Abstract
Invasive Aspergillosis (IA) is a life-threatening infectious disease caused by fungi from the genus Aspergillus, with an associated mortality as high as 90% in certain populations. IA-associated pulmonary lesions are characteristically depleted in oxygen relative to normal lung tissue, and it has been shown that the most common causal agent of IA, Aspergillus fumigatus, must respond to low-oxygen environments for pathogenesis and disease progression. Previous studies have demonstrated marked alterations to the Aspergillus fumigatus transcriptome in response to low oxygen environments that induce a “hypoxia response.” Consequently, we hypothesized that these transcriptomic changes would alter the volatile metabolome and generate a volatile hypoxia signature.
In the present study, we analyzed the volatile molecules produced by A. fumigatus in both oxygen replete (normoxia) and depleted (hypoxia) environments via headspace solid-phase microextraction coupled to two-dimensional gas chromatography-time-of-flight mass spectrometry (HS-SPME-GC×GC-TOFMS). Using the machine learning algorithm Random Forest, we identified 19 volatile molecules that were discriminatory between the four growth conditions assessed in this study (i.e., early hypoxia (1 h), late hypoxia (8 h), early normoxia (1 h), and late normoxia (8 h)), as well as a set of 19 that were discriminatory between late hypoxia cultures and all other growth conditions in aggregate. Nine molecules were common to both comparisons, while the remaining 20 were specific to only one of two.
We assigned putative identifications to 13 molecules, of which six were most highly abundant in late hypoxia cultures. Previously acquired transcriptomic data identified putative biochemical pathways induced in hypoxia conditions that plausibly account for the production of a subset of these molecules, including 2,3-butanedione and 3-hydroxy-2-butanone. These two molecules may represent a novel hypoxia fitness pathway in A. fumigatus, and could be useful in the detection of hypoxia-associated A. fumigatus lesions that develop in established IA infections.
Keywords: Aspergillus fumigatus, Fungal metabolites, Hypoxia, Invasive Aspergillosis, Volatile organic compounds (VOCs), Two-dimensional gas chromatography-time-of-flight mass spectrometry (GC×GC-TOFMS)
Introduction
Aspergillus fumigatus is an environmental mold responsible for severe pulmonary and extrapulmonary disease in many individuals [1]. In those with airway hyperresponsiveness or cystic fibrosis, inhalation of A. fumigatus conidia can result in allergic bronchopulmonary aspergillosis (ABPA) that often necessitates treatment with oral corticosteroids and clinical monitoring [2]. On the other end of the immunological spectrum, immune compromised individuals are at risk for invasive aspergillosis (IA), defined by invasion of Aspergillus into the bloodstream from the respiratory tract (with possible further invasion into other tissues and organs). IA is of particular concern in individuals with quantitative or qualitative neutrophil defects, including high dose corticosteroid therapy for graft versus host disease, chemotherapy, and recent organ transplantation [3]. Mortality associated with IA can be as high as 90%, for example, in individuals with recent hematopoietic stem-cell transplantation [4, 5].
Histopathologic and cytologic examination of lung tissue accompanied by fungal culture is currently recommended for the diagnosis of IA [6]. However, the process of obtaining tissue for examination can result in potentially severe complications, such as pneumothorax or pulmonary hemorrhage [7]. Immunoassays conducted using blood or bronchoalveolar lavage (BAL) fluid can be used for the detection of the Aspergillus-associated surface markers galactomannan and 1,3 β-D-glucan, although these assays often suffer from unacceptably low sensitivities (average of approximately 0.71 for galactomannan [8] and 0.77 for β-D-glucan [9]). Alternatively, nucleic acid amplification of fungal ribosomal RNA (rRNA) in either whole blood or serum yields a higher average sensitivity (0.84) relative to either galactomannan or β-D-glucan-based assays, but at the cost of specificity (average of 0.76) [10]. Culturing of sputum, an expectorated sample from the respiratory tract that cannot be produced by all patients, is the least invasive option for the detection of A. fumigatus, and while this technique is poorly-suited for discriminating between colonization and infection in the setting of an immunocompetent individual, isolation of A. fumigatus from an immunocompromised host yields a positive predictive value of upwards of 0.90 [3]. Taken together, there remains a clear need for a less invasive diagnostic assay for IA with sensitivities and specificities that approach the current diagnostic gold-standard of lung tissue histopathology [11].
There is evidence to suggest that the presence of A. fumigatus in a human lung can be discerned by measuring the small molecules in their exhaled breath, a biofluid that can be collected non-invasively. For example, Koo and colleagues have demonstrated that Aspergillus-derived sesquiterpenes (α- and β-trans-bergamotene), identified via in vitro culture experiments, were found in higher abundance in the breath of individuals with IA relative to those without, with a sensitivity and specificity of 0.94 and 0.93, respectively [12]. Further, Chambers and colleagues have reported that 2-pentylfuran, which they identified in cultures of different Aspergillus species, was also present in the exhaled breath of individuals who cultured A. fumigatus from either sputum or BAL, with a sensitivity and specificity of 0.77 and 0.78, respectively [13]. Given the suite of volatile monoterpenes, sesquiterpenes, and heterocycles that have been reported for A. fumigatus, the development of a breath-based diagnostic of IA seems achievable. However, one important consideration that remains to be addressed is how volatile molecules can discriminate between Aspergillus colonization and the development of pathogenic processes such as IA.
