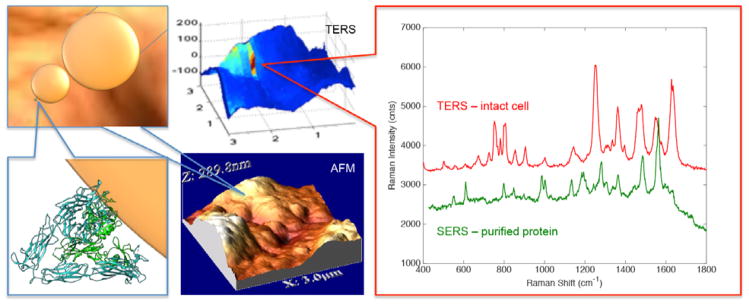
Abstract
Surface enhanced Raman scattering (SERS) has become a powerful technique for trace analysis of biomolecules. The use of SERS-tags has evolved into clinical diagnostics, the enhancement of the intrinsic signal of biomolecules on SERS active materials shows tremendous promise for the analysis of biomolecules and potential biomedical assays. The detection of the de novo signal from a wide range of biomolecules has been reported to date. In this review, we examine different classes of biomolecules for the signals observed and experimental details that enable their detection. In particular, we survey nucleic acids, amino acids, peptides, proteins, metabolites, and pathogens. The signals observed show that the interaction of the biomolecule with the enhancing nanostructure has a significant influence on the observed spectrum. Additional experiments demonstrate that internal standards can correct for intensity fluctuations and provide quantitative analysis. Experimental methods that control the interaction at the surface are providing for reproducible SERS signals. Results suggest that combining advances in methodology with the development of libraries for SERS spectra may enable the characterization of biomolecules complementary to other existing methods.
INTRODUCTION
The detection and identification of biomolecules is fundamental to biomedical diagnostics, a challenge which surface enhanced Raman scattering (SERS) is poised to make an impact.1–3 Recent advances in systems biology have shown that monitoring the levels of multiple molecular species can provide a holistic perspective on the health or disease of an organism. Understanding the interconnected biochemical pathways requires assays that assess multiple molecules in complex biofluids with high sensitivity and selectivity.4–6 The intrinsic molecular fingerprint, or de novo Raman signal of a molecule, combined with the improved sensitivity of SERS, provides an attractive approach for identifying and quantifying molecules for diagnostic purposes.
SERS provides molecular identification on the basis of the structure of the analyte, providing complementary analysis to other technologies currently used for biomolecular identification. The current technologies for molecular identification include fluorescence, nuclear magnetic resonance (NMR), infrared (IR), Raman, and mass spectrometry (MS); each with distinct advantages and disadvantages. Fluorescence can enable trace detection, down to the single molecule level; however, the number of natively fluorescent molecules is limited and requires extra sample preparation steps to attach chromophores. Additionally, the fluorescence spectra of biomolecules are quite indistinct and it is complicated to distinguish similar fluorophores within a sample. The sensitivity of fluorescence enables quick measurements of the kinetic activity of proteins, such as receptor-ligand binding and folding, but is of limited use for identification of analytes.7–8 In contrast, NMR provides exquisite structural information that can be deconvoluted into the absolute biomolecule structures.6, 9–10 However, NMR characterization is limited by the concentrations needed, requiring longer analysis times, and larger samples relative to other analytical techniques.6, 10 MS has become the analysis method of choice. Current high-resolution mass spectrometers can provide empirical formulas of molecules for identification, and tandem (MS-MS) experiments can sequence peptides and proteins. Additionally, MS has excellent sensitivity and the ability to couple to separation techniques such as liquid chromatography (LC), gas chromatography (GC) and capillary electrophoresis (CE) for separation and detection of biomolecules in complex mixtures.6, 10–13 Despite the many advantages of MS, identification of certain classes of molecules are still problematic. Poorly ionizing molecules, structural isomers (i.e. same empirical formula but different structures), and difficulties with small molecule identification in LC-MS experiments can challenge MS-based experiments. It was noted that less than 2% of the MS spectra recorded in an LC-MS metabolomics experiment can be associated with a specific molecule.14 Vibrational spectroscopy (IR and Raman) also provides structural information that can be used for molecular identification.15–16 IR, however, shows poor sensitivity when detecting low concentration analytes in aqueous environments due to the strong water interference. Spontaneous Raman spectroscopy, while free from water interference, suffers from low intrinsic signals. While Raman, in various forms, has seen increasing use in biomedical microscopy,17 small molecule identification typically requires some form of signal enhancement.
Since its discovery, SERS has advanced to become a promising bioanalytical technique, combining the structural information present in the Raman signal with ultrasensitive detection. Raman scattering arises from the inelastic scattering of monochromatic light (i.e. laser) as the frequency of photons change upon interaction with a molecule. This frequency shift correlates to the vibrational modes of the molecule, which gives rise to a molecular fingerprint. The intensity of Raman scattering is proportional to the analyte concentration in the detection volume, which allows both qualitative and quantitative measurements.3, 17–18 Interactions with nanomaterials provide sufficient Raman signal enhancements for bioanalytical studies, in which the analytes concentrations are within micro to nanomolar range. It has been shown that enhancements arise from near-field effects between molecules and nanostructured noble metal surfaces, commonly gold and silver.19–20 When an electromagnetic wave interacts with metal nanostructures, it excites the localize surface plasmon resonance (LSPR) on the surface.18 The SERS intensity is distance dependent, and the target analyte has to be in very close proximity to the metal surface in order to generate enhanced scattering.21 There are reports of the Raman scattering being enhanced as much as 1011.22–23 Additional enhancements can be achieved when the incident laser frequency and nanomaterial plasmon resonance match an electronic transition state of the absorbed molecule, similar to resonance Raman scattering (RRS), resulting in surface-enhanced resonance Raman scattering (SERRS).24–25 Under appropriate conditions, the Raman scattering from a single molecule can be detected.20, 26–27 SERS, like Raman, is relatively insensitive to water, making it a suitable detection method for samples in aqueous environments such as biological materials. The electromagnetic field enhancement associated with increased local fields around nanomaterials, suggests a generic method for increasing the Raman signal of a wide range of molecules.
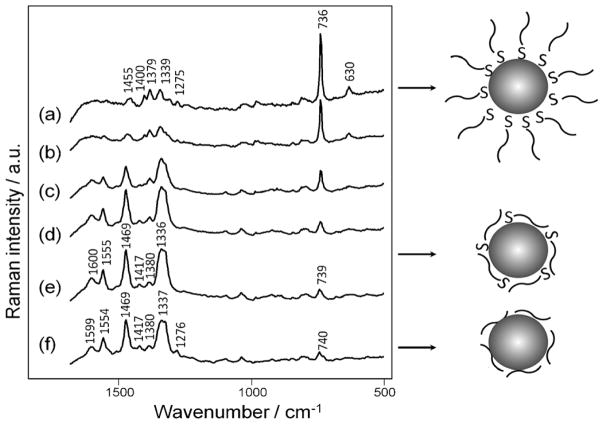
SERS detection can be divided into two categories for molecular detection. Figure 1 illustrates the common approaches to SERS detection. Perhaps the most common method is to prepare nanoparticles (NPs) functionalized with specific molecules that give a distinct Raman spectrum and can be used with affinity reagents to “tag” specific molecules. Thus, indirect detection is based on the signal of the SERS “labels” or “tags” attached to the target analytes instead of the analytes itself. The advantage of SERS tags versus fluorophores is the distinctive Raman spectrum associated with different reporter molecules would facilitate multiplex analysis.28
Figure 1.
