Abstract
Cnidarians harbor a variety of small regulatory RNAs that include microRNAs (miRNAs) and PIWI-interacting RNAs (piRNAs), but detailed information is limited. Here, we report the identification and expression of novel miRNAs and putative piRNAs, as well as their genomic loci, in the symbiotic sea anemone Anemonia viridis. We generated a draft assembly of the A. viridis genome with putative size of 313 Mb that appeared to be composed of about 36% repeats, including known transposable elements. We detected approximately equal fractions of DNA transposons and retrotransposons. Deep sequencing of small RNA libraries constructed from A. viridis adults sampled at a natural CO2 gradient off Vulcano Island, Italy, identified 70 distinct miRNAs. Eight were homologous to previously reported miRNAs in cnidarians, whereas 62 appeared novel. Nine miRNAs were recognized as differentially expressed along the natural seawater pH gradient. We found a highly abundant and diverse population of piRNAs, with a substantial fraction showing ping–pong signatures. We identified nearly 22% putative piRNAs potentially targeting transposable elements within the A. viridis genome. The A. viridis genome appeared similar in size to that of other hexacorals with a very high divergence of transposable elements resembling that of the sea anemone genus Exaiptasia. The genome encodes and expresses a high number of small regulatory RNAs, which include novel miRNAs and piRNAs. Differentially expressed small RNAs along the seawater pH gradient indicated regulatory gene responses to environmental stressors.
Keywords: coastal ecology, CO2 seep, ocean acidification, miRNA, piRNA, transposable elements
Introduction
Two major classes of small regulatory RNAs in eumetazoans are microRNAs (miRNAs) and PIWI-interacting RNAs (piRNAs). These classes are distinct in terms of their sizes, biogenesis, biological function, and origin (Ghildiyal and Zamore 2009). Compared with Bilateria, knowledge about cnidarian small RNAs remains scarce. To date, small RNAs have only been reported in four cnidarians; the non-symbiotic sea anemone Nematostella vectensis, the stony corals Stylophora pistillata and Acropora digitifera, and the hydroid Hydra magnipapillata (Grimson et al. 2008; Wheeler et al. 2009; Chapman et al. 2010; Krishna et al. 2013; Liew et al. 2014; Moran et al. 2014; Gajigan and Conaco 2017).
miRNAs represent a well-studied class of small RNAs that usually range in size from 20 to approximately 24 nt (miRBase, http://mirbase.org; last accessed January 14, 2018). In animals, miRNAs are initially transcribed as RNA polymerase II transcripts (pri-miRNAs), which are further processed by the RNases Drosha and Dicer into stem–loop precursor miRNAs (pre-miRNAs) and mature miRNAs, respectively. One strand of the mature miRNA duplex is usually incorporated into the RNA-induced silencing complex (RISC) (Schwarz et al. 2003; Gregory et al. 2005), and it guides the whole complex to complementary mRNA for post-transcriptional gene silencing. In plants, the miRNA biogenesis pathway involves a Dicer-like 1 (DCL-1) protein that is responsible for both cropping and slicing miRNA precursors in the nucleus (Voinnet 2009). The miRNA silencing mechanism is fundamentally different in animals and plants. Although animal miRNAs usually perform translational repression through partial base-pairing to target mRNAs, plant miRNAs mostly bind with full or nearly full complementarity leading to targeted mRNA cleavage (Bartel 2009). Cnidarian miRNAs appear to contain plant-like features in their biogenesis and the post-transcriptional gene silencing follows a plant-like regulatory pathway (Moran et al. 2013, 2014). Among cnidarians, Nematostella was reported to express 87 distinct miRNAs, compared with 26, 31, and 126 miRNAs in Acropora, Stylophora, and Hydra, respectively (Grimson et al. 2008; Wheeler et al. 2009; Chapman et al. 2010; Krishna et al. 2013; Liew et al. 2014; Moran et al. 2014; Gajigan and Conaco 2017). Interestingly, only one miRNA (miR-100) was found conserved between the bilaterian and the cnidarian species. miRNAs have several important regulatory roles in plants and animals (Bartel 2004; Ghildiyal and Zamore 2009; Vashisht and Nodine 2014). Expression profiling indicated the cnidarian miRNAs to be involved in developmental regulation, regeneration and thermal stress resilience (Krishna et al. 2013; Moran et al. 2014; Gajigan and Conaco 2017). However, their roles in other biological processes, including other environmental stress responses, have not been investigated in detail.
piRNAs are usually between 23 and 30 nt in size (Krishna et al. 2013; Liew et al. 2014; Moran et al. 2014). The single-stranded piRNA precursors are either derived from transposable elements or from specific piRNA genomic clusters, and they do not require Dicer nuclease activity for their processing (Vagin et al. 2006; Houwing et al. 2007; Das et al. 2008). piRNAs represent a highly diverse class of small regulatory RNAs, reaching several thousand distinct members within a single organism (Aravin et al. 2006; Kawamura et al. 2008). The uniqueness of piRNAs arises from phased production of primary piRNAs (Han et al. 2015; Mohn et al. 2015). A secondary piRNA pathway serves for piRNA amplification by a “ping–pong loop” mechanism (Brennecke et al. 2007; Gunawardane et al. 2007). The two distinct piRNA populations (primary and secondary) show opposite orientation and complementarity in their first 10 nt positions (Brennecke et al. 2007; Gunawardane et al. 2007). piRNAs lack universal sequence conservation, except that the primary and secondary piRNAs show a preference for an uracil residue at the 5′end (1 U) and an adenine residue at the 10th position (10 A), respectively. In all cnidarians investigated so far, piRNAs appear to be highly abundant compared with miRNAs and short-interfering RNAs (siRNAs) (Grimson et al. 2008; Krishna et al. 2013; Juliano et al. 2014; Moran et al. 2014; Gajigan and Conaco 2017; Praher et al. 2017). The biological role of piRNAs is not well understood, but their most important function seems to be guiding PIWI proteins to suppress transposon activity in animal germ cells (Brennecke et al. 2007; Gunawardane et al. 2007). piRNA profiling in cnidarians (Nematostella and Hydra) suggested a similar role in transposon silencing, but proposed broader silencing functionalities as well (Grimson et al. 2008; Juliano et al. 2014; Praher et al. 2017).
The sea anemone Anemonia viridis exposed to natural ocean acidification conditions appears to be physiologically acclimatized to low pH and optimizes its energy utilization under elevated pCO2 through an increased autotrophic input (Suggett et al. 2012, Horwitz et al. 2015). Our recent transcriptome sequencing from the same sampling site indicates increased expression of stress-related transcripts, repression of global synthesis and boost in certain retrotransposon elements at low pH in A. viridis (Urbarova et al., unpublished results). In plants, it is known that small RNAs can regulate species tolerance to stress via post-transcriptional gene silencing (Sunkar et al. 2007). We therefore wanted to elucidate if acclimatization responses of A. viridis that we observe at the transcriptome level could be caused by small RNA-mediated post-transcriptional regulation. Here, we report whole genome and small RNA library sequencing of the symbiotic sea anemone A. viridis, sampled at a natural seawater pH gradient off Vulcano Island, Sicily—Italy. We mainly aimed to identify novel small RNA species in A. viridis, but also elucidate their possible involvement in the acclimatization responses to low pH conditions. We detected 70 distinct miRNA species, and assessed differentially expressed small RNAs. Most of the putative piRNAs contained features typical of primary piRNAs and a large fraction showed ping–pong signatures. Our study indicates possible regulatory gene responses of small RNAs to low pH.
Materials and Methods
Sampling
The temperate symbiotic sea anemone A. viridis (the Snakelocks Anemone) was collected at Levante Bay, North Vulcano Island, Sicily—Italy. Acidification conditions are created here by the release of CO2 into the seawater from a natural vent site at −1 m depth (Boatta et al. 2013; Johnson et al. 2013). Sampling was performed on May 13 and 14, 2013, at the depth of 1–2 m at >350 m from the vent site along a gradient of decreasing pH (∼ pH 7.6 and 7.9), and at a control location at ∼800 m from the vent site with pH corresponding to ambient seawater levels (∼pH 8.2). For simplicity, we are referring to average pH values throughout this work as reported in Johnson et al. (2013). A total of nine individuals of A. viridis were sampled in 2 days (three from each location). Small pieces of tissue (≤0.5 cm) from body wall, tentacles and oral disc of each individual were collected and stored separately at 4 °C in RNAlater (ThermoFisher Scientific, Waltham, MA, USA) during transport from the sampling site to laboratory. Then, RNAlater solution was removed and all samples were frozen at −80 °C before further processing steps.