One potential approach to discriminate between these clinical presentations is through the use of biomarkers associated with the metabolic state of A. fumigatus during IA. Previous work has demonstrated that the progression of invasive Aspergillus lesions is accompanied by the development of severely low-oxygen microenvironments [14]. Response to a low-oxygen environment is critical for IA during the development of invasive lesions [15], where necrosed, poorly-perfused tissue is a common pathological finding [16]. A. fumigatus responds to low-oxygen conditions by changing its gene expression profile and metabolic enzyme production [17, 18], altering its production of virulence factors, and increasing its utilization of low oxygen (hypoxia response) respiratory pathways [19]. It is therefore likely that the metabolic profile of the organism, including its volatile metabolome, also changes in response to alterations in oxygen tension that occur throughout the course of disease. In the present study, we explore the volatile metabolome of A. fumigatus, with a specific interest in the hypoxia-associated volatile metabolome, by comparing the volatile molecules produced in vitro under both normoxia and hypoxia conditions. The identification of biomarkers specific to the fungal hypoxia response would aid in the establishment of breath-based IA diagnostic method development, assist in monitoring the temporal progression of IA in at risk patient populations, and provide fundamental insight into mechanisms of hypoxia adaptation and virulence.
Materials and Methods
Fungal Strains and Culturing
Aspergillus fumigatus strain CEA10 was used for all experiments, as this clinical isolate has been extensively studied in the context of hypoxia fitness [14, 15, 18, 20]. Conidial suspensions were stored in 50% glycerol at -80 °C and maintained on 1% glucose minimal media, as previously described [21]. Conidia for experimental cultures were collected in 0.01% Tween-80. Fungal cultures of 106 conidia/mL were grown in 50 mL glucose minimal medium (GMM; 6 g/L NaNO3, 0.52 g/L KCl, 0.52 g/L MgSO4•7H2O, 1.52 g/L KH2PO4, 2.2 mg/L ZnSO4•7H2O, 1.1 mg/L H3BO3, 0.5 mg/L MnCl2•4H2O, 0.5 mg/L FeSO4•7H2O, 0.16 mg/L CoCl2•5H2O, 0.16 mg/L CuSO4•5H2O, 0.11 mg/L (NH4)6Mo7O24•4H2O, 5 mg/L Na4EDTA, 1% glucose; pH 6.5) at 37 °C with shaking at 250 rpm for 18 h. Mycelia were collected with vacuum filtration and transferred to 50 mL fresh liquid GMM. Cultures were incubated at 37 °C with 200 rpm shaking in either normoxia (ambient air; ∼21% O2, ∼0.04% CO2) or hypoxia (0.2% O2, 5% CO2) for either 1 h (early cultures) or 8 h (late cultures). Hypoxia was achieved using an INVIVO2 400 hypoxia chamber (The Baker Company (Ruskinn), Sanford, ME). Culture conditions were chosen in an attempt to model the gaseous environment that A. fumigatus is exposed to from initiation to progression of disease. Ten biological replicates were prepared for each of the four experimental conditions.
Sample Preparation and Concentration of Volatile Molecules
Culture supernatants were filtered through Miracloth (Merck Millipore, Billerica, MA) to remove mycelia, and 4 mL were transferred to 20 mL headspace vials that were immediately sealed with a PTFE/silicone cap (Sigma-Aldrich, St. Louis, MO). Samples were stored at -20 °C prior to analysis of volatile molecules, and all samples were analyzed within one week of collection. Headspace volatile molecules were concentrated using a 2 cm Divinylbenzene/Carboxen/Polydimethylsiloxane (DVB/CAR/PDMS) solid-phase micro-extraction (SPME) fiber (film thickness: 50/30 μm) (Supelco, Bellenfonte, PA). The SPME fiber was suspended in the headspace for 30 min at 50 °C with 250 rpm agitation. Sterile media controls were prepared identically.
Gas Chromatographic and Mass Spectrometric Methods
The GC×GC-TOFMS (Pegasus 4D, LECO Corporation, St. Joseph, MI) was equipped with a rail autosampler (MPS, Gerstel, Linthicum Heights, MD) and fitted with a two-dimensional column set consisting of an Rxi-624Sil MS (60 m × 250 μm × 1.4 μm (length × internal diameter × film thickness); Restek Corporation, Bellefonte, PA) first column followed by a Stabilwax (Crossbond Carbowax polyethylene glycol; 1 m × 250 μm × 0.5 μm; Restek) second column. The primary oven ramped at 5.0 °C/min from 35 °C to 230 °C, with a 5 min hold at 230 °C. The secondary oven, and quad-jet modulator (2.0 s modulation period, 0.5 s alternating hot and cold pulses), were heated in step with the primary oven with +5 °C and +25 °C offsets, respectively. The helium carrier gas flow rate was 2 mL/min. A splitless injection was used, with a 180 s desorption time. The inlet and transfer line temperatures were set to 270 °C and 250 °C, respectively. Time-of-flight mass spectra were acquired over the range of 30 to 500 m/z, with an acquisition rate of 200 spectra/s, and a 70 eV electron ionization. Data acquisition and analysis were performed using ChromaTOF software, version 4.50 (LECO Corp.).
Determination of Retention Indices
Retention indices (RIs) were calculated using external alkane standards (C6 – C17). The SPME fiber was exposed to a vial containing a pure retention index mixture (Sigma-Aldrich) for 10 min at 50 °C and desorbed at a 40:1 split. Reported RIs are between the literature values for polar and nonpolar column sets, owing to the midpolarity of the Rxi-624Sil MS stationary phase. Retention indices less than 600 (corresponding to C6) or greater than 1700 (corresponding to C17) were not extrapolated.
Data Processing, Chromatographic Alignment, and Compound Reporting
Chromatographic data was processed and aligned using the Statistical Compare feature of ChromaTOF. For peak identification, a signal-to-noise (S/N) cutoff was set at 50:1, and resulting peaks were identified by a forward search of the NIST 2011 library. Subpeaks within a chromatogram were combined if their second dimension retention time shift was ≤ 0.1 s between subsequent modulation periods, and their mass spectral match score was ≥ 600 (out of 1000). For peaks detected at a S/N threshold of ≥ 50:1 in at least one chromatogram, a reduced threshold of 5:1 was employed to assess low-level compound abundance in other chromatograms. For the alignment of peaks across chromatograms, maximum allowed first and second-dimension retention time deviations were set at 6.0 s and 0.15 s, respectively.
A forward match score of at least 850/1000, accompanied by affirmation via linear retention indices, was required for putative compound identification. For four of the five hypoxia-associated compounds that could be linked to putative metabolic pathways (2,3-butanedione, 2-methylpropanal, 3-hydroxy-2-butanone, and 3-methylbutanal) standard solutions (Sigma-Aldrich) were analyzed using the same analytical method described above (Supplementary Table S1). The solutions (ranging from 0.1 to 2.0 ppm) allowed for the validation of compound identifications, and assessment of linearity under the analytical conditions employed in this study.
Statistical Analyses
Statistical analyses were performed using R version v3.2.2 (R Foundation for Statistical Computing, Vienna, Austria). Atmospheric gases (e.g., carbon dioxide and argon), as well as suspected chromatographic artifacts or contaminants [22], were eliminated prior to statistical analyses. In addition, peaks eluting prior to 300 s were eliminated due to inefficient modulation of low molecular weight compounds [23]. The relative abundance of compounds across chromatograms was normalized using Probabilistic Quotient Normalization [24]. The machine learning algorithm Random Forest (RF) was used to identify volatile molecules that were discriminatory between different experimental conditions [25]. For each statistical model, 100 iterations of RF were performed, and volatile molecules were ranked according to their average mean decrease in accuracy (MDA), a measure of variable importance. Molecules with an MDA > 0 across all 100 RF iterations were selected as being discriminatory. Principal component analysis (PCA) was performed to reduce data dimensionality and visualize variance amongst samples using the molecules selected from RF [26]. Hierarchical clustering analysis (HCA) was used to assess relatedness between samples, using Euclidean distance as the distance metric.
Supplementary Figure S1 summarizes the methods employed in this study.
Results and Discussion
Aspergillus fumigatus produces volatile molecules differentially across growth conditions
We hypothesized that A. fumigatus would differentially produce volatile molecules as a function of its growth environment, particularly between growth in either oxygen replete or depleted environments. The A. fumigatus volatile metabolome is influenced by factors including iron availability, culture aeration, and exposure to inhibitory small molecules such as simvastatin, pravastatin, and alendronate [27], but to the best of our knowledge, the influence of low oxygen conditions that induce a hypoxia response has not been assessed to-date. Headspace volatile molecules from A. fumigatus culture supernatants were concentrated and analyzed using headspace solid-phase microextraction coupled to comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry (HS-SPME-GC×GC-TOFMS). This approach has been employed previously for the analysis of bacterial volatile metabolites [23, 28-32], owing to its increased resolution and sensitivity relative to traditional gas chromatography-mass spectrometry (GC-MS) [33]. Cultures of A. fumigatus were collected and analyzed following incubation under either normoxia or hypoxia conditions for either 1 h or 8 h (i.e., early hypoxia (H1), late hypoxia (H8), early normoxia (N1), or late normoxia (N8)). After chromatographic alignment, peak finding, and elimination of environmental contaminants and chromatographic artifacts (see Materials and Methods), a total of 652 peaks were identified across all samples.
Using the machine learning algorithm Random Forest (RF) [25], we achieved a classification accuracy of 97.5% for the discrimination between the four culture conditions, and identified 19 volatile molecules that were highly discriminatory (see Materials and Methods). We generated a principal components (PC) scores plot using these molecules to visualize variance in the data (Figure 1a), which revealed four distinct sample clusters corresponding to the four experimental conditions of interest (H1, H8, N1, and N8). PC1 (50%) primarily accounted for the variance between hypoxia and normoxia, while PC2 (29%) primarily accounted for the variance between early and late cultures.
Figure 1.
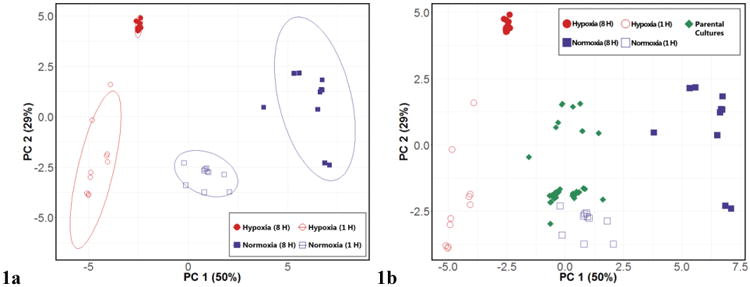
Differential production of volatile molecules by Aspergillus fumigatus across four experimental conditions. 1a, left: PC scores plot generated using 19 discriminatory volatile molecules for the differentiation between H8 (red, filled), H1 (red, hollow), N8 (blue, filled), and N1 (blue, hollow). Ellipses represent 95% confidence intervals. 1b, right: PC scores plot with parental culture samples (green) projected, using same 19 discriminatory molecules. PC 1: 50%, PC 2: 29% for both scores plots.