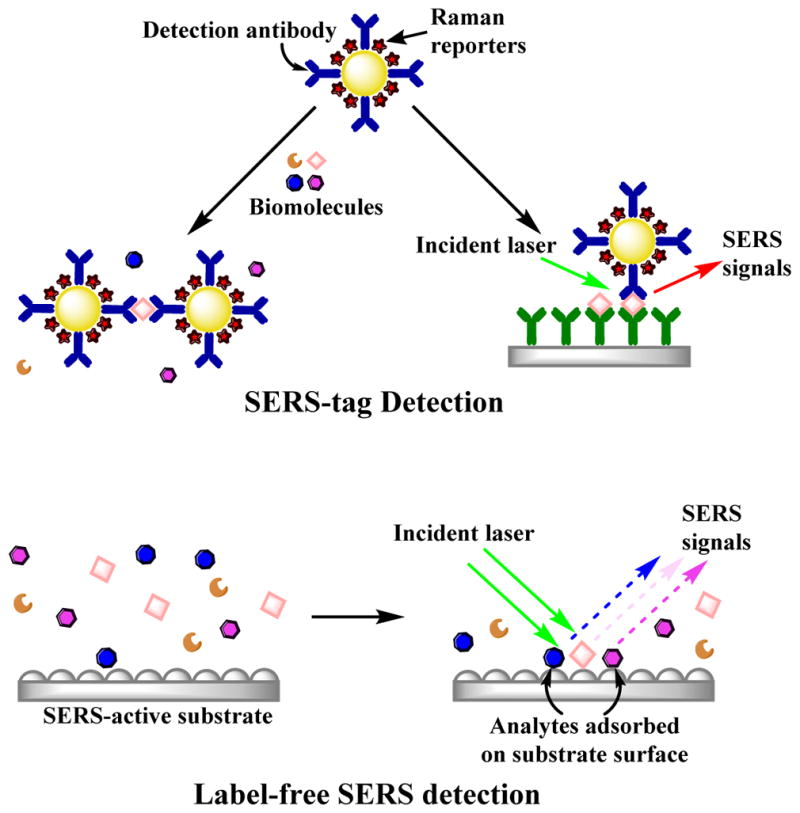
The diagram illustrates the differences between the label-free SERS detection and the SERS approaches using dye labels as reporter molecules. SERS-tags provide the SERS signal from a reporter molecule when the analyte interacts via some form of molecular recognition chemistry (e.g. antibodies or nucleic acid sequence). In contrast, label-free SERS directly detects the signal originating from the conformation and the orientation of the analytes adsorbed on the SERS-active substrate (direct detection). Label free SERS can also be accomplished using nanoparticles.
SERS tags have been used with flow cytometry29–30, SERS based immunoassays31–33, and analyte-induced SERS tags aggregation assays 34–35 to detect DNA, proteins, cells, and other biomarkers. The Vo-Dinh group was the first to utilize SERS-based molecular sentinel nanoprobes for multiplex detection of DNA.36 Two molecular sentinel probes tagged with Cy3 and TAMRA as Raman reporters were applied to detect two genes characteristic of breast cancers. The nanoprobe consisted of a Raman reporter attached to a hairpin DNA probe strand with a silver NP at the other end, which led to a decrease or disappearance of the SERS signal upon hybridization by disrupting the hairpin structure and distancing the reporter from the silver NP. Instead of obtaining ‘turn off’ SERS signal upon hybridization, positive signal enhancement was demonstrated by Graham et al. by using resonant dye-coated silver NPs functionalized with thiolated DNA probe sequences.37 Hybridization with complementary DNA strands facilitates NP aggregation, which substantially increases the SERS signal. Recent advances have further increased the sensitivity of SERS-based ELISAs to a level comparable with fluorescence-based immunoassay techniques for multiplex protein biomarkers detection over a large dynamic range.38–40
The basis for SERS-tags is the adsorption of specific molecules that give rise to distinctive Raman signals. In principle, any molecule at the NP surface should give rise to increased scattering. This suggests a powerful method of molecular identification. Direct label-free SERS detection originates from the conformation and the orientation of molecules on the metal surface. While this can provide heterogeneous signals, it also provides label-free detection that does not require additional sample preparation or affinity reagents. Indeed, direct SERS detection has been utilized for a number of different biomolecules.
This review focuses on the aspects of direct, label-free SERS analysis and identification of various biomolecules ranging from nucleic acids, proteins, carbohydrates, and nucleotides to small molecule metabolites. While direct detection may seem straightforward, complications have been attributed to low Raman cross-sections of certain analytes as well as interferences in complex biofluids. Quantitative SERS has been notoriously difficult, with challenges attributed to subtle differences in nanostructure that affect the observed Raman enhancements.41 All of these listed factors remain challenges for SERS detection and identification of biomolecules. Here we focus on which molecules have been detected, looking for trends in the classes of molecules investigated and signals observed and surveying the advances that are facilitating de novo detection of biomolecules by SERS.
NUCLEIC ACIDS AND NUCLEOTIDES
Nucleic acid detection, the components of DNA and RNA, has attracted much attention over the years for identifying pathogens, infectious agents, and cancerous diseases. Common conventional detection methods for nucleic acids, include polymerase chain reaction (PCR) and fluorescence; however, SERS detection offers simple procedures and intrinsically narrow bands that facilitate multiplexed analysis.
Molecular Orientation Affects the SERS Spectra DNA
Early SERS experiments with DNA focused on understanding how the orientation of each DNA base on the metal surface affected the SERS spectra. Otto et al. used a Raman microprobe system to detect DNA bases of adenine, guanine, thymine, and cytosine that adsorbed on a roughen silver working electrode.42 SERS spectra showed significant frequency shifts and changes in the relative intensities and widths of peaks when compared to the spontaneous Raman spectra observed from each base in solution, indicating the molecular orientation at the metal surface is an important factor. For example, they reported differences in behavior of the carbonyl groups attached to the pyrimidine ring in DNA bases at approximately 1640 cm−1. The SERS spectra of cytosine and thymine show strong carbonyl stretching modes; however, in the case of thymine, two carbonyl bands were expected but only one was observed.42 Moreover, guanine’s carbonyl peak intensity was very weak and sometimes could not be seen, which indicates not all DNA bases orient in the same direction to the metal surface. Improvements in SERS-active nanostructures preparation remarkably improved the detection range of nucleic acids. NIR excitation of SERS from colloidal clusters provided enhancement that was capable of detecting a single DNA base.43 Sub-micromolar label-free SERS detection of mononucleotides was done by Bell and Sirimuthu using silver colloids aggregated with MgSO4.44 They reported the SERS spectra of the five major nucleotides (adenine, guanine, thymine, cytosine, and uracil) along with their nucleosides, which had similar spectra that suggest they bind to the surface in the same way, but with enough distinct Raman bands to be easily distinguished. Moreover, the SERS spectra of adenine, adenosine, and dAMP showed differences associated with the phosphate and sugar groups that indicate slight changes in the orientation at metal surface can lead to significant changes in SERS spectra and signal intensity.44 In a later study, Lo et al. were able to achieve label free detection of sub-picomolar DNA bases and ds-DNA using AgNPs decorated Au-nanotip SERS arrays.45
Thiol-Modified Oligonucleotides
Thiol modification of the DNA can bond the DNA to the SERS substrate, bring the DNA in close proximity to the strong enhancing field, and facilitate uniform orientation of the DNA at the metal surface. Barhoumi and Halas have reported on DNA hybridization and the adsorption of thiolated single stranded (ssDNA) and double stranded DNA (dsDNA) to Au nanoshells.46–48 They observed that the Au nanoshells provide highly reproducible SERS signals of DNA oligomers following thermal treatment of the DNA to achieve extended linear conformations.48 Despite the different contributions of each base in ssDNA and dsDNA, the SERS spectra was dominated by the adenine vibrational modes at 736 cm−1, which obscures other features.46, 48 The dominating adenine signal, however, can be used as a marker for DNA hybridization. In a further study, the adenine on the DNA probe sequence was substituted with 2-aminopurine, which is an artificial adenine isomer.46 The adenine signal at 736 cm−1, which was not observed with adenine free DNA probe, was strongly enhanced upon hybridization with the adenine containing target sequence. The peak ratio between adenine and 2-aminopurine was used to determine DNA hybridization efficiency. Papadopoulou and Bell confirmed that DNA adsorbed through thiol linkers on AuNPs aggregated with MgSO4 orient perpendicular to the metal surface and the SERS spectra are dominated by the adenine ring mode at 736 cm−1, as shown in Figure 2.49 However, unthiolated DNA can reduce the overwhelming adenine signal by promoting non-specific binding of the DNA nucleobases, which reorients the DNA on metal surface with weaker adenine bands in the SERS spectra.49 The spectra changes were applied to detect DNA hybridization of a molecular beacon.
Figure 2.
SERS spectra of thiolated poly A adsorbed on on AuNPs aggregated with MgSO4 at concentrations (a)–(e) 10−4 – 10−8 M and unthiolated poly A at 10−5 M (f). The diagram represents that unthiolated DNA can reduce the overwhelming adenine signal at 736 cm−1 through the reorientation of the DNA on metal surface, with weaker adenine bands shown in the SERS spectra. Reproduced from Ref. 49 with permission of The Royal Society of Chemistry.
Quantitative Analysis of Oligonucleotides
Overall, the surface chemistry of different NP colloids affects the SERS signals of adsorbed DNA.50 Also, aggregating agents serve a critical role in the affinity between the DNA and the metal surface resulting in varied SERS spectra of ssDNA and dsDNA.50 Quantitative SERS analysis of nucleotides is often challenging since the strands are made up by similar bases that can all contribute equally to the SERS spectra. Moreover, the reproducibility and sensitivity of SERS studies on DNA rely heavily on the experimental conditions and the orientation of DNA strands on the metal surface.