Reference Genome Assembly
DNA from one individual of A. viridis (pH 8.2) was extracted using Wizard(R) Genomic DNA Purification Kit (Promega, Madison, Wisconsin, USA). Two whole genome paired-end libraries (2 × 150 bp) were constructed and sequenced on Illumina HiSeq2500 at Eurofins MWG Operon (Germany). The paired-end reads were processed using Trimmomatic (Bolger et al. 2014). Adapters were removed and reads were trimmed and quality filtered using sliding window with Phred score > 20. A bias at the first nine nucleotides was removed by trimming these bases, and reads with length <40 bp were discarded. SGA preqc tool was run pooling the forward and reverse reads from the two libraries together (Simpson 2014). Platanus, a de novo genome assembler for highly heterozygous diploidic organisms, was then used to assemble reads with k-mer length of 51 (Kajitani et al. 2014). To be able to assess our A. viridis genome assembly for repeat-enriched regions using RepeatMasker (Smit et al. 2013–2015), we first filtered out short reads from our reference assembly using N75 statistics, resulting in 210,233 sequences with sizes larger than 173 bp. The filtered assembly was then assessed for repeat-enriched regions using RepeatMasker (Smit et al. 2013–2015), with a custom library created by RepeatModeler, which integrates RECON, RepeatScout, and Tandem Repeats Finder (TRF) de novo repeat finding tools to build a repeat library for an assembly (Smit and Hubley 2008–2015). In addition to RepeatMasker annotation, the repeat-enriched regions were extracted from the assembly and transposable element annotation was performed as described previously (Chapman et al. 2010; Baumgarten et al. 2015). The annotation pipeline included then also a TBLASTX run using RepBase database (Bao et al. 2015), version 22.09 (e-value < 10−20), and a BLASTX search (e-value < 10−10) against a custom-made non-redundant database of proteins encoded by transposable elements (TEs; NCBI keywords: retrotransposon, transposase, reverse transcriptase, gypsy, and copia). These two databases were separately queried against our reference genome assembly and the best annotation was chosen based on alignment coverage and score. A combined tabular output from the searches was further run through two Perl scripts, “blast92gff3.pl” with additional options -lowscore 0.0001 -alignmax 9999 -exonType exon (http://arthropods.eugenes.org/EvidentialGene/evigene/scripts/blast92gff3.pl; last accessed January 14, 2018) and the “overbestgene2.pl” (http://iubio.bio.indiana.edu/gmod/tandy/perls/overbestgene2.perl; last accessed January 14, 2018) to create a gff file from blast results and to remove overlapping blast hits, respectively. The results were imported into IBM SPSS Statistics software (version 23), where counting of transposable elements was performed. Sequence regions corresponding to transposable elements in our reference genome assembly were then extracted from our scaffolds using BLAST fastacmd tool (Altschul et al. 1990, 1997) and used as a reference for piRNA analyses.
RNA Extraction
Each tissue sample (without excess RNAlater solution) was immediately transferred from −80 °C to 1 ml cold TRIzol reagent (ThermoFisher Scientific, Waltham, MA, USA). The tissue was then crushed using Precellys tissue homogenizer at 6,000 rpm for 30 s (Stretton Scientific, Stretton, UK) to minimize degradation of RNA. RNA was twice extracted by chloroform, and subsequently precipitated in isopropanol at 4 °C overnight, washed with 70% ethanol, and rehydrated in Nuclease-Free Water (ThermoFisher Scientific, Waltham, MA, USA). The RNA quality was examined using the Agilent 2100 Bioanalyzer (Agilent technologies, Santa Clara, CA, USA) and quantity of the samples was measured using Qubit 2.0 fluorometer (ThermoFisher Scientific, Waltham, MA, USA). Only high quality samples with RNA integrity number (RIN) equal to 7 or higher were used in library constructions.
Small RNA Sequencing
Nine individuals of A. viridis representing three different pH conditions (8.2, 7.9, and 7.6) were included in small RNA sequencing. Total RNA from three different tissue samples of an individual was pooled at equal amounts. The small RNA fraction was enriched using PureLink miRNA Isolation Kit (ThermoFisher Scientific, Waltham, MA, USA). Libraries were prepared only from high quality RNA samples (RIN ≥ 7) following the SOLiD Total RNA-Seq Kit protocol (ThermoFisher Scientific, Waltham, MA, USA). Different A. viridis small RNA libraries were barcoded and sequencing was performed on three lanes of a SOLiD™ 6-Lane FlowChip using SOLiD 5500xl sequencer at the Nord University (Bodø, Norway).
Discovery of Novel miRNA
After removing low quality (quality score < 18) and less complex sequences from our raw small RNA sequencing data set, adapter sequences were trimmed away using trimSOLiDAdaptor.pl Perl script keeping only sequences equal to or longer than 18 nt. The filtered reads were mapped in color space using Bowtie (Langmead et al. 2009) to our A. viridis genome reference with parameters –integer-quals -l 18 -M 20 –best –strata -e 150 –nomaqround –maxbts 800 –tryhard -a –col-cqual –col-keepends –mapq 20 –threads 14 –chunkmbs 200. These options select the best alignments based on the seed mismatches only and mismatches outside the seed region are ignored. Therefore, we needed to perform additional filtering using processBowtieAlignments.pl Perl script to select alignments with the minimum mismatches along the whole reads. Both Perl scripts are available at https://github.com/patelhardip/bitx.git (last accessed January 14, 2018). Mapped reads from each condition were then preprocessed by bwa_sam_converter.pl Perl script (Friedländer et al. 2012), outputting two files essential for running miRDeep2 software tool for the novel miRNA predictions (Friedländer et al. 2012). The two files were used as input into miRDeep2.pl Perl script (Friedländer et al. 2012) that was run for identification of novel miRNAs in A. viridis. In addition, known miRNAs of three related species, N. vectensis, S. pistillata, and H. magnipapillata (Krishna et al. 2013; Liew et al. 2014; Moran et al. 2014) that were available at the time of the analysis, have been used in the prediction pipeline. These miRNA sequences were downloaded from miRBase, release 21 (Kozomara and Griffiths-Jones 2014). Output from miRDeep2 software was then inspected manually, keeping only predicted miRNAs with miRDeep2 score larger than ten and with significant randfold value (p value < 0.05). Small RNA sequencing data from each individual were assessed separately for the presence of novel miRNAs. Only miRNAs identified in at least two individuals were considered further. Possible tRNA contamination was examined by running tRNAscan on the reference genome (Lowe and Eddy 1997). Further, presence of rRNA sequences in predicted hairpin structures was tested by querying against a custom database combining known rRNA sequences from N. vectensis and A. viridis. No contamination was found in either case. In addition, to ensure that our miRNA candidates come from the host, assembled scaffolds of A. viridis were screened for their possible contamination by symbiont DNA using genomes of Symbiodinium minutum, Symbiodinium microadriaticum, and Symbiodinium kawaguti (Shoguchi et al. 2013; Lin et al. 2015; Aranda et al. 2016). 1,642 scaffolds (mainly short ones with length ∼100 bp) that were highly similar (e-value < 10−20) to Symbiodinium genomes in the A. viridis genome assembly were filtered out prior to the small RNA alignment. Genomic setting of aligned putative miRNAs was inspected for overlapping regions corresponding to open reading frames (ORFs). Our scaffolds were searched for ORFs using OrfPredictor (http://bioinformatics.ysu.edu/tools/OrfPredictor.html; last accessed January 14, 2018) (Min et al. 2005).
miRNA Analyses
Expression of selected miRNAs was confirmed by quantitative PCR (qPCR). Six Locked Nucleid Acid (LNA) probes targeting the predicted miRNAs were designed using online miRNA qPCR designer tool from Exiqon (Vedbaek, Denmark). cDNA was synthesized from three individuals (10 ng of total RNA each) per condition using miRCURY LNA™ Universal RT microRNA PCR, Polyadenylation, and cDNA synthesis kit II (Exiqon, Vedbaek, Denmark) following the instruction manual. Small RNA for the qPCR analysis was isolated from the same samples that were used for preparation of small RNA libraries. qPCR was performed in duplicates using miRCURY LNA microRNA PCR, ExiLENT SYBR Green master mix (Exiqon, Vedbaek, Denmark) in 10 µl. miRNAs were assessed for differential expression among the sampling sites with differing pH by edgeR (FDR < 0.05) (Robinson et al. 2010). Mature miRNAs were aligned to their precursor sequences. miRNAs with <20 counts in less than three conditions were not considered. We searched for putative animal-like miRNA targets by Probability of Interaction by Target Accessibility (PITA) software, based on target complementarity and site accessibility (Kertesz et al. 2007). Coding regions were predicted from the A. viridis transcriptome (Urbarova et al., unpublished results) by TransDecoder, version 2.0.1 (Haas et al. 2013) and used as input into PITA software. The results were filtered based on the change in free energy (ddG) of miRNAs binding to its targets (ddG < −10 kcal/mol), seed length of 8 nt with no mismatches and no wobble pairs. Only targets fulfilling these criteria were considered further. We then checked the predicted targets from PITA for more extensive complementarity to the miRNAs using FASTA v36 (Pearson and Lipman 1988) as previously described (Moran et al. 2014), and scored the alignments accordingly. Blast hits were obtained using NCBI nr database (e-value < 10−5) and GO terms were assigned to the potential mRNA targets using B2G4Pipe Blast2GO pipeline (Götz et al. 2008).
