantification was carried out using Fiji (Schindelin et al. 2012). Briefly, all images were split into RGB channels, and the area stained by Oil Red O was quantified using the identical thresholds for all images based on the blue channel. For each well, at least 10 images from randomly picked locations were analyzed.
Genome-wide studies in human mesenchymal stem cells
mRNA-seq was performed according to a standard protocol as previously described (Schmidt et al. 2016). Briefly, human mesenchymal stem cells, treated or differentiated as indicated in individual figures, were harvested in TRIzol (Thermo Fisher Scientific), and the RNA was purified using Econo Spin columns (Epoch Life Sciences). Library preparation, including RNA fragmentation and cDNA synthesis, was performed according to the manufacturer's instructions (TruSeq 2, Illumina). Sequencing data were mapped to the hg19 assembly of the human genome using STAR (Dobin et al. 2013). (The major difference between hg19 and hg38 is the addition of alternative loci, which more accurately captures variation in the human genome. Since IMAGE does not analyze human variation, the choice of genome will not significantly impact the results. Therefore, hg19 was chosen to make the data as easy to use as possible, since currently, the majority of data sets are mapped using hg19.) Gene counts were quantified using iRNA-seq (Madsen et al. 2015). Differential expression analysis was performed using edgeR (Robinson et al. 2010). Gene ontology enrichment analysis was performed using GOseq (Young et al. 2010) against the Reactome database (Croft et al. 2014) or the KEGG database (Kanehisa et al. 2016). Metabolic flux estimations were performed using SPOT (Kim et al. 2016) and Recon v2 (Thiele et al. 2013).
MED1 ChIP-seq was performed essentially as previously described (Siersbaek et al. 2011, 2012). Briefly, human mesenchymal stem cells, treated or differentiated as indicated in individual figures, were cross-linked for 20 min in 0.5 M DSG (Proteochem), followed by 10 min in 1% formaldehyde. Chromatin was sheared using a Bioruptor (Diagenode), immunoprecipitation was performed using MED1 antibody (SC-8998, Santa Cruz), and ChIP'ed DNA was prepared for sequencing according to the manufacturer's instructions (Illumina). Sequencing data was mapped to the human genome (hg19) using STAR (Dobin et al. 2013). Peaks were identified and tags were counted using HOMER (Heinz et al. 2010). Differential occupancy analysis was performed using edgeR (Robinson et al. 2010).
Data access
The high-throughput sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE104537. The complete R code for analyses of 3T3-L1 and hMSC data has been submitted to GitHub at https://github.com/JesperGrud/IMAGE and is in the Supplemental Material. The IMAGE pipeline used for this paper is available in the Supplemental Material. For the newest version, as well as instructions and examples, go to the Bioinformatics Tools pane at http://sdu.dk/mandrupgroup.
Competing interest statement
M.W. is an employee of Beiersdorf AG.
Supplementary Material
Acknowledgments
We thank present and former members of the Mandrup laboratory for helpful comments and discussions and Moustapha Kassem (Odense University Hospital, Denmark) for providing the hMSC-TERT4 cells. This work was supported by grants from the Danish Independent Research Council | Natural Science, the Novo Nordisk Foundation, the Lundbeck Foundation, the EMBO Long-Term Fellowship program, the Danish Diabetes Academy supported by the Novo Nordisk Foundation, and a grant from the VILLUM Foundation to the VILLUM Center for Bioanalytical Sciences at University of Southern Denmark.
Author contributions: J.G.S.M., S.F.S., and S.M. conceptualized the method. J.G.S.M. designed and implemented the tool, and A.R. assisted testing the code. The method comparison framework was designed by J.G.S.M., and M.W. provided data for method comparison. A.R. and E.L.V.H. performed experiments with hMSCs, and J.G.S.M. analyzed the data. J.G.S.M. prepared the figures with input from S.M. and A.R., and J.G.S.M. and S.M. wrote the paper with input from A.R.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.227231.117.