Smaller nucleotide sequences of microRNA (miRNA) can be analyzed using appropriate statistical analysis.51–52 Abell et al. demonstrated label free detection of miRNAs absorbed on Ag nanorods coated onto a multi-well chip.51 Characteristic bands of the four nucleotides were deconvoluted from the SERS spectra, which allowed quantitative analysis of the ratios of bases comprising the mixture. Prado et al. previously demonstrated that least squares analysis could quantify oligoribonucleotides polyadenosine (pA), polyuridine (pU), polycytosine (pC) and polyguanosine (pG) using Ag colloids with the limit of detection (LOD) varying from 2 pM to 20 pM.53 In general, the surface area of the mixed Raman spectrum is broken down into the sum of the surface areas of each RNA bases’ spectrum, which is then fit to the linear spectrum using least square analysis to calculate the percentage of each nucleotide in the mixture.51, 53 There are significant differences between the miRNAs spectra after treatment of noncomplementary and complementary targets which denote hybridization. Least square analysis was also performed to estimate the composition of A, C, G, and U after hybridization and better determine the hybridization efficiency.51
Internal standards are employed to correct for any variations associated with SERS substrate and instrumental conditions. Zhang et al. utilized magnetic microspheres AgNPs as an SERS substrate coated with probe DNA and treated with mercaptobenzoic acid (MBA) solution.54 The SERS intensity of DNA increased with the increase in MBA signals. Therefore, by detecting MBA and plotting the intensity of MBA as function of DNA concentrations, a calibration curve was generated to quantify the DNA concentration in a mixture with a LOD of 10 nM. This work, however, was not truly a direct detection of DNA since the SERS spectra originated from MBA instead of DNA strands. Other studies have treated the backbone phosphate stretch at 1087 cm−1 as an intrinsic internal standard for a more direct, quantitative analysis of the fraction of each base in DNA.55–56
Direct Detection of Oligonucleotides and Modifications
ssDNA and dsDNA can spontaneously adsorb directly to Ag silver colloids through the nucleotide side chains without the need for labeling or thiolation. The observed SERS spectra of unthiolated DNA arose from each constituent base, which identified nucleotide components in the DNA sequence.57–58 Digital subtraction produced difference spectrum that contained positive and negative features attributed to changes in the DNA sequence, and even detecting single base mismatches in short DNA strands.57 Recently, Guerrini et al. introduced the used of spermine bound to AgNPs for label free and direct analysis of DNA duplexes.59 Spermine acts as a stabilizer to promote controlled aggregation between positively charge AgNPs and negatively charged DNAs into small clusters without the need of external aggregation agents. Single base mismatches and base methylations (5-methylated cysteine and N6-methylated adenine) in DNA duplexes were detected for the first time by digital subtraction of the SERS spectrum of original dsDNA sequence from other samples as demonstrated in Figure 3.59 Spermine stabilized AgNPs were further combined with chemometrics and microfluidics to reduce the amount of sample used and accurately quantify chemical modifications in DNA sequences.60
Figure 3.
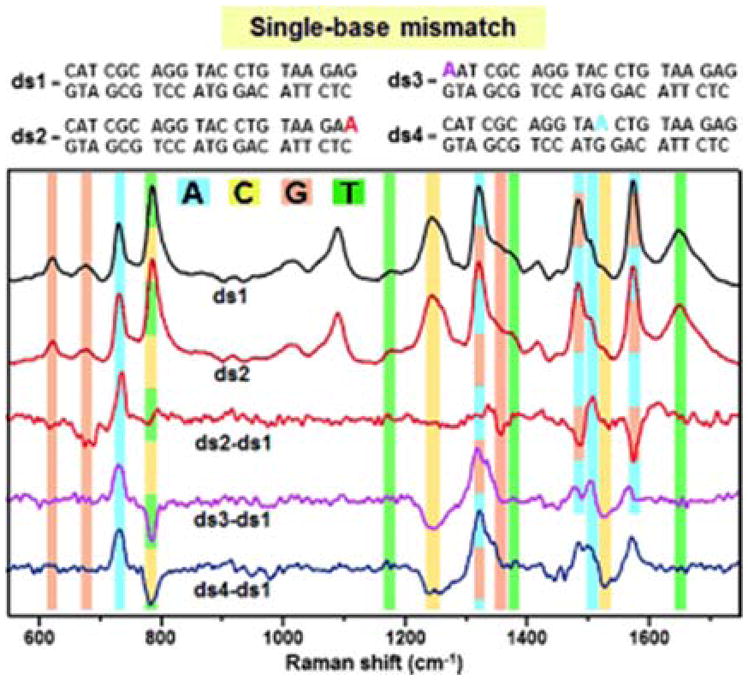
The detection single-base mismatches in DNA was demonstrated by digital subtraction between the SERS spectrum of the original dsDNA and from mismatched duplexes containing one adenine base (A) replacing one guanine (G) (ds2), one terminal cytosine (C) (ds3), and one internal C (ds4). Vibrational modes associated with nucleotide bands are highlighted in: blue (A), yellow (C), orange (G), and green (T). Reproduced from Ref. 59 with permission from John Wiley & Sons, Inc.
Beyond identification of the base composition of DNA and RNA strands, the conformation changes of nucleic acids sequences can provide important information for diagnosis.28, 58, 61 The Bell group demonstrated the ability to successfully separate and identify five DNA strains of Escherichia coli (E. coli) without the need for labeling or multivariate analysis.58 Changes in SERS spectra of each E. coli strain were dependent on the nucleobase composition of DNA sequences. For example, the ring breathing and carbonyl stretching modes of cytidine and thymidine at 793 cm−1 and 1636 cm−1 are more intense in the SERS spectrum of specific strains. Similarly, the characteristic bands of adenosine at 737 cm−1 and 1329 cm−1 are more intense in the spectrum of other strains, which contained more adenosine nucleotides. The SERS spectra show changes with respect to the order of the bases, indicating SERS is indeed a valuable technique to extract DNA information from biological samples.58 Chen et al. developed methodology to detect and differentiate RNAs in the serum of colorectal cancer patients.62 RNA was extracted from serum at concentrations ranging from 18.8 to 56.3 ng/μL. The differences between SERS spectra of normal and cancer groups can be tentatively assigned to different vibrational modes that are associated with the molecular changes and transformation of RNAs in cancerous serum. Further, a label-free multifunctional probe was fabricated and coupled to SERS through dual circular-strand displacement for targeted miRNAs detection with significant signal amplification.63 miRNA 203 was selected as a model analyte. They found that the SERS intensity increased with the increases in concentration of miRNA enabling quantitative analysis of miRNA in a mixture. The LOD was determined to be 6.3 fM. This method was also tested on miRNA 203 extracted from human epithelial cancer cell lines and shows good agreement with qRT-PCR results.63
AMINO ACIDS, PEPTIDES, AND PROTEINS
SERS has been used to identify proteins based on the contributions of the amino acids in the protein sequence and resonance enhancement of intrinsic chromophores. Because the vibrational modes that give rise to the SERS spectrum are sensitive to structure, conformational changes can be monitored as well.