Search for Putative piRNAs
Raw reads from SOLiD sequencing were quality filtered and adapter sequences were trimmed, as described previously for miRNA discovery. However, only sequences equal to or longer than 23 nt were kept for the piRNA analyses. Sequences were further filtered for reads mapping to miRNA precursors identified in the present study and reads mapping to rRNAs using rRNA databases according to Praher et al. (2017). Sequences in color space were aligned using Bowtie with the same parameters as for the miRNA alignment, but allowing for maximum three mismatches with the “seed length” of 23 nt (-l 23 -n 3). We refrained from mapping our reads to unique locations in the A. viridis reference genome due to presence of many sequence stretches in assembled scaffolds that most probably correspond to the same genomic locations. The aligned sequences were filtered for the best matches using the same custom Perl script as for filtering the miRNA alignments and filtered for reads with 1 U or 10 A sequence signatures. TE-targeting potential of putative piRNAs was then assessed by including only putative piRNAs mapping antisense to the transposable elements. Overlap probabilities of the putative piRNA sequences with opposite orientation were analyzed using signature.py script (Antoniewski 2014). The script computes the probability of an antisense read overlapping a sense read with defined length and assigns each overlap length a z-score. The overlapping signatures of the putative piRNAs were inspected in more detail by running PingPongPro v1.0 software (http://sourceforge.net/projects/pingpongpro/; last accessed January 14, 2018). This software also served for inspection of transposon silencing by putative piRNAs. Silencing of transposable elements by the putative piRNAs was only considered if FDR (q value) < 0.01 and if it was supported by at least ten putative piRNA reads with ping–pong signatures normalized to the transposon length. To inspect data for presence of piRNA clusters, the putative piRNA reads were mapped onto the masked genome reference produced by RepeatMasker (Smit et al. 2013–2015) as described previously in color space using Bowtie, reporting at maximum five valid alignments. Finally, our sorted alignment files were submitted to piClust software (Jung et al. 2014). Here, the Eps parameter was set to 1000 and MinReads to 50.
Results
Genome Reference Assembly and Search for Repeat-Enriched Regions
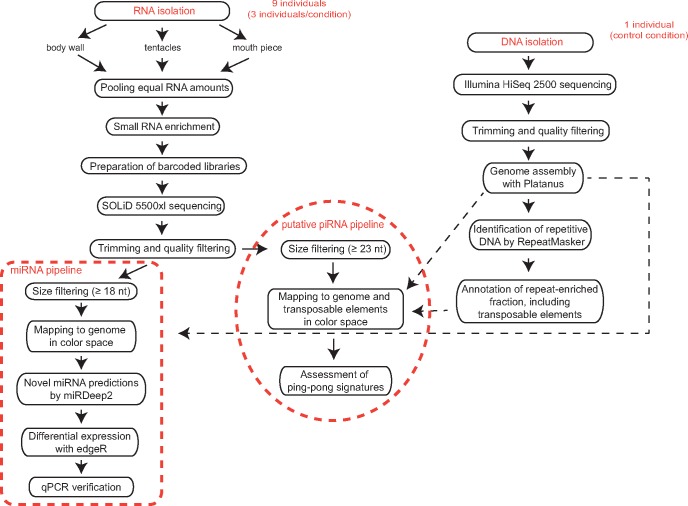
Total DNA from a single A. viridis polyp (normal seawater conditions, pH 8.2) was extracted and subjected to whole genome sequencing on the Illumina HiSeq2500 platform (fig. 1). Sequencing generated 43 billion nucleotides (nt) of genomic data, which corresponded to 144 million paired-end reads (table 1). The basic genome characteristics were determined using the SGA preqc software tool (Simpson 2014), showing an estimated genome size of 313 Mb (∼140x coverage). Adapters were trimmed and the reads were quality filtered before de novo genome assembly, which created about 1.1 million short scaffolds with N50 = 2,087. This genome assembly is highly fragmented, but it was sufficient for the mapping of small RNAs and the identification of transposable elements (fig. 1).
Fig. 1.
—Data analysis overview. DNA and RNA were isolated from A. viridis adult polyps sampled from a natural seawater pH gradient (at normal seawater pH 8.2, and at low seawater pH 7.9 and 7.6) off Vulcano Island, Sicily—Italy. Only one polyp (pH 8.2) was used for DNA extraction and was subjected to paired-end sequencing on the Illumina HiSeq2500 platform. Sequencing reads were assembled into a draft genome reference. Subsequently, repeat-enriched regions, including transposable elements were identified and annotated in this assembly. Nine polyps were used for small RNA library preparation and sequencing on the SOLiD 5500xl platform. Sequencing reads were further used for novel miRNA discovery and description of putative piRNA reads.
Table 1.
The Amount of Reads Gained from Genome Sequencing of Anemonia viridis
| Sequencing Index | No. of Paired-End Raw Readsa | No. of Trimmed and Quality Filtered Reads | % Paired-End Reads Kept After Filtering |
|---|---|---|---|
| CTTGTA | 82,428,617 | 71,676,503 | 87.0 |
| GCCAAT | 61,246,666 | 52,649,432 | 86.0 |
Sequencing of barcoded genome libraries was performed in one lane of Illumina HiSeq2500 sequencing machine in 2 × 150 bp mode. The amount of sequences presented here is the number of raw paired-end sequences obtained after the run.
The A. viridis genome assembly was inspected for repeat and low complexity regions using the RepeatMasker software tool (Smit et al. 2013–2015), and about 36% of the genome reference was found to contain repetitive regions. About 27.5% of the repetitive sequences could be assigned to previously known repetitive elements, but most of these sequences (25% of the genome) could not be classified into any assigned category (supplementary table S1, Supplementary Material online). Repeat annotation identified only about 8.3% of genome to be comprised of transposable elements (TEs). These are similar observations as made previously for symbiotic sea anemone Exaiptasia sp. (formerly known as Aiptasia sp.; Grajales and Rodríguez 2014; Baumgarten et al. 2015). From the identified TE fraction, about half (44.4%) were retrotransposons, and amongst them, non-long terminal repeat (non-LTR) retrotransposons were predominating (supplementary table S1, Supplementary Material online).
miRNA Discovery
Small RNA libraries from nine individual polyps of A. viridis, sampled at three different seawater pH conditions (at normal seawater pH 8.2, as well as at pH 7.9 and 7.6) were prepared and subjected to sequencing on the SOLiD 5500xl platform (fig. 1). The sequencing generated approximately 116 million reads (≥18 nt) after adapter trimming and quality filtering (table 2 and fig. 2). Despite the fragmented nature of our genome draft assembly, a high proportion of the small RNA reads mapped to the genomic reference (between 88% and 92%, table 2). This indicates that even a very preliminary genome assembly is sufficient for the discovery of small RNAs.
Table 2.
The Amount of Reads Gained from Small RNA Sequencing of Anemonia viridis
| Individuals | Raw Readsa | Filtered Small RNA Reads (≥18 nt)b | % Reads Aligned to the Genome (≥18 nt) | Filtered Small RNA Reads (≥23 nt)b | Reads Aligned to Genome (≥23 nt) | Putative piRNA Reads Aligned to Genomec | % Putative piRNAs Aligned to Genome |
|---|---|---|---|---|---|---|---|
| pH 7.6–1 | 16,111,517 | 12,117,933 | 88.5 | 9,831,771 | 7,681,304 | 6,643,549 | 86.5 |
| pH 7.6–2 | 12,492,090 | 11,175,766 | 89.5 | 9,465,543 | 7,721,730 | 6,795,032 | 88.0 |
| pH 7.6–3 | 13,794,761 | 10,609,870 | 89.1 | 8,921,147 | 7,238,217 | 6,310,624 | 87.2 |
| pH 7.9–1 | 18,815,344 | 15,837,619 | 91.5 | 13,235,131 | 11,175,365 | 10,001,436 | 89.5 |
| pH 7.9–2 | 20,094,253 | 17,369,794 | 90.0 | 16,247,704 | 13,837,596 | 11,960,594 | 86.4 |
| pH 7.9–3 | 16,794,089 | 15,567,040 | 89.3 | 14,958,985 | 12,687,556 | 11,353,938 | 89.5 |
| pH 8.2–1 | 13,066,319 | 12,109,753 | 88.0 | 11,317,833 | 9,184,500 | 8,163,612 | 88.9 |
| pH 8.2–2 | 17,379,705 | 10,309,153 | 90.3 | 8,124,195 | 6,661,625 | 5,543,870 | 83.2 |
| pH 8.2–3 | 13,492,380 | 11,225,436 | 90.3 | 8,769,865 | 7,189,261 | 6,271,324 | 87.2 |
Small RNA libraries from each individual were barcoded, pooled, and sequencing was performed on three lanes of a SOLiD™ 6-Lane FlowChip using the SOLiD 5500xl sequencer. The amount of sequences presented here is the sum of raw reads from the three lanes.
These reads had adapter removed and were quality filtered. They differ only according to the size filtering.
Reads (≥23 nt) aligned to the reference genome and filtered for piRNA sequence signatures (1 U and 10 A).
Fig. 2.
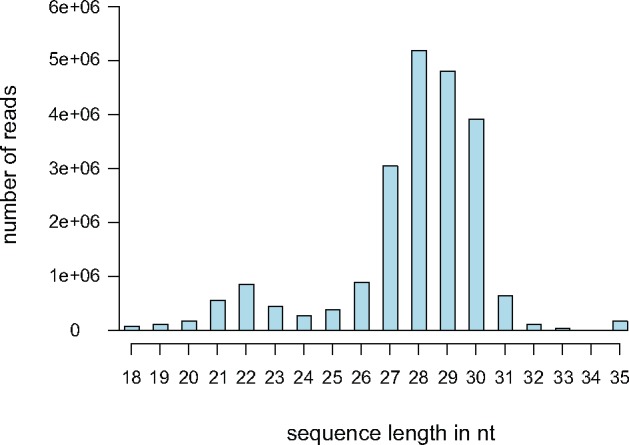
—Sequence length representation of small RNAs in A. viridis. Distribution of small RNA reads after adapter trimming and quality filtering in one individual sampled from pH 8.2. Two distinct peaks could be observed; first around 22 nt representing both miRNA and siRNA reads, and second around 28 nt representing putative piRNA reads.
