Abstract
Nucleic acid editing holds promise for treating genetic disease, particularly at the RNA level, where disease-relevant sequences can be rescued to yield functional protein products. Type VI CRISPR-Cas systems contain the programmable single-effector RNA-guided RNases Cas13. Here, we profile Type VI systems to engineer a Cas13 ortholog capable of robust knockdown and demonstrate RNA editing by using catalytically-inactive Cas13 (dCas13) to direct adenosine to inosine deaminase activity by ADAR2 to transcripts in mammalian cells. This system, referred to as RNA Editing for Programmable A to I Replacement (REPAIR), has no strict sequence constraints, can be used to edit full-length transcripts containing pathogenic mutations. We further engineer this system to create a high specificity variant, REPAIRv2, that is 919 times more specific than REPAIRv1 as well as minimize the system to ease viral delivery. REPAIR presents a promising RNA editing platform with broad applicability for research, therapeutics, and biotechnology.
Introduction
Precise nucleic acid editing technologies are valuable for studying cellular function and as novel therapeutics. Current editing tools, based on programmable nucleases such as the prokaryotic clustered regularly interspaced short palindromic repeats (CRISPR)-associated nucleases Cas9 (1–4) or Cpf1(5), have been widely adopted for mediating targeted DNA cleavage which in turn drives targeted gene disruption through non-homologous end joining (NHEJ) or precise gene editing through template-dependent homology-directed repair (HDR) (6). NHEJ utilizes host machineries that are active in both dividing and post-mitotic cells and provides efficient gene disruption by generating a mixture of insertion or deletion (indel) mutations that can lead to frame shifts in protein coding genes. HDR, in contrast, is mediated by host machineries whose expression is largely limited to replicating cells. Accordingly, the development of gene-editing capabilities for post-mitotic cells remains a major challenge. DNA base editors, consisting of a fusion between Cas9 nickase and cytidine deaminase can mediate efficient cytidine to uridine conversions within a target window and significantly reduce the formation of double-strand break induced indels (7, 8). However the potential targeting sites of DNA base editors are limited by the requirement of Cas9 for a protospacer adjacent motif (PAM) at the editing site (9). Here, we describe the development of a precise and flexible RNA base editing technology using the type VI CRISPR-associated RNA-guided RNase Cas13 (10–13).
Cas13 enzymes have two Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) endoRNase domains that mediate precise RNA cleavage with a preference for targets with protospacer flanking site (PFS) motif observed biochemically and in bacteria (10, 11). Three Cas13 protein families have been identified to date: Cas13a (previously known as C2c2), Cas13b, and Cas13c (12, 13). We recently reported that Cas13a enzymes can be adapted as tools for nucleic acid detection (14) as well as mammalian and plant cell RNA knockdown and transcript tracking (15). Interestingly, the biochemcial PFS was not required for RNA interference with Cas13a (15). The programmable nature of Cas13 enzymes makes them an attractive starting point to develop tools for RNA binding and perturbation applications.
The adenosine deaminase acting on RNA (ADAR) family of enzymes mediates endogenous editing of transcripts via hydrolytic deamination of adenosine to inosine, a nucleobase that is functionally equivalent to guanosine in translation and splicing (16, 17). There are two functional human ADAR orthologs, ADAR1 and ADAR2, which consist of N-terminal double stranded RNA-binding domains and a C-terminal catalytic deamination domain. Endogenous target sites of ADAR1 and ADAR2 contain substantial double stranded identity, and the catalytic domains require duplexed regions for efficient editing in vitro and in vivo (18, 19). Importantly, the ADAR catalytic domain is capable of deaminating target adenosines without any protein cofactors in vitro (20). ADAR1 has been found to target mainly repetitive regions whereas ADAR2 mainly targets non-repetitive coding regions (17). Although ADAR proteins have preferred motifs for editing that could restrict the potential flexibility of targeting, hyperactive mutants, such as ADAR2(E488Q) (21), relax sequence constraints and increase adenosine to inosine editing rates. ADARs preferentially deaminate adenosines mispaired with cytidine bases in RNA duplexes (22), providing a promising opportunity for precise base editing. Although previous approaches have engineered targeted ADAR fusions via RNA guides (23–26), the specificity of these approaches has not been reported and their respective targeting mechanisms rely on RNA-RNA hybridization without the assistance of protein partners that may enhance target recognition and stringency.
Here we assay a subset of the family of Cas13 enzymes for RNA knockdown activity in mammalian cells and identify the Cas13b ortholog from Prevotella sp. P5–125 (PspCas13b) as the most efficient and specific for mammalian cell applications. We then fuse the ADAR2 deaminase domain (ADAR2DD) to catalytically inactive PspCas13b and demonstrate RNA editing for programmable A to I (G) replacement (REPAIR) of reporter and endogenous transcripts as well as disease-relevant mutations. Lastly, we employ a rational mutagenesis scheme to improve the specificity of dCas13b-ADAR2DD fusions to generate REPAIRv2 with more than 919-fold higher specificity.
Comprehensive Characterization of Cas13 Family Members in Mammalian Cells
We previously developed LwaCas13a for mammalian knockdown applications, but it required an monomeric superfolder GFP (msfGFP) stabilization domain for efficient knockdown and, although the specificity was high, knockdown levels were not consistently below 50% (15). We sought to identify a more robust RNA-targeting CRISPR system by characterizing a genetically diverse set of Cas13 family members to assess their RNA knockdown activity in mammalian cells (Fig. 1A). We generated mammalian codon-optimized versions of multiple Cas13 proteins, including 21 orthologs of Cas13a, 15 of Cas13b and 7 of Cas13c, and cloned them into an expression vector with N- and C-terminal nuclear export signal (NES) sequences and a C-terminal msfGFP to enhance protein stability (Supplementary Table 1). To assay interference in mammalian cells, we designed a dual reporter construct expressing the independent Gaussia (Gluc) and Cypridinia (Cluc) luciferases under separate promoters, which allows one luciferase to function as a measure of Cas13 interference activity and the other to serve as an internal control. For each Cas13 ortholog, we designed protospacer flanking site (PFS)-compatible guide RNAs, using the Cas13b PFS motifs derived from an ampicillin interference assay (fig S1; Supplementary Table 2; Supplementary Information) and the 3’ H (not G) PFS from previous reports of Cas13a activity (10).
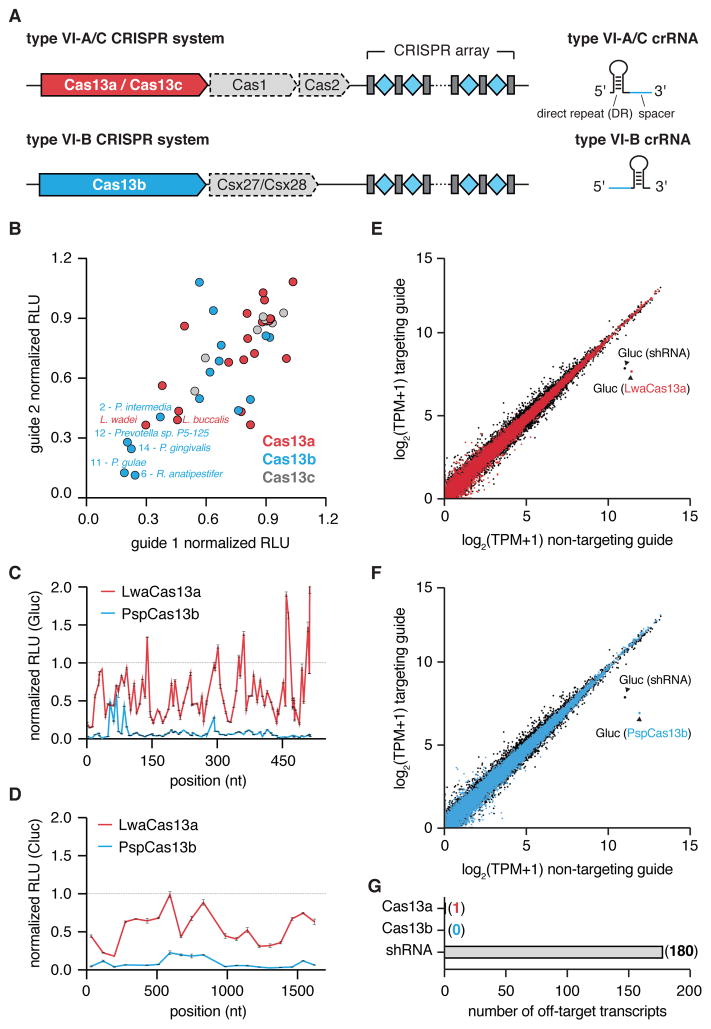
Figure 1. Characterization of a highly active Cas13b ortholog for RNA knockdown.
A) Schematic of stereotypical Cas13 loci and corresponding crRNA structure.
B) Evaluation of 19 Cas13a, 15 Cas13b, and 7 Cas13c orthologs for luciferase knockdown using two different guides. Orthologs with efficient knockdown using both guides are labeled with their host organism name. Values are normalized to a non-targeting guide with designed against the E. coli LacZ transcript, with no homology to the human transcriptome.
C) PspCas13b and LwaCas13a knockdown activity (as measured by luciferase activity) using tiling guides against Gluc. Values represent mean +/− S.E.M. Non-targeting guide is the same as in Fig. 1B.
D) PspCas13b and LwaCas13a knockdown activity (as measured by luciferase activity) using tiling guides against Cluc. Values represent mean +/− S.E.M. Non-targeting guide is the same as in Fig. 1B.
E) Expression levels in log2(transcripts per million (TPM+1)) values of all genes detected in RNA-seq libraries of non-targeting control (x-axis) compared to Gluc-targeting condition (y-axis) for LwaCas13a (red) and shRNA (black). Shown is the mean of three biological replicates. The Gluc transcript data point is labeled. Non-targeting guide is the same as in Fig1B.
F) Expression levels in log2(transcripts per million (TPM+1)) values of all genes detected in RNA-seq libraries of non-targeting control (x-axis) compared to Gluc-targeting condition (y-axis) for PspCas13b (blue) and shRNA (black). Shown is the mean of three biological replicates. The Gluc transcript data point is labeled. Non-targeting guide is the same as in Fig. 1B.
G) Number of significant off-targets from Gluc knockdown for LwaCas13a, PspCas13b, and shRNA from the transcriptome wide analysis in E and F.
We transfected HEK293FT cells with Cas13-expression, guide RNA, and reporter plasmids and then quantified levels of Cas13 expression and the targeted Gluc 48 hours later (Fig. 1B, fig. S2A). Testing two guide RNAs for each Cas13 ortholog revealed a range of activity levels, including five Cas13b orthologs with similar or increased interference across both guide RNAs relative to the recently characterized LwaCas13a (Figure 1B), and we observed only a weak correlation between Cas13 expression and interference activity (fig. S2B–D). We selected the top five Cas13b orthologs, as well as the top two Cas13a orthologs for further engineering.
We next tested Cas13-mediated knockdown of Gluc without msfGFP, to select orthologs that do not require stabilization domains for robust activity. We hypothesized that Cas13 activity could be affected by subcellular localization, as we previously reported for optimization of LwaCas13a (15). Therefore, we tested the interference activity of the seven selected Cas13 orthologs C-terminally fused to one of six different localization tags without msfGFP. Using the luciferase reporter assay, we identified the top three Cas13b designs with the highest level of interference activity: Cas13b from Prevotella sp. P5–125 (PspCas13b) and Cas13b from Porphyromonas gulae (PguCas13b) C-terminally fused to the HIV Rev gene NES and Cas13b from Riemerella anatipestifer (RanCas13b) C-terminally fused to the MAPK NES (fig. S3A). To further distinguish activity levels of the top orthologs, we compared the three optimized Cas13b constructs to the optimal LwaCas13a-msfGFP fusion and to shRNA for their ability to knockdown the endogenous KRAS transcript using position-matched guides (fig. S3B). We observed the highest levels interference for PspCas13b (average knockdown 62.9%) and thus selected this for further comparison to LwaCas13a.
To more rigorously define the activity of PspCas13b and LwaCas13a, we designed position-matched guides tiling along both Gluc and Cluc transcripts and assayed their activity using our luciferase reporter assay. We tested 93 and 20 position-matched guides targeting Gluc and Cluc, respectively, and found that PspCas13b had consistently increased levels of knockdown relative to LwaCas13a (average of 92.3% for PspCas13b vs. 40.1% knockdown for LwaCas13a) (Fig. 1C,D).