References
- Alleyne TM, Pena-Castillo L, Badis G, Talukder S, Berger MF, Gehrke AR, Philippakis AA, Bulyk ML, Morris QD, Hughes TR. 2009. Predicting the binding preference of transcription factors to individual DNA k-mers. Bioinformatics 25: 1012–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya-Kudo M, Shimano H, Hasty AH, Yahagi N, Yoshikawa T, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, et al. 2002. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J Lipid Res 43: 1220–1235. [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32: D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A, Agius P, Noble WS, Leslie C. 2012. Sequence and chromatin determinants of cell-type–specific transcription factor binding. Genome Res 22: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. 2014. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res 24: 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard B, Thorsson V, Rovira H, Shmulevich I. 2012. Increasing coverage of transcription factor position weight matrices through domain-level homology. PLoS One 7: e42779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Christie KR, Dolan ME, Drabkin HJ, Hill DP, Ni L, Sitnikov D, Burgess S, Buza T, Gresham C, et al. 2015. Gene Ontology Consortium: going forward. Nucleic Acids Res 43: D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 5: 1538–1552. [DOI] [PubMed] [Google Scholar]
- Chen X, Yu B, Carriero N, Silva C, Bonneau R. 2017. Mocap: large-scale inference of transcription factor binding sites from chromatin accessibility. Nucleic Acids Res 45: 4315–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al. 2014. The Reactome pathway knowledgebase. Nucleic Acids Res 42: D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowski M, Dojer N, Krystkowiak I, Kaminska B, Wilczynski B. 2015. Optimally choosing PWM motif databases and sequence scanning approaches based on ChIP-seq data. BMC Bioinformatics 16: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium & Riken Omics Science Center. 2009. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet 41: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JH, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33: 1–22. [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, Salgado H, Santos-Zavaleta A, Ledezma-Tejeida D, Muniz-Rascado L, Garcia-Sotelo JS, Alquicira-Hernandez K, Martinez-Flores I, Pannier L, Castro-Mondragon JA, et al. 2016. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 44: D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubelmann C, Schwalie PC, Raghav SK, Röder E, Delessa T, Kiehlmann E, Waszak SM, Corsinotti A, Udin G, Holcombe W, et al. 2014. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. eLife 3: e03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg L, Zuk O, Getz G, Domany E. 2005. Finding motifs in promoter regions. J Comput Biol 12: 314–330. [DOI] [PubMed] [Google Scholar]
- Jankowski A, Tiuryn J, Prabhakar S. 2016. Romulus: robust multi-state identification of transcription factor binding sites from DNase-seq data. Bioinformatics 32: 2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kähärä J, Lähdesmäki H. 2015. BinDNase: a discriminatory approach for transcription factor binding prediction using DNase I hypersensitivity data. Bioinformatics 31: 2852–2859. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Löytynoja A. 2007. MATLIGN: a motif clustering, comparison and matching tool. BMC Bioinformatics 8: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. 2003. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 31: 3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P, Gburcik V, Petrovic N, Gallagher IJ, Nedergaard J, Cannon B, Timmons JA. 2011. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr Disord 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Kellis M. 2014. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res 42: 2976–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Lane A, Kelley JJ, Lun DS. 2016. E-Flux2 and SPOT: validated methods for inferring intracellular metabolic flux distributions from transcriptomic data. PLoS One 11: e0157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, Chiang C-y, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y, et al. 2007. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods 4: 847–849. [DOI] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Soboleva AV, Kasianov AS, Ashoor H, Ba-alawi W, Bajic VB, Medvedeva YA, Kolpakov FA, et al. 2016. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res 44: D116–D125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JGS, Schmidt SF, Larsen BD, Loft A, Nielsen R, Mandrup S. 2015. iRNA-seq: computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res 43: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier E, Droit A, Li L, Robertson G, Zhang X, Gottardo R. 2011. An integrated pipeline for the genome-wide analysis of transcription factor binding sites from ChIP-seq. PLoS One 6: e16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, et al. 2015. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol 33: 555–562. [DOI] [PubMed] [Google Scholar]
- Nakata M, Nagasaka S, Kusaka I, Matsuoka H, Ishibashi S, Yada T. 2006. Effects of statins on the adipocyte maturation and expression of glucose transporter 4 (SLC2A4): implications in glycaemic control. Diabetologia 49: 1881–1892. [DOI] [PubMed] [Google Scholar]