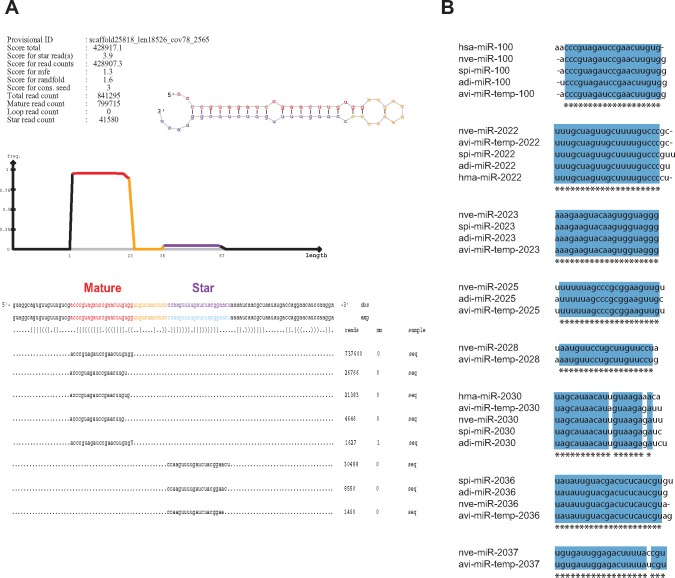
A. viridis miRNAs were identified by the miRDeep2 software tool (Friedländer et al. 2012), and a representative analysis result is shown in figure 3A. We predicted in total 70 high-confidence miRNA candidates (20 to 25 nt) for A. viridis, including 61, 60, and 65 distinct miRNA species at pH 8.2, 7.9, and 7.6, respectively (table 3). Most miRNAs were detected in all pH conditions studied (n = 51), 14 miRNAs in two different pH conditions, and five miRNAs were detected in one pH condition only (table 3). Eight candidate miRNAs in A. viridis were apparently homologous to those reported in other cnidarian species (avi-miR-temp-100, 2022, 2023, 2025, 2028, 2030, 2036, and 2037) (fig. 3B). The predicted miRNA with the highest miRDeep2 score (avi-miR-temp-100), and which was highly expressed in all pH conditions, was identical in sequence to miR-100 in N. vectensis and S. pistillata (fig. 3B) (Grimson et al. 2008; Liew et al. 2014; Moran et al. 2014). Similarly, avi-miR-temp-2022, 2023, and 2025 were identical to the corresponding miRNAs in N. vectensis (Moran et al. 2014). Other A. viridis miRNAs (avi-miR-temp-2028, 2030, 2036, and 2037) have one or two nucleotides substitution compared with that of the N. vectensis homolog (fig. 3B). Multiple precursor sequences for some of the predicted miRNAs were found by miRDeep2 (table 3). All mature, star, and precursor sequences are presented in supplementary table S2, Supplementary Material online, with read counts for each pH condition in supplementary table S3, Supplementary Material online.
Fig. 3.
—Identified miRNAs in A.viridis with similarity to known miRNAs. (A) A typical prediction result from miRDeep2 software tool showing the miRNA precursor, mature and star sequence and their abundances in the sample. Shown is avi-miR-temp-100 precursor with top sequence alignments in the sample for each strand. (B) Alignments of novel miRNAs from A. viridis to known miRNAs from other species. Sequences of our predicted miRNAs from A. viridis (denoted as avi-miR-temp) were aligned to known miRNA sequences from H. sapiens (hsa-miR), N. vectensis (nve-miR), H. magnipapillata (hma-miR), S. pistillata (spi-miR), and A. digitifera (adi-miR). (temp = temporary; miRNAs that are not yet registered in the miRBase).
Table 3.
The List of 70 Predicted miRNAs in Anemonia viridis from Various pH Conditions
| Temporary miRNA Name | Mature Sequence | Length | Stem–Loop Lengtha | Present in Condition | Similarity to Known miRNAs |
|---|---|---|---|---|---|
| avi-miR-temp-100 | acccguagauccgaacuugugg | 22 | 56 | All | nve-miR-100-5p |
| avi-miR-temp-2022 | uuugcuaguugcuuuugucccgc | 23 | 52; 53 | All | nve-miR-2022-3p |
| avi-miR-temp-2023 | aaagaaguacaagugguaggg | 21 | 53 | All | nve-miR-2023-3p |
| avi-miR-temp-2025 | uuuuuuagcccgcggaaguugu | 22 | 53 | All | nve-miR-2025-3p |
| avi-miR-temp-2028 | aaauguuccugcuuguuccug | 21 | 48 | All | nve-miR-2028-5p |
| avi-miR-temp-2030 | uagcauaacauaguaagagauu | 22 | 52 | All | nve-miR-2030-5p |
| avi-miR-temp-2036 | uauauuguacgacucucaucguag | 24 | 54 | All | nve-miR-2036-3p |
| avi-miR-temp-2037 | ugugauuggagacuuuuaucgu | 22 | 54 | All | nve-miR-2037-3p |
| avi-miR-temp-1 | gaucaagucaaauacaucucu | 21 | 48 | All | |
| avi-miR-temp-2 | uaucaaggcagucuuaccauau | 22 | 54 | All | |
| avi-miR-temp-3 | uacaaauguuacgcagcagaac | 22 | 55 | All | |
| avi-miR-temp-4 | ugacauugcugcccgaaucucc | 22 | 88 | All | |
| avi-miR-temp-5 | uuuaauguuacugcucguucc | 21 | 53 | All | |
| avi-miR-temp-6 | aauuucaaauauccacugauuga | 23 | 55 | All | |
| avi-miR-temp-7 | uugagcaucuguugcaugucua | 22 | 53 | All | |
| avi-miR-temp-8 | aucaucgccacuagcaucguca | 22 | 55 | All | |
| avi-miR-temp-9 | aagggcaagacaauagaauuuca | 23 | 59 | All | |
| avi-miR-temp-10 | cuugauaguacuuuugccuugc | 22 | 52 | pH 7.9, pH 8.2 | |
| avi-miR-temp-11 | uaguagguucuuauaagcuauu | 22 | 55 | All | |
| avi-miR-temp-12 | uauaagucuaggcugguuaaga | 22 | 56 | All | |
| avi-miR-temp-13 | auacugaacuugaaagaagugau | 23 | 55 | All | |
| avi-miR-temp-14 | aaacgcuguucuugguaguca | 21 | 55 | All | |
| avi-miR-temp-15 | uaacaaagcaguuuggcuguau | 22 | 55 | All | |
| avi-miR-temp-16 | ucuggcugauuugaagaaaga | 21 | 51 | All | |
| avi-miR-temp-17 | acaucaaacaaagcaguuug | 20 | 53 | All | |
| avi-miR-temp-18 | auuacccguaaauaaauucaau | 22 | 54 | All | |
| avi-miR-temp-19 | aaccccaacgcgggccucugg | 21 | 51 | All | |
| avi-miR-temp-20 | uuaguuugcacucauuugcugg | 22 | 55 | All | |
| avi-miR-temp-21 | auuacccagaauggggccuuu | 21 | 55 | All | |
| avi-miR-temp-22 | uauucuccaaaaauucacaagg | 22 | 52 | All | |
| avi-miR-temp-23 | uaaacuaguugauaggauugu | 21 | 51 | All | |
| avi-miR-temp-24 | acagauugcggcaaccgugcag | 22 | 72; 88 | All | |
| avi-miR-temp-25 | ucaaauguugcgcagcagaac | 21 | 55 | All | |
| avi-miR-temp-26 | ugcugcaguuuagacugaccuc | 22 | 53 | All | |
| avi-miR-temp-27 | uccucaaguuuugauuguaauac | 23 | 51; 52 | All | |
| avi-miR-temp-28 | uucuuaaguuuugauuguaauac | 23 | 52 | All | |
| avi-miR-temp-29 | aucuacugauacuaaguauccg | 22 | 54 | All | |
| avi-miR-temp-30 | uuucuguaguacuuuauccuggc | 23 | 54 | All | |
| avi-miR-temp-31 | uauucaaucagucuggcuguua | 22 | 52 | All | |
| avi-miR-temp-32 | ucuuuugauaaauaccaccaaca | 23 | 56 | All | |
| avi-miR-temp-33 | uacucugaaguguacuuagugu | 22 | 53; 54 | All | |
| avi-miR-temp-34 | gauaugauauaauauguaugug | 22 | 57 | All | |
| avi-miR-temp-35 | uauacauauuuaguaucgauaucag | 25 | 57; 58 | All | |
| avi-miR-temp-36 | uaaauacacaauaucuauagcagu | 24 | 55 | All | |
| avi-miR-temp-37 | uaugguagugauguuuagaaa | 21 | 49 | pH 7.6, pH 7.9 | |
| avi-miR-temp-38 | ccggacaaugagaauagcuga | 21 | 56 | All | |