Detection of Individual Amino Acids
The basic amino acids have been detected by SERS. O’Neal et al. demonstrated a quantitative analysis of glutamic acid (Glu) and aspartic acid (Asp) at concentrations ranging between 0.4 to 5 μM.64 Differences in the observed Raman bands indicate chemical fluctuations in traumatized rat brain tissues.64 Nascimento et al. studied the effect of salts (NaCl, MgCl2, KBr, CaSO4, K2SO4, MgSO4, KI, NH4Cl, SrCl2, CaCl2, Na2SO4) on sodium and borohydride citrate colloids to detect amino acids in prebiotic Earth conditions.65 The amino acids investigated were alanine (Ala), glycine (Gly), cysteine (Cys), and 2-amianoisobutyric acid, and it was reported that the amino acids could not be characterized by the SERS spectra in the presence of higher concentration salts.65
Similar to nucleotides, the interactions between the amino acids and the surface has a significant effect on the observed vibrational bands. Other amino acids have been investigated such as D-penicillamine, an amino acid related to Cys and valine (Val). D-penicillamine was detected at various pH levels.66 López-Ramírez et al. reported that the major vibrational bands were associated with the thiol, amine, and the carboxylic acids groups of the amino acid. However, from the enhanced intensity and red shift of the C-S stretch it was concluded that a chemical bond forms between the NP and the sulfur group.66 With the knowledge that thiol groups act as anchors between desired analytes and NPs, Du et al. investigated the use of Ag@oxide nanoplates to detect biomolecules with and without thiol groups.67 The Ag nanoplates were coated with ultrathin layers of SiO2 or TiO2. 4-aminothiophenol (4-ATP) was used to prove that the modified NPs can detect model analytes containing thiols. 4-ATP showed characteristic peaks of the benzene and amine group. The amino acid, Gly, was used to model a carboxyl containing analyte. They showed that the carboxylic group can strongly absorb to the oxide layer of the NP, making it possible to detect a wider range of biomolecules that do not contain thiol moieties.67
Individual amino acids can be detected by separating the components with techniques such as capillary electrophoresis (CE). He et al. used a post-column CE-SERS to detect amino acids.68 The SERS-active substrate was located on the moving collection plate. The CE elution trail was then retraced with a Raman microscope to collect the desired SERS spectra. They showed the biological relevance of their set-up detecting two amino acids: tyrosine (Tyr) and tryptophan (Trp).68 Negri and Schultz demonstrated capillary zone electrophoresis (CZE) with sheath flow SERS detection was able to separate and identify all 20 amino acids through characteristic vibrational bands.69 From this study, it was reported that the amino, carboxylate, or side chain groups can interact with the metal NP and therefore contribute to the SERS spectra.69
Peptides
In addition to individual amino-acids, peptides have also been investigated. Wei et al. used Au nanoshells to obtains highly reproducible SERS spectra of cysteine-containing aromatic di-peptides: tryptophan-cysteine, tyrosine-cysteine, and phenylalanine-cysteine, and penetratin (peptide oligomer containing 19 amino acids).70 This study suggested that the SERS spectra were dominated by the aromatic residues and the peptide backbone, which may help predict major features in more complex peptide and protein spectra, such as penetratin. The predicted and experimental spectra of penetratin showed high agreement except in the backbone region.70
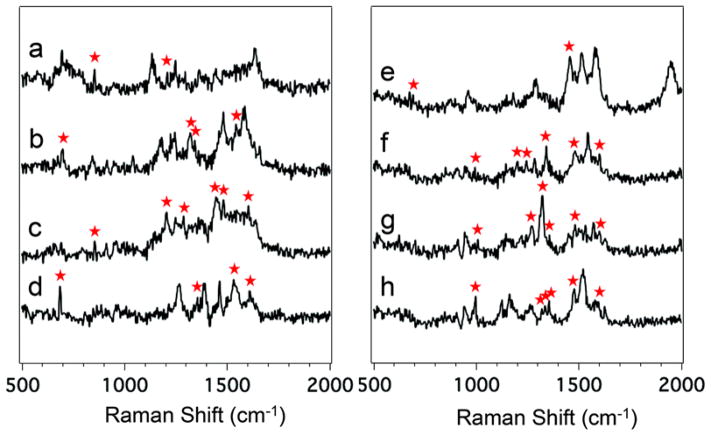
Using the CZE-sheath flow SERS method mentioned above, Negri et al. also detected and identified eight biologically-active peptides.71 The peptides were chosen to have aromatic and/or sulfur-containing amino acid residues: amyloid β-protein, Angiotensin I, Angiotensin II, Angiotensin III, bombesin, laminin pentapeptide, somatostatin, and substance P.71 Figure 4 shows the average SERS spectra of the eight different peptides. The aromatic, sulfur-containing, or side chain vibrations are denoted to show diversity between the peptides.
Figure 4.
Average SERS spectra of the eight detected peptides: Laminin Pentapeptide (a), Bombesin (b), Angiotensin III (c), Somatostatin (d), Amyloid beta-protein (e), Angiotensin I (f), Angiotensin II (g), and Substance P (h). The differentiating vibrational bands are labeled with a red asterisk. Reproduced from Ref. 71 with permission from The Royal Society of Chemistry.
Amide I Mode
The amide I mode is commonly used to assign backbone structure. SERS characterization of the amide backbone has generated conflicting results. As noted above, Wei et al’s study of penetratin showed discrepancies in the backbone region.70 A tip-enhanced Raman scattering (TERS) study of model peptides exhibited only vibrational bands associated with aromatic amino acids with a distinct lack of signals from the amide backbone.72
The sporadic observation of the amide I mode in SERS and TERS spectra was investigated by Kurouski et al. with insulin fibrils and model peptides.73 They explored native insulin along with the homo-peptides containing Gly-, Ala-, Tyr-, and Trp- rich side chains to verify the side change size dependence. For insulin, the observed vibrational bands were associated the amino acid residues: Tyr, Cys, and Phe. Only 50% of the insulin spectra showed peaks assignable to an amide I band whereas all the penta-Gly homo-peptide spectra showed intense amide I bands. The short peptides also showed intense CH2 and C-C stretching bands. Since the side chains in the insulin fibrils are so bulky the peptide bond is further away from the metal NP decreasing the SERS signal of the amide I vibration. Kurouski et al. hypothesize that the absence of the amide I band is dependent on the size of the amino acid side chains.73
Nonetheless SERS has been used to investigate the structure of other proteins. Dou et al. used NIR SERS (1064 nm) to detect protein antigens without the need for bound/free antigen separation.74 In this study, they were able to detect anti-mouse IgG and IgG-anti-IgG complex at 0.22 nM and 19 nM, respectively. The β-sheet rich structure was identified in the SERS spectrum by the amide I and III vibrational bands at 1673 cm−1 and 1239 cm−1, respectively. However, these bands were weak compared to the multiple vibrational bands from the Tyr and Trp residues.74
De Novo SERRS of Proteins
Single protein detection was demonstrated independently by Xu et al. and Bizzarri and Cannistraro using SERRS. Xu et al. used 514.5 nm laser excitation to detect 100 nM hemoglobin (Hb).75 While, Bizzarri and Cannistraro detected myoglobin (Mb) at 100 nM at the excitation wavelength of 633 nm.76 The single molecule SERRS experiments showed temporal fluctuations common with single molecule experiments. The detection of native met-Mb at concentrations as low as 1×10−7 M was also achieved with the use of a T-shaped flow system.77 The vibrational bands associated with the in-plane porphyrin ring of the heme group dominate the SERRS spectra of Mb because of the resonance enhancement.76–77 Other proteins, such as green fluorescent protein (GFP), have been detected at single molecule concentrations by SERRS, in the case of GFP using an excitation wavelength of 488 nm.78 The prominent vibrational modes observed were from the delocalized imidazolinone/exocyclic C=C modes of the chromophore,78 consistent with previously performed SERRS experiments. Delfino et al. detected single molecule yeast cytochrome c at a concentration of 1.7×10−12 M for both suspended and immobilized samples at the excitation wavelength of 514.5 nm.79 The main vibrations are associated with the pyrrole breathing mode CalphaCm mode.79
SERRS has also been used to detect proteins in various studies other than single molecule detection. Zheng et al. used SERRS to detect mutants of iso-1-cytochrome c at the excitation wavelength of 413 nm.80 In this investigation two mutants were studied: the wild type protein, where the cysteine-102 was replaced with serine (Ser) to avoid dimerization, and where the phenylalanine-82 (Phe) was replaced with histidine (His).80 Han et al. used the layer-by-layer technique (Au/Ag) with SERRS to quantify Atto610-biotin with 568 nm excitation.81 The detection limit was 10 pg/mL. They reported that the amino acids with high hydrophobicity bind to the metal surface causing those residues to dominate the spectra.81
Proteins with chromophores have also been detected without the use of a resonant excitation source. By exciting at multiple wavelengths (488, 514, and 633 nm), Etchegoin et al. monitored the oxygenation of the Hb with the Fe spin vibration at 1640 cm−1. It was reported that the size difference between the oxygenated and deoxygenated Hb comes from the absorbed or released oxygen.82
Nanoparticle-Protein Interactions
Proteins without chromophores have also been detected, identified, and quantified in recent years. With the use of an adaptive substrate, recombinant human insulin and insulin lispro were detected at a surface density of 80 fmol/mm2.83 The adaptive substrate allowed for the restructuring of the NPs in the presence of the protein; it is suggested that the proteins acted like “glue” for the NPs therefore, decreasing the distance between NPs and increasing the SERS signal. The major vibrational bands were associated Phe, Tyr, the amide I and amide III bands of the peptide backbone. The spectral difference between the two insulins came from the Phe B1 displacement and the alpha-helical N-terminus of the B-chain in human insulin.83
Wang et al. utilized a graphene-Au nano-pyramid substrate to detect lysozyme at nanomolar concentrations, achieving single protein detection in the absence of a chromophore.84 They reported that the lysozyme vibrational band associated with C-N mode showed a greater enhancement compared to the CH2 bending mode,84 which suggests how the protein associated with the nanopyramid substrate.