- Nicholson AC, Hajjar DP, Zhou X, He W, Gotto AM Jr, Han J. 2007. Anti-adipogenic action of pitavastatin occurs through the coordinate regulation of PPARγ and Pref-1 expression. Br J Pharmacol 151: 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YPS. 2008. Guide to threshold selection for motif prediction using positional weight matrix. IMECS 1: 19–21. [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. 2011. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SF, Madsen JGS, Frafjord KØ, Poulsen Ll, Salö S, Boergesen M, Loft A, Larsen BD, Madsen MS, Holst JJ, et al. 2016. Integrative genomics outlines a biphasic glucose response and a ChREBP-RORγ axis regulating proliferation in β cells. Cell Rep 16: 2359–2372. [DOI] [PubMed] [Google Scholar]
- Schupp M, Cristancho AG, Lefterova MI, Hanniman EA, Briggs ER, Steger DJ, Qatanani M, Curtin JC, Schug J, Ochsner SA, et al. 2009. Re-expression of GATA2 cooperates with peroxisome proliferator-activated receptor-γ depletion to revert the adipocyte phenotype. J Biol Chem 284: 9458–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O'Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. 2014. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol 32: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. 2011. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J 30: 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk MS, Loft A, Aagaard MM, Nielsen R, Schmidt SF, Petrovic N, Nedergaard J, Mandrup S. 2012. Genome-wide profiling of peroxisome proliferator-activated receptor γ in primary epididymal, inguinal, and brown adipocytes reveals depot-selective binding correlated with gene expression. Mol Cell Biol 32: 3452–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, La Cour Poulsen L, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. 2014. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep 7: 1443–1455. [DOI] [PubMed] [Google Scholar]
- Siersbaek R, Madsen JGS, Javierre BM, Nielsen R, Bagge EK, Cairns J, Wingett SW, Traynor S, Spivakov M, Fraser P, et al. 2017. Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol Cell 66: 420–435.e5. [DOI] [PubMed] [Google Scholar]
- Simonsen JL, Rosada C, Serakinci N, Justesen J, Stenderup K, Rattan SIS, Jensen TG, Kassem M. 2002. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol 20: 592–596. [DOI] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rhos R, Honig B, Bussemaker HJ, et al. 2011. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147: 1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söhle J, Machuy N, Smailbegovic E, Holtzmann U, Grönniger E, Wenck H, Stäb F, Winnefeld M. 2012. Identification of new genes involved in human adipogenesis and fat storage. PLoS One 7: e31193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Lazar MA. 2010. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 24: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs L, Sun Y, Caetano-Anolles D. 2011. Function and evolution of C2H2 zinc finger arrays. Subcell Biochem 52: 75–94. [DOI] [PubMed] [Google Scholar]
- Tan G, Lenhard B. 2016. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32: 1555–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I, Swainston N, Fleming RMT, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, et al. 2013. A community-driven global reconstruction of human metabolism. Nat Biotechnol 31: 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. 1994. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79: 1147–1156. [DOI] [PubMed] [Google Scholar]
- Touzet H, Varré J-S. 2007. Efficient and accurate P-value computation for Position Weight Matrices. Algorithms Mol Biol 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, Liu XS. 2013. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc 8: 2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zang C, Xiao T, Fan J, Mei S, Qin Q, Wu Q, Li XL, Xu K, He HH, et al. 2016. Modeling cis-regulation with a compendium of genome-wide histone H3K27ac profiles. Genome Res 26: 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote A, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Schoeps T, Dönitz J. 2013. TFClass: an expandable hierarchical classification of human transcription factors. Nucleic Acids Res 41: D165–D170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, Stark A. 2012. Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res 22: 2018–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL. 1995. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 9: 168–181. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanighomi M, Lin Z, Wang Y, Jiang R, Wong WH. 2017. Predicting transcription factor binding motifs from DNA-binding domains, chromatin accessibility and gene expression data. Nucleic Acids Res 45: 5666–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Liao H, Liu Y, Lee T-S, Zhu M, Wang X, Stemerman MB, Zhu Y, Shyy JY-J. 2004. Sterol-responsive element-binding protein (SREBP) 2 down-regulates ATP-binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism. J Biol Chem 279: 48801–48807. [DOI] [PubMed] [Google Scholar]
- Zhang J-W, Klemm DJ, Vinson C, Lane MD. 2004. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein β gene during adipogenesis. J Biol Chem 279: 4471–4478. [DOI] [PubMed] [Google Scholar]
- Zhang H-M, Liu T, Liu C-J, Song S, Zhang X, Liu W, Jia H, Xue Y, Guo A-Y. 2015. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Research 43: D76–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, He X, Bar-Joseph Z. 2013. Predicting tissue specific transcription factor binding sites. BMC Genomics 14: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.