| avi-miR-temp-39 | ugaucaauaaaagaaacaucguu | 23 | 54 | All | |
| avi-miR-temp-42 | uaucacauuuaaaacacucaug | 22 | 53 | All | |
| avi-miR-temp-43 | ucauacgauauuuuucacuagu | 22 | 55; 56 | All | |
| avi-miR-temp-44 | aaccucaugucagagaucaaa | 21 | 53 | pH 7.6, pH 8.2 | |
| avi-miR-temp-45 | acagagccuccuuuaaccuccu | 22 | 60 | pH 7.6, pH 8.2 | |
| avi-miR-temp-47 | ugguagaacaaguaacuugcugc | 23 | 55 | pH 7.6, pH 8.2 | |
| avi-miR-temp-48 | gaaaaagacauuuagagacuug | 22 | 56 | pH 7.6 | |
| avi-miR-temp-49 | aaugucaccaaguuucgacca | 21 | 50 | pH 7.6, pH 8.2 | |
| avi-miR-temp-50 | aggcccuggggaaacaaugga | 21 | 54 | pH 7.6, pH 7.9 | |
| avi-miR-temp-52 | uggaugcucaauuugccaauugc | 23 | 75 | pH 7.9, pH 8.2 | |
| avi-miR-temp-53 | aacuuaaaacaaaaaucucccu | 22 | 53 | pH 7.6, pH 8.2 | |
| avi-miR-temp-54-1 | aucuauucacugugggcguccagu | 24 | 54 | All | |
| avi-miR-temp-54-2 | aucuauucauugugggcguccagu | 24 | 55 | All | |
| avi-miR-temp-55 | uacuacuuugacaaugugaugg | 22 | 53; 77 | pH 7.6, pH 8.2 | |
| avi-miR-temp-56 | aggucagucuaaacugcagca | 21 | 54; 55 | pH 7.6, pH 8.2 | |
| avi-miR-temp-57 | gcuuugaaaauguaaagaaca | 21 | 50 | pH 7.6, pH 7.9 | |
| avi-miR-temp-58 | ugcaguauucaguaugcacua | 21 | 65 | pH 7.9 | |
| avi-miR-temp-59 | ucggcgccggucacgcgauaga | 22 | 52 | pH 7.9 | |
| avi-miR-temp-60 | caagcuauaaauuccaacuga | 21 | 50 | pH 7.6 | |
| avi-miR-temp-61 | ucgaguaaaauauuacagaaaug | 23 | 54 | pH 7.6, pH 8.2 | |
| avi-miR-temp-64 | ucaucucuuguggcuugacauu | 22 | 51 | pH 7.6, pH 7.9 | |
| avi-miR-temp-65 | uggugcaguuuagacugacccuu | 23 | 54 | pH 7.9 | |
| avi-miR-temp-66 | cuagauuaugagagcuuaugu | 21 | 53 | All | |
| avi-miR-temp-67 | ugugugaaaacaugacaagaucu | 23 | 50 | All |
Presence of two numbers in this column indicates that two different miRNA precursors (pre-miRNAs) of the same mature miRNAs have been detected in our genome assembly. All nucleotide sequences of mature and star miRNAs and pre-miRNA precursors are listed in supplementary table S2, Supplementary Material online.
Genome Context of Identified miRNAs
In sea anemones, little is known about the genomic miRNA clusters that generate pri-miRNAs. Therefore, we searched the A. viridis reference genome sequence for the presence of putative miRNA clusters. We identified four clusters, three contained two miRNA sequences (avi-miR-temp-11 and 66; avi-miR-temp-28 and 27; and avi-miR-temp-64 and 67) and one cluster contained three miRNAs (avi-miR-temp-2, 13, and 39) (supplementary fig. S1, Supplementary Material online). No open reading frames (ORFs) spanning cluster regions could be predicted, implying that the miRNAs were transcribed as independent transcription units. Further clustering of miRNA loci could not be assessed due to the fragmented nature of the genome assembly.
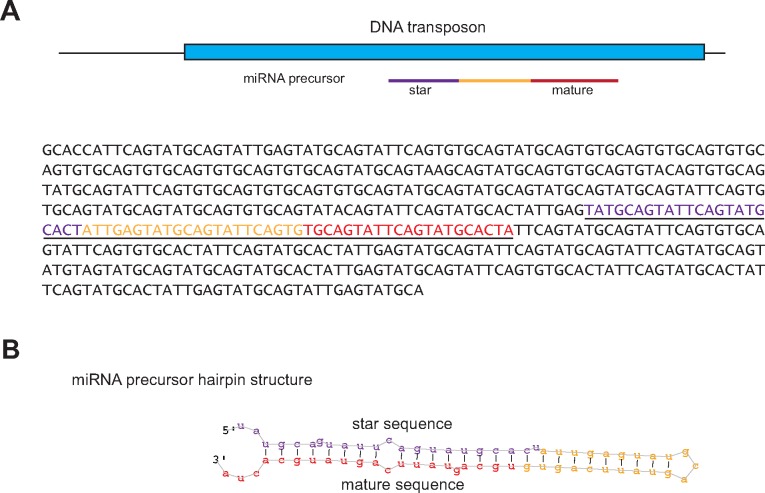
We then asked if any of the expressed miRNAs were colocalized with predicted transposable elements in the A. viridis genome reference. One miRNA (avi-miR-temp-58) was found encoded within a DNA transposon (fig. 4A). avi-miR-temp-58 was detected only at pH 7.9 in the small RNA sequencing experiment (but in all three individuals inspected). However, we detected avi-miR-temp-58 in all conditions studied by a quantitative PCR (qPCR) approach (see below), though in higher abundance at pH 7.9 (supplementary fig. S2, Supplementary Material online). avi-miR-temp-58 and its precursor was predicted to create a 1 nt 3′overhang (fig. 4B). The latter feature suggests a group II pre-miRNA that requires a 3′-end mono-uridylation for further Dicer processing (Heo et al. 2012).
Fig. 4.
—Precursor of avi-miR-temp-58 and its DNA transposon localization. (A) The miRNA precursor of avi-miR-temp-58 was found localized in a DNA transposon. A schematic representation of the scaffold region is depicted above the DNA sequence. Only the part of the scaffold with similarity to the DNA transposon is shown. The DNA transposon has homology to a transposable element from Crassostrea gigas. The miRNA precursor is marked in color and the whole sequence is underlined. Mature miRNA is indicated in red and star sequence in violet. (B) Hairpin structure of avi-miR-temp-58 precursor with 1 nt 3′ overhang.
Differential miRNA Expression upon Seawater pH Gradient
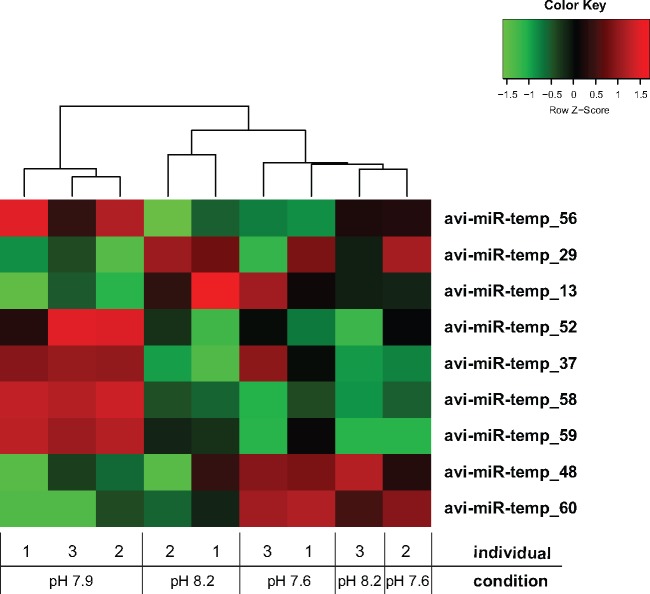
All high-confidence miRNAs were included in differential expression analyses. Despite that 19 candidate miRNAs could not be detected in all conditions studied, only nine miRNAs were recognized as differentially expressed between conditions by edgeR (FDR < 0.05) (fig. 5; supplementary tables S4 and S5, Supplementary Material online) (Robinson et al. 2010). Here, avi-miR-temp-37, 52, 56, 58, and 59 appeared up-regulated at pH 7.9, whereas avi-miR-temp-13, 29, 48, and 60 appeared down-regulated at pH 7.9 (fig. 5). Six miRNAs were then selected for verification analysis by qPCR (avi-miR-temp-37, 58, 60, 100, 2023, and 2028), where three miRNAs homologous to Nematostella with apparently unaffected expression levels in the different pH conditions served as controls (supplementary fig. S2, Supplementary Material online). The control miRNAs (avi-miR-temp-100, 2023, and 2028) were detected by qPCR in all pH conditions at similar expression levels, and thus are in good agreement with results generated from small RNA sequencing. The miRNA avi-miR-temp-37 was detected only at pH 7.9 and only in one individual at pH 7.6, and avi-miR-temp-60 was detected only at pH 8.2. However, in contrast with the observation from small RNA sequencing, we detected the presence of avi-miR-temp-58 in all conditions studied, though in higher abundance at pH 7.9 (supplementary fig. S2, Supplementary Material online).
Fig. 5.