Chowdhury et al. were able to quantify two purified human integrins: α5β1 and αVβ3.85 From the observed peaks, it was suggested that the α5β1 integrin adsorbs to the NP at the alpha-helix region. Peaks from aromatic amino acid residues were also observed: Tyr, Phe, His, and Trp. The peptide backbone was observed through the amide I and III vibrational modes. They reported the LOD to be 30 nM and 60 nM for α5β1 and αVβ3, respectively.85
Yang et al. used a tip-coated multimode fiber (TCMMF) probe to create a NP-protein-NP sandwich to quantify lysozyme and cytochrome c with a detection limit of 0.2 μg/mL.86 The major peaks for lysozyme were associated with Trp (765 and 1359 cm−1), CH2 scissoring (1442 cm−1), Phe, His, and Trp (1508 cm−1), and the amide I band (1646 cm−1). The main SERS peaks for cytochrome c, a hemeprotein, come from the NH3 deformation (1121 cm−1), CH2 (1313 cm−1), Trp (1363 and 1554 cm−1), carboxylic acid (1392 cm−1), and amide I (1622 cm−1). The proteins have very similar spectra however the lysozyme obtains additional vibrational bands from Tyr, Trp, and Phe.86
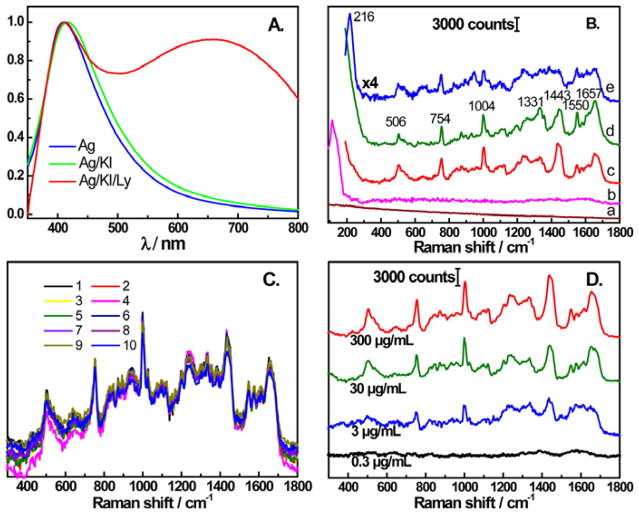
Modulating the protein nanoparticle interaction can significantly alter the observed SERS spectrum proteins. In Figure 5, Xu et al. used iodide modified citrate reduced AgNPs to obtain very reproducible SERS signals, that resembled spontaneous Raman signals, from lysozyme, avidin, BSA, cytochrome c, Hb, Trp, and Phe.87 It is believed that the iodide enables a reproducible signal by maintaining the native states of the proteins. The LOD for lysozyme was 3 μg/mL. Avidin and BSA showed characteristics of disulfide bonds, Phe, Trp, CH2, and amide. Cytochrome c and Hb were dominated by the vibrations associated with the heme. Lysozyme showed peaks relating to disulfide bonds, Trp, Phe, CH2, and the peptide backbone (amide I and III).87
Figure 5.
(A) Comparison of UV-vis spectra of Ag NPs, Ag iodide-modified NPs, and Ag iodide-modified NPs with lysozyme. (B) SERS spectra of Ag NPs (a), Ag iodide-modified NPs (b), Ag iodide-modified NPs with 300 μg/mL of lysozyme (c), normal Raman spectra of 100 mg/mL of lysozyme (d), and SERS spectra of Ag NPs with 300 μg/mL of lysozyme (e). (C) Ten consecutive SERS spectra of lysozyme with the iodide-modified NPs. (D) SERS spectra of lysozyme with the iodide-modified NPs at concentrations ranging between 0.3 μg/mL to 300 μg/mL. Reprinted with permission from Ref. 87. Copyright 2014 American Chemical Society.
Ivleva et al. employed label-free in situ SERS to image biofilms. Reference samples of BSA, l-phenylalanine, cellulose, dextran, xanthan, gellan, and alginic acid were collected.88 The glucosidic ring mode vibrations were very strong and the prominent carboxylic bands suggest that this is where the proteins adsorb to the surface of the NP.88 Binary mixtures of cytochrome c, lysozyme, Hb, HSA, apotransferrin, IgG, fibrinogen, and IgA were discriminated using principal component analysis on the observed SERS spectra.89 The binary mixtures were based on the size of the proteins: Lys-apoTrf, Cyt c-Hb, HSA-Fb, and IgG-IgA. They reported that the affinity between the protein and NP was significantly dependent on the charge of the protein. The apoTrf showed stronger affinity (than lysozyme) to the NP through the functional groups: COO, SH, and OH. A larger protein may not be able to adhere to the NP through weak ionic interactions unless a protein has covalent interactions to the NP.89
Limits of Detection and Quantification of Proteins
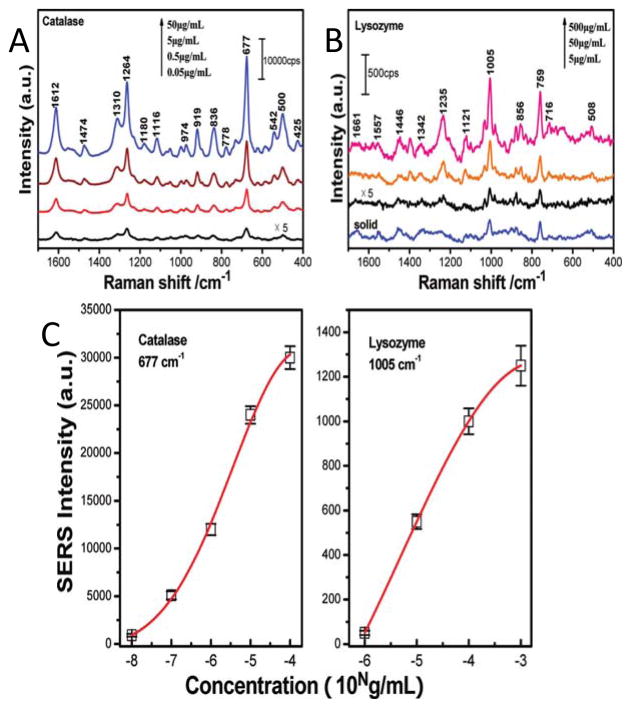
Han et al. investigated the quantification of lysozyme and catalase in aqueous solutions as shown in Figure 6.90 The LOD was calculated to be 5 μg/mL and 50 ng/mL for lysozyme and catalase, respectively. Lysozyme showed vibrational bands from the amide and aromatic amino acid residues (Phe, Tyr, and Trp). They reported the detection of ribonuclease B, avidin, Hb, and cytochrome c in aqueous solutions was also achievable.90 The hemeproteins were dominated by the heme group vibrations as previously reported.
Figure 6.
The SERS spectra of aqueous catalase (A) and lysozyme (B) are shown at concentrations ranging between 0.05 μg/mL to 500 μg/mL. (C) The SERS intensity of characteristic bands shows a concentration dependence for catalase (677 cm−1) and lysozyme (1005 cm−1). The red line is the Gaussian fit. Adapted with permission from Ref. 90. Copyright 2009 American Chemical Society.