We next sought to characterize the volatile molecular profile of the parental cultures that were used to prepare cultures belonging to each of the four experimental conditions of interest. The parental cultures had been incubated for 18 h under normoxia conditions; however, growth of filamentous fungi in liquid culture often results in pellet formation, where oxygen consumption inside the pellet may result in hypoxic microenvironments, even in well aerated cultures [34]. Therefore, we hypothesized that their volatile molecular profiles would potentially share attributes with both hypoxia and normoxia cultures. Using the 19 molecules that were discriminatory between our four experimental conditions, we projected the 40 parental cultures onto the PC scores plot, and observed that these samples projected approximately half-way between the N8 and H8 cultures along PC1, the axis primarily accounting for the variance between normoxia and hypoxia (Figure 1b). Therefore, these parental cultures appear to possess an intermediate volatile molecular signature between those of late hypoxia and late normoxia cultures, consistent with the development of heterogeneous oxygen tension and hypoxic microenvironments.
A volatile molecular signature of adaptation to hypoxia in A. fumigatus
Next, we sought to compare the volatile metabolic profile of late hypoxia A. fumigatus cultures with all other culture conditions assessed in this study (late normoxia, early hypoxia, and early normoxia) in aggregate, in order to identify the volatile molecules that are associated with the fungal response to hypoxia. These late cultures represent an environment with significant metabolic changes due to the hypoxia and hypercapnia response of the fungus, similar to what is found in an established infection microenvironment [14]. This culture condition therefore represents a hypoxia-associated metabolic profile distinct from the three other experimental conditions, potentially reflective of fungal metabolism in the setting of in vivo hypoxia lesions. A two-class RF model comparing late hypoxia with the other three experimental conditions aggregated into a single class (“all others”) resulted in a perfect 100% classification accuracy for the differentiation between late hypoxia cultures versus all others. From this model, we identified 19 volatile molecules that were discriminatory between late hypoxia cultures and all others, and again generated a PC scores plot using these 19 molecules, which revealed two distinct sample clusters corresponding to late hypoxia versus all others (Figure 2). Of note, within the “all others” group, the late normoxia cultures were observed to form a sub-cluster away from the early hypoxia and early normoxia samples (Supplementary Figure S2).
Figure 2.
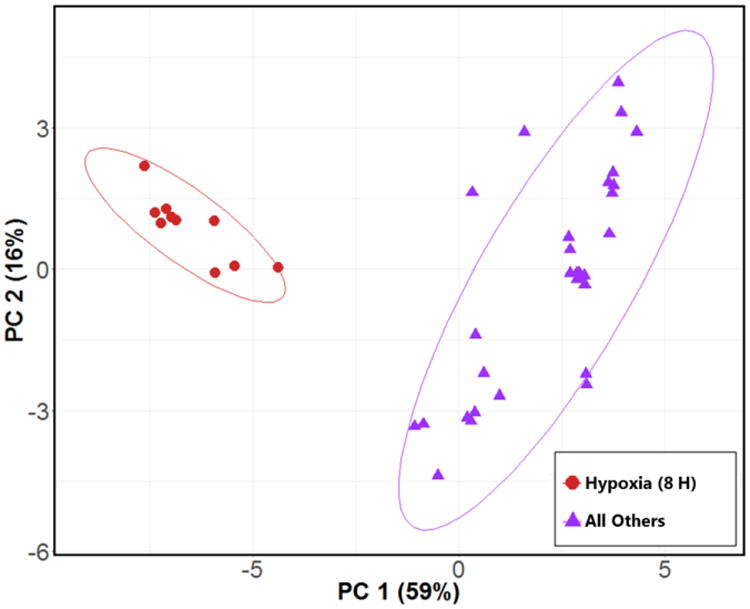
PC scores plot generated using 19 top discriminatory volatile molecules for the differentiation between H8 (red circles) and all other experimental conditions (purple triangles). Ellipses represent 95% confidence intervals. PC 1: 59%, PC 2: 16%. A PC scores plot depicting “all other” samples labeled by experimental condition is presented in Supplementary Figure S2.
Nine of the identified discriminatory molecules were common to both the four-class and two-class models, while the remainder were specific to a single model. All 29 discriminatory volatile molecules were evaluated using hierarchical clustering analysis (HCA), and are visualized via heat map (Figure 3). Samples were observed to cluster as a function of growth condition, with the late hypoxia cultures exhibiting the most distinct volatile molecular profile. For the nine discriminatory molecules identified in both the four-class and two-class RF models, six were most highly abundant in the late hypoxia cultures and three most highly abundant in the late normoxia cultures. For all 29 discriminatory molecules identified, 11 were most highly abundant in late hypoxia, 14 in late normoxia, 2 in early hypoxia, and 2 in early normoxia. Of the molecules assigned putative identifications, 3-hydroxy-2-butanone was most dramatically overabundant (72-fold higher) in late hypoxia cultures relative to all other experimental conditions, followed by 2,3-butanedione (7.1-fold higher), with the remainder ranging from 5.3-fold higher in late hypoxia (benzonitrile) to not detectable under late hypoxia conditions (2,3-hexanedione). 2,3-butanedione was also the most highly abundant peak detected in late hypoxic cultures (14.4 × 106 total ion chromatogram (TIC)), followed by benzaldehyde (2.4 × 106 TIC), and 3-hydroxy-2-butanone (7.2 × 105 TIC).