—Differentially expressed miRNAs under low pH conditions. Nine miRNAs were found differentially expressed among the sampling sites (edgeR, FDR < 0.05). This included two miRNAs detected in all pH conditions studied (avi-miR-temp-13 and 29) and seven miRNAs that could be detected in only one or two different pH conditions. Five miRNAs were differentially expressed between pH 7.6 and 7.9, three downregulated (avi-miR-temp-52, 58, and 59) and two upregulated (avi-miR-temp-48 and 60) at pH 7.6 compared with pH 7.9. Only one miRNA was detected differentially expressed between pH 7.6 and 8.2 (avi-miR-temp-37), and it was upregulated at pH 7.6 compared with pH 8.2. Eight miRNAs were found differentially expressed between pH 7.9 and 8.2, three downregulated (avi-miR-temp-13, 29, and 48) and five upregulated (avi-miR-temp-37, 52, 56, 58, and 59) at pH 7.9 compared with pH 8.2. Differentially expressed miRNAs were hierarchically clustered into heatmap based on counts per million (cpm) and scaled by row.
We then searched for putative mRNA targets of 13 selected miRNAs that were differentially expressed along the pH gradient (avi-miR-temp-13, 29, 37, 48, 52, 56, 58, 59, and 60) (fig. 5), detected only in one pH condition (avir-miR-temp-48, 58, 59, 60, and 65), or detected only at low pH, that is, at pH 7.6 and/or pH 7.9 (avi-miR-temp-37, 48, 50, 57, 58, 59, 60, 64, and 65) (table 3). Differentially expressed mRNAs previously identified in A. viridis in the same individuals from the same sampling experiment (Urbarova et al., unpublished results) were assessed as potential targets. After stringent filtering criteria, including full seed matching with extended pairing, we identified 9 out of the 13 selected miRNAs that could potentially target 13 of the differentially expressed mRNAs along the low pH gradient (supplementary table S6, Supplementary Material online). Although we could not consistently detect miRNA upregulation and its mRNA target downregulation, we made one interesting observation. We detected avi-miR-temp-50, present only at pH 7.6 and pH 7.9, to target an RNase HI domain of a DIRS1 retrotransposon. The corresponding transcript was found downregulated both at pH 7.6 and 7.9 compared with pH 8.2, which could mean that this domain is inactivated and reverse transcription is therefore inhibited.
Search for Putative piRNAs and Their Characteristics
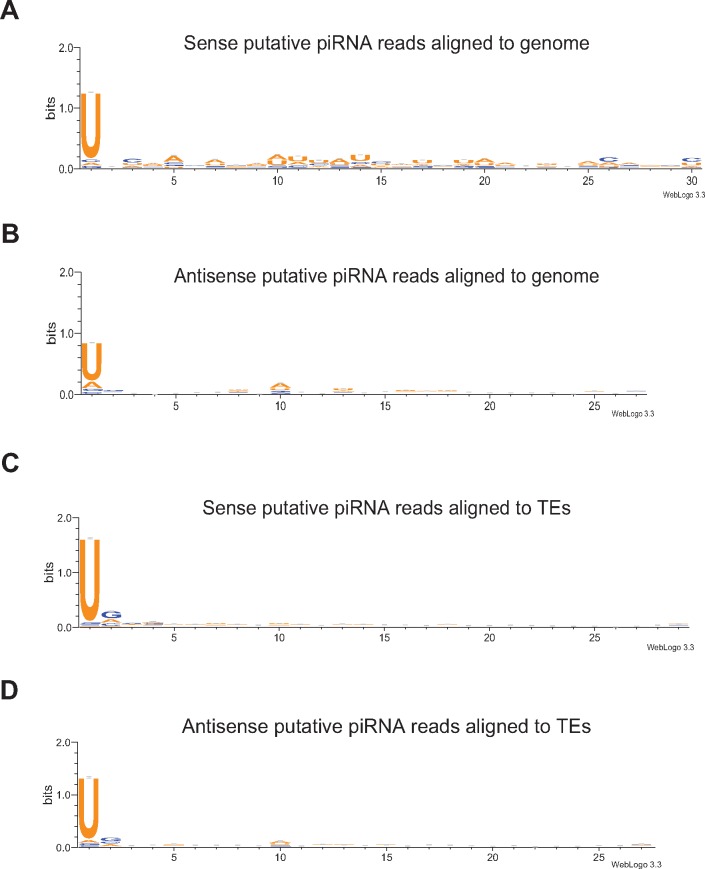
Most small RNA sequences present in our data set showed a distinct peak at 27–29 nt in the small RNA size distribution plot (fig. 2), and most likely represent PIWI-interacting RNAs (piRNAs). Based on an earlier report on piRNA signatures in cnidarians (Moran et al. 2014), we explored the trimmed and quality filtered small RNA reads with minimum length of 23 nt and with the typical base preference signatures (hereafter called putative piRNAs) in our data set. The putative piRNAs were aligned to two different reference data sets; the A. viridis reference genome, and the transposable elements identified in the genome. In total, about 83–90% of reads ≥23 nt aligned to the reference genome were putative piRNAs (table 2), with about 14–42% mapping to transposable elements (supplementary table S7, Supplementary Material online), including unclassified fraction of repeats. Most of the putative piRNA reads mapped to the reference genome and transposable elements showed strong preference for 1 U (fig. 6), a feature consistent with the primary piRNA population. Most of the putative piRNAs are found in genomic clusters and the majority of piRNA cluster loci appear unistranded (61–68%), where piRNAs are transcribed from one strand of the piRNA locus (supplementary fig. S3, Supplementary Material online).
Fig. 6.
—Base preferences of putative piRNA reads mapped to various data sets. Shown are base preferences of putative piRNA reads mapping to sense (A, C) and antisense (B, D) strand of genome (A, B) and transposable elements (TEs; C, D). Base preferences did not significantly differ at various pH conditions. Depicted is always one sequence set of specific length from one condition. The Y-axis represents the entropy score for the base bias.
About one third of the genome scaffolds contained expressed piRNA loci (supplementary table S8, Supplementary Material online). We observed more expressed piRNA loci at pH 7.9 than at pH 8.2 or 7.6. Putative piRNAs were found to map to about 24–30% of the identified transposable elements in all conditions and in all individuals (supplementary table S9, Supplementary Material online). These included both DNA transposons and retrotransposons (supplementary fig. S4, Supplementary Material online). Interestingly, retrotransposons appeared more frequently targeted by piRNAs than DNA transposons (supplementary table S9, Supplementary Material online).
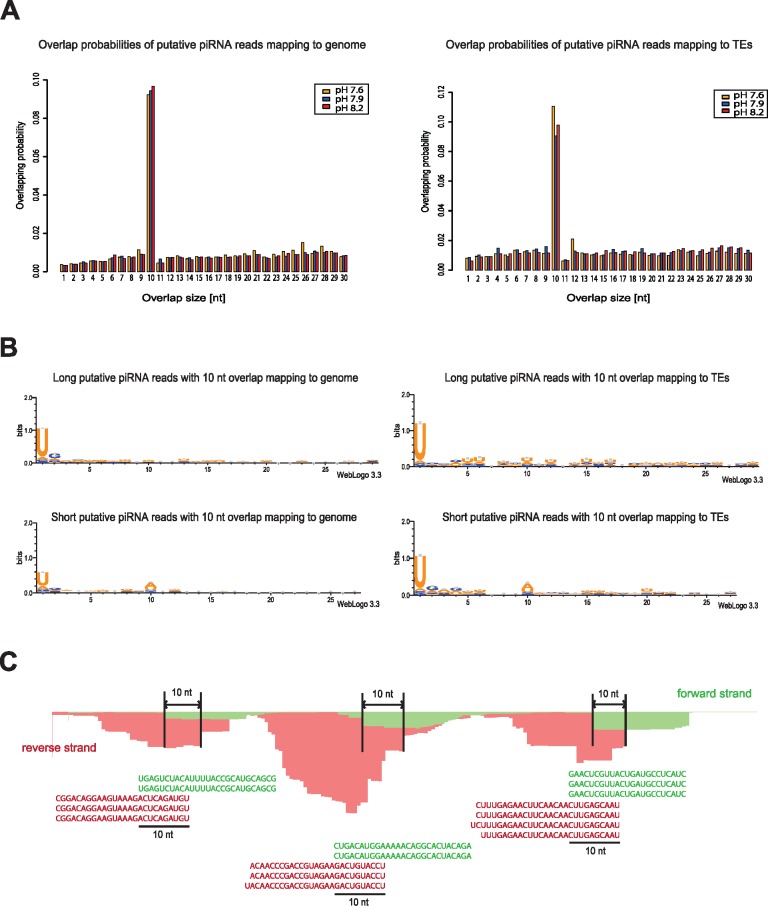
Ping–Pong piRNA Amplification Signature in A. viridis
We further investigated if ping–pong signatures, that is, 10 nt overlaps of putative piRNAs with opposite direction, were common in our data set. The probability of overlap by 1–30 nt of putative piRNAs with opposite orientation was assessed. The data exhibited strong ping–pong signatures, since most reads showed preference for 10 nt 5′overlaps of putative piRNAs with opposite orientation in all conditions, in all individuals, and for all reference data sets (z-score > 5). Probability of other overlaps was much lower (z-score < 1) (fig. 7A). We could detect ∼14–15% putative piRNAs mapping to identified transposable elements, and up to 42% (supplementary table S7, Supplementary Material online) when including the unclassified fraction of identified repeats (supplementary table S1, Supplementary Material online). Only around 10% of putative piRNAs mapped to transposable elements showed ping–pong signatures. A slightly higher proportion of putative piRNA reads with ping–pong signatures was found to map to the genome reference outside the repeat-enriched regions (∼17%). Majority of these most probably represent protein-coding genes. When assessing the base-preferences of piRNAs with ping–pong signatures, we found that a substantial amount of the putative piRNAs had 1 U preference (primary piRNAs) (fig. 7B). Sequences mapping to transposable elements showed preference for 1 U in both sense and antisense orientation (figs. 7B and C). Shorter antisense reads (27 nt) mapped to transposable elements showed minor preference for 10 A (secondary piRNAs) (fig. 7B). Only around 8–9% of the putative piRNAs appeared to be targeting transposable elements in all conditions studied (supplementary table S7, Supplementary Material online). This fraction further increased nearly up to 22% when including the unclassified fraction of the identified repeats in the genome (supplementary table S7, Supplementary Material online). These might potentially represent very divergent transposable elements, as reported also for Exaiptasia sp. (Baumgarten et al. 2015). Higher fraction of putative piRNAs targeting transposable elements is also expected to be found during development or when extracting specifically germline cells from A. viridis, as observed in N. vectensis (Praher et al. 2017).