Protein A and IgG were quantified with the use of Ag NPs arranged on a silicon substrate.91 The LOD for protein A and IgG were determined to be 5 nM and 500 fM, respectively, with this substrate. Protein A has high concentrations of lysine (Lys), glutamine (Gln), and asparagine (Asn), which are observed in the SERS spectra at 1150 and 1507 cm−1 (Lys) and 1272 cm−1 (Gln and Asn). IgG showed vibrational bands associated with Trp and Tyr. For IgG the 1649 cm−1 peak was used follow the adsorption process.91 Silver nanostructured surfaces on stainless steel were used to quantify albumin and globulin.92 The LOD was reported at 400 μg/mL.92 Hughes et al. used antibody-functionalized gold NP to detect mouse monoclonal anti-Erythropoietin (EPO) and caffeine.93 The lower limits of quantification for EPO and caffeine were 3.5×10−13 M and 1×10−10 M, respectively. The EPO concentration was comparable to the human urine concentration. Caffeine showed a band associated with the imidazole trigonal ring stretch that was used for quantification. The EPO was quantified using the CH2 vibrational band.93 Buividas et al. developed a gold coated grating-like ripple structure for fast and reliable detection of amyloid-β 40 oligomers (Aβ-40).94 The roughened surface allowed for the detection of Aβ-40 with concentrations ranging between10 nM and 10 μM. Most vibrational bands were associated with the L-Phe but vibrational bands related with L-Val and L-Tyr amino acids were also observed.94
The use of convective assembly between proteins and NPs allowed for the quantification of BSA, catalase, pepsin, cytochrome c, avidin, and lysozyme.95 The LOD for all the proteins was 0.5 μg/mL. The vibrational bands were mainly associated with amino acids residues: Cys, Trp, Phe, and Tyr. However, the peptide backbone was observed from the amide I and III vibrational modes.95 Convective assembly was also used detect proteins in binary and ternary mixtures such as: IgA with Hb and avidin with cytochrome c, and insulin with human serum albumin (HSA) and IgA.96 The use of Euclidian distance plots allowed for the distinguish between proteins based on their associated bands: IgA (928 and 950 cm−1), Hb (971 and 1326 cm−1), and cytochrome c (1254 and 1366 cm−1).96 Another study investigated the fabrication of a highly sensitive chip-based substrate for the quantification of BSA, mouse IgG, and cardiac troponin T (CTnT) where the detection limit of 3 ng/mL was achieved.97 The signal for all three proteins were dominated by the amino acid residues: Trp, Tyr, Cys, His, and Phe.97 The suspended droplet technique was used to quantify HSA, transferrin, Mb, cytochrome c, avidin, and lysozyme with a detection limit of 0.5 μg/mL.98 From the obtained spectra it is observed that the phenylalanine residue and the C-O, C-N stretch dominate. Other amino acid residues and vibrational bands associated with the peptide backbone were also noted.98 Kahraman et al. also observed a LOD of 0.5 μg/mL for BSA, Hb, thrombin, avidin, cytochrome c, and lysozyme by using hydrophobicity-driven self-assembly.99 For quantification, a reference peak was chosen for each protein: lysozyme (760 cm−1), cytochrome c (1005 and 1262 cm−1), and avidin (945 cm−1).99
The fabrication of a 3D plasmonic silver nanovoid structure was reported to obtain label-free protein detection by SERS at the excitation wavelength of 785 nm.100 The protein solutions were drop coated and naturally dried on the nanovoid structure. BSA, Hb, thrombin, avidin, cytochrome c, and lysozyme were detected with a LOD of approximately 0.05 μg/mL. The fabricated nanovoid structure provided more reproducible spectra than colloidal NPs. The band assignments for all the proteins are mainly associated with the amino acid residues: Cys, Phe, Tyr, and Trp. The peptide backbone was also observed though the amide I and III bands.100
Nano- and Microfluidic Approaches to In Situ Protein Characterization
Nano- and microfluidic devices have been investigated to characterize proteins in solution environments. Studies have shown that the interaction between the NP and protein denatures or changes the protein conformation. To address this challenge nanofluidic devices have been implemented obtain high signal without interfering with the naturally structure of the protein. β-amyloid 40, a protein critically involved in Alzheimer’s disease, was detected with the use of a nanofluidic device.101 The device allowed the mixture of NPs and proteins to be transported through a microfluidic channel to the entrance of a nanochannel where the analyte and NP were concentrated and confined, leading to an enhanced SERS signal. Other proteins such as bovine serum albumin (BSA) and bovine pancreas insulin were also detected with this nanofluidic device. The spectra were mainly dominated by the amino acids containing aromatic side chains (Phe, Tyr, and His). However, it was reported that protein-NP interaction occurs at the His residue.101 Zhou et al. used a low-cost T-shape poly(dimethylsiloxane) microfluidic chip to mix NPs with proteins to quantify the amount of BSA.102 They reported a LOD of 1 pM. The BSA vibrational bands were associated with Tyr, Trp, Phe, and amide I. A mixture of BSA and cytochrome c was also investigated; showing different proteins could be distinguished based on the vibrational bands observed.102
Protein Kinetics and Conformational Changes
Because SERS can be obtained in solution, it is possible to monitor enzymatic reactions and changes in protein conformation. Double-Cys mutants of FynSH3 (K105C/S115C and A95C/V141C) were expressed by pET vector in E. coli and detected at concentrations as low as 5×10−9 M.103 The Cys residues promoted ‘self-assembly’ with the NPs creating a ‘hot spot’. Strong C-S vibrational mode was observed.103 Das et al. employed off-resonant SERS (λex=830 nm) for lysozyme, ribonuclease-B, BSA, and Mb at various temperatures.104 BSA’s spectrum is dominated by the amide I and III, Phe, Trp, Tyr, and Cys vibrational bands at room temperature. The vibrational bands remained constant over the various temperatures besides at the 2 extreme temperatures. The lysozyme spectrum was dominated by the amide I band however bands associated with the amide II and III, Cys (C-S), Trp (indole ring), and Phe were also observed.104
Keskin et al. used a hydrophobic glass surface to execute the suspended droplet technique of ternary mixtures under different temperature conditions.105 The conformational changes of the proteins were investigated for the temperature gradient of 30 to 70 °C. The two different mixtures were HSA-transferrin-Hb and HSA-fibrinogen-Hb. They concluded that the proteins do show different behaviors based on the interactions in the mixtures and with the NPs. At the different temperatures, the human serum transferrin denatured, as evidenced by increased Phe peaks at 676 cm−1 and 1001 cm−1 that suggest the Phe residues and the NPs interact more. Fibrinogen (critical for hemostasis) was also studied at various temperatures. Peaks associated with S-S stretching, Trp, proline (Pro), and Ala were observed. However, at high temperatures, 60 and 70°C, an intense Tyr (640 cm−1) and C-C stretching (671 cm−1) vibrational modes were observed, suggesting a change in structure at 50°C.105
Li et al. monitored protein tyrosine nitration (PTN) for BSA and three different peptide sequences.106 The peptide sequences were IEDNE(Y)TARQGGC, FSA(Y)LER, and I(Y)GEFKKKAAC. They also studied the hemin-catalyzed PTN process for BSA and human blood serum samples. This process allows for the in-situ monitoring of in-vitro hemin-catalyzed protein tyrosine nitration. Nitration was monitored by the increase in the 330–400 cm−1 vibrational bands.106
Biorecognition
Biorecognition by proteins has been studied by SERS. Li et al. investigated carbohydrate-protein binding, relevant to human influenza hemagglutinins, by incubating gold NPs with various solutions of galectin 1, galectin 3, and BSA.107 Wang and Xiao further investigated protein-ligand binding interactions by comparing the SERS signal of ligand functionalized NPs mixed with soluble proteins against TERS experiments of immobilized proteins on surfaces binding the ligand functionalized NPs.108–111 On the basis of the amino acids detected, it is believed that the SERS spectrum can identify the amino acid residues of, for example, an integrin receptor binding to an RGD peptide ligand.110 Figure 7 shows how the SERS spectrum from purified αVβ3 integrins mixed with RGD functionalized NPs strongly resembles the spectrum observed from TERS detection of the same functionalized NPs interacting on a cell membrane with TERS detection.110 The amino acid selectivity of this recognition assay was probed by mutating a tryptophan near the binding pocket in streptavidin, which dramatically altered the observed SERS spectrum and thereby identified the binding site.108
Figure 7.
The spectrum from an RGD-functionalized nanoparticle bound to a cellular membrane and detected with a Au-ball TERS tip gives rise to a spectrum that is statistically similar to that observed mixing purified αvβ3 integrin with the functionalized nanoparticles. The features observed in these spectra correspond to features known to be in the binding pocket of the protein.
Towards Clinical Applications
One application of SERS for protein detection is the “Western SERS” demonstrated by Han et al., which allows for the label-free multi-protein detection on a nitrocellulose membrane.112 With the use of SERRS they were able to quantify bovine Mb with an estimated LOD of 4 ng.112 The Mb showed vibrational bands associated with the heme group. They were also able to detect BSA, HSA, and lysozyme on the nitrocellulose membrane. These proteins exhibited bands from the amide I and III structures along with the Tyr, Phe, and Trp residues.112
Further development of such assays is leading toward clinical applications of SERS based protein detection. Wang et al. detected purified albumin and globulin from healthy and colorectal cancer patients.113 PCA-LDA analysis was used to provide a “yes/no” assessment of colorectal cancer. They assigned the major peaks of albumin and globulin to: Trp, Phe, Tyr, methionine (Met), amide I (alpha helix: 1646 cm−1 and beta sheet: 1706 cm−1) and III, S-S stretching, C-S stretch, C-C and CH2. Changes in amide I vibrations suggest that the cancer patients’ samples contained less alpha helix regions and more beta sheets than the normal samples.113
METABOLITES
Increased interest in characterizing metabolites, the small molecules involved in metabolic pathways, has resulted in the new detection methods complimentary to the gold standards: MS and NMR. SERS has been used in various studies to detect and quantify metabolites related to pesticides, natural biological pathways, and drug metabolism.
Vitamins
The various B vitamins have been detected by SERS. Bailey et al. used the combination of microfluidics, sheath-flow SERS, and amperometry to quantify vitamin B2, also known as riboflavin.114 Silver was electrodeposited onto a gold electrode to act as a SERS active electrode. The SERS detection limit was observed at 1 nM while the electrochemical detection limit was 89 nM. They reported the orientation of the analyte relative to the SERS electrode contributes to the enhancement of certain vibrational bands.114 Stokes et al. quantified folic acid (pterin based-vitamin B9) in water and detected it in human serum.115 The LOD was calculated to be 0.018 μM. The main vibrational modes observed in the SERS spectrum were associated with the coupled phenyl ring deformations.115 Other pterin containing compounds such as tetrahydropterin and dihydropterin were identified and quantified at the optimal excitation wavelength of 514.5 nm in water and rat serum.116 The LOD was reported as 1 nM for the pterins.116
Folic acid and the cancer drug, methotrexate, were also quantified in complex mixtures.117 A sandwich substrate technique was implemented by drop coating the target analyte/NP mixture onto the planar substrate. The planar substrate was fabricated by galvanic displacement of AgNPs onto copper foils. The LOD for this technique was reported to be 100 pM. However, the LOD of folic acid in PBS buffer with 1% BSA was 1nM. The characteristic Raman bands for quantification for folic acid and methotrexate were 1502 and 1563 cm−1, respectively.117
β-carotene, a precursor to vitamin A, was detected in whole blood using SERS.118 The SERS signal selectively probed either β-carotene or hemoglobin with various excitations wavelength: 785 nm and 514 nm. At 514 nm, the SERS spectra of whole blood was dominated by the vibrational bands associated with heme group while the normal Raman spectra were dominated with β-carotene bands.118
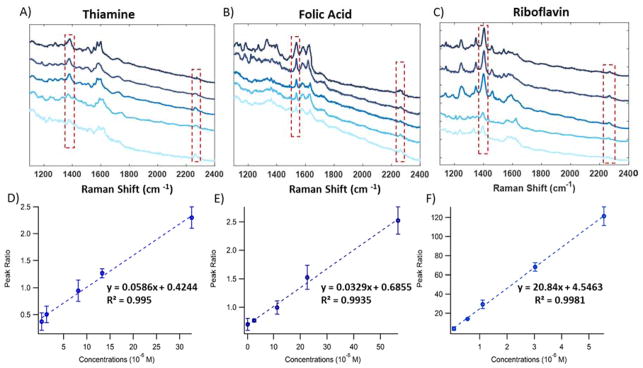
DeVault and Sepaniak used capillary electrophoresis with off -column SERS to separate and detect riboflavin from benzyoxyresorufin, and resorufin.119 An electrofilament was used to deposit the CE elution onto a silver colloid coated aluminum block where the SERS detection would later be performed. With this system, the three analytes were detected in the micromolar regime.119 Another study used LC-sheath flow SERS to separate and detect thiamine (vitamin B1), folic acid, and riboflavin.120 Quantification of the analytes was achieved using acetonitrile from the mobile phase as an internal standard. Figure 8, shows the average SERS spectra of the three analytes at various concentrations. The peak area ratio between the internal standard and analyte shows a linear relationship with concentration. The SERS LOD for thiamine, folic acid, and riboflavin were reported as 1.02 μM, 0.94 μM, and 84 nM, respectively. The unique vibrational bands allowed for the identification of the three vitamins. Observed vibrational bands for thiamine were associated with the protonated pyrimidine ring stretch. The SERS spectrum of folic acid was dominated by the C-H rocking, C=N asymmetric stretch, and the C=O stretch + N-H bending modes. While, riboflavin showed peaks associated with the C=O bending, C-N stretch of the pyrazine ring, C-C stretch, C-CH3 bending, C-N stretching, and C-C stretching of the benzene.120
Figure 8.
SERS spectra of various concentrations of thiamine (a), folic acid (b), and riboflavin (c) with a constant acetonitrile peak present. Peak area ratio between acetonitrile and thiamine (d), folic acid (e), and riboflavin (f) all show a linear dependence with concentration. Adapted from Ref.120 with permission of The Royal Society of Chemistry.
Neurotransmitters
Wang et al. used a graphene-gold tip hybrid platform to detect single molecule dopamine and serotonin (10−10 M).121 They also reported the detection of these neurotransmitters at concentrations as low as 10−9 M in simulated body fluids supplemented with background serum proteins. The dominant vibrational band for dopamine was associated with the C=C stretching mode. Indole ring vibrations dominated the serotonin spectrum. It was reported that the π-π interactions between the ring structures and the graphene contributed to the chemical enhancement.121 Bailey et al. detected the neurotransmitters: dopamine, serotonin, norepinephrine, epinephrine, and catechol to investigate the role of surface absorption in SERS.122 The dominant vibrational bands were associated with phenol moiety, which suggest that this area of the neurotransmitter adsorbs to the SERS active-surface. It was reported that the affinity and orientation is highly correlated to the SERS enhancement.122 Li et al. fabricated a stationary nanoporous gold disk in a microfluidic device to quantify dopamine and urea.123 The LOD for dopamine and urea was 32.4 nM and 0.67 mM, respectively.123
Metabolites in Biological Matrices
Other small molecules have also been identified and quantified by SERS in biological fluids. Isotope dilution was used to quantify serum creatinine; ultrafiltration and ion exchange treatment was used to remove any interfering substances from the serum.124 The LOD was reported to be 0.1 μg/mL.124 However, Premasiri et al. used gold NPs to detect creatinine, urea (the major component in urine), and uric acid in urine samples.125 They show it is possible to detect nitrogen compounds in the presence of other compounds without separation. The major vibrational band near 1000 cm−1 corresponds to the symmetric C-N stretch of urea. The SERS spectrum of uric acid was dominated by the peaks at 464, 783, and 1745 cm−1.125
Li et al. used stamping SERS with a nanoporous gold disk as SERS substrates to detect creatinine.126 A droplet of the desired analyte was dried on a PDMS surface before the SERS substrate was stamped against the PDMS film creating contact between the NP and the analyte. It was reported that creatinine in water had a LOD of 13.2 nM. Creatinine was also studied in the presence of urea and mouse urine. The calculated LOD for creatinine in mouse urine was 0.68 mg/mL. From this study, it was reported that creatinine can be observed in the presence of 100× higher concentration of urea.126
Lussier et al. used gold nanoraspberries as a plasmonic nanosensor to detect low level metabolites excreted from living Madin-Darby canine (MDCK11) kidney cells.127 PCA was used to extract SERS spectra of pyruvate, lactate, ATP, glucose, and urea while dynamic SERS (DSERS) was used to monitor the single molecule fluctuations of the metabolites.127 Glucose sensing was investigated by Yonzon et al. with the use of (1-mercaptoundeca-11-yl)tri(ethylene glycol) modified silver film over nanospheres (AgFON) on copper substrates.128 A small volume flow cell was used to minimize environmental interferences at the SERS active surface. The major vibrational bands for glucose were associated with the C-C-H bend (1320 cm−1), C-C+C-O stretch (1260, 1168, 1124 cm−1), and C-OH stretch (1076 cm−1). To prove biological relevance, glucose was also detected in the presence of serum albumin at physiological concentrations (3.9–5.5 mM).128
Drug Metabolites
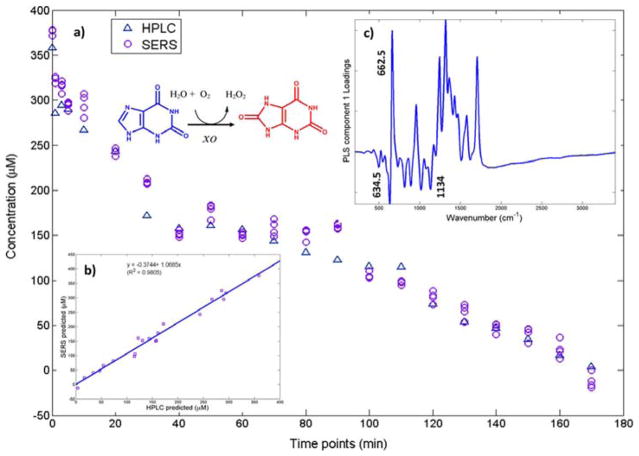
Oxypurinol, the active metabolite of the pharmaceutical drug allopurinol, was investigated for potential drug screening.129 Oxypruinol is the xanthine oxidase inhibitor. It was reported that this SERS technique was comparable to the standard HPLC method, as shown in Figure 9b. Hydroxylamine reduce silver colloids were used as the SERS-active substrate to monitor the chemical cascade between hypoxanthine and the enzyme xanthine oxidase.129 Hypoxanthine, xanthine, uric acid, and oxypurinol were all detected using SERS.
Figure 9.
(a) Partial least-squares regression results of SERS and HPLC concentration verses time results. Reaction present is xanthine to uric acid. (b) Plot of predicted SERS concentrations verses HPLC concentrations. (c) Partial least-squares loadings plot showing characteristics of the xanthine (662.5 cm−1) to uric acid (634.5 and 1134 cm−1) reaction. Reprinted with permission from Ref. 129. Copyright 2016 American Chemical Society.
Farquharson et al. investigated the SERS detection of the cancer medication, 5-fluorouracil (5-FU), and the two major metabolites of 5-FU, 5-fluorouridine (5-FUrd) and 5-fluoro-2′-deoxyuridine (5-FdUrd).130 5-FU was detected in saliva within the physiological range, 1–30 μg/mL. The 5-FU spectrum is dominated by the pyrimidine ring, trigonal ring plus C-F stretching mode, a ring plus C-H waging mode, a ring plus N-H wagging mode, and the symmetric C=O stretch. The 5-FUrd and 5-FdUrd spectra are dominated by the same vibrational modes as 5-FU, however, the 5-FdUrd had an additional peak corresponding to the combination of ring and ribose vibrational modes. It is suggested that the analytes interact with the metal NPs through the N3 giving the analyte an upright geometry relative to the NP.130
Han et al. used core-shell silver coated gold NPs to identify the antitumor drug, 6-mercaptopurine (6MP), along with the drug metabolites, 6-thioinosine monophosphate (6TIMP) and 6-mercaptopurine-ribose (6MPR).131 They monitored the real-time metabolism of 6-mercaptopurine in living tumor cells. It was reported that the 6MP and the 6MPR absorbed to the NP surface through the thiol group and the lone pair on the N atom. The dominate vibrational bands were associated with the purine ring therefore there was significant spectral overlap between the different analytes.131 Aqueous silver colloids have also been used to detect other low level anticancer drugs such as sunitinib, SN-38, irinotecan, paclitaxel, and doxorubicin at 633 nm excitation.132 The colloids and analyte mixtures were drop coated onto aluminum. Paclitaxel showed no SERS signal while the other anticancer drugs were quantifiable. The LOD for sunitinib, SN-38, and irinotecan were calculated as 11–15 ng/mm2 (18–26 ng), 11–28 ng/mm2 (20–50 ng), and 34–40 ng/mm2 (60–70 ng), respectively. This technique allowed for the detection of these analytes at medically relevant concentrations (102–103 ng).132 Yuen et al. earlier reported detecting paclitaxel in blood plasma using microwave-treated Au-polystyrene beads as the SERS-active substrate.133 Concentrations varying from 1.0 μM to 10 nM were detected using this method. The major vibrational bands for paclitaxel were associated with the C-C and C=C stretching modes.133
Other prescription drugs have been quantified by SERS. A study investigated the quantification of tricyclic antidepressants in plasma, imipramine and desipramine with Ag colloids.134 The LOD for imipramine was reported as 9.8×10−7 M. While the LOD for desipramine was not reported, it was stated that the pharmaceutical was undetectable under 10 μM. The high enhancement of the 1432 cm−1 band suggests that aliphatic chain was near the silver colloids. The 1206 and 1228 cm−1 bands corresponding to the lone pair of electrons on the N, suggest another interaction with the Ag colloids through the lone pair of electrons.134 Choi et al. detected promethazine (pain medication) and urea with a SERS active silver coated nanodome sensor.135 The sensor can be place in line with biomedical tubing for the real-time detection of biomolecules in flow. The LOD for promethazine and urea are 2.32 mg/mL and 13 mM, respectively. Urea was identified by the symmetric C-N stretch at 1000 cm−1. Whereas, promethazine showed dominate vibrational bands associated with the ring-breathing mode of the aromatic ring (1030 cm−1) and the aromatic C=C stretching mode (1567 and 1589 cm−1).135 Levene et al. tested different metal colloids to detect the β blocker, propranolol.136 The largest enhancement and most consistent spectra occurred with citrate reduction of HAuCl4 at the excitation wavelength 785 nm. Propranolol showed a well-defined naphthalene ring stretch at 1375 cm−1. The LOD was reported as 7.97 nM, which is within physiological concentrations in human blood.136
Clarke et al. used SERS to detect multiple different antibiotics such as: carbenicillin, ampicillin, penicillin V, penicillin G, and 6-aminopenicillanic acid.137 The antibiotics were detected at concentrations as low as 62.5 mM in aqueous solutions. A quantitative analysis of penicillin G in fermentation broth had a detection limit of 20 mg/mL. The core structure of penicillin consists of a thiazolidine ring, four-membered beta-lactam ring, and a variable side chain that usually consist of a benzene ring. The five different antibiotics showed distinctions in their SERS spectra based off the variability of the side chain. Quantifiable peaks for penicillin G were associated with the aromatic residues, benzene, and peaks that were combination of multiple vibrations.137
Recreational drugs have also been investigated for medical diagnostics and law enforcement monitoring. SERS detection suggests a fast, reliable, and inexpensive detection method.
The metabolites of caffeine were detected in tertiary mixtures using citrate reduced silver colloid based SERS.138 Caffeine and its metabolites, theobromine and paraxanthine, were quantified at concentrations relevant to human fluids. Alternating least squares and neural network modelling were able to quantify the metabolites without the need for separation. The caffeine and paraxanthine spectra showed vibrational bands associated with the C=O deformation. For theobromine, the vibrational band for v(C-N)+p(CH3) was observed in the tertiary mixture.138
Nicotine, cotinine, and anabasine were detected with the use of gold colloids at the excitation wavelength 785 nm.139 With the use of standard addition, the nicotine metabolites were detected in pure solutions, mixtures, and human urine. They used multivariate curve resolution-alternating least squares (MCR-ALS) to compensate for the matrix affects from the urine to be able to quantify the nicotine without separation. Vibrational bands were associated with the symmetrical breathing (1032 cm−1) and the trigonal deformation (1052 cm−1) of the pyridine. Since the three metabolites all consist of the same backbone, the SERS spectra contained significant overlap.139 Another study reported the simultaneous detection of a tertiary mixture of nicotine, cotinine, and trans-3′-hydroxycotinine by SERS.140 Here, they investigated the pH dependence of these analytes on the SERS signal. They analyzed the SERS spectra of the tertiary mixture with the artificial neural network.140 An additional study explored, magnetically optimized SERS with the use of gold dotted magnetic nanocomposites to detect cotinine and benzoylecgonine (BZE) in saliva.141 The LOD of cotinine and BZE were reported as 8.8 ppb and 29 ppb, respectively, which is below the 50 ppb detection limit currently set by SAMHSA for drugs in saliva. They reported the spectral difference between cotinine and BZE could lead to the distinction between drug addicts and smokers.141
Sanles-Sobrido et al. showed the direct and indirect detection of BZE with silver coated carbon nanotubes.142 The label-free indirect detection of BZE with mouse mAB anti-BZE was quantifiable at physiological concentrations. The BZE spectrum showed vibrational bands associated with the C=C stretching, C-H bending, NC-H stretching, C-N stretching, ring breathing, pyrrolidine C-C stretching, C-H deformation, and piperidine C-C stretching. The antibody complex shows an enhancement in the 693 cm−1 band.142 BZE in human urine was detected with the combination of CE-sheath flow SERS and with DSERS.143 Direct detection of BZE was observed in urine at a physiologically relevant concentration of 30 ng/mL.143
SERS was employed as a detection technique for HPLC separations for other recreational drugs detection such as cocaine, heroin and amphetamine or the pharmaceuticals (Nor-) papaverine and procaine.144 The LC fraction of specific analytes was collected and deposited into a microplate well containing a SERS substrate. The off-line SERS detection of these drugs varied from 2 μg/well to 75 ng/well.144 Psychoactive drugs such as dihydrocodeine, doxepine, citalopram, trimipramine, carbamazepine, methadone were analyzed in human blood and urine.145 By collecting HPLC fractions and depositing them onto gelatin-based silver halide dispersion wells, the detection limit in the1 μg/sample regime was achieved.145
Gold and silver-doped sol-gel SERS substrates incorporated inside capillaries were used to detect and quantify 80 different drugs and metabolites.146 Cocaine and five different drugs were spiked into saliva and successfully detected at concentrations as low as 1 ppm.146 A rapid, SERS detection was developed for multiplex screenings of drugs with LOD ranging from 0.09 – 0.39 μg/mL.147 The binding between drugs and HSA can lead to changes in conformation of HSA when drug-protein complexes form that are evident in the SERS spectra. HSA was employed as a model tethering protein to specifically bind four different classes of drug molecules: barbiturate, benzodiazepine, amphetamine and cocaine with the use of silver core-gold shell NPs. The analytes quantified were butalbital (barbiturate), alpha-hydroxyalprazolam (benzodiazepine, used to enhance heroin and cocaine), pseudoephedrine (precursor of methamphetamine) and BZE (metabolite of cocaine). Thus, demonstrating SERS as a promising tool to monitor drug binding efficiency and quantify the amount of bound drugs in a complex mixture.147 Fabriciova et al. also investigated the interaction between anthraquinone drugs and HSA.148 The defatted HSA was used to understand the influence of fatty acids in the binding event by comparing the SERS spectra of the various complexes. It was reported that emodin, an antitumor drug, can interact with HSA through the I and II Sudlow binding sites.148
Pesticides
Pesticide metabolites have also been investigated with SERS. The major metabolite of the pesticide cyromazine is melamine. Melamine has been investigated in complex mixtures without using a separation technique. The FDA set detection limit for melamine in food is 2.5 ppm. Lin et al. quantified melamine in gluten, chicken feed, cake, and noