Summary
Arc/Arg3.1 is required for synaptic plasticity and cognition and mutations in this gene are linked to autism and schizophrenia. Arc bears a domain resembling retroviral/retrotransposon Gag-like proteins, which multimerize into a capsid that packages viral RNA. The significance of such a domain in a plasticity molecule is uncertain. Here we report that the Drosophila Arc1 protein forms capsid-like structures that bind darc1 mRNA in neurons and is loaded into extracellular vesicles that are transferred from motorneurons to muscles. This loading and transfer depends on the darc1-mRNA 3′-untranslated region, which contains retrotransposon-like sequences. Disrupting transfer blocks synaptic plasticity, suggesting that transfer of dArc1 complexed with its mRNA is required for this function. Notably, cultured cells also release extracellular vesicles containing the Gag region of the Copia retrotransposon complexed with its own mRNA. Taken together, our results point to a trans-synaptic mRNA transport mechanism involving retrovirus-like capsids and extracellular vesicles.
Keywords: Arc/Arg3.1, plasticity, synapse, retrotransposon, exosomes, Gag domain, RNA trafficking, extracellular vesicles, copia, gypsy retrotransposon, trans-synaptic RNA transport, RNA-binding protein
In brief
Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons

Introduction
Mammalian Activity-Regulated Cytoskeleton-Associated protein (Arc/Arg3.1) is pivotal for synapse maturation, synaptic plasticity, and learning and memory (Shepherd and Bear, 2011). Arc is an activity-dependent immediate early gene, and its mRNA is translocated to dendrites via sequences in its 3′-Untranslated Region (UTR) (Dynes and Steward, 2007). Following plasticity-inducing stimulation arc mRNA moves into active dendritic spines, where it is translated and regulates trafficking of AMPA receptors by engaging the endocytic machinery (Chowdhury et al., 2006). Arc is also involved in regulating dendritic spine morphology during plasticity (Peebles et al., 2010). Arc protein is composed of Group-specific antigen (Gag)-like amino acid sequences typically found in retroviruses such as HIV and in retrotransposons (Campillos et al., 2006). Beyond its binding to Tarpγ2, an AMPA-receptor binding protein (Zhang et al., 2015), the physiological significance of the Gag-like sequences in Arc is unknown. During retroviral replication, Gag proteins multimerize into capsids, which bind and package viral RNA (Chen, 2016). Capsids then undergo secondary envelopment by membranes and exit the host cell being competent to infect other cells (Alenquer and Amorim, 2015). There is a growing aggregate of evidence that suggest some viruses, commandeer host exosomal pathways suggesting a connection between viruses and exosomes (Meckes, 2015).
Exosomes and other extracellular vesicles (EVs) such as microvesicles have recently emerged as a novel trans-cellular communication strategy in the healthy and diseased brain (Basso and Bonetto, 2016). For instance, glutamate release by neurons induces oligodendroglial secretion of exosomes, which are taken up and regulate the physiology of recipient neurons (Fruhbeis et al., 2013). At Drosophila neuromuscular synapses, Wnt signaling is mediated by trans-synaptic transfer of the Wnt, Wingless, via exosomes in vivo (Koles et al., 2012; Korkut et al., 2009). In mammals, the propagation of neurodegenerative disorders, such as ALS, appears to be partly mediated by the transfer of prion-like proteins across cells via exosomes (Basso and Bonetto, 2016).
In an expression profile of Drosophila cultured cell EVs we found that one of the most abundant and enriched mRNAs was the Drosophila darc1 (also see (Lefebvre et al., 2016). At the NMJ, darc1 mRNA and protein were enriched at synaptic boutons of the larval NMJ and both were transferred from presynaptic boutons to postsynaptic muscles, likely via EVs. Notably, this transfer was dependent on a gypsy retrotransposon-like sequence fragment in the 3′UTR of darc1. Evocative of retroviral Gags, dArc1 protein physically associated with its own mRNA. In support of this observation, a spliced fragment of the Copia retrotransposon mRNA and protein were also found within EVs. This retroviral-like mechanism of transfer is required for dArc1 function, as blocking the transfer resulted in aberrations in both synapse maturation and activity-dependent plasticity. We propose that in Drosophila, dArc1 influences synaptic plasticity by utilizing a retroviral-like mechanism for transport between synaptic partners.
Results
darc1 mRNA is enriched in S2 cell exosome-like EVs at the Drosophila larval NMJ
To elucidate the expression profile of Drosophila S2 cell EVs, we collected EV fractions by differential centrifugation and sucrose gradient sedimentation, treated the EV fraction with RNase to digest non-specifically associated extravesicular cytoplasmic RNA and then constructed poly(A)+ libraries for deep sequencing using RNA collected from treated EVs and total cell mRNA. While most mRNAs in the EV fraction (~96.6% of cellular mRNAs) were equal to or under-represented when compared to total cellular mRNAs, approximately 100 transcripts were enriched in the EV fraction by at least twofold. Among the most abundant and enriched mRNAs was darc1 (Fig. 1A,B,D). The large enrichment of darc1 in EV preps was confirmed by qPCR (Fig. 1C). To determine if darc1 mRNA was present in vivo at the Drosophila larval NMJ, we conducted fluorescent in situ hybridization (FISH) and double labeled the preparation with anti-HRP, an antibody that recognizes neuronal membrane antigens in insects (Jan and Jan, 1982), to label the presynaptic compartment. darc1 mRNA was present in a punctate pattern both inside presynaptic boutons and muscles, but was particularly enriched at the muscle postsynaptic junctional region, the region of the muscle immediately adjacent to the HRP label (Fig. 2A). The FISH signal was largely specific since it was severely reduced in a predicted darc1 null mutant, darc1esm113 (Mattaliano et al., 2007), although some residual signal remained (Fig. 2B).
Figure 1. darc1 mRNA is enriched in EVs.
(A) Comparison of RNA expression levels in EV versus cellular RNA from S2 cells determined by deep sequencing, and enrichment of darc1 (red) in the EV fraction. Note that darc2 mRNA (blue) levels are not statistically different from cellular levels.
(B) Volcano Plot of RNA-Seq data from 4 replicates, where the x-axis represents fold change in transcripts from EVs vs. total cellular mRNA levels (a positive score represents enrichment, a negative score represents depletion). The y-axis represents statistical confidence for each x-axis point. Green circles are transcripts that are significantly enriched. darc1 is encircled by a red marker.
(C) Normalized quantitative PCR confirming that darc1 is enriched in the exosome fraction compared to the total cell. N= 3 biological replicates.
(D) Raw number of RNA-seq reads for darc1 from 4 biological RNA-Seq replicates.
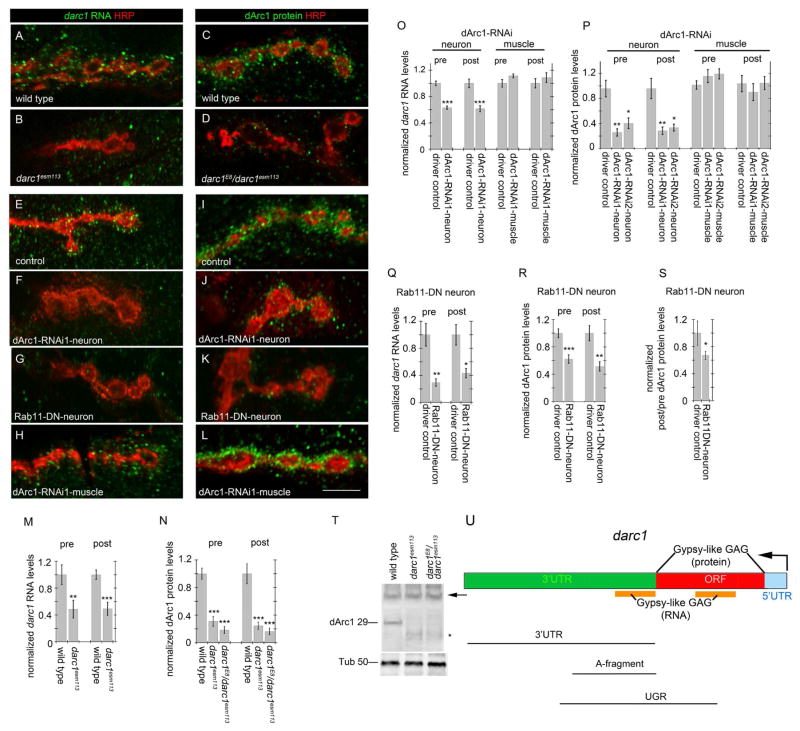
Figure 2. darc1 mRNA and protein are present at the Drosophila NMJ, and are transferred from pre- to postsynaptic sites.
(A–D) Confocal slices of NMJ branches in preparations double labeled with anti-HRP and either (A,B) a darc1 FISH probe, or (C,D) anti-dArc1 in (A,C) wild type and (B) darc1esm113 and (D) darc1E8/darc1esm113 mutants.
(E–L) Confocal slices of NMJ branches in preparations double labeled with anti-HRP and either (E–H) a darc1 FISH probe, or (I–L) anti-dArc1, in (E,I) C380-Gal4/+ control, upon expression of (F,J) dArc1-RNAi-neuron (C380-Gal4>UAS-RNAi1), (G,K) Rab11-DN-neuron (C380-Gal4>UAS-Rab11DN), and (H,L) dArc1-RNAi1-muscle (C57-Gal4> dArc1-RNAi1).
(M–R) Quantification of (M,O,Q) normalized darc1 FISH signal and (N,P,R) normalized dArc1 immunocytochemical signal at the NMJ in the indicated genotypes.
(S) Normalized ratio of postsynaptic to presynaptic signal in the indicated genotypes
(T) Western blot of body wall muscle protein extracts derived from the indicated genotypes probed sequentially with anti-dArc1 (top) and Tubulin (Tub; bottom). Numbers at the left of the blots represent molecular weight in kilodaltons. Arrow points to an unspecific band labeled by the dArc1 antibody. Tub=tubulin.
(U) Diagram of darc1 mRNA showing the 5′ UTR (blue), the open reading frame (ORF; red), and the 3′ UTR (green). Black lines underneath represent different portions of the darc1 transcript. Orange bars represent regions of the darc1 mRNA resembling Gypsy-like Gag sequences. Note that the entire ORF encodes a Gypsy-like Gag protein.
Calibration bar is 6 μm; N=(from left to right; animals/arbors) M(6/11, 6/10), N(12/16, 6/10, 6/10), O(8/14, 8/13, 8/15, 8/16), P(21/44, 12/24, 9/14, 18/29, 9/17, 9/14), Q(8/14, 8/13), R(15/28, 15/28), S(15/28,15/28); Data are represented as mean and error bars represent SEM; statistical analysis was conducted using student’s t-test for M and one-way ANOVA with Tukey Post Hoc test for the rest of the graphs. *= p < 0.05; **= p < 0.001; ***= p < 0.0001.
To establish if dArc1 protein was also observed at these sites, we generated a polyclonal anti-dArc1 antibody and found that dArc1 was also present both pre- and postsynaptically at the NMJ, in a pattern very similar to the RNA localization (Fig. 2C). We also generated an independent, cleaner darc1 null mutant via Crispr/Cas9, darc1E8, because the darc1esm113 deletion spans into the putative darc2 promoter region (see methods). In darc1E8/darc1esm113 and darc1wsm113 mutants there was a similar drastic reduction in dArc1 RNA and protein (Fig. 2B,D), but residual signal was still observed. The residual signal might correspond to either maternal contribution of darc1 RNA, which is quite large, or to unspecific antibody binding. In support to the former possibility, darc1 RNA could still be detected in extracts from the mutants by PCR, although the amount was very low compared to wild type (Fig. S1C). The anti-dArc1 antibody recognized a band of the expected molecular weight in Western blots of body wall muscle protein extracts (Fig. 2T) and in immunoprecipitations of dArc1 (Fig. S1A). This band was absent in the darc1esm113 and darc1E8/darc1esm113 mutant, demonstrating the specificity of the antibody (Fig. 2T;S1A). However, the antibody also recognized higher molecular weight bands (arrow in Fig. 2T) that were not altered by the darc1 mutation. This indicates that at least some of the residual signal observed in the mutants might be unspecific. Unlike the specific signal, the residual signal was distributed throughout the muscle and was not concentrated at the NMJ. Thus, darc1 mRNA is enriched in S2 cell EVs, and both darc1 mRNA and dArc1 protein are present at pre- and postsynaptic sites of the NMJ.
A postsynaptic muscle darc1 RNA pool is derived from motorneurons and RNA transfer depends on darc1 3′UTR
The ORF of darc1 is mostly represented by the remnant of a Gypsy-superfamily of transposons (Fig. 2U), specifically dArc1 has strong similarities to the GAG region of a Gypsy-like transposon. The presence of darc1 in EVs together with the observation that darc1 encodes a retroviral Gag-like protein raised the possibility that the mRNA, similar to retroviral genomic RNA (gRNA), could be transferred across synaptic partners, as we have previously shown for two transmembrane proteins, Evi and Syt4 (Korkut et al., 2009; Korkut et al., 2013). To test this possibility, we downregulated darc1 exclusively in motorneurons, using the Gal4 driver C380-Gal4 to express UAS-dArc1-RNAi, and examined the pre-and postsynaptic levels of dArc1 mRNA via FISH. Expressing dArc1-RNAi in motorneurons resulted in a significant decrease in darc1 FISH signal, not only at presynaptic boutons, but also at the muscle postsynaptic region (Fig. 2E,F,O, Fig. S1B). Given that the C380-Gal4 driver does not express Gal4 in muscles (Budnik et al., 1996) and that we have not previously observed transfer of RNAi between NMJ synaptic partners (e.g., (Ataman et al., 2006)), the most parsimonious explanation is that much of the postsynaptic darc1 mRNA is derived from a presynaptic pool, presumably via EVs (see Discussion). Similarly to the FISH results, expressing dArc1-RNAi in motorneurons resulted in a significant decrease in dArc1 protein immunoreactive signal, both in pre- and postsynaptic compartments (Fig. 2I,J,P).
In contrast, expressing dArc1-RNAi with a muscle Gal4 driver, C57-Gal4, did not affect dArc1 RNA nor dArc1 protein levels (Fig. 2H,L,O,P). This raises the possibility that postsynaptic dArc1 RNA is somehow protected from the RNAi machinery (see Discussion). Taken together, the above results suggest that postsynaptic darc1 transcript and/or protein is transported from presynaptic boutons to the postsynaptic compartment in a unidirectional manner.
To eliminate the possibility that dArc1 RNAi could, unlike other RNAi species, be transferred from neuron to muscles, we used an approach independent from RNAi to block EV release from presynaptic boutons. We have previously demonstrated that Rab11 is required for the release of exosome-like EVs at the NMJ (Koles et al., 2012; Korkut et al., 2013). We therefore examined whether expressing Rab11 dominant-negative (Rab11-DN) specifically in neurons induced a change in the levels of darc1 RNA or protein in the postsynaptic region. We observed that this manipulation resulted in a highly significant reduction in darc1 RNA and protein in the postsynaptic region (Fig. 2G,K,Q,R). Interestingly, when expressing Rab11DN in neurons, we observed a decrease in dArc1 levels in both the pre- and postsynaptic compartments, suggesting that Rab11 could be required both for the transport and release of dArc1. To distinguish between these possibilities, we calculated the proportion postsynaptic dArc1 levels relative to presynaptic levels (postsynaptic dArc1 levels/presynaptic dArc1 levels). We found that this ratio was significantly decreased upon expressing Rab11DN in neurons (Fig. 2S). Together, these data provide additional evidence for the trans-synaptic transfer of darc1 mRNA and/or protein.
To further support the above conclusion, we expressed a dArc1 transgene in neurons, in a darc1E8/darc1esm113 mutant background. The transgene was composed of the darc1 open reading frame, plus the untranslated regions (UTR). This resulted in the presence of dArc1, not only in presynaptic boutons, but also at the postsynaptic region (Fig. 3A). Interestingly, expressing a dArc1 transgene lacking the 3′UTR resulted in accumulation of the dArc1 signal in presynaptic terminals, but virtually no signal was observed at the postsynaptic region (Fig. 3B,I). Notably, the distribution of dArc1 lacking the 3′UTR was variable, being highly enriched at some synaptic boutons, and less enriched in others (Fig. 3C). The quantification of the dArc1 signal in Figure 3I omitted the highly-enriched boutons because in order for the quantification to be informative, the fluorescence levels must be in a linear, non-saturating range. Those boutons with very high expression were orders of magnitude brighter, such that the signal became saturated with confocal acquisition parameters that allowed us detect the postsynaptic signal. Notably, expressing the dArc1 construct including the 3′UTR in the muscles of the null mutant resulted in diffuse distribution of dArc1 signal throughout the muscle, and no postsynaptic enrichment was observed (Fig. 3D). Together, the above results provide compelling evidence that dArc1 protein and/or RNA are transferred from synaptic boutons to muscles, and that a normal dArc1 localization at postsynaptic sites depends on such transfer.
Figure 3. darc1 mRNA and protein transfer across synaptic boutons depends on darc1 3′UTR (see also Supplemental Fig. 1).
(A–H,L–M) Single confocal slices of NMJ branches
(A–D) from darc1 mutant larvae expressing dArc1 transgenes either (A,D) containing, or (B,C) lacking the 3′UTR, double labeled with antibodies to dArc1 and HRP. Transgenes were expressed in (A–C) neurons (C380-Gal4>transgene)or (D) muscles (C57-Gal4>transgene).
(E–H) in preparations double labeled with GFP and HRP from larvae expressing (E) darc1 3′UTR-GFP-neuron (C380> darc1 3′UTR-GFP), (F) darc1 3′UTR A-fragment-GFP-neuron (C380-Gal4> darc1 3′UTR A-fragment-GFP, (G) UGR-GFP (C380>UAS-UGR-GFP), (H) GFP neuron (C380-Gal4>UAS-GFP).
(L,M) in preparations double labeled with GFP and HRP from darc1 3′UTR-GFP-neuron (C380> darc1 3′UTR-GFP) in a (L) wild type and (M) darc1E8/darc1esm113 mutant background.
(I–K) Quantification of (I) dArc1 immunoreactive and (J,K) GFP immunoreactive signal in the indicated genotypes.
(L) Diagram of darc1 mRNA showing the 5′ (blue), the ORF encoding (red), and 3′ (green) UTR. The black lines underneath represent different portions of the darc1 transcript testing the darc1 localization signal.
Calibration bar is 8 μm; N=(from left to right; animals/arbors) I(10/13, 10/13, 10/23, 9/17, 9/15, 9/13, 9/19, 9/17, 9/15), J(9/18, 15/27, 9/18, 9/16), K(10/29, 10/25). Data are represented as mean and error bars represent SEM; statistical analysis was conducted using one-way ANOVA with Tukey post hoc test for I,J, and Student’s t-test for K. *= p < 0.05; **= p < 0.001; ***= p < 0.0001.
Yet, an alternative possibility is that alterations in darc1 RNA and/or protein in presynaptic boutons might limit a signaling mechanism that induces darc1 transcription or enrichment of darc1 RNA and/or protein at the postsynaptic compartment. As RNA localization often depends on the RNA 3′UTR (Berkovits and Mayr, 2015), and as the lack of the darc1 3′UTR prevented the transfer of darc1, we determined if expressing darc1 3′UTR alone in neurons was sufficient to allow the transfer. In these experiments darc1-3′UTR was fused to GFP (Fig. 2U), expressed in neurons, and the levels of GFP signal at the postsynaptic compartment was assessed. As a control, we expressed GFP alone. Expressing darc1-3′UTR-GFP (Fig. 3E), but not GFP alone (Fig. 3H,J), resulted in the presence of GFP signal in the muscle. Furthermore, expressing a shorter fragment of the 3′UTR (A-fragment [Fig. 2U]) fused to GFP in motorneurons was also sufficient for the transport of GFP (Fig. 3F,J). Interestingly, transfer of GFP was also observed when fusing it to a duplication of a darc1 gene fragment (UGR [Fig. 2U]) that includes part of the 3′UTR (Fig. 3G,J; see below for further analysis of the UGR). Thus, darc1 mRNA can be transferred from neurons to postsynaptic muscles in a manner dependent on its 3′UTR.