Fig. 7.
—Ping–pong pathway signature. (A) Overlap probabilities of sense and antisense reads mapping to the genome and transposable elements (TEs). Overlap probabilities in all the different pH conditions are shown. (B) Depicted are base preferences of putative piRNA reads with ping–pong signatures mapping to the genome and TEs. The Y-axis represents the entropy score for the base bias. (C) Shown are putative piRNA reads aligned to a TE. Three regions with identified ping–pong signatures are highlighted. Green reads correspond to sense strand, and red reads to antisense strand.
Ping–pong activity is mainly linked to transposon silencing (Brennecke et al. 2007; Gunawardane et al. 2007). Therefore, we investigated if ping–pong activity changes could be observed at different pH conditions. Transposable elements were only considered silenced if a significant ping–pong activity feature could be detected within the transposon region, with FDR (q value) < 0.01. We found that a possible ping–pong dependent suppression varied among individuals in each condition, and examples from the BEL and Gypsy LTR retrotransposons are shown in supplementary figures S5 and S6, Supplementary Material online. However, only a small fraction of the identified transposable elements appeared silenced by the ping–pong pathway in all conditions studied (< 1%; supplementary table S10, Supplementary Material online).
Discussion
Here, we report a preliminary draft genome reference sequencing of the symbiotic sea anemone A. viridis, with an estimated genome size of approximately 313 Mb. The partially assembled reference genome was used to assess transposable element and small RNA loci. We also performed small RNA sequencing along a natural seawater pH gradient and identified differentially expressed RNA candidates. In A. viridis, we found 70 distinct miRNAs and thousands of putative piRNAs, suggesting that small RNAs are widespread regulators in the control of gene expression and transposable element silencing in this species.
The estimated genome size of A. viridis appears intermediate compared with the sea anemones Exaiptasia sp. (260 Mb) and Nematostella vectensis (329/450 Mb), slightly less than the stony coral Acropora digitifera (420 Mb), and substantially smaller than the freshwater hydroid Hydra magnipapillata (1.3 Gb) (Putnam et al. 2007; Chapman et al. 2010; Shinzato et al. 2011; Baumgarten et al. 2015). Thus, a general trend is that hexacorals harbor relatively small genomes. We found that about 36% of the A. viridis genome contains repeated sequences, which is a higher fraction than Exaiptasia and Nematostella (both 26%) and Acropora (13%), but less than Hydra (57%) (Putnam et al. 2007; Chapman et al. 2010; Shinzato et al. 2011; Baumgarten et al. 2015). There is a significant heterogeneity in the distribution of classes and subclasses of transposable elements among the investigated cnidarians. Whereas Hydra contains approximately equal fractions of DNA transposons and retrotransposons, Acropora harbors four times as many retrotransposons than DNA transposons. There are also significant differences between the sea anemones Nematostella and Exaiptasia. The non-symbiotic Nematostella was reported to carry about four times more DNA transposons than retrotransposons (Putnam et al. 2007), which contrasts that of the symbiotic Exaiptasia with slightly more retrotransposons than DNA transposons (Baumgarten et al. 2015). Our data from the symbiotic A. viridis does not appear to resemble any previously sequenced cnidarian in terms of the transposable element distribution, even though it contains approximately equal fractions of DNA transposons and retrotransposons. It is interesting to note that the Gypsy element is the most frequent LTR retrotransposon in all the cnidarian species, including A. viridis.
We identified 70 distinct miRNAs in A. viridis, and 61 of these were detected in normal seawater conditions at pH 8.2. Only eight miRNAs were similar to previously known miRNAs in Nematostella (Grimson et al. 2008; Moran et al. 2014), six in Acropora (Gajigan and Conaco 2017), five in Stylophora (Liew et al. 2014) and two in Hydra (Krishna et al. 2013). These results support that taxonomically restricted miRNAs are common to cnidarians, including A. viridis—an observation seen mainly in plants, and which could be explained by high sequence turnover rates of miRNAs, as suggested by Moran et al. (2017). In agreement with other reports in cnidarians (Grimson et al. 2008; Wheeler et al. 2009; Liew et al. 2014; Moran et al. 2014, Gajigan and Conaco 2017), we detected only one miRNA in A. viridis (avi-miR-temp-100) to be conserved with miRNAs in bilaterians. This miRNA belongs to the miR-100 family, and it was found identical in sequence to nve-miR-100 and spi-miR-100 in Nematostella and Stylophora, respectively (Grimson et al. 2008; Wheeler et al. 2009; Liew et al. 2014; Moran et al. 2014). In bilaterians, including nematodes and humans, miR-100 makes a cluster in the genome together with let-7 and miR-125, and regulates transcripts involved in multiple cellular and developmental processes, as well as cancer progression (Christodoulou et al. 2010; Sokol 2012; Li et al. 2015). The absence of miR-51/miR-100 family was first reported in nematodes to result in lethality during development (Shaw et al. 2010). In A. viridis, as well as in other cnidarians, miR-100 appears to be transcribed from an individual gene locus, but its biological role in gene repression is not well established. In the coral Stylophora, Liew and coworkers speculated that miR-100 could be involved in the calcification process (Liew et al. 2014). However, since sea anemones lack any sort of calcified skeleton, other processes have to be regulated by miR-100 in sea anemones.
We identified and described four miRNA clusters within the A. viridis genome. However, a more detailed analysis in regard to clustering of individual miRNAs was not possible due to insufficient contiguity of our draft assembly. Therefore, we cannot exclude that some miRNAs predicted in our study form additional miRNA clusters, where miRNA pairs are located further apart. Interestingly, one of the identified miRNA locus localized inside a DNA transposon (fig. 4), and this miRNA (avi-miR-temp-58) appeared expressed mostly at seawater pH 7.9. The formations of small RNAs from transposable element loci are not unusual among animals, and dozens of publications inspecting various species have reported miRNAs originating from transposable elements (reviewed in Roberts et al. 2014). However, to our knowledge avi-miR-temp-58 is the first example of a TE-encoded miRNA reported in any cnidarian. We found nine miRNAs to be differentially expressed in A. viridis along the seawater pH gradient, indicating that miRNA-based gene repression might be involved in compensating environmental stressors. Here, we identified few potential mRNA targets, including stress-related and mobile element proteins.
PIWI-interacting RNAs (piRNAs) have previously been reported in Nematostella (Grimson et al. 2008; Praher et al. 2017). This sea anemone contains two piRNA classes, where class I possesses an unknown function during germline development and class II is involved in gene silencing, including transposons, by the ping–pong mechanism. In A. viridis, we found a high number of expressed piRNA candidates, apparently representing both piRNA classes, even though we did not specifically extract and analyze germline cells in our study. However, we were not able to characterize piRNA gene loci at high resolution in A. viridis due to presence of many short scaffolds in the genome assembly.
A relatively high proportion of our piRNA reads showed a strong enrichment for uridine at 5′ends (1 U) and a higher probability to carry an adenine at the nucleotide 10 (10 A). In addition, the majority of sense and antisense putative piRNA reads showed an overlap by exactly ten nucleotides. This bidirectional production of piRNA reads with 10 nt offset indicated a ping–pong dependent piRNA biogenesis. However, only a small fraction of the putative piRNA reads that mapped to transposable elements showed ping–pong signatures. Although we could detect nearly 22% putative piRNAs potentially targeting transposons, only around 10% showed ping–pong signatures in all pH conditions studied. This might be caused by very high divergence of transposable elements in A. viridis, an observation made previously in Exaiptasia sp. (Baumgarten et al. 2015). Another possible explanation is that piRNAs may fulfil various functions mainly during development or in female adults. Here, TE-targeting piRNAs could be connected to the process of oogenesis and serve the maintenance of the germline genome, as recently reported by Praher et al. (2017). However, it indicates that piRNAs in cnidarians may also have additional function to that of transposable element silencing, a notion supported by observations in Hydra (Krishna et al. 2013; Juliano et al. 2014). More detailed characteristics of the putative piRNA population remain to be elucidated once better genome assembly and gene predictions are available for A. viridis. One important aim of our study was to identify and assess differentially expressed small RNAs along a natural seawater pH gradient. We found high amounts of putative piRNA reads in all the different pH conditions. Although it was difficult to detect any significantly differentially expressed piRNAs or piRNA clusters at this point, we noted an increase in putative piRNA expression at pH 7.9 compared with pH 7.6 and 8.2. One possible biological implication could be less restricted transposon activities at seawater pH 7.6 compared with pH 7.9.