Figure 3.
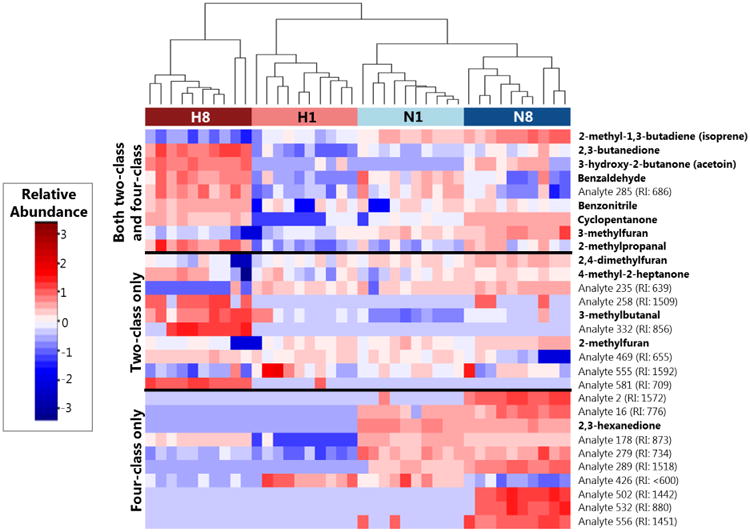
Heat map depicting the relative abundance of discriminatory volatile molecules across samples from either late hypoxia (H8, dark red), early hypoxia (H1, light red), early normoxia (N1, light blue), or late normoxia (N8, dark blue) after log10-transformation, mean centering, and unit scaling. Molecules (rows) are divided into those that were identified in both the four-class and two-class RF models (top), the two-class model only (middle), or the four-class model only (bottom). Cell colors correspond to relative abundance of molecules across samples, ranging from low abundance (blue) to high abundance (red). Dendrogram was calculated using Euclidean distance as a measure of similarity between samples. “Analytes” correspond to molecules for which putative identifications could not be determined, and are accompanied by an experimentally-determined retention index (RI).
Putative metabolic origins of the hypoxia-associated volatile molecular signature
In the present study, we report on 29 discriminatory molecules identified via RF, of which 13 could be assigned putative identifications based on mass spectral matching combined with affirmation via linear retention indices. Of these, six were most highly abundant in late hypoxia cultures relative to all other experimental conditions: one dione (2,3-butanedione), one hydroxy ketone (3-hydroxy-2-butanone), three aldehydes (2-methylpropanal, 3-methylbutanal, and benzaldehyde), and one benzene-derivative (benzonitrile). All six of these molecules were also significantly more abundant in late hypoxia cultures relative to sterile media controls incubated under identical conditions (Supplementary Table S2). To the best of our knowledge, none of the reported molecules have previously been measured in the headspace of A. fumigatus cultures, likely because the hypoxia-associated volatile metabolome of this organism has never been measured. Five of the six molecules (excluding benzonitrile) have, however, been measured in the headspace of either Aspergillus flavus or Aspergillus niger cultures [35, 36].
The putative metabolic pathways that contribute to the production of nearly all hypoxia-associated volatile molecules (except for benzonitrile) can be inferred based on prior studies evaluating the metabolism of other microorganisms in combination with prior transcriptomic data obtained from A. fumigatus CEA10 cultured under identical conditions [20]. For example, in the yeast Saccharomyces cerevisiae, production of 2,3-butanedione and 3-hydroxy-2-butanone results from the activity of at least two pathways, one involving the conversion of pyruvate to acetolactate via acetolactate synthase, and another involving the conversion of pyruvate to acetaldehyde via pyruvate decarboxylase [37]. Prior evaluation of the A. fumigatus CEA10 transcriptome following adaptation to hypoxia under identical growth conditions revealed marked increases in transcript abundance for two of three putative pyruvate decarboxylase complexes (pdcC and pdcA), which were upregulated by 140-fold and 63-fold, respectively, after two hours of growth under hypoxia conditions [20]. The two putative acetolactate synthase genes, however, were not significantly upregulated under these conditions. We therefore hypothesize that the production of 2,3-butanedione and 3-hydroxy-2-butanone results either solely from pyruvate decarboxylase activity or from the combined activities of pyruvate decarboxylase with other presently uncharacterized enzymes that contribute to acetolactate production. Alternatively, it is possible that the expression of genes encoding acetolactate synthase were increased after 2 h post-inoculation, and were therefore not captured by the previously-acquired transcriptomic data.
Although our current knowledge of A. fumigatus metabolism cannot account for the generation of acetolactate in the culture conditions described in this study, the high abundance of 2,3-butanedione in our samples supports the hypothesis that acetolactate is produced by A. fumigatus. Indeed, activity of the pyruvate decarboxylase pathway results in the direct production of 3-hydroxy-2-butanone only, and production of 2,3-butanedione from this molecule is a NAD+-consuming step. In the setting of fermentative metabolism, NAD+ recycling is critical for cellular fitness, and consumption of NAD+ via the conversion of 3-hydroxy-2-butanone to 2,3-butanedione represents a seemingly maladaptive process. In addition to the utility of this pathway for NAD+ regeneration, a model has been proposed relating the synthesis of these molecules to oxygen tension and pH, whereby microorganisms undergoing hypoxia fermentation produce these pH neutral metabolites instead of metabolites that acidify the microenvironment in order to prevent lethal acidification [38]. We hypothesize that A. fumigatus may be leveraging this mechanism during fermentation under hypoxia conditions to eliminate reductive stress and potentially regenerate NAD+ levels, a process that has not previously been reported for this organism. In addition, the terminally-reduced product of this pathway, 2,3-butanediol (which we also detect in the headspace of our late hypoxia cultures), is hypothesized to play a role in the host-pathogen interaction, potentially in the context of immune evasion [39] and enhanced virulence [40].
The three aldehydes reported in this study (2-methylpropanal, 3-methylbutanal, and benzaldehyde) are known volatile metabolic byproducts of valine, leucine, and phenylalanine catabolism, respectively [41]. It is also possible that benzonitrile production results from phenylalanine metabolism, although the mechanism for this has not been well-characterized in A. fumigatus. Of note, it has been observed that in Aspergillus nidulans, production of branched-chain amino acids (e.g., valine and leucine) increases under hypoxia, likely for the purpose of regenerating NAD+ and NADP+ [42]. Catabolism of valine and leucine to 2-methylpropanal and 3-methylbutanal, respectively, is initiated by a branched-chain amino acid transaminase [43]. Of the four putative branched-chain amino acid aminotransferases in A. fumigatus, one (AFUB_092450) was overexpressed by 6.1-fold following adaptation to hypoxia, while the expression of the other three (AFUB_002080, 026220, and 063280) were reduced by between 2.0-fold and 10-fold [20]. In contrast to valine and leucine, catabolism of phenylalanine in A. fumigatus involves at least three distinct pathways, one involving conversion to trans-cinnamate (via phenylalanine ammonia-lyase), another involving conversion to phenylethylamine (via aromatic-L-amino acid decarboxylase), and a third involving conversion to phenylpyruvate (via aromatic amino acid aminotransferase) [43]. Transcriptomic data reveals that phenylalanine ammonia-lyase, aromatic-L-amino acid decarboxylase, and aromatic amino acid aminotransferase were downregulated by 8.8-fold, 8.3-fold, and 1.3-fold after 2 h in hypoxia, a result that seeming contradicts our present findings [20]. It is possible, however, that temporal differences between the collection of transcriptomic and volatile metabolomic data is responsible for the apparent discrepancy between expression levels of the genes responsible for the catabolism of these amino acids and the production of these volatile molecules. Alternatively, the identity of the enzymes responsible for the production of these molecules may not yet be annotated in A. fumigatus.
Study strengths and limitations
This study represents the first application of GC×GC-TOFMS for the analysis of A. fumigatus headspace volatiles, as well as the first direct comparison of the volatile molecules produced by this organism in a hypoxia physiological state versus standard normoxia culture growth conditions. The hypoxia-associated volatile molecular signature was investigated due to its clinical relevance in the context of IA. We were able to identify a possible novel hypoxia adaptation mechanism of A. fumigatus involving the production of 2,3-butanedione and 3-hydroxy-2-butanone, likely for the purpose of regenerating NAD+ and preventing lethal acidification. As this study was conducted as a proof-of-concept to assess the contribution of oxygen tension to the volatile molecular profile of A. fumigatus, future studies must take into account several points in order to validate these preliminary results. First, based on previous studies, we hypothesize that pH may have an impact the production of volatile molecules by A. fumigatus, as well as their partitioning between the liquid culture and headspace. This parameter should be monitored in further investigations. Second, filamentous fungal batch cultures often result in pellet formation in which a hypoxia response may develop in late normoxia cultures [34]. Consequently, our experimental design using batch culture may not have captured the full extent of the volatilomic differences that exist between A. fumigatus cultured under normoxia versus hypoxia conditions. Finally, the relationship between catabolic pathways and the hypoxia-associated volatile metabolites described in this study are presently only putative. Assessment of the volatile metabolic profiles of A. fumigatus mutants lacking the genes that we have putatively linked to the production of hypoxia-associated metabolites is essential for confirming biological origins.
Conclusions
In conclusion, we present the first profiling of the A. fumigatus volatile metabolome using GC×GC-TOFMS, with a focus on the differential production of volatile molecules under low-oxygen and normoxia growth conditions. We report on 19 molecules that were discriminatory between the four experimental conditions assessed in this study (early hypoxia, late hypoxia, early normoxia, and late normoxia), as well as 19 that were discriminatory in the comparison of late hypoxia cultures versus all others experimental groups in aggregate. Nine molecules were shared between the two comparisons, while 10 were specific to each statistical model. Using previously acquired transcriptomic data, we were able to elucidate metabolic pathways that might account for the production of a subset of putatively-identified hypoxia-associated volatile molecules. Two of these molecules, namely 2,3-butanedione and 3-hydroxy-2-butanone, reveal a potential novel hypoxia adaptation strategy in A. fumigatus that could simultaneously allow for the regeneration of NAD+ while preventing lethal acidification via the production of pH neutral catabolic byproducts.
Supplementary Material
Acknowledgments
Dartmouth College holds an Institutional Program Unifying Population and Laboratory Based Sciences award from the Burroughs Wellcome Fund, and C. Rees was supported by this grant (Grant#1014106). P-H. Stefanuto is a Marie-Curie COFUND postdoctoral fellow co-funded by the European Union and the University of Liège. R. Cramer holds an Investigator in the Pathogenesis of Infectious Diseases award from the Burroughs Wellcome Fund (BWF) and is also supported by a National Institute of Allergy and Infectious Diseases (NIAID) award 2R01AI081838, National Institute of General Medicine Sciences (NIGMS) award P30GM106394 (Bruce Stanton, principal investigator), and a Cystic Fibrosis Foundation award (Bruce Stanton, principal investigator). S. Beattie has been supported NIGMS T32GM008704 (Compton, D PI) and NIAID T32AI007519 (O'Toole, G PI).
References
- 1.Park SJ, Mehrad B. Innate immunity to Aspergillus species. Clin Microbiol Rev. 2009;22:535–51. doi: 10.1128/CMR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal R. Allergic bronchopulmonary aspergillosis. Chest. 2009;135:805–26. doi: 10.1378/chest.08-2586. [DOI] [PubMed] [Google Scholar]
- 3.Kousha M, Tadi R, Soubani AO. Pulmonary aspergillosis: a clinical review. Eur Respir Rev. 2011;20:156–74. doi: 10.1183/09059180.00001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeghen T, Kibbler CC, Prentice HG, Berger LA, Wallesby RK, McWhinney PH, Lampe FC, Gillespie S. Management of invasive pulmonary aspergillosis in hematology patients: a review of 87 consecutive cases at a single institution. Clin Infect Dis. 2000;31:859–68. doi: 10.1086/318133. [DOI] [PubMed] [Google Scholar]
- 5.Fukuda T, Boeckh M, Carter RA, Sandmaier BM, Maris MB, Maloney DG, Martin PJ, Storb RF, Marr KA. Risks and outcomes of invasive fungal infections in recipients of allogeneic hematopoietic stem cell transplants after nonmyeloablative conditioning. Blood. 2003;102:827–33. doi: 10.1182/blood-2003-02-0456. [DOI] [PubMed] [Google Scholar]
- 6.Patterson TF, et al. Executive Summary: Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;63:433–42. doi: 10.1093/cid/ciw444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu CC, Maher MM, Shepard JA. Complications of CT-guided percutaneous needle biopsy of the chest: prevention and management. AJR Am J Roentgenol. 2011;196:W678–82. doi: 10.2214/AJR.10.4659. [DOI] [PubMed] [Google Scholar]
- 8.Pfeiffer CD, Fine JP, Safdar N. Diagnosis of invasive aspergillosis using a galactomannan assay: a meta-analysis. Clin Infect Dis. 2006;42:1417–27. doi: 10.1086/503427. [DOI] [PubMed] [Google Scholar]
- 9.Karageorgopoulos DE, Vouloumanou EK, Ntziora F, Michalopoulos A, Rafailidis PI, Falagas ME. beta-D-glucan assay for the diagnosis of invasive fungal infections: a meta-analysis. Clin Infect Dis. 2011;52:750–70. doi: 10.1093/cid/ciq206. [DOI] [PubMed] [Google Scholar]
- 10.Arvanitis M, Ziakas PD, Zacharioudakis IM, Zervou FN, Caliendo AM, Mylonakis E. PCR in diagnosis of invasive aspergillosis: a meta-analysis of diagnostic performance. J Clin Microbiol. 2014;52:3731–42. doi: 10.1128/JCM.01365-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ascioglu S, et al. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin Infect Dis. 2002;34:7–14. doi: 10.1086/323335. [DOI] [PubMed] [Google Scholar]
- 12.Koo S, Thomas HR, Daniels SD, Lynch RC, Fortier SM, Shea MM, Rearden P, Comolli JC, Baden LR, Marty FM. A breath fungal secondary metabolite signature to diagnose invasive aspergillosis. Clin Infect Dis. 2014;59:1733–40. doi: 10.1093/cid/ciu725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers ST, Syhre M, Murdoch DR, McCartin F, Epton MJ. Detection of 2-Pentylfuran in the breath of patients with Aspergillus fumigatus. Med Mycol. 2009;47:468–476. doi: 10.1080/13693780802475212. [DOI] [PubMed] [Google Scholar]
- 14.Grahl N, Puttikamonkul S, Macdonald JM, Gamcsik MP, Ngo LY, Hohl TM, Cramer RA. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog. 2011;7:e1002145. doi: 10.1371/journal.ppat.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA., Jr A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denning DW, Ward PN, Fenelon LE, Benbow EW. Lack of vessel wall elastolysis in human invasive pulmonary aspergillosis. Infect Immun. 1992;60:5153–6. doi: 10.1128/iai.60.12.5153-5156.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vodisch M, Scherlach K, Winkler R, Hertweck C, Braun HP, Roth M, Haas H, Werner ER, Brakhage AA, Kniemeyer O. Analysis of the Aspergillus fumigatus proteome reveals metabolic changes and the activation of the pseurotin A biosynthesis gene cluster in response to hypoxia. J Proteome Res. 2011;10:2508–24. doi: 10.1021/pr1012812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barker BM, Kroll K, Vodisch M, Mazurie A, Kniemeyer O, Cramer RA. Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter. BMC Genomics. 2012;13:62. doi: 10.1186/1471-2164-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wezensky SJ, Cramer RA., Jr Implications of hypoxic microenvironments during invasive aspergillosis. Med Mycol. 49. 2011;(1):S120–4. doi: 10.3109/13693786.2010.495139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Losada L, Barker BM, Pakala S, Pakala S, Joardar V, Zafar N, Mounaud S, Fedorova N, Nierman WC, Cramer RA. Large-scale transcriptional response to hypoxia in Aspergillus fumigatus observed using RNAseq identifies a novel hypoxia regulated ncRNA. Mycopathologia. 2014;178:331–9. doi: 10.1007/s11046-014-9779-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellors T, Rees C, Wieland-Alter W, von Reyn CF, Hill JE. The volatile molecule signature of four mycobacteria species. J Breath Res. 2017 doi: 10.1088/1752-7163/aa6e06. [DOI] [PubMed] [Google Scholar]
- 23.Bean HD, Rees CA, Hill JE. Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates. J Breath Res. 2016;10:047102. doi: 10.1088/1752-7155/10/4/047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78:4281–90. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 25.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 26.Hotelling H. Analysis of a complex of statistical variables into principal components. J Educ Psychol. 1933;24:417–441. [Google Scholar]
- 27.Heddergott C, Calvo AM, Latge JP. The volatome of Aspergillus fumigatus. Eukaryot Cell. 2014;13:1014–25. doi: 10.1128/EC.00074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bean HD, Dimandja JM, Hill JE. Bacterial volatile discovery using solid phase microextraction and comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;901:41–6. doi: 10.1016/j.jchromb.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nizio KD, Perrault KA, Troobnikoff AN, Ueland M, Shoma S, Iredell JR, Middleton PG, Forbes SL. In vitro volatile organic compound profiling using GCxGC-TOFMS to differentiate bacteria associated with lung infections: a proof-of-concept study. J Breath Res. 2016;10:026008. doi: 10.1088/1752-7155/10/2/026008. [DOI] [PubMed] [Google Scholar]
- 30.Rees CA, Smolinska A, Hill JE. The volatile metabolome of Klebsiella pneumoniae in human blood. J Breath Res. 2016;10:027101. doi: 10.1088/1752-7155/10/2/027101. [DOI] [PubMed] [Google Scholar]
- 31.Rees CA, Franchina FA, Nordick KV, Kim PJ, Hill JE. Expanding the Klebsiella pneumoniae volatile metabolome using advanced analytical instrumentation for the detection of novel metabolites. J Appl Microbiol. 2016;122:785–795. doi: 10.1111/jam.13372. [DOI] [PubMed] [Google Scholar]
- 32.Rees CA, Shen A, Hill JE. Characterization of the Clostridium difficile volatile metabolome using comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2016;1039:8–16. doi: 10.1016/j.jchromb.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Blomberg J, Schoenmakers PJ, Beens J, Tijssen R. Comprehensive two-dimensional gas chromatography (GCxGC) and its applicability to the characterization of complex (petrochemical) mixtures. J High Resolut Chromatogr. 1997;20:539–544. [Google Scholar]
- 34.Krull R, Cordes C, Horn H, Kampen I, Kwade A, Neu TR, Nortemann B. Morphology of filamentous fungi: linking cellular biology to process engineering using Aspergillus niger. Adv Biochem Eng Biotechnol. 2010;121:1–21. doi: 10.1007/10_2009_60. [DOI] [PubMed] [Google Scholar]
- 35.Sun D, Wood-Jones A, Wang W, Vanlangenberg C, Jones D, Gower J, Simmons P, Baird RE, Mlsna TE. Monitoring MVOC profiles over time from isolates of Aspergillus flavus using SPME GC-MS. J Agric Chem Environ. 2014;3:48–63. [Google Scholar]
- 36.Costa CP, Silva DG, Rudnitskaya A, Almeida A, Rocha SM. Shedding light on Aspergillus niger volatile exometabolome. Sci Rep. 2016;6 doi: 10.1038/srep27441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng CY, Jung MY, Lee J, Oh MK. Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb Cell Fact. 2012;11:68. doi: 10.1186/1475-2859-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whiteson KL, Meinardi S, Lim YW, Schmieder R, Maughan H, Quinn R, Blake DR, Conrad D, Rohwer F. Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2,3-butanedione fermentation. ISME J. 2014;8:1247–58. doi: 10.1038/ismej.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh SC, Lu CC, Horng YT, Soo PC, Chang YL, Tsai YH, Lin CS, Lai HC. The bacterial metabolite 2,3-butanediol ameliorates endotoxin-induced acute lung injury in rats. Microbes Infect. 2007;9:1402–9. doi: 10.1016/j.micinf.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Venkataraman A, Rosenbaum MA, Werner JJ, Winans SC, Angenent LT. Metabolite transfer with the fermentation product 2,3-butanediol enhances virulence by Pseudomonas aeruginosa. ISME J. 2014;8:1210–20. doi: 10.1038/ismej.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz S, Dickschat JS. Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 2007;24:814–42. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu M, Fujii T, Masuo S, Takaya N. Mechanism of de novo branched-chain amino acid synthesis as an alternative electron sink in hypoxic Aspergillus nidulans cells. Appl Environ Microbiol. 2010;76:1507–15. doi: 10.1128/AEM.02135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