Notably, expressing darc1-3′UTR-GFP in neurons in a darc1 mutant background partly but significantly inhibited the transfer of GFP (Fig. 3K, L,M). The potential implication of this result is explored below.
dArc1 resembles a fragment of a retrotransposon, and a spliced form of copia RNA is also loaded into EVs
In addition to the presence of retroviral Gag-like sequences (Abrusan et al., 2013b; Campillos et al., 2006) (Fig. 7A), structural analyses have revealed that the Arc Gag region resembles the three-dimensional structure of the HIV virus Gag region (Zhang et al., 2015). Given that retroviruses are also similar to retrotransposons, which are present throughout the mammalian and fly genomes, we analyzed the S2 deep sequencing data using an algorithm designed to identify transposable elements, which are typically masked by the standard genome browsing algorithms. Strikingly, we found that copia mRNA, a common Drosophila retrotransposon, was highly enriched in S2 EVs (Fig. 4A). Closer examination of the sequence reads revealed that the copia RNA sequences present in EVs corresponded to a predicted spliced short copia (copiaS) species consisting primarily of the Gag region (Fig. 4B). This is in contrast to the cellular Copia, which has RNA-seq reads representing the whole transcript (Fig. S2). The copiaS isoform appeared to be selectively loaded into EVs as the ratio of copiaS/full length copia (copiaL) was 12.4/1 in the in the EV fraction, compared to 1.4/1 in the cell. To ascertain if Copia protein was also present in EVs we performed a mass spectrometry-based proteomic analysis of the S2 cell EV fraction. Notably, we found that the short Copia protein isoform was enriched in EVs (Fig. 4C).
Figure 7. Consequences of the UGR for darc1 expression and NMJ morphology, and proposed model of dArc1 transfer (see also Supplemental Fig. 2).
(A) Diagram of the darc1 genomic region in OR showing the UGR element between the darc1 and darc2 genes.
(B,C) Quantification in the indicated genotypes of (B) anti-dArc1 signal and (C) number of synaptic boutons.
(D,E) Merged confocal Z-stacks of NMJ arbors labeled with antibodies to HRP and DLG in (D) Canton-S and (E) Oregon-R.
(F) Diagram depicting a larval NMJ, in which exosome-like vesicles (EV) containing dArc1 protein and transcript are packaged then released from non-synaptic sites. It is still unclear whether these EVs contain multiple enveloped capsid-like particles (a) or a single capsid-like structure (b). These capsid-like particles (c) are taken up by the postsynaptic muscle, either through EV fusion with the muscle membrane, or endocytosis and further fusion with the endosome membrane. We propose that this transfer serves to stimulate synaptic maturation, as downregulation of presynaptic dArc1 leads to accumulation of ghost boutons. (MVB = multivesicular body, SV = synaptic vesicle, AZ = active zone, SSR = subsynaptic reticulum)
Calibration bar is 26 μm; N=(from left to right; animals/arbors) B(12/12,12/24), C(6/10,6/12); data are represented as mean and error bars represent SEM. Statistical analysis was conducted using student’s T-test. *= p < 0.05; **= p < 0.001; ***= p < 0.0001.
Figure 4. Enrichment of Copia-retrotransposon RNA and protein in S2 cell EVs and darc1 mRNA association withdarc1 protein.
(A) Enrichment of copia mRNA in the S2 EV fraction.
(B) Long (L) and short (S) copia isoforms, predicted to be generated by alternative RNA splicing, and enrichment of copiaS, encoding the Gag region, in EVs.
(C) Selected proteins showing enrichment in S2 cell EVs and their abundance in the EV vs cellular fractions, highlighting dArc1, dArc2 and Copia.
(D,E) Immunoprecipitation of darc1 RNA using anti-dArc1 antibodies from extracts of (D) S2 cells and (E) body wall muscles.
(F) Immunoprecipitation of GFP RNA using anti-dArc1 antibodies from extracts of body wall muscles with neurons expressing either GFP alone or GFP upstream of the darc1 3′UTR.
(G) Biotinylated RNA pull down of dArc1, using biotinylated darc1 3′UTR RNA or control RNA. Both pull down dArc1 protein, while beads or RNA alone do not.
N=3 biological repeats for D,E,F; data are represented as mean and error bars represent SEM; statistical analysis was conducted using student’s T-test; *= p < 0.05; **= p < 0.001; ***= p < 0.0001.
dArc1 and dArc2 proteins are also present in EVs, and dArc1 protein binds mRNA
Given that both CopiaS protein and copiaS RNA were found in S2 EVs, we also determined if dArc1 protein, in addition to darc1 mRNA, was similarly enriched in S2 EVs. To determine the relative abundance and enrichment of EV proteins compared to those in the cell, we performed mass spectrometry-based proteomic analysis of EVs compared to total cell proteins. Both dArc1 and dArc2 proteins were highly enriched in EVs compared to cellular proteins (Fig. 4C). Interestingly, our mRNA expression analysis did not reveal any enrichment of darc2 mRNA in the EV fraction. The dArc2 ORF has 52.2% identity and 71.6% similarity with the dArc1 ORF, but lacks the putative C-terminal zinc finger domain, as well as the long darc1 3′UTR mRNA sequences present in darc1.
The Gag region of retroviruses binds to and packages retroviral gRNA. Thus, the enrichment of both dArc1 protein and mRNA in EVs and their similarity to Gag sequences raised the question as to whether dArc1 protein could bind its own mRNA in vivo. This was addressed by conducting RNA immunoprecipitation (RIP) experiments using S2 cell and larval body wall muscle extracts, gel shift assays, and in vitro binding assays. In the RIP experiments, anti-dArc1 was used to immunoprecipitate dArc1 protein and the immunoprecipitate was analyzed by qRT-PCR to determine if darc1 mRNA was present in this fraction. The dArc1 antibody co-immunoprecipitated darc1 mRNA in both S2 cells and body wall muscle preparations (Fig. 4D,E). In contrast, no co-precipitation of other RNAs, such as 18s, eEf1a1, and copia was observed (Fig. 4E), showing that the binding of dArc1 with its own mRNA has some degree of specificity in vivo. This is consistent with the idea that like retroviral Gag proteins, dArc1 exists in a complex with its own mRNA in vivo.
To further explore the binding of dArc1 to its own transcript in vivo, we expressed darc1-3′UTR-GFP or GFP alone in larval neurons, immunoprecipitated dArc1 from extracts of body wall muscles derived from the above larvae, and tested the precipitate for the presence of GFP mRNA by qRT-PCR. Anti-dArc1 antibodies immunoprecipitated GFP RNA only when GFP was fused to the darc1 3′UTR, but not when GFP was expressed alone (Fig. 4F), in agreement with the model that dArc1 protein binds its own 3′UTR in vivo.
We next tested whether purified dArc1 is capable of directly binding RNA. We found, by co-incubating dArc1 with biotinylated control mRNA or biotinylated darc1 mRNA, that dArc1 protein is immobilized on streptavidin-labelled beads (Fig. 4G). We next attempted electrophoretic mobility assays with both biotinylated or radiolabeled mRNAs but were unable to convincingly demonstrate specificity of dArc1 for its own mRNA versus controls. This is consistent with published observations showing that HIV-1 Gag proteins associate specifically with HIV gRNA in vivo, but that this binding specificity is lost in vitro (Comas-Garcia et al., 2016).
dArc1 protein assembles into capsid like structures
We noted that cleaving the HIS-MBP tag after affinity purification of bacterially produced dArc1 protein resulted in the formation of a precipitate. Given that dArc1 resembles a Gag protein, and that retroviral Gag proteins have the ability to auto-assemble into capsids, we examined the precipitate at the EM level by negative staining. Notably, we observed the presence of round structures of about 39.3±5.2 nm (Fig. 5A, B), which resembled HIV capsids. Thus, dArc1 appears to auto-assemble, and together with its ability to bind its own RNA in vivo, it is reasonable to suggest that has some properties similar to retroviruses.
Figure 5. Purified dArc1 protein assembles into capsid-like structures and these structures are contained in EVs.
(A,B) Negatively stained capsid-like structures (white arrow) formed by purified dArc1 protein shown at (A) low and (B) high magnification.
(C,D) Capsid-like structures (white arrow) observed after EV lysis, shown at (A) low and (B) high magnification.
(E–G) Anti-dArc1 ImmunoEM labeling of capsid-like structures (black arrows) derived from lysed EVs, shown at (E) low and (F,G) high magnification. White arrows point to unlabeled capsid like structures. Calibration bar is 120 nm for A,C,E and 40 nm for B,D,F,G.
Owing to the viral properties of dArc1 and we next sought to determine whether we could observe the dArc1 capsid-like structures in our EV preparations (Fig. S3A). In these experiments, the EV preparation was treated with saponin, to lyse EVs, and the resulting preparation examined by EM. Remarkably, we did observe the presence of ~40 nm round structures resembling the capsid-like elements obtained from purified dArc1 (Fig. 5C,D). Unlike those in purified protein, however, the capsid-like structures derived from EVs appeared electron dense, perhaps indicating the presence of RNA within them. To confirm if the ~40 nm structures observed in EVs contained dArc1 protein, we immunolabeled the grids with anti-dArc1 antibodies and 10nm gold conjugated secondary antibody. We found that many (Fig. 5E,F,G; black arrows;), but not all (Fig. 5E; white arrows) of the capsid-like structures contained gold granules (Fig. S3 for controls). This might be due to gold-free secondary, to the stringency of the washes during antibody staining, or to the presence of other capsid-like structures formed by alternative Gags.
dArc1 is involved in synaptic plasticity at the NMJ
We then examined the role of dArc1 during NMJ expansion, a process occurring throughout larval development, involving the addition of new synaptic boutons (Budnik et al., 1990). In addition, we tested its role in rapid activity-dependent synaptic bouton formation (Ataman et al., 2008). We found a drastic reduction in bouton numbers in 3rd instar (the last stage of larval development prior to metamorphosis) darc1 null mutant larvae compared to controls (Fig. 6A,B,C,E,F,G,I). A similar result was observed upon expressing dArc1-RNAi in motorneurons (Fig. 6D,H,I). This suggests that a reduction of dArc1 in motorneurons inhibits synaptic expansion during larval development. Consistent with this model, presynaptic expression of a genomic darc1 rescue construct in motorneurons completely rescued the reduced number of synaptic boutons resulting from eliminating or decreasing dArc1 levels (Fig. 6I). In contrast, expressing the darc1 rescue construct in postsynaptic muscles, or eliminating its 3′UTR, did not rescue the mutant phenotype (Fig. 6I).
Figure 6. dArc1 influences developmental and activity-dependent plasticity at the NMJ.
(A–D) Merged confocal Z-stacks of NMJ arbors labeled with antibodies against HRP and DLG in (A) C380-Gal4/+ control, (B) darc1esm113 mutant, (C) darc1E8/darc1esm113 mutant, and (D) motorneuron expression of dArc1-RNAi1.
(E–H) High magnification view of single confocal slices through NMJ branches in the same genotypes as (A–D).
(I,J,O) Quantification of third instar larval (I) synaptic boutons, (J) ghost boutons, and (O) activity induced ghost boutons in the indicated genotypes and conditions.
(K–M) Single confocal slices of NMJs in preparations with antibodies against HRP and DLG in (K,L) unstimulated NMJs and (M,N) after stimulating NMJs with a spaced stimulation protocol in the indicated genotypes.
Calibration bar is 46 μm in A–H and 6 μm in K–N; N=(from left to right; animals/arbors) I,J(14/27, 8/15, 6/12, 6/12, 6/10, 6/12, 12/21, 6/12, 6/12, 12/21, 8/16, 6/10, 6/10, 14/27, 10/20, 9/16, 6/10), O(12/19,12/22,6/12,6/11,7/14,7/12). Data are represented as mean and error bars represent SEM. Statistical analysis was conducted using one-way ANOVA with Tukey post hoc test for I,J and Student’s t-test for O. *= p < 0.05; **= p < 0.001; ***= p < 0.0001.
NMJ expansion involves the transient formation of immature synaptic boutons (ghost boutons) devoid of neurotransmitter release sites and postsynaptic proteins and specializations (Ataman et al., 2008; Koon et al., 2011). At the postsynaptic side, this is followed by the recruitment of postsynaptic proteins and formation of postsynaptic structures. Several mutations in genes required for NMJ development result in a reduced number of synaptic boutons and an accumulation of ghost boutons (Ataman et al., 2006; Harris et al., 2016). We found that, in addition to a reduction in bouton numbers, transallelic darc1 mutations or darc1 downregulation in neurons resulted in an accumulation of ghost boutons (Fig. 6F,G,H,J). As with the number of synaptic boutons, this phenotype was completely rescued by expressing the genomic darc1 rescue construct in neurons, but not in muscles (Fig. 6J). Moreover, expressing a darc1 transgene lacking the 3′UTR in neurons or muscles did not result in rescue.
NMJ expansion can be stimulated by increased synaptic activity during larval development (Ataman et al., 2008; Budnik et al., 1990). In addition, acute spaced stimulation of motorneurons gives rise to the rapid formation of ghost boutons, some of which subsequently undergo maturation (Ataman et al., 2008). To determine if dArc1 was required for this activity-dependent new bouton formation, we reduced dArc1 levels by expressing dArc1-RNAi in neurons. While the formation of ghost boutons upon spaced stimulation was normal in control animals, it was significantly reduced upon dArc1 downregulation (Fig. 6K,L,M,N,O). Thus, like mammalian Arc, darc1 is required for both developmental and acute forms of synaptic plasticity.
The darc1 genomic region is polymorphic and duplications of darc1 sequences in this region downregulate darc1 expression
It has been previously reported that, in contrast to our observations, dArc1 is not involved in synaptic plasticity (Mattaliano et al., 2007). To determine the potential reason for this discrepancy, we first sequenced the darc1 genomic region of the wild type strain Canton-S (CS). Surprisingly, a neighboring gene annotated as pseudo-gene (referred to here as UGR) in the fly genome and located between darc1 and darc2 was absent in CS (Fig. 7A). The annotated Drosophila genome sequence was derived from a wild type strain different than CS, Oregon-R (OR). Thus, there is a polymorphism in the darc1-darc2 intergenic region locus between CS and OR wild type strain. Closer analysis of the UGR revealed that it was a duplication of a fragment of the darc1 open reading frame (ORF) and part of its 3′UTR (Fig. 7A). To determine the potential impact of the UGR element in the OR strain, we compared darc1 mRNA and protein levels in CS and OR. Notably, darc1 RNA and protein levels were drastically reduced in OR compared to CS (Fig. 7B, Fig. S4). In addition, the number of synaptic boutons was significantly reduced in OR (Fig. 7C,D,E). We propose that the difference between our darc1 mutant analysis and the previous report might arise from this difference between strains. Interestingly, upon sequencing the darc1-darc2 intergenic region from S2 cells, which were generated from OR approximately 40 years ago (Schneider, 1972), we found that it lacks the UGR which raises the possibility that the UGR is a relatively recent divergence.
Discussion
Here we report a mechanism of trans-synaptic communication with several properties resembling retroviruses and retrotransposons, for the trans-synaptic transport of a Gag-related endogenous protein, dArc1, and its mRNA. dArc1 protein associates with the 3′UTR of its own transcript in vivo, and is transported, likely through EVs, from presynaptic boutons to the postsynaptic region of the Drosophila NMJ (Fig. 7F). This mechanism is required for proper synapse maturation during development and for activity-dependent synapse formation. We also provide evidence that, similarly, both Gag protein and RNA sequences from the retrotransposon Copia are loaded into EVs and released by cells, indicating either the domestication of viral mechanisms to shuttle material across cells, or the co-option of an endogenous cellular mechanism for viral infection.
Retroviruses and trans-synaptic transport of darc1 RNA and protein
Multiple lines of evidence support the idea that dArc1 uses certain retroviral-like mechanisms to transfer a signal from the presynaptic compartment to the postsynaptic site required for NMJ expansion during development and for acute activity-dependent synaptic plasticity (Fig. 7F). (1) much like a viral capsid binds its own transcript, dArc1 protein interacts with the darc1 mRNA, specifically its 3′UTR. It remains to be determined if dArc1 directly binds to its own transcript and if this binding is needed for EV loading of transcript and how dArc1 protein itself is loaded into EVs. (2) Like retroviruses, darc1 RNA and protein are transmitted from cell to cell as determined by our observations of trans-synaptic transfer of wild type dArc1 protein and mRNA. (3) Arc protein appears to self-assemble forming capsid-like structures that are released from cells and can be extracted from exosome preparations. (4) The transfer appeared to be unidirectional, as postsynaptic muscle darc1 mRNA and dArc1 protein levels were decreased when expressing dArc1-RNAi in neurons but not in muscles. The inability of dArc1-RNAi to downregulate darc1 mRNA might be due to darc1 RNA, at the postsynaptic region, being inaccessible to the RNAi machinery (e.g. D2 bodies (Nishida et al., 2013)). In our experiments dArc1-RNAi was expressed with the UAS/Gal4 system, which requires its transcription and export from the nucleus. In contrast, our experiments suggest that postsynaptic darc1 mRNA and protein are locally derived from the presynaptic neuron, which likely determines their postsynaptic localization. Indeed, expressing a dArc1 transgene in muscle did not result in synaptic localization of the transgenic protein. Another possible explanation of the resistance of postsynaptic Arc mRNA to postsynaptic RNAi is the ability of Arc protein to multimerize (Myrum et al., 2015) to form a capsid which protects dArc1 RNA. Future studies will determine how darc1 mRNA is unpackaged post-transfer from its Gag capsid to carry out its function and what processes are regulated by dArc1 leading to synapse growth and maturation.
Supporting the model that dArc1 takes advantage of viral properties for trans-synaptic signaling is our observation with a common endogenous fly retrotransposon, Copia. Retrotransposons like Copia contain the entire set of genes present in a retroviral genome except for envelope genes (Nefedova and Kim, 2017). Thus, unlike retroviruses, retrotransposons are thought not to be transferred between cells. Nevertheless, we found that both copia RNA and protein were released from cells via EVs which likely allows them to spread to neighboring cells. Strikingly, the Gag-encoding truncated region of copia RNA and protein were the predominant forms in S2 cell EVs. This short form does not encode the proteins necessary for Copia replication and integration into the genome, supporting the idea that this type of transcellular communication simply uses the RNA-binding properties of Gags to transport RNAs across cells.
The ORF of mammalian Arc and darc1 are largely comprised of regions derived of viral-like Gag sequences, likely the remnants of an earlier transposon insertion, and previous work showed that the Gag region of Arc can fold like a viral capsid (Zhang et al., 2015). There are over 30 other proteins in Drosophila that have significant portions of their coding region comprised of Gag like sequences (Abrusan et al., 2013a). Both the fly and mammalian genomes contain a large proportion (~40%) of transposable elements (Lander et al., 2001), so far referred to as “junk DNA”. Our results raise the provocative idea that other Gag-related proteins and Gag-containing transposons might have a physiological function in cell-cell communications. This would explain the retention of retrotransposons through evolution. In addition, it is possible that the Gag-containing proteins encoded in the fly genome could represent the serendipitous integration of a retrotransposon into a functional gene, similarly to how dArc1 was likely created. Future studies geared to understanding the function of these Gag proteins as well as Gag-encoding transposons or transposon fragments will be needed to address these possibilities.
darc1-3′UTR as a mechanism of dArc1 loading into EVs
Drosophila Arc1 and Arc2 appear to result from a genomic duplication event and are solely composed of a Gypsy transposon-derived Gag domain. While the ORF of darc1 and darc2 are highly conserved, they differ vastly in their 3′UTR. Our studies with a GFP reporter show that the 3′UTR of darc1 mRNA is necessary and sufficient for the transport and accumulation of darc1 postsynaptically. This suggests that the 3′UTR of darc1 imparts some function needed to load darc1 mRNAs into EVs. In this regard, it’s interesting to note that the dArc2 protein, but not its mRNA, is enriched in EVs. While the protein and mRNA sequences of darc1 and darc2 are very similar, they differ dramatically in the 3′UTR, which in the case of darc2 is much shorter. We hypothesize that this difference might explain the absence of darc2 mRNA in EVs. Our data suggests that darc1 mRNA may prove to be a powerful model to understand EV RNA loading in vivo. Alternatively, as reported in the companion paper (Day et al.), RNA binding to Gag proteins might be required to assemble a capsid, with might be needed for EV loading. The rat Arc 3′UTR contains Gypsy-like sequences, transposon sequences similar to those of darc1. Since these genes most likely evolved independently (Abrusan et al., 2013b) (also see the companion paper; Day et al.) the similarity of the mammalian and fly Arc proteins and mRNAs indicates the possibility of convergent evolution of this mechanism of trans-cellular communication.
Trans-synaptic dArc1 transfer in Drosophila synapse development and plasticity
Our studies of darc1 mutants, dArc1-RNAi, and expression of transgenic dArc1 variants suggest that the transfer of dArc1 is required for normal expansion of the NMJ, synaptic bouton maturation, and activity-dependent synaptic bouton formation. For example, expressing a dArc1 transgene lacking the 3′UTR in neurons, while resulting in the localization of the transgenic protein at presynaptic boutons, it is not transferred to the postsynaptic region and fails to rescue mutant phenotypes at the NMJ. In contrast, expressing a transfer-competent dArc1 transgene, containing the 3′UTR, results in complete rescue. Therefore, it is not just the presence of dArc1 in presynaptic terminals, but the actual transfer to the postsynaptic region which is required for dArc1 function at the NMJ. This is also supported by our finding that expressing dArc1 containing the 3′UTR in muscles alone did not result in normal postsynaptic dArc1 localization, nor did it rescue mutant phenotypes at the NMJ. Thus, both normal postsynaptic localization of dArc1 and its function in synaptic development and plasticity requires dArc1 transfer from the presynaptic terminus. The requirement of darc1-3′UTR also provides support to the idea that darc1 mRNA, and not just dArc protein is transferred, which is also supported by our finding of both dArc1 protein and RNA in EVs.
Previous studies show that the transfer or release of EV proteins, such as Evi and Wg, is enhanced by electrical activity (Ataman et al., 2006), and similar observations have been made with cultured mammalian neurons and glia (Faure et al., 2006; Fruhbeis et al., 2013). This opens the possibility that dArc1 might be delivered to postsynaptic sites in an activity-dependent fashion. This would allow the functional modification of specific postsynaptic sites.
Trans-synaptic transfer as a general mechanism of Arc/dArc1 function
In mammals Arc is a master regulator of synaptic plasticity, being involved in many aspects of synapse formation, maturation and plasticity, as well as in learning and memory (Shepherd and Bear, 2011). Arc expression is induced by synaptic activity and its mRNA becomes localized to active dendritic spines, where it contributes to local translation during synaptic plasticity (Farris et al., 2014). Not surprisingly, in humans, mutations in Arc are associated with multiple neurological disorders affecting synapses, including autism spectrum disorders (Alhowikan, 2016), Angelman syndrome (Cao et al., 2013), and schizophrenia (Fromer et al., 2014). While some mechanisms of Arc function, such as its involvement in trafficking of glutamate receptors during plasticity (Chowdhury et al., 2006) are beginning to be elucidated, the extent of its roles remain to be deciphered. In this and the companion paper (Day et al.), we reveal a highly significant Arc/dArc1 role in trans-synaptic signaling. The studies reveal the significance of a long-noted but mysterious feature of Arc/dArc1 protein, its resemblance to retroviral Gags. Together with the companion paper we show that, like retroviruses, Arc/dArc1 proteins can form capsids capable of packaging RNAs. These capsids are loaded into EV-like vesicles that can be released from synaptic sites and taken up by synaptic partners. While a functional role in synaptic development and plasticity is documented here at the Drosophila NMJ, the significance of this transfer at mammalian synapses remains to be determined. In Drosophila dArc1 protein and mRNA are present both inside presynaptic boutons and at the postsynaptic muscle region (this report). In contrast, Arc has been reported to localize exclusively in dendrites, and not at presynaptic sites (Lyford et al., 1995). The finding that mammalian Arc is also released in EVs (Day et al) raises the possibility that this release might serve as a signaling mechanism between dendritic spines. However, if this signaling process plays a role in synaptic plasticity, it would call into question the synapse-specificity of synaptic plasticity documented in the mammalian brain (Viola et al., 2014). An alternative possibility is that mammalian Arc, while primarily being localized at postsynaptic sites, it is also present in lesser amounts at presynaptic terminals. Indeed, in the fly, most of the dArc1 protein and RNA is present at the postsynaptic region. Studies of Arc downregulation in presynaptic neurons and its effect in the localization of Arc at dendritic spines, may serve to distinguish between these possibilities.
STAR Methods
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vivian Budnik (Vivian.Budnik@umassmed.edu).
Experimental Model and Subject Details
The following fly lines were used, UAS-dArc1-RNAi1 (31122, Vienna Drosophila Research Center), UAS-Rab11DN[N124I] (Satoh et al., 2005). UAS-dArc1-RNAi2 (see below), the wild type strain, CantonS (CS) (1, Bloomington Drosophila stock center, BDSC), the wild type strain, OregonR (OR) (6362, BDSC), UAS-GFP (5431, BDSC), UAS-darc1-3′UTR-GFP (see below), UAS-darc1-A-fragment-GFP (see below), UAS-darc1-UGR-GFP (see below), C380-Gal4 and C57-Gal4 (Budnik, 1996). Unless otherwise mentioned in the text, for constiency, female 3rd Instar larvae were dissected for NMJ preparations.
Method Details
Fly husbandry
All flies were raised on standard molasses formulation food at either 25°C (most crosses) or 29°C (RNAi crosses).
Generation of a dArc1null allele
A mutant allele of darc1 was generated by Crispr/Cas9-mediated genomic engineering as previously described (Port and Bullock, 2016). Briefly, sequences encoding two single-guide RNAs (sgRNA1 and sgRNA2) targeting darc1 gene flanking sites which were predicted to have no off-targets (flyCRISPR target finder; (http://tools.flycrispr.molbio.wisc.edu/targetFinder/) (Gratz et al., 2014) were cloned into the pCFD4 plasmid. This construct was injected into Actin-Cas9 expressing embryos and flies containing potentially mutant chromosomes were captured over a balancer chromosome and made into stocks. The stocks were subsequently screened for the desired deletion by PCR, employing deletion-flanking primers sets. Sequencing of PCR products generated from the darc1E8 line revealed the expected deletion.
Constructs
The sequence of constructs made for this paper can be found in Table S1 DNA sequences of constructs used. Related to STAR Methods.
The UAS-darc1-3′UTR-GFP constructs were cloned into pAWG vector using the Gateway system (ThermoFisher). The GFP with resulting 3′UTR fragments were amplified out of this construct, and cloned into pENTR/DTOPO. The resulting construct was finally cloned into the pTWM construct, containing an attB site for targeted genomic insertion. The darc1 sequences are in Table S1.
The UAS-dArc1-RNAi2 construct was cloned by first amplifying a dArc1 fragment, that was non-overlapping with the VDRC 31122 line, using RNAi2 forward and reverse primers.
The PCR product was cloned into PENTR-DTOPO (ThermoFisher), and then, using the Gateway system (ThermoFisher), it was cloned into pWalium10. All constructs were then injected into flies and integrated at site attP2 on the third chromosome, through targeted integration by BestGene.
The UAS-dArc1 rescue transgene was synthesized (Genscript) and using the Gateway system (ThermoFisher), cloned into the pTWM vector. The sequence of the darc1 rescue construct is included in Table S1.
Immunocytochemistry and antibodies
Body wall muscles from third instar larva were dissected in low calcium (0.1mM Calcium) HL3 saline (Stewart et al., 1994) and fixed in either Bouin’s fixative (0.9% picric acid, 5% acetic acid, 9% formaldehyde, 2.5–5% methanol) or 4% paraformaldehyde in 0.1 M phosphate buffer. Fixed larvae were washed and permeabilized in PBT (0.1 M phosphate buffer; 0.2% (v/v) Triton X-100) and incubated in primary antibody overnight at 4°C. Samples were washed three times with PBT and incubated in secondary antibodies for 2 hours at room temperature. After incubation with secondary antibodies, samples were washed thrice with PBT and mounted in Vectashield (Vector Laboratories). The following antibodies were used: anti-dArc1 (see below 1:500 for immunocytochemistry (ICC), 1:1000 for Western blots), anti-GFP (Developmental Studies Hybridoma Bank (DSHB), 4C9, 1:200), and rabbit anti-DLG, 1:40,000 (Koh et al., 1999). DyLight-conjugated secondary antibodies were from Jackson ImmunoResearch (DyLight-594-conjugated goat anti-HRP, DyLight-488-conjugated anti-mouse or anti-rabbit or, DyLight-594-conjugated anti-mouse or anti-rabbit) and were used at 1:200 dilution.
dArc1 antibodies were generated against the first 56 amino acids of dArc1 by immunizing rabbits and rats with purified 6X His tagged peptides (Pocono Rabbit Farm and Laboratory). A synthetic gene representing the first 56 amino acids was synthesized into pET151 (ThermoFisher), transfected into BL21(DE3) bacterial cells (ThermoFisher) and His tagged dArc1 peptide was purified on a nickel column (Pierce).
Fluorescent in situ hybridization (FISH)
In situ hybridization was performed as described previously (Speese et al., 2012) with minor modifications. Briefly larval body wall muscles were dissected as described above and fixed with 4% paraformaldyhyde for 30min. Preparations were then washed three times, 10 min each, with 0.2%PBT with RNasin (Promega). Samples were equilibrated with hybridization buffer (2XSSC, 10% dextran sulfate, RNasin (Promega), 50% formamide). Gene specific probes (125ng probe, 1.25μg salmon sperm DNA, 1.25μg yeast tRNA) were heated to 80°C for 5min and chilled on ice immediately. Probes were then combined with equal volumes of 2X hybridization buffer (final concentration of 2.5ng/μL gene specific probes). Samples were incubated with probes for 3 hours at 37°C. From this point onward, the protocol was the same as in Speese et al 2012 (Speese et al., 2012). To visualize synaptic boutons, samples were colabeled with goat anti-HRP-Dylight594 (Jackson ImmunoResearch).
Probe preparation
Probe preparation was the same as described previously(Speese et al., 2012). Briefly, probes were designed based on the cDNA sequences of dArc1 and amplified using dArc1 probe primers. The probes were produced by nick translation of the PCR product (Bionick; ThermoFisher) with digoxigenin-11-dUTP (Roche) for 2.5hrs at 18°C, and the reaction was inactivated by heating to 65°C for 10min. Probes were precipitated and resuspended in formamide and stored at −80°C. Blocking probe against shibire was prepared using the same method except dTTP was used instead of digoxigenin-labeled dUTP.
Confocal Microscopy and Signal Intensity Measurements
Images shown in figures were acquired on Zeiss LSM 700 or LSM 800 confocal microscope equipped with a Zeiss 63X Plan-Apochromat 1.4 NA DIC oil immersion objective. For quantification of signal intensity using Volocity Software (version 6.3.1), NMJs were imaged at identical settings for control and experimental groups processed simultaneously on a Zeiss Axioplan microscope equipped with a Yokogawa CSU10 spinning disk confocal scanning unit and a Hamamatsu 9100 EM-CCD camera (512 × 512) and a 40X EC-Plan-NeoFluar 1.3 NA objective. Briefly, after image acquisition, the bouton volume bounded by HRP staining was selected using Volocity, and fluorescence intensity inside was measured. To calculate the postsynaptic area, the presynaptic bouton selection was dilated by 8 iterations, the HRP containing volume was subtracted and the intensity within the remaining volume was measured. Intensity was determined as the sum of total pixel intensity in each volume (pre- and postsynaptic) and normalized to bouton volume, as described previously (Ramachandran et al., 2009).
Exosome Preparation
Exosomes were prepared from S2 cells that were cultured in serum-free medium to avoid contamination from Bovine serum exosomes. Cultures were grown in spinner flasks (BellCo Glass Inc.) at 22 °C and harvested at 1–1.5 × 106 cells/mL density. Exosomes were further purified as described previously (Koles et al., 2012) but detailed as follows. The cells from 500 ml of culture were pelleted at 300 × g for 5 min, the supernatant was then spun for10 min at 2000 × g, to clear dead cells. To remove large cellular debris the samplers were then centrifuged at 10,000 × g for 30 min to remove. To collect exosomes the samples were spun at 100,000 × g for 70 min, the pellet was resuspended in PBS. Tris/sucrose/D2O (200 mM, 30% (w/v), 50% (v/v)) was mixed together and put into a centrifuge tube to form a sucrose cushion on top of which the exosome pellet was layered, then centrifuged in an SW28 ultracentrifuge rotor at 100,000 × g for 75 min. Using a syringe through the side of the tube 3.5ml of the Tris/sucrose/PBS interface was collected. To remove the sucrose and D2O 60 ml of PBS was added to samples which were then centrifuged at100,000 × g for 70 min. The final pellet representing the purified exosomes were resuspended in 500ul of PBS and either processed for future experiments or stored at −80 °C.
RNA sequencing
Exosomes were treated with micrococcal nuclease (NEB) for 1 hour at 37°C, and then both exosomes and cell pellets we re treated with RLT buffer (Qiagen) and RNA was extracted using RNeasy micro Kit (Qiagen). Libraries were prepared using NEBNext Ultra directional RNA library prep kit for Illumina sequencing. Library 1 was subjected to single end sequencing with 18934428 and 13513146 reads for cell and exosomes respectively. Libraries 2, 3 and 4 were pair end-sequenced using miSeq illumina. While library 2 had 1745707 exosome and 2188190 cell reads, library 3 had 2921143 exosome and 1823805 cell reads and library 4 had 3649518 exosome and 5047915 cell reads. Reads from each library were sorted by barcode, and adapter sequences removed. Reads were then mapped using TopHat2 (Kim et al., 2013) to the Drosophila genome. Transcript expression was measured using Cufflinks (Trapnell et al., 2012), and differential expression was determined using DeSeq2 (Love et al., 2014). For transposon mapping reads, reads from each library were mapped to common Drosophila transposon sequences using Bowtie2 (Langmead et al., 2009). Expression was then measured using eXpress (Roberts and Pachter, 2013) and DeSeq2.
Biotinylated RNA pull down assay
Reagents used for this assay were from the LightShift™ Chemiluminescent RNA EMSA Kit (ThermoFisher, Catalog number: 20158). For dArc1 protein, the dArc1 ORF was cloned into the pDEST-His-MBP(Nallamsetty et al., 2005) vector using the Gateway system (ThermoFisher). Protein was then expressed in Bl21(DE3) E. coli (ThermoFisher) and purified by nickel-chelation chromatography (New England Biolabs). The 3′UTR of darc1 was cloned into pDEST17 (ThermoFisher) using the Gateway system (ThermoFisher). The resulting vector was then digested with EcoRV, and then incubated overnight with T7 RNA polymerase (ThermoFisher). The subsequent RNA was then purified through lithium chloride precipitation, and then end labeled using 3′ Biotinylation (ThermoFisher). For competition assays, the specific competitor was the unlabeled 3′UTR transcript, while the unspecific competitor was generated from an empty pDEST17 vector linearized with SmaI, incubated overnight with T7 RNA polymerase, and purified through lithium chloride precipitation as above. Each binding assay was carried out using 80ng of labelled darc1 3′UTR and 9μg of dArc1 protein. After incubating for 30 minutes, 25μl of washed Steptavidin magnetic beads (ThermoFisher) was added. The beads were washed 3X with binding buffer, then protein loading buffer was added, the samples were boiled, run on a stain-free 4–20% acrylamide gel (Bio-Rad), and imaged in a ChemiDoc Imaging System (BioRad).
Sample preparation for LC/MS/MS
Purified exosomes and cell pellets were diluted in loading buffer (see below), incubated at 95°C for 1 5 min, and resolved by SDS-PAGE in a 4–20% gradient gel under reducing and denaturing conditions. Proteins were run 2cm into the resolving gel, excised, and processed for mass spec analysis.
LC/MS/MS
Gel slices were cut into 1×1 mm pieces and placed in 1.5ml Eppendorf tubes with 1ml of water for 30 min. The water was removed and 150μL of 250mM ammonium bicarbonate was added. For reduction, 20μL of a 45mM solution of 1,4 dithiothreitol (DTT) was added and the samples were incubated at 50°C for 30 min. The samples were cooled to room temperature and then, for alkylation, 20μL of a 100mM iodoacetamide solution was added and allowed to react for 30 min. The gel slices were washed 2 X with 1ml water aliquots. The water was removed, 1ml of 50:50 (50mM ammonium: bicarbonate: acetonitrile) was added to each tube, and samples were incubated at room temperature for 1hr. The solution was then removed and 200μL of acetonitrile was added to each tube at which point the gels slices turned opaque white. The acetonitrile was removed and gel slices were further dried in a Speed Vac. Gel slices were rehydrated in 100μL of 4ng/μL trypsin (Sigma or Promega sequencing grade) in 0.01% ProteaseMAX Surfactant (Promega): 50mM ammonium bicarbonate. Additional bicarbonate buffer was added to ensure complete submersion of the gel slices. Samples were incubated at 37°C for 21hrs. The supernatant of each sample was then removed and placed in a separate 1.5mL eppendorf tube. Gel slices were further dehydrated with 100μL of 80:20 (acetonitrile: 1% formic acid). The extract was combined with the supernatants of each sample. The samples were then dried down in a Speed Vac. Samples were dissolved in 25μL of 5% acetonitrile in 0.1% trifluroacetic acid prior to injection on LC/MS/MS. A 3.0μl aliquot was directly injected onto a custom packed 2cm × 100μm C18 Magic 5μ particle trap column. Peptides were then eluted and sprayed from a custom packed emitter (75μm × 25cm C18 Magic 3μm particle) with a linear gradient from 95% solvent A (0.1% formic acid in water) to 35% solvent B (0.1% formic acid in acetonitrile) in 90 minutes at a flow rate of 300 nl/min on a Waters Nano Acquity UPLC system. Data-dependent acquisitions were performed on a Q Exactive mass spectrometer (Thermo Scientific) according to an experiment where full MS scans from 300–1750 m/z were acquired at a resolution of 70,000 followed by 10 MS/MS scans acquired under HCD fragmentation at a resolution of 17,500 with an isolation width of 1.6 Da. Raw data files were processed with Proteome Discoverer (version 1.4) prior to searching with Mascot Server (version 2.5) against the Uniprot database. Search parameters utilized were fully tryptic with 2 missed cleavages, parent mass tolerances of 10 ppm and fragment mass tolerances of 0.05 Da. A fixed modification of carbamidomethyl cysteine and variable modifications of acetyl (protein N-term), pyro glutamic for N-term glutamine, oxidation of methionine and serine/threonine phosphorylation were considered. Search results were loaded into the Scaffold Viewer (Proteome Software, Inc.) for assessment of protein identification probabilities and label free quantitation.
RNA IP with Real time quantitative PCR
For S2 cell preparations, cells were raised in serum free medium, as above, and then centrifuged at 300 × g to pellet the cells. Once pelleted, cells were resuspended in RIPA buffer (Abcam), and homogenized using 0.5mm glass beads at 4°C using a BBX24B Bullet Blen der Blue homogenizer (Next Advance Inc.). Larval body wall muscles from wild type animals were homogenized in RIPA buffer (Abcam), and homogenized as above. Supernatants were cleared against magnetic beads (Pierce), and then incubated with either anti-dArc1, or equal amounts of preimmune serum. Samples were incubated overnight with serum and magnetic beads, and washed several times with RIPA buffer. Finally, beads were either incubated directly with 5X loading buffer (4% SDS, 250mM Tris, Bromphenol blue, 30% glycerol, and 2-Mercaptoethanol) for Western blot, or RNA was eluted from the beads with RLT buffer (Qiagen) and then purified using the RNeasy micro kit (Qiagen). RNA samples from both conditions were DNase treated with TurboDNase (ThermoFisher) and then equal volumes were reverse transcribed into cDNA using Superscript III (ThermoFisher). The RT-quantitative PCRs were performed in triplicate in a 96-well plate (BioRad) using a CFX96 system (BioRad). For the reactions, a Sybr green master mix (ThermoFisher) was used with the gene specific primer sets for dArc1, Copia, GFP, dArc2 and 18S rRNA. RpL32 was used as a reference gene. All transcript levels were normalized to RpL32 transcript level, using the same cDNA template. Data were expressed in graphs as ΔCT. For RTq-PCR on body wall preparations, 5 larvae per genotype were dissected and RNA was purified from Brains and body wall muscles separately using RNeasy kit (Qiagen). All cDNA for body wall preparation analysis was synthesized with 100ng of RNA, and then samples were treated as above for qRT-PCR.
Western Blotting
Extracts from RIP experiments were incubated at 95 °C for 15 min, and resolved by SDS-PAGE in a 4–20% gradient gel under reducing and denaturing conditions. Proteins were transferred onto nitrocellulose membrane (Bio-Rad) and blocked in 5% instant nonfat dry milk in TBST (50 mM Tris (pH 7.4), 150 mM NaCl, 0.05% Tween 20) and incubated with primary antibodies (diluted to working concentration in blocking solution) overnight at 4 °C. After washing in TBST, blots were incubated with HRP-conjugated secondary antibodies diluted to 1:3000 in blocking solution for 1 h at room temperature. Western blots were visualized using the SuperSignal West Femto Maximum Sensitivity Substrate kit (ThermoFisher). Blots were imaged using Chemidoc Touch imaging system (BioRad).
Activity Paradigm
Potassium stimulations were performed as in (Ataman et al., 2008). Briefly, larva were dissected in low calcium HL3 (Stewart et al., 1994) saline, and then pulsed with high potassium saline for 2 minutes, 3 times, each separated by 15 minutes of rest in normal saline. The final two stimulations were done for 4 and 6 minutes separated by 15 minutes of rest. Samples were then fixed and processed for immunocytochemistry.
dArc1 Capsid Formation
The darc1 open reading frame was cloned into pENTR (Thermo Fisher), its sequence confirmed, and pENTR-Arc was subsequently recombined using LR Clonase (Thermo Fisher) with the pDEST-HisMbp Destination vector (Nallamsetty et al., 2005) to generate His-MBP-dArc1. Expression induction and purification under native conditions in a Ni-NTA column (Qiagen) were performed following standard procedures. To generate soluble protein for ultrastructural studies, protein was diluted to ~ 1 mg/ml and cleaved at 30 degrees C with AcTEV (Thermo-Fisher), followed by removal of the His-MBP tag and the His tagged TEV by binding to a Ni-NTA column and dialysis against PBS.
Electron Microscopy of dArc1 capsid
After formation of dArc1 capsids, the capsids were examined using negative staining. Capsids were fixed in 2% paraformaldehyde overnight at 4°C. 5μL of fixed capsids were spotted onto formvar coated grids. After 20 min absorption, grids were rinsed with PBS and then fixed with 1% glutaraldehyde for 5 min. Finally, samples were washed with water and then counter-stained with 2% w/v uranyl acetate, and blotted dry. Samples were imaged on an FEI EM 10 electron microscope at 80 kv.
ImmunoEM of Exosomes
Exosome preparations were fixed overnight in a final concentration of 2% paraformaldehyde (EM grade) at 4°C. After fixation, solution was gently pipetted up and down several times to resuspend exosomes. Formvar coated grids (EMS) were glow discharged, spotted with 4μl of exosomes, and incubated for 10 min at room temperature. Excess solution was removed by gently wicking liquid off of the grid on #50 filter paper. Grids were washed 2 times, 3 min each, in 100mM Tris, followed by 4 washes, 3 min each, in 100mM Tris + 50mM Glycine. Grids were blocked for 10 min with 100mM Tris + 0.1% BSA. After block, exosomes were either incubated in 100mM Tris (control) or lysed with 0.05% saponin in 100mM Tris. Grids were washed for 2 min with 100mM Tris followed by an anti-dArc1 antibody (1:500 in 100mM Tris) incubation for 1hr. Grids were then washed 5 times, 3 min each, in 100mM Tris, followed by a 30 min incubation with goat anti-rabbit conjugated to 10nm gold (Electron Microscopy Sciences) secondary (1:60 dilution in 100mM Tris). Grids were washed 8 times, 2 min each, in 100mM Tris, and then fixed with 1% glutaraldehyde in 100mM Tris for 1 min at room temperature. Finally, grids were washed again 8 times, 2 min each, with distilled water, and then negative stained with 1% uranyl acetate for 30 seconds. Grids were imaged on an FEI EM 10 as above.
Quantification and Statistical Analysis
Statistical analysis was performed using a Student’s t test when a single experimental sample was compared with control. For comparison of multiple experimental groups, a one-way analysis of variance (ANOVA) was used followed by a Tukey’s post hoc test. *, p < 0.05; **, p < 0.001; ***, p < 0.0001. Excel (Microsoft) and Kaleidograph (Synergy Software) were used to measure, standard deviations, standard errors and averages, as well as create graphs. Each figure contains relevant information as to the number of experimental repeats and other data needed to assess the accuracy of the data.
Data and Software Availability
Data Resources
The accession number for the raw and analyzed RNA-sequencing data reported in this paper is GEO:GSE104972
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Deutsch et al., 2017) via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD008136 and 10.6019/PXD008136.
Supplementary Material
Figure S1.dArc1 Immunoprecipitation anddarc1RNA levels (see also Figure 2)
(A) Western blot showing dArc1 immunoprecipitation from body wall muscle extracts derived from wild type (CS) and darc1esm113 mutants labeled with anti-dArc1. Number at the left of the blot represents molecular weight in kilodaltons.
(B) darc1 qRT-PCR performed on RNA isolated from either dissected body wall muscles or brains in the indicated genotypes.
(C) darc1 RT-qPCR performed on RNA isolated from wild type larval body wall and darc1esm113.
N (biological replicates per genotype from left to right) is B:6,5,4,4; C 3 for each genotype; Graphs represent mean, and error bars SEM; statistical analysis was performed using a Student’s t-test; *= p < 0.05; **= p < 0.001; ***= p < 0.0001.
Figure S2.Full-lengthcopiaLtranscript is found in the cellular fraction (see also Figure 4)
Raw number of RNA-seq reads for cellular Copia showing reads across the entire predicted transcript.
Figure S3.Controls for EVs and capsid-like structures (see also Figure 5)
(A) Ultrastructure of negatively stained exosomal fraction without saponin treatment
(B) No primary antibody control for lysed EVs. Grids containing this fraction were processed for immunocytochemistry, but the dArc1 primary antibody was omitted. No 10nm gold granules are observed.
Calibration bar is 70 nm in A and 350 nm in B.
Figure S4.dArc1 transcript levels in Canton-S vs Oregon-R. (see also Fig. 7)
qRT-PCR results from body wall preparations from the indicated genotypes.
N (biological replicates; from left to right) = 10,9; Graphs represent mean, and error bars SEM; statistical analysis was performed using a Student t-test; *= p < 0.05; **= p < 0.001; ***= p < 0.0001.
Highlights.
arc1 mRNA/protein traffic across synapses via exovesicles, requiring its 3′UTR
dArc-protein Gag binds darc1 mRNA, and the protein forms capsid-like structures
Exovesicles contain GAG-protein and Gag-encoding-mRNA of the retrotransposon Copia
Transfer of darc1 protein and/or mRNA is required for synaptic plasticity
Acknowledgments
We thank the Bloomington Drosophila Stock Center and flybase (NIH P40OD018537) for providing stocks and managing the fly database, and the Vienna Drosophila Resource Center (VDRC). We thank Drs. Melissa Moore for valuable discussions and methodological guidance, Emiliano Riccio for help in library preparation in the initial stages of this project, and Franz Wendler for exosome preparation. We also thank Dr. Jason Shepherd for helpful discussions. This work was supported by NIH Grant R37MH070000 to VB.
Footnotes
Author contributions (CRediT taxonomy)
Conceptualization: VB and TT; Methodology: JA, BC, LF, VB, and TT; Software: JA, TT; Validation: VB, TT, JA; Formal Analysis: TT, JA, VB, BC; Investigation: JA, BC, DL, LF and TT; Resources: VB, EM core, Neurobiology Department, Deep Sequencing Core; Data Curation: TT, JA, VB; Writing – Original Draft: VB, TT; Writing – Review & Editing: VB, TT, JA, BC, LF; Visualization: VB; Supervision: VB, JA, TT; Project Administration: VB, JA, TT; Funding Acquisition: VB.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrusan G, Szilagyi A, Zhang Y, Papp B. Turning gold into ‘junk’: transposable elements utilize central proteins of cellular networks. Nucleic Acids Res. 2013a;41:3190–3200. doi: 10.1093/nar/gkt011. doi:3110.1093/nar/gkt3011. Epub 2013 Jan 3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrusan G, Szilagyi A, Zhang Y, Papp B. Turning gold into ‘junk’: transposable elements utilize central proteins of cellular networks. Nucleic Acids Res. 2013b;41:3190–3200. doi: 10.1093/nar/gkt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenquer M, Amorim MJ. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses. 2015;7:5066–5083. doi: 10.3390/v7092862. doi:5010.3390/v7092862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhowikan AM. Activity-Regulated Cytoskeleton-Associated Protein Dysfunction May Contribute to Memory Disorder and Earlier Detection of Autism Spectrum Disorders. Med Princ Pract. 2016;25:350–354. doi: 10.1159/000445351. doi:310.1159/000445351. Epub 000442016 Mar 000445311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataman B, Ashley J, Gorczyca D, Gorczyca M, Mathew D, Wichmann C, Sigrist SJ, Budnik V. Nuclear trafficking of Drosophila Frizzled-2 during synapse development requires the PDZ protein dGRIP. Proc Natl Acad Sci U S A. 2006;103:7841–7846. doi: 10.1073/pnas.0600387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataman B, Ashley J, Gorczyca M, Ramachandran P, Fouquet W, Sigrist SJ, Budnik V. Rapid activity-dependent modifications in synaptic structure and function require bidirectional Wnt signaling. Neuron. 2008;57:705–718. doi: 10.1016/j.neuron.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso M, Bonetto V. Extracellular Vesicles and a Novel Form of Communication in the Brain. Front Neurosci. 2016;10:127. doi: 10.3389/fnins.2016.00127. eCollection 02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. doi:310.1038/nature14321. Epub 12015 Apr 14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V. Synapse maturation and structural plasticity at Drosophila neuromuscular junctions. Curr Opin Neurobiol. 1996;6:858–867. doi: 10.1016/s0959-4388(96)80038-9. [DOI] [PubMed] [Google Scholar]
- Budnik V, Koh YH, Guan B, Hartmann B, Hough C, Woods D, Gorczyca M. Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron. 1996;17:627–640. doi: 10.1016/s0896-6273(00)80196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V, Zhong Y, Wu CF. Morphological plasticity of motor axons in Drosophila mutants with altered excitability. J Neurosci. 1990;10:3754–3768. doi: 10.1523/JNEUROSCI.10-11-03754.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillos M, Doerks T, Shah PK, Bork P. Computational characterization of multiple Gag-like human proteins. Trends Genet. 2006;22:585–589. doi: 10.1016/j.tig.2006.09.006. Epub 2006 Sep 2018. [DOI] [PubMed] [Google Scholar]
- Cao C, Rioult-Pedotti MS, Migani P, Yu CJ, Tiwari R, Parang K, Spaller MR, Goebel DJ, Marshall J. Impairment of TrkB-PSD-95 signaling in Angelman syndrome. PLoS Biol. 2013;11:e1001478. doi: 10.1371/journal.pbio.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. HIV Capsid Assembly, Mechanism, and Structure. Biochemistry. 2016;55:2539–2552. doi: 10.1021/acs.biochem.6b00159. doi:2510.1021/acs.biochem.2536b00159. Epub 02016 Apr 00126. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas-Garcia M, Davis SR, Rein A. On the Selective Packaging of Genomic RNA by HIV-1. Viruses. 2016;8 doi: 10.3390/v8090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes JL, Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol. 2007;500:433–447. doi: 10.1002/cne.21189. [DOI] [PubMed] [Google Scholar]
- Farris S, Lewandowski G, Cox CD, Steward O. Selective localization of arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. J Neurosci. 2014;34:4481–4493. doi: 10.1523/JNEUROSCI.4944-13.2014. doi:4410.1523/JNEUROSCI.4944-4413.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, Blot B, Grange J, Schoehn G, Goldberg Y, Boyer V, et al. Exosomes are released by cultured cortical neurones. Molecular and cellular neurosciences. 2006;31:642–648. doi: 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. doi:110.1038/nature12929. Epub 12014 Jan 12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruhbeis C, Frohlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Mobius W, Goebbels S, Nave KA, et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. doi:1001610.1001371/journal.pbio.1001604. Epub 1002013 Jul 1001609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KP, Akbergenova Y, Cho RW, Baas-Thomas MS, Littleton JT. Shank Modulates Postsynaptic Wnt Signaling to Regulate Synaptic Development. J Neurosci. 2016;36:5820–5832. doi: 10.1523/JNEUROSCI.4279-15.2016. doi:5810.1523/JNEUROSCI.4279-5815.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc Natl Acad Sci U S A. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh YH, Popova E, Thomas U, Griffith LC, Budnik V. Regulation of DLG localization at synapses by CaMKII-dependent phosphorylation. Cell. 1999;98:353–363. doi: 10.1016/s0092-8674(00)81964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koles K, Nunnari J, Korkut C, Barria R, Brewer C, Li Y, Leszyk J, Zhang B, Budnik V. Mechanism of evenness interrupted (Evi)-exosome release at synaptic boutons. J Biol Chem. 2012;287:16820–16834. doi: 10.1074/jbc.M112.342667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koon AC, Ashley J, Barria R, DasGupta S, Brain R, Waddell S, Alkema MJ, Budnik V. Autoregulatory and paracrine control of synaptic and behavioral plasticity by octopaminergic signaling. Nat Neurosci. 2011;14:190–199. doi: 10.1038/nn.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut C, Ataman B, Ramachandran P, Ashley J, Barria R, Gherbesi N, Budnik V. Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell. 2009;139:393–404. doi: 10.1016/j.cell.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut C, Li Y, Koles K, Brewer C, Ashley J, Yoshihara M, Budnik V. Regulation of postsynaptic retrograde signaling by presynaptic exosome release. Neuron. 2013;77:1039–1039. doi: 10.1016/j.neuron.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre FA, Benoit Bouvrette LP, Perras L, Blanchet-Cohen A, Garnier D, Rak J, Lecuyer E. Comparative transcriptomic analysis of human and Drosophila extracellular vesicles. Sci Rep. 2016;6:27680. doi: 10.1038/srep27680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Mattaliano MD, Montana ES, Parisky KM, Littleton JT, Griffith LC. The Drosophila ARC homolog regulates behavioral responses to starvation. Molecular and cellular neurosciences. 2007;36:211–221. doi: 10.1016/j.mcn.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG., Jr Exosomal communication goes viral. J Virol. 2015;89:5200–5203. doi: 10.1128/JVI.02470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrum C, Baumann A, Bustad HJ, Flydal MI, Mariaule V, Alvira S, Cuellar J, Haavik J, Soule J, Valpuesta JM, et al. Arc is a flexible modular protein capable of reversible self-oligomerization. Biochem J. 2015;468:145–158. doi: 10.1042/BJ20141446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S, Austin BP, Penrose KJ, Waugh DS. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 2005;14:2964–2971. doi: 10.1110/ps.051718605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefedova L, Kim A. Mechanisms of LTR-Retroelement Transposition: Lessons from Drosophila melanogaster. Viruses. 2017;9 doi: 10.3390/v9040081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Miyoshi K, Ogino A, Miyoshi T, Siomi H, Siomi MC. Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Mol Cell. 2013;49:680–691. doi: 10.1016/j.molcel.2012.12.024. [DOI] [PubMed] [Google Scholar]
- Peebles CL, Yoo J, Thwin MT, Palop JJ, Noebels JL, Finkbeiner S. Arc regulates spine morphology and maintains network stability in vivo. Proc Natl Acad Sci U S A. 2010;107:18173–18178. doi: 10.1073/pnas.1006546107. doi:18110.11073/pnas.1006546107. Epub 1006542010 Oct 1006546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Bullock SL. Creating Heritable Mutations in Drosophila with CRISPR-Cas9. Methods Mol Biol. 2016;1478:145–160. doi: 10.1007/978-1-4939-6371-3_7. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh AK, O’Tousa JE, Ozaki K, Ready DF. Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development. 2005;132:1487–1497. doi: 10.1242/dev.01704. [DOI] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. doi:210.1038/nn.2708. Epub 2011 Jan 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola H, Ballarini F, Martinez MC, Moncada D. The tagging and capture hypothesis from synapse to memory. Prog Mol Biol Transl Sci. 2014;122:391–423. doi: 10.1016/B1978-1010-1012-420170-420175.400013-420171. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu J, Ward MD, Yang S, Chuang YA, Xiao M, Li R, Leahy DJ, Worley PF. Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron. 2015;86:490–500. doi: 10.1016/j.neuron.2015.03.030. doi:410.1016/j.neuron.2015.1003.1030. Epub 2015 Apr 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.dArc1 Immunoprecipitation anddarc1RNA levels (see also Figure 2)
(A) Western blot showing dArc1 immunoprecipitation from body wall muscle extracts derived from wild type (CS) and darc1esm113 mutants labeled with anti-dArc1. Number at the left of the blot represents molecular weight in kilodaltons.
(B) darc1 qRT-PCR performed on RNA isolated from either dissected body wall muscles or brains in the indicated genotypes.
(C) darc1 RT-qPCR performed on RNA isolated from wild type larval body wall and darc1esm113.
N (biological replicates per genotype from left to right) is B:6,5,4,4; C 3 for each genotype; Graphs represent mean, and error bars SEM; statistical analysis was performed using a Student’s t-test; *= p < 0.05; **= p < 0.001; ***= p < 0.0001.
Figure S2.Full-lengthcopiaLtranscript is found in the cellular fraction (see also Figure 4)
Raw number of RNA-seq reads for cellular Copia showing reads across the entire predicted transcript.
Figure S3.Controls for EVs and capsid-like structures (see also Figure 5)
(A) Ultrastructure of negatively stained exosomal fraction without saponin treatment
(B) No primary antibody control for lysed EVs. Grids containing this fraction were processed for immunocytochemistry, but the dArc1 primary antibody was omitted. No 10nm gold granules are observed.
Calibration bar is 70 nm in A and 350 nm in B.
Figure S4.dArc1 transcript levels in Canton-S vs Oregon-R. (see also Fig. 7)
qRT-PCR results from body wall preparations from the indicated genotypes.
N (biological replicates; from left to right) = 10,9; Graphs represent mean, and error bars SEM; statistical analysis was performed using a Student t-test; *= p < 0.05; **= p < 0.001; ***= p < 0.0001.