Specificity of Cas13 mammalian interference activity
To characterize the interference specificities of PspCas13b and LwaCas13a we designed a plasmid library of luciferase targets containing single mismatches and double mismatches throughout the target sequence and the three flanking 5’ and 3’ base pairs (fig. S3C). We transfected HEK293FT cells with either LwaCas13a or PspCas13b, a fixed guide RNA targeting the unmodified target sequence, and the mismatched target library corresponding to the appropriate system. We then performed targeted RNA sequencing of uncleaved transcripts to quantify depletion of mismatched target sequences. We found that LwaCas13a and PspCas13b had a central region that was relatively intolerant to single mismatches, extending from base pairs 12–26 for the PspCas13b target and 13–24 for the LwaCas13a target (fig. S3D). Double mismatches were even less tolerated than single mutations, with little knockdown activity observed over a larger window, extending from base pairs 12–29 for PspCas13b and 8–27 for LwaCas13a in their respective targets (fig. S3E). Additionally, because there are mismatches included in the three nucleotides flanking the 5’ and 3’ ends of the target sequence, we could assess PFS constraints on Cas13 knockdown activity. Sequencing showed that almost all PFS combinations allowed robust knockdown, indicating that a PFS constraint for interference in mammalian cells likely does not exist for either enzyme tested. These results indicate that Cas13a and Cas13b display similar sequence constraints and sensitivities against mismatches.
We next characterized the interference specificity of PspCas13b and LwaCas13a across the mRNA fraction of the transcriptome. We performed transcriptome-wide mRNA sequencing to detect significant differentially expressed genes. LwaCas13a and PspCas13b demonstrated robust knockdown of Gluc (Fig. 1E,F) and were highly specific compared to a position-matched shRNA, which showed hundreds of off-targets (Fig. 1G), consistent with our previous characterization of LwaCas13a specificity in mammalian cells (15).
Cas13-ADAR fusions enable targeted RNA editing
Given that PspCas13b achieved consistent, robust, and specific knockdown of mRNA in mammalian cells, we envisioned that it could be adapted as an RNA binding platform to recruit RNA modifying domains, such as the deaminase domain of ADARs (ADARDD) for programmable RNA editing. To engineer a PspCas13b lacking nuclease activity (dPspCas13b, referred to as dCas13b hereafter), we mutated conserved catalytic residues in the HEPN domains and observed loss of luciferase RNA knockdown (fig. S4A). We hypothesized that a dCas13b-ADARDD fusion could be recruited by a guide RNA to target adenosines, with the hybridized RNA creating the required duplex substrate for ADAR activity (Fig. 2A). To enhance target adenosine deamination rates we introduced two additional modifications to our initial RNA editing design: we introduced a mismatched cytidine opposite the target adenosine, which has been previously reported to increase deamination frequency, and fused dCas13b with the deaminase domains of human ADAR1 or ADAR2 containing hyperactivating mutations to enhance catalytic activity (ADAR1DD(E1008Q) (27) or ADAR2DD(E488Q) (21)).
Figure 2. Engineering dCas13b-ADAR fusions for RNA editing.
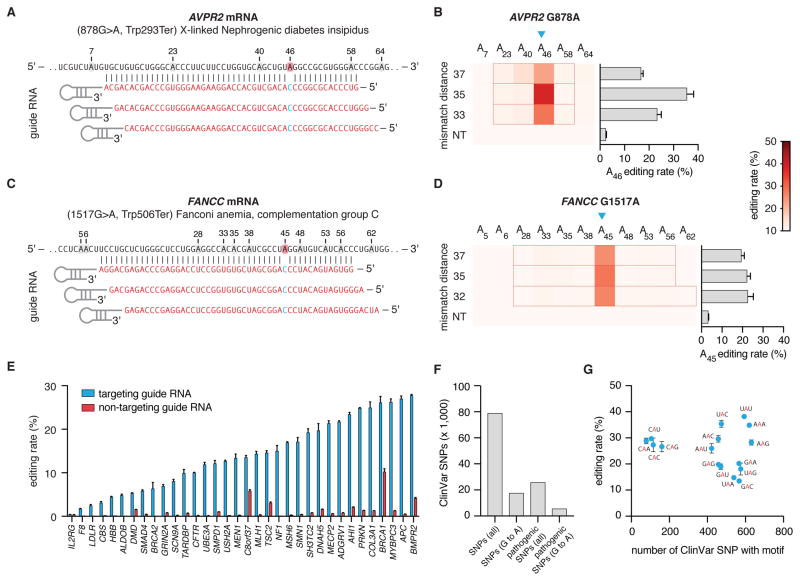
A) Schematic of RNA editing by dCas13b-ADARDD fusion proteins. Catalytically dead Cas13b (dCas13b) is fused to the deaminase domain of human ADAR (ADARDD), which naturally deaminates adenosines to insosines in dsRNA. The crRNA specifies the target site by hybridizing to the bases surrounding the target adenosine, creating a dsRNA structure for editing, and recruiting the dCas13b-ADARDD fusion. A mismatched cytidine in the crRNA opposite the target adenosine enhances the editing reaction, promoting target adenosine deamination to inosine, a base that functionally mimics guanosine in many cellular reactions.
B) Schematic of Cypridina luciferase W85X target and targeting guide design. Deamination of the target adenosine restores the stop codon to the wildtype tryptophan. Spacer length is the region of the guide that contains homology to the target sequence. Mismatch distance is the number of bases between the 3’ end of the spacer and the mismatched cytidine. The cytidine mismatched base is included as part of the mismatch distance calculation.
C) Quantification of luciferase activity restoration for dCas13b-ADAR1DD(E1008Q) (left) and dCas13b-ADAR2DD(E488Q) (right) with tiling guides of length 30, 50, 70, or 84 nt. All guides with even mismatch distances are tested for each guide length. Values are background subtracted relative to a 30nt non-targeting guide that is randomized with no sequence homology to the human transcriptome.
D) Schematic of the sequencing window in which A to I edits were assessed for Cypridinia luciferase W85X.
E) Sequencing quantification of A to I editing for 50-nt guides targeting Cypridinia luciferase W85X. Blue triangle indicates the targeted adenosine. For each guide, the region of duplex RNA is outlined in red. Values represent mean +/− S.E.M. Non-targeting guide is the same as in Fig. 2C.
To test the activity of dCas13b-ADARDD we generated an RNA-editing reporter on Cluc by introducing a nonsense mutation (W85X (UGG->UAG)), which could functionally be repaired to the wildtype codon through A->I editing (Fig. 2B) and then be detected as restoration of Cluc luminescence. We evenly tiled guides with spacers of 30, 50, 70 or 84 nucleotides in length across the target adenosine to determine the optimal guide placement and design (Fig. 2C). We found that dCas13b-ADAR1DD required longer guides to repair the Cluc reporter, while dCas13b-ADAR2DD was functional with all guide lengths tested (Fig. 2C). We also found that the hyperactive E488Q mutation improved editing efficiency, as luciferase restoration with the wildtype ADAR2DD was reduced (fig. S4B). From this demonstration of activity, we chose dCas13b-ADAR2DD(E488Q) for further characterization and designated this approach as RNA Editing for Programmable A to I Replacement version 1 (REPAIRv1).
To validate that restoration of luciferase activity was due to bona fide editing events, we directly measured REPAIRv1-mediated editing of Cluc transcripts via reverse transcription and targeted next-generation sequencing. We tested 30- and 50-nt spacers around the target site and found that both guide lengths resulted in the expected A to I edit, with 50-nt spacers achieving higher editing percentages (Fig. 2D,E, fig. S4C). We also observed that 50-nt spacers had an increased propensity for editing at non-targeted adenosines within the sequencing window, likely due to increased regions of duplex RNA (Fig. 2E, fig. S4C).
We next targeted an endogenous gene, PPIB. We designed 50-nt spacers tiling PPIB and found that we could edit the PPIB transcript with up to 28% editing efficiency (Fig. S4D). To test if REPAIR could be further optimized, we modified the linker between dCas13b and ADAR2DD(E488Q) (fig. S4E, Supplementary Table 3) and found that linker choice modestly affected luciferase activity restoration. Additionally, we tested the ability of dCas13b and guide alone to mediate editing events, finding that the ADAR deaminase domain is required for editing (fig. S5A–D).
Defining the sequence parameters for RNA editing
Given that we could achieve precise RNA editing at a test site, we wanted to characterize the sequence constraints for programming the system against any RNA target in the transcriptome. Sequence constraints could arise from dCas13b targeting limitations, such as the PFS, or from ADAR sequence preferences (28). To investigate PFS constraints on REPAIRv1, we designed a plasmid library carrying a series of four randomized nucleotides at the 5’ end of a target site on the Cluc transcript (Fig. 3A). We targeted the center adenosine within either a UAG or AAC motif and found that for both motifs, all PFSs demonstrated detectable levels of RNA editing, with a majority of the PFSs having greater than 50% editing at the target site (Fig. 3B). Next, we sought to determine if the ADAR2DD in REPAIRv1 had any sequence constraints immediately flanking the targeted base, as has been reported previously for ADAR2DD (28). We tested every possible combination of 5’ and 3’ flanking nucleotides directly surrounding the target adenosine (Fig. 3C), and found that REPAIRv1 was capable of editing all motifs (Fig. 3D). Lastly, we analyzed whether the identity of the base opposite the target A in the spacer sequence affected editing efficiency and found that an A–C mismatch had the highest luciferase restoration, in agreement with previous reports of ADAR2 activity, with A–G, A–U, and A-A having drastically reduced REPAIRv1 activity (fig. S5E).
Figure 3. Measuring sequence flexibility for RNA editing by REPAIRv1.
A) Schematic of screen for determining Protospacer Flanking Site (PFS) preferences of RNA editing by REPAIRv1. A randomized PFS sequence is cloned 5’ to a target site for REPAIR editing. Following exposure to REPAIR, deep sequencing of reverse transcribed RNA from the target site and PFS is used to associate edited reads with PFS sequences.
B) Distributions of RNA editing efficiencies for all 4-N PFS combinations at two different editing sites
F) Quantification of the percent editing of REPAIRv1 at Cluc W85 across all possible 3 base motifs. Values represent mean +/− S.E.M. Non-targeting guide is the same as in Fig. 2C.
C) Heatmap of 5’ and 3’ base preferences of RNA editing at Cluc W85 for all possible 3 base motifs.
Correction of disease-relevant human mutations using REPAIRv1
To demonstrate the broad applicability of the REPAIRv1 system for RNA editing in mammalian cells, we designed REPAIRv1 guides against two disease relevant mutations: 878G>A (AVPR2 W293X) in X-linked Nephrogenic diabetes insipidus and 1517G>A (FANCC W506X) in Fanconi anemia. We transfected expression constructs for cDNA of genes carrying these mutations into HEK293FT cells and tested whether REPAIRv1 could correct the mutations. Using guide RNAs containing 50-nt spacers, we were able to achieve 35% correction of AVPR2 and 23% correction of FANCC (Fig. 4A–D). We then tested the ability of REPAIRv1 to correct 34 different disease-relevant G>A mutations (Supplementary Table 4) and found that we were able to achieve significant editing at 33 sites with up to 28% editing efficiency (Fig. 4E). The mutations we chose are only a fraction of the pathogenic G to A mutations (5,739) in the ClinVar database, which also includes an additional 11,943 G to A variants (Fig. 4F and fig. S6). Because there are no sequence constraints (Fig. 3), REPAIRv1 is capable of potentially editing all these disease relevant mutations, especially given that we observed editing regardless of the target motif (Fig. 3C and Fig. 4G).
Figure 4. Correction of disease-relevant mutations with REPAIRv1.
A) Schematic of target and guide design for targeting AVPR2 878G>A.
B) The 878G>A mutation (indicated by blue triangle) in AVPR2 is corrected to varying levels using REPAIRv1 with three different guide designs. For each guide, the region of duplex RNA is outlined in red. Values represent mean +/− S.E.M. Non-targeting guide is the same as in Fig. 2C.
C) Schematic of target and guide design for targeting FANCC 1517G>A.
D) The 1517G>A mutation (indicated by blue triangle) in FANCC is corrected to varying levels using REPAIRv1 with three different guide designs. For each guide, the region of duplex RNA is outlined in red. The heatmap scale bar is the same as in panel B. Values represent mean +/− S.E.M. Non-targeting guide is the same as in Fig. 2C.
E) Quantification of the percent editing of 34 different disease-relevant G>A mutations selected from ClinVar using REPAIRv1. Non-targeting guide is the same as in Fig. 2C.
F) Analysis of all the possible G>A mutations that could be corrected using REPAIR as annotated in the ClinVar database.
G) The distribution of editing motifs for all G>A mutations in ClinVar is shown versus the editing efficiency by REPAIRv1 per motif as quantified on the Gluc transcript. Values represent mean +/− S.E.M.
Delivering the REPAIRv1 system to diseased cells is a prerequisite for therapeutic use, and we therefore sought to design REPAIRv1 constructs that could be packaged into therapeutically relevant viral vectors, such as adeno-associated viral (AAV) vectors. AAV vectors have a packaging limit of 4.7kb, which cannot accommodate the large size of dCas13b-ADARDD (4,473 bp) along with promoter and expression regulatory elements. To reduce the size, we tested a variety of N-terminal and C-terminal truncations of dCas13 fused to ADAR2DD(E488Q) for RNA editing activity. We found that all C-terminal truncations tested were still functional and able to restore luciferase signal (fig. S7), and the largest truncation, C-terminal Δ984-1090 (total size of the fusion protein 4,152bp) was small enough to fit within the packaging limit of AAV vectors.
Transcriptome-wide specificity of REPAIRv1
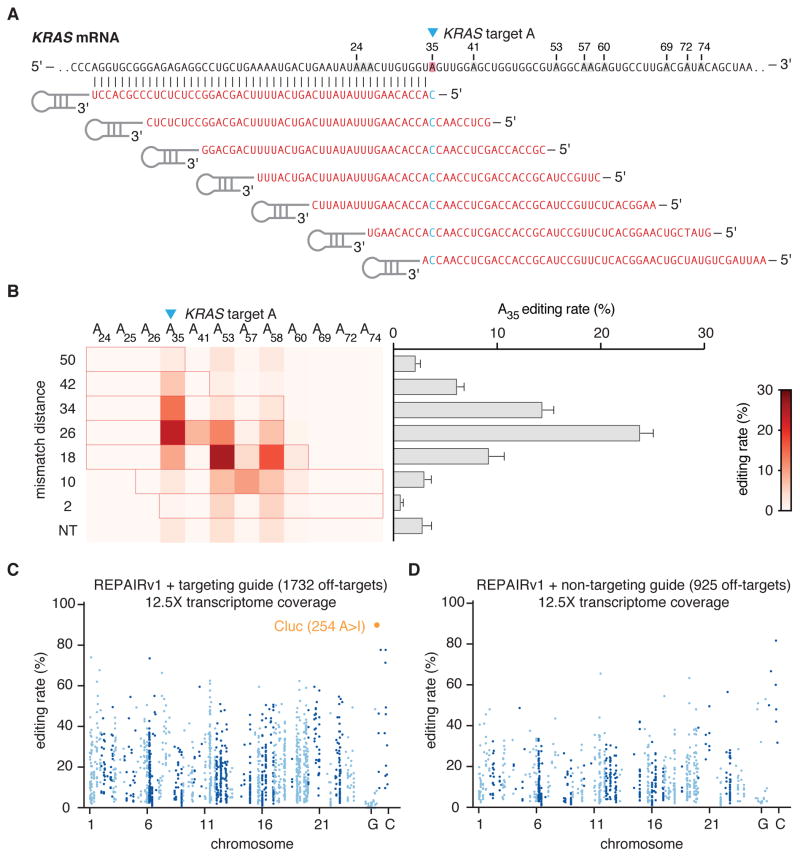
Although RNA knockdown with PspCas13b was highly specific in our luciferase tiling experiments, we observed off-target adenosine editing within the guide:target duplex (Fig. 2E). To see if this was a widespread phenomenon, we tiled an endogenous transcript, KRAS, and measured the degree of off-target editing near the target adenosine (Fig. 5A). We found that for KRAS, while the on-target editing rate was 23%, there were many sites around the target site that also had detectable A to I edits (Fig. 5B).
Figure 5. Characterizing specificity of REPAIRv1.
A) Schematic of KRAS target site and guide design.
B) Quantification of percent A to I editing for tiled KRAS-targeting guides. Editing percentages are shown for the on-target (blue triangle) and neighboring adenosine sites. For each guide, the region of duplex RNA is outlined in red. Values represent mean +/− S.E.M.
C) Transcriptome-wide sites of significant RNA editing by REPAIRv1 (150ng REPAIR vector transfected) with Cluc targeting guide. The on-target site Cluc site (254 A>I) is highlighted in orange.
D) Transcriptome-wide sites of significant RNA editing by REPAIRv1 (150ng REPAIR vector transfected) with non-targeting guide. Non-targeting guide is the same as in Fig. 2C.
Because of the observed off-target editing within the guide:target duplex, we initially evaluated transcriptome-wide off-targets by performing RNA sequencing on all mRNAs with 12.5X coverage. Of all the editing sites across the transcriptome, the on-target editing site had the highest editing rate, with 89% A to I conversion. We also found that there was a substantial number of A to I off-target events, with 1,732 off-targets in the targeting guide condition and 925 off-targets in the non-targeting guide condition, with 828 off-targets shared between the targeting and non-targeting guide conditions (Fig. 5C,D). Given the high number of overlapping off-targets between the targeting and non-targeting guide conditions, we reasoned that the off-targets may arise from ADARDD. To test this hypothesis, we repeated the Cluc targeting experiment, this time comparing transcriptome changes for REPAIRv1 with a targeting guide, REPAIRv1 with a non-targeting guide, REPAIRv1 alone, or ADARDD(E488Q) alone (fig. S8). We found differentially expressed genes and off-target editing events in each condition (fig. S8B,C). Interestingly, there was a high degree of overlap in the off-target editing events between ADARDD(E488Q) and all REPAIRv1 off-target edits, supporting the hypothesis that REPAIR off-target edits are driven by dCas13b-independent ADARDD(E488Q) editing events (fig. S8D).
Next, we sought to compare two RNA-guided ADAR systems that have been described previously (fig. S9A). The first utilizes a fusion of ADAR2DD to the small viral protein lambda N (ƛN), which binds to the BoxB-ƛ RNA hairpin (24). A guide RNA with double BoxB-ƛ hairpins guides ADAR2DD to edit sites encoded in the guide RNA (25). The second design utilizes full-length ADAR2 (ADAR2) and a guide RNA with a hairpin that the double strand RNA binding domains (dsRBDs) of ADAR2 recognize (23, 26). We analyzed the editing efficiency of these two systems compared to REPAIRv1 and found that the BoxB-ADAR2 and full-length ADAR2 systems demonstrated 50% and 34.5% editing rates, respectively, compared to the 89% editing rate achieved by REPAIRv1 (fig. S9B–E). Additionally, the BoxB and full-length ADAR2 systems created 1,814 and 66 observed off targets, respectively, in the targeting guide conditions, compared to the 2,111 off targets in the REPAIRv1 targeting guide condition. Notably, all the conditions with the two ADAR2DD-based systems (REPAIRv1 and BoxB) showed a high percentage of overlap in their off-targets whereas the full-length ADAR2 system had a largely distinct set of off-targets (fig. S9F). The overlap in off-targets between the targeting and non-targeting conditions and between REPAIRv1 and BoxB conditions suggests ADAR2DD drives off-targets independent of dCas13 targeting (fig. S9F).
Improving specificity of REPAIR through rational protein engineering
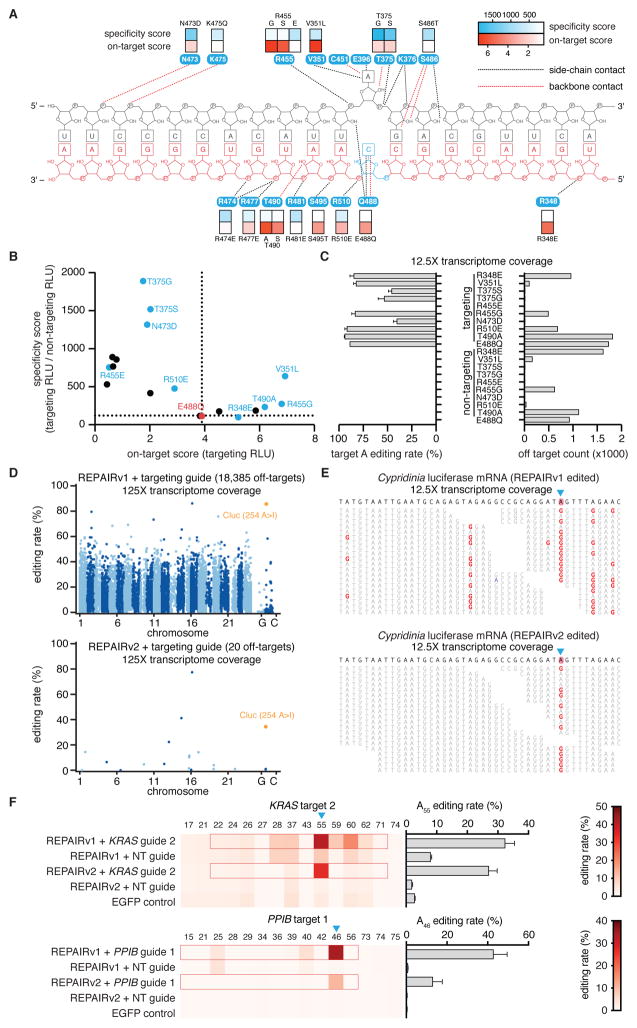
To improve the specificity of REPAIRv1, we employed structure-guided protein engineering of ADAR2DD(E488Q). Because of the guide-independent nature of the off-targets, we hypothesized that destabilizing ADAR2DD(E488Q)-RNA binding would selectively decrease off-target editing, but maintain on-target editing due to increased local concentration from dCas13b tethering of ADAR2DD(E488Q) to the target site. We mutated residues in ADAR2DD(E488Q) previously determined to contact the duplex region of the target RNA (Fig. 6A) (19). To assess efficiency and specificity, we tested 17 single mutants with both targeting and non-targeting guides, under the assumption that background luciferase restoration in the non-targeting condition would be indicative of broader off-target activity. We found that mutations at the selected residues had significant effects on the luciferase activity for targeting and non-targeting guides (Fig. 6A,B, fig. S10A). A majority of mutants either significantly improved the luciferase activity for the targeting guide or increased the ratio of targeting to non-targeting guide activity, which we termed the specificity score (Fig. 6A,B).
Figure 6. Rational mutagenesis of ADAR2 to improve the specificity of REPAIRv1.
A) Quantification of luciferase signal restoration (on-target score, red boxes) by various dCas13-ADAR2DD mutants as well as their specificity score (blue boxes) plotted along a schematic of the contacts between key ADAR2 deaminase residues and the dsRNA target (target strand shown in gray; the non-target strand is shown in red). All deaminase mutations were made on the dCas13-ADAR2DD(E488Q) background. The specificity score is defined as the ratio of the luciferase signal between targeting guide and non-targeting guide conditions. Schematic of ADAR2 deaminase domain contacts with dsRNA is adapted from ref (20).
B) Quantification of luciferase signal restoration by various dCas13-ADAR2 mutants versus their specificity score. Non-targeting guide is the same as in Fig. 2C.
C) Quantification of on-target editing and the number of significant off-targets for each dCas13-ADAR2DD(E488Q) mutant by transcriptome wide sequencing of mRNAs. Values represent mean +/− S.E.M. Non-targeting guide is the same as in Fig. 2C.
D) Transcriptome-wide sites of significant RNA editing by REPAIRv1 (top) and REPAIRv2 (bottom) with a guide targeting a pretermination site in Cluc. The on-target Cluc site (254 A>I) is highlighted in orange. 10 ng of REPAIR vector was transfected for each condition.
E) Representative RNA sequencing reads surrounding the on-target Cluc editing site (254 A>I; blue triangle) highlighting the differences in off-target editing between REPAIRv1 (top) and REPAIRv2 (bottom). A>I edits are highlighted in red; sequencing errors are highlighted in blue. Gaps reflect spaces between aligned reads. Non-targeting guide is the same as in Fig. 2C.
F) RNA editing by REPAIRv1 and REPAIRv2 with guides targeting an out-of-frame UAG site in the endogenous KRAS and PPIB transcripts. The on-target editing fraction is shown as a sideways bar chart on the right for each condition row. For each guide, the region of duplex RNA is outlined in red. Values represent mean +/− S.E.M. Non-targeting guide is the same as in Fig. 2C.
We selected a subset of these mutants (Fig. 6B) for transcriptome-wide specificity profiling by next generation sequencing. As expected, off-targets measured from transcriptome-wide sequencing correlated with our specificity score (fig. S10B) for mutants. We found that with the exception of ADAR2DD(E488Q/R455E), all sequenced REPAIRv1 mutants could effectively edit the reporter transcript (Fig. 6C), with many mutants showing reduction in the number of off-targets (Fig. 6C, fig S10C, S11). We further explored the surrounding motifs of off-targets for the various specificity mutants, and found that REPAIRv1 and most of the engineered variants exhibited a strong 3’ G preference for their off-target edits, in agreement with the characterized ADAR2 motif (fig. S12A) (28).
We focused on the mutant ADAR2DD(E488Q/T375G), as it had the highest percent editing of the four mutants with the lowest numbers of transcriptome-wide off targets and termed it REPAIRv2. Compared to REPAIRv1, REPAIRv2 exhibited increased specificity, with a reduction from 18,385 to 20 transcriptome-wide off-targets by high-coverage sequencing (125X coverage, 10ng DNA transfection) (Fig. 6D). In the region surrounding the targeted adenosine in Cluc, REPAIRv2 also had reduced off-target editing, visible in sequencing traces (Fig. 6E). In motifs derived from the off-target sites, REPAIRv1 presented a strong preference towards 3’ G, but showed off-targeting edits for all motifs (fig. S12B); by contrast, REPAIRv2 only edited the strongest off-target motifs (fig. S12C). The distribution of edits on transcripts was heavily skewed for REPAIRv1, with highly-edited genes having over 60 edits (fig. S13A), whereas REPAIRv2 only edited one transcript (EEF1A1) multiple times (fig. S13B). REPAIRv1 off-target edits were predicted to result in numerous variants, including 1000 missense base changes (fig. S13C) with 93 events in genes related to cancer processes (fig. S13D). In contrast, REPAIRv2 only had 6 predicted base changes (fig. S10E), none of which were in cancer-related genes (fig. S13F). Analysis of the sequence surrounding off-target edits for REPAIRv1 or v2 did not reveal homology to guide sequences, suggesting that off-targets are likely dCas13b-independent (fig. S14), consistent with the high overlap of off-targets between REPAIRv1 and the ADAR deaminase domain (fig. S8D). To directly compare REPAIRv2 against other programmable ADAR systems, we repeated our Cluc targeting experiments with all systems at two different dosages of ADAR vector, finding that REPAIRv2 had comparable on-target editing to BoxB and ADAR2 but with significantly fewer off-target editing events at both dosages (fig S15). REPAIRv2 had enhanced specificity compared to REPAIRv1 at both dosages (fig. S15B), a finding that also extended to two guides targeting distinct sites on PPIB (fig. S16A–D). It is also worth noting that, in general, the lower dosage condition (10 ng) had fewer off-targets than the higher dosage condition (150 ng) (fig. S5).
To assess editing specificity with greater sensitivity, we sequenced the low dosage condition (10 ng of transfected DNA) of REPAIRv1 and v2 at significantly higher sequencing depth (125X coverage of the transcriptome). Increased numbers of off-targets were found at higher sequencing depths corresponding to detection of rarer off-target events (fig. S17). Furthermore, we speculated that different transcriptome states could also potentially alter the number of off-targeting events. Therefore, we tested REPAIRv2 activity in the osteosarcoma U2OS cell line, observing 6 and 7 off-targets for the targeting and non-targeting guide, respectively (fig. S18).
We targeted REPAIRv2 to endogenous genes to test if the specificity-enhancing mutations reduced nearby edits in target transcripts while maintaining high-efficiency on-target editing. For guides targeting either KRAS or PPIB, we found that REPAIRv2 had no detectable off-target edits, unlike REPAIRv1, and could effectively edit the on-target adenosine at efficiencies of 27.1% (KRAS) or 13% (PPIB) (Fig. 6F). This specificity extended to additional target sites, including regions that demonstrate high-levels of background in non-targeting conditions for REPAIRv1, such as other KRAS or PPIB target sites (fig. S19). Overall, REPAIRv2 eliminated off-targets in duplexed regions around the edited adenosine and showed dramatically enhanced transcriptome-wide specificity.
Discussion
We show here that the RNA-guided RNA-targeting type VI-B CRISPR effector Cas13b is capable of highly efficient and specific RNA knockdown, providing the basis for improved tools for interrogating essential genes and non-coding RNA as well as controlling cellular processes at the transcript level. Catalytically inactive Cas13b (dCas13b) retains programmable RNA binding capability, which we leveraged here by fusing dCas13b to the adenosine deaminase domain of ADAR2 to achieve precise A to I edits, a system we term REPAIRv1 (RNA Editing for Programmable A to I Replacement version 1). Further engineering of the system produced REPAIRv2, which has dramatically higher specificity than previously described RNA editing platforms (25, 29) while maintaining high levels of on-target efficacy.
Although Cas13b exhibits high fidelity, our initial results with dCas13b-ADAR2DD(E488Q) fusions revealed a substantial number of off-target RNA editing events. To address this, we employed a rational mutagenesis strategy to vary the ADAR2DD residues that contact the RNA duplex, identifying a variant, ADAR2DD(E488Q/T375G), capable of precise, efficient, and highly specific editing when fused to dCas13b. Editing efficiency with this variant was comparable to or better than that achieved with two currently available systems, BoxB-ADAR2DD(E488Q) or ADAR2 editing. Moreover, the REPAIRv2 system created only 20 observable off-targets in the whole transcriptome, at least an order of magnitude better than both alternative editing technologies. While it is possible that ADAR could deaminate adenosine bases on the DNA strand in RNA-DNA heteroduplexes (20), it is unlikely to do so in this case as Cas13b does not bind DNA efficiently and that REPAIR is cytoplasmically localized. Additionally, the lack of homology of off-target sites to the guide sequence and the strong overlap of off-targets with the ADARDD(E488Q)-only condition suggest that off-targets are not mediated by off-target guide binding. Deeper sequencing and novel inosine enrichment methods could further refine our understanding of REPAIR specificity in the future.
The REPAIR system offers many advantages compared to other nucleic acid editing tools. First, the exact target site can be encoded in the guide by placing a cytidine within the guide extension across from the desired adenosine to create a favorable A–C mismatch ideal for ADAR editing activity. Second, Cas13 has no targeting sequence constraints, such as a PFS or PAM, and no motif preference surrounding the target adenosine, allowing any adenosine in the transcriptome to be potentially targeted with the REPAIR system. The lack of motif for ADAR editing, in contrast with previous literature, is likely due to the increased local concentration of REPAIR at the target site due to dCas13b binding. We do note that DNA base editors can target either the sense or anti-sense strand, while the REPAIR system is limited to transcribed sequences, thereby constraining the total number of possible editing sites. However, due to the less constrained nature of targeting with REPAIR, this system can effect more edits within ClinVar (Fig. 4C) than Cas9-DNA base editors. Third, the REPAIR system directly deaminates target adenosines to inosines and does not rely on endogenous repair pathways, such as base-excision or mismatch repair, to generate desired editing outcomes. Therefore, REPAIR should be able to mediate efficient RNA editing even in post-mitotic cells such as neurons. Fourth, in contrast to DNA editing, RNA editing is transient and can be more easily reversed, allowing the potential for temporal control over editing outcomes. The temporary nature of REPAIR-mediated edits will likely be useful for treating diseases caused by temporary changes in cell state, such as local inflammation and could also be used to treat disease by modifying the function of proteins involved in disease-related signal transduction. For instance, REPAIR editing would allow the re-coding of some serine, threonine and tyrosine residues that are the targets of kinases (fig. S20). Phosphorylation of these residues in disease-relevant proteins affects disease progression for many disorders including Alzheimer’s disease and multiple neurodegenerative conditions (30). REPAIR might also be used to transiently or even chronically change the sequence of expressed, risk-modifying G to A variants to decrease the chance of entering a disease state for patients. For instance, REPAIR could be used to functionally mimic A to G alleles of IFIH1 that protect against autoimmune disorders such as type I diabetes, immunoglobulin A deficiency, psoriasis, and systemic lupus erythematosus (31, 32).
The REPAIR system provides multiple opportunities for additional engineering. Cas13b possesses pre-crRNA processing activity (13), allowing for multiplex editing of multiple variants, any one of which alone may not affect disease, but together might have additive effects and disease-modifying potential. Extension of our rational design approach, such as combining promising mutations and directed evolution, could further increase the specificity and efficiency of the system, while unbiased screening approaches could identify additional residues for improving REPAIR activity and specificity.
Currently, the base conversions achievable by REPAIR are limited to generating inosine from adenosine; additional fusions of dCas13 with other catalytic RNA editing domains, such as APOBEC, could enable cytidine to uridine editing. Additionally, mutagenesis of ADAR could relax the substrate preference to target cytidine, allowing for the enhanced specificity conferred by the duplexed RNA substrate requirement to be exploited by C to U editors. Adenosine to inosine editing on DNA substrates may also be possible with catalytically inactive DNA-targeting CRISPR effectors, such as dCas9 or dCpf1, either through formation of DNA-RNA heteroduplex targets (20) or mutagenesis of the ADAR domain.
We have demonstrated the use of the PspCas13b enzyme as both an RNA knockdown and RNA editing tool. The dCas13b platform for programmable RNA binding has many applications, including live transcript imaging, splicing modification, targeted localization of transcripts, pull down of RNA-binding proteins, and epitranscriptomic modifications. Here, we used dCas13 to create REPAIR, adding to the existing suite of nucleic acid editing technologies. REPAIR provides a new approach for treating genetic disease or mimicking protective alleles, and establishes RNA editing as a useful tool for modifying genetic function.
Supplementary Material
Acknowledgments
We would like to thank R. Munshi, A. Baumann, A. Philippakis, and E. Banks for computational guidance, C. Muus for sequencing assistance, A. Smargon for gene synthesis of a Cas13b orthologs 1–8, V. Verdine for assistance with RNA sequencing library preparation, R. Macrae, R. Belliveau, and the entire Zhang lab for discussions and support. D.B.T.C. is supported by award number T32GM007753 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the NIH. O.O.A. is supported by a Paul and Daisy Soros Fellowship and a NIH F30 NRSA 1F30-CA210382. F.Z. is a New York Stem Cell Foundation–Robertson Investigator. F.Z. is supported by the NIH grants (1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201); the Howard Hughes Medical Institute; the New York Stem Cell, Simons, Paul G. Allen Family, and Vallee Foundations; and J. and P. Poitras, R. Metcalfe, and D. Cheng. D.B.T.C., J.S.G., O.O.A., and F.Z. are co-inventors on U.S. provisional patent application no. 62/525,181 relating to REPAIR and other applications relating to CRISPR and CRISPR-Cas13 technology filed by the Broad Institute. Sequencing data are available at Sequence Read Archive under BioProject accession number PRJNA414340. The authors plan to make the reagents widely available to the academic community through Addgene and to provide software tools via the Zhang lab website (www.genome-engineering.org) and GitHub (github.com/fengzhanglab).
Footnotes
References
- 1.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 7.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishida K, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 9.Kim YB, et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abudayyeh OO, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov S, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmakov S, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smargon AA, et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell. 2017;65:618–630 e617. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gootenberg JS, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abudayyeh OO, et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan MH, et al. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017;550:249–254. doi: 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 19.Matthews MM, et al. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat Struct Mol Biol. 2016;23:426–433. doi: 10.1038/nsmb.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y, Lorenzo C, Beal PA. DNA editing in DNA/RNA hybrids by adenosine deaminases that act on RNA. Nucleic Acids Res. 2017;45:3369–3377. doi: 10.1093/nar/gkx050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuttan A, Bass BL. Mechanistic insights into editing-site specificity of ADARs. Proc Natl Acad Sci U S A. 2012;109:E3295–3304. doi: 10.1073/pnas.1212548109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong SK, Sato S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuda M, et al. Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing. Sci Rep. 2017;7:41478. doi: 10.1038/srep41478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, Rosenthal JJ. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci U S A. 2013;110:18285–18290. doi: 10.1073/pnas.1306243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montiel-Gonzalez MF, Vallecillo-Viejo IC, Rosenthal JJ. An efficient system for selectively altering genetic information within mRNAs. Nucleic Acids Res. 2016;44:e157. doi: 10.1093/nar/gkw738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wettengel J, Reautschnig P, Geisler S, Kahle PJ, Stafforst T. Harnessing human ADAR2 for RNA repair - Recoding a PINK1 mutation rescues mitophagy. Nucleic Acids Res. 2017;45:2797–2808. doi: 10.1093/nar/gkw911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Havel J, Beal PA. A Phenotypic Screen for Functional Mutants of Human Adenosine Deaminase Acting on RNA 1. ACS Chem Biol. 2015;10:2512–2519. doi: 10.1021/acschembio.5b00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 29.Stafforst T, Schneider MF. An RNA-deaminase conjugate selectively repairs point mutations. Angew Chem Int Ed Engl. 2012;51:11166–11169. doi: 10.1002/anie.201206489. [DOI] [PubMed] [Google Scholar]
- 30.Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, et al. Carriers of rare missense variants in IFIH1 are protected from psoriasis. J Invest Dermatol. 2010;130:2768–2772. doi: 10.1038/jid.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira RC, et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat Genet. 2010;42:777–780. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- 33.Joung J, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picardi E, D'Erchia AM, Montalvo A, Pesole G. Using REDItools to Detect RNA Editing Events in NGS Datasets. Curr Protoc Bioinformatics. 2015;49:12 12 11–15. doi: 10.1002/0471250953.bi1212s49. [DOI] [PubMed] [Google Scholar]
- 36.Picardi E, Pesole G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013;29:1813–1814. doi: 10.1093/bioinformatics/btt287. [DOI] [PubMed] [Google Scholar]
- 37.Glusman G, Caballero J, Mauldin DE, Hood L, Roach JC. Kaviar: an accessible system for testing SNV novelty. Bioinformatics. 2011;27:3216–3217. doi: 10.1093/bioinformatics/btr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson JD. Molecular biology of the gene. 7. Pearson; Boston: 2014. p. xxxiv.p. 872. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.