Conclusion
The A. viridis genome appears similar in size and in transposable element divergence to that of Exaiptasia sp., a related sea anemone with a symbiotic lifestyle resembling that of Anemonia spp. The A. viridis genome encodes and expresses a high number of small regulatory RNAs, and when compared with the sea anemone Nematostella, a large fraction (89%) of miRNAs appears taxonomically restricted. A. viridis expresses a high amount of candidate piRNA sequences with putative functions in transposable element silencing and in other still unknown cellular functions. Some small RNAs appeared differentially expressed along a seawater pH gradient, suggesting a regulatory role in the response to environmental stressors.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank to Sebastian Uhrig, Johannes Gutenberg University of Mainz, for advices using PingPongPro software tool, Inuk Jung, Seoul National University, for assistance in running piClust software tool, and Professor Don Gilbert, Indiana University, for advices on filtering of the transposable element search output. We also thank members of the RAMP research group at UiT and the Genomics group at Nord University for practical support and discussions. We also thank two anonymous reviewers for their valuable comments and suggestions that helped us to improve the manuscript. This work was supported by grants from the Research Council of Norway (CoralSeq; to S.D.J.); and Tromsø Research Foundation (to S.D.J.). The publication charges for this article have been funded by a grant from the publication fund of UiT The Arctic University of Norway.
Author Contributions
I.U. and S.D.J. designed the study; I.U. collected, analyzed and interpreted the data; S.F., H.P., B.O.K., and I.U. designed workflows for the data analyses; H.P. wrote two Perl scripts for processing of miRNA sequencing data; I.U. and J.M.H.-S. organized and performed the fieldwork; T.E.J. sequenced the small RNA libraries; all authors helped I.U. and S.D.J. prepare the manuscript for publication. All authors (except S.F.) reviewed, commented and approved the final manuscript for publication. S.F. recently passed away; S.F. reviewed, commented and approved an earlier version of the final manuscript.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402.http://dx.doi.org/10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewski C. 2014. Computing siRNA and piRNA overlap signatures. Methods Mol Biol. 1173:135–146. [DOI] [PubMed] [Google Scholar]
- Aranda M, et al. 2016. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci Rep. 6:39734.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442(7099):203–207. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297.http://dx.doi.org/10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233.http://dx.doi.org/10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci U S A. 112(38):11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatta F, et al. 2013. Geochemical survey of Levante Bay, Vulcano Island (Italy), a natural laboratory for the study of ocean acidification. Mar Pollut Bull. 73(2):485–494.http://dx.doi.org/10.1016/j.marpolbul.2013.01.029 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120.http://dx.doi.org/10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, et al. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128(6):1089–1103.http://dx.doi.org/10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Chapman JA, et al. 2010. The dynamic genome of Hydra. Nature 464(7288):592–596.http://dx.doi.org/10.1038/nature08830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou F, et al. 2010. Ancient animal microRNAs and the evolution of tissue identity. Nature 463(7284):1084–1088.http://dx.doi.org/10.1038/nature08744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 31(1):79–90.http://dx.doi.org/10.1016/j.molcel.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N.. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40(1):37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajigan AP, Conaco C.. 2017. A microRNA regulates the response of corals to thermal stress. Mol Ecol. 26(13):3472–3483.http://dx.doi.org/10.1111/mec.14130 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD.. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet. 10(2):94–108.http://dx.doi.org/10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, et al. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36(10):3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales A, Rodríguez E.. 2014. Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actiniaria, Metridioidea). Zootaxa 3826(1):55–100. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R.. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123(4):631–640. [DOI] [PubMed] [Google Scholar]
- Grimson A, et al. 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455(7217):1193–1197.http://dx.doi.org/10.1038/nature07415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, et al. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315(5818):1587–1590. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512.http://dx.doi.org/10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD.. 2015. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348(6236):817–821.http://dx.doi.org/10.1126/science.aaa1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, et al. 2012. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151(3):521–532.http://dx.doi.org/10.1016/j.cell.2012.09.022 [DOI] [PubMed] [Google Scholar]
- Horwitz R, Borell EM, Yam R, Shemesh A, Fine M.. 2015. Natural high pCO2 increases autotrophy in Anemonia viridis (Anthozoa) as revealed from stable isotope (C, N) analysis. Sci Rep. 5:8779.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129(1):69–82.http://dx.doi.org/10.1016/j.cell.2007.03.026 [DOI] [PubMed] [Google Scholar]
- Johnson VR, et al. 2013. Responses of marine benthic microalgae to elevated CO2. Mar Biol. 160(8):1813–1824. [Google Scholar]
- Juliano CE, et al. 2014. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc Natl Acad Sci U S A. 111(1):337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I, Park JC, Kim S.. 2014. piClust: a density based piRNA clustering algorithm. Comput Biol Chem. 50:60–67.http://dx.doi.org/10.1016/j.compbiolchem.2014.01.008 [DOI] [PubMed] [Google Scholar]
- Kajitani R, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395.http://dx.doi.org/10.1101/gr.170720.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, et al. 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453(7196):793–797.http://dx.doi.org/10.1038/nature06938 [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E.. 2007. The role of site accessibility in microRNA target recognition. Nat Genet. 39(10):1278–1284. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S.. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42(Database issue):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S, et al. 2013. Deep sequencing reveals unique small RNA repertoire that is regulated during head regeneration in Hydra magnipapillata. Nucleic Acids Res. 41(1):599–616.http://dx.doi.org/10.1093/nar/gks1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, et al. 2015. Multiple roles of microRNA-100 in human cancer and its therapeutic potential. Cell Physiol Biochem. 37(6):2143–2159.http://dx.doi.org/10.1159/000438572 [DOI] [PubMed] [Google Scholar]
- Liew YJ, et al. 2014. Identification of microRNAs in the coral Stylophora pistillata. PLoS One 9(3):e91101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, et al. 2015. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350(6261):691–694.http://dx.doi.org/10.1126/science.aad0408 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.http://dx.doi.org/10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min XJ, Butler G, Storms R, Tsang A.. 2005. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33(Web Server issue):W677–W680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J.. 2015. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348(6236):812–817.http://dx.doi.org/10.1126/science.aaa1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Praher D, Fredman D, Technau U.. 2013. The evolution of miRNA pathway protein components in Cnidaria. Mol Biol Evol. 30(12):2541–2552.http://dx.doi.org/10.1093/molbev/mst159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, et al. 2014. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 24(4):651–663.http://dx.doi.org/10.1101/gr.162503.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Agron M, Praher D, Technau U.. 2017. The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol. 1(3):27..http://dx.doi.org/10.1038/s41559-016-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ.. 1988. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 85(8):2444–2448.http://dx.doi.org/10.1073/pnas.85.8.2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praher D, et al. 2017. Characterization of the piRNA pathway during development of the sea anemone Nematostella vectensis. RNA Biol. 7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317(5834):86–94.http://dx.doi.org/10.1126/science.1139158 [DOI] [PubMed] [Google Scholar]
- Roberts JT, Cardin SE, Borchert GM.. 2014. Burgeoning evidence indicates that microRNAs were initially formed from transposable element sequences. Mob Genet Elements. 4:e29255.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140.http://dx.doi.org/10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, et al. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115(2):199–208.http://dx.doi.org/10.1016/S0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- Shaw WR, Armisen J, Lehrbach NJ, Miska EA.. 2010. The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics 185(3):897–905.http://dx.doi.org/10.1534/genetics.110.117515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476(7360):320–323.http://dx.doi.org/10.1038/nature10249 [DOI] [PubMed] [Google Scholar]
- Shoguchi E, et al. 2013. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 23(15):1399–1408.http://dx.doi.org/10.1016/j.cub.2013.05.062 [DOI] [PubMed] [Google Scholar]
- Simpson JT. 2014. Exploring genome characteristics and sequence quality without a reference. Bioinformatics 30(9):1228–1235.http://dx.doi.org/10.1093/bioinformatics/btu023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R.. 2008–2015. RepeatModeler Open-1.0. Available from: http://www.repeatmasker.org (last accessed January 14, 2018).
- Smit AFA, Hubley R, Green P.. 2013–2015. RepeatMasker Open-4.0. Available from: http://www.repeatmasker.org (last accessed January 14, 2018).
- Sokol NS. 2012. Small temporal RNAs in animal development. Curr Opin Genet Dev. 22(4):368–373.http://dx.doi.org/10.1016/j.gde.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suggett DJ, et al. 2012. Sea anemones may thrive in a high CO2 world. Glob Chang Biol. 18(10):3015–3025.http://dx.doi.org/10.1111/j.1365-2486.2012.02767.x [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK.. 2007. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12(7):301–309.http://dx.doi.org/10.1016/j.tplants.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Vagin VV, et al. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313(5785):320–324.http://dx.doi.org/10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- Vashisht D, Nodine MD.. 2014. MicroRNA functions in plant embryos. Biochem Soc Trans. 42(2):352–357.http://dx.doi.org/10.1042/BST20130252 [DOI] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136(4):669–687.http://dx.doi.org/10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Wheeler BM, et al. 2009. The deep evolution of metazoan microRNAs. Evol Dev. 11(1):50–68.http://dx.doi.org/10.1111/j.1525-142X.2008.00302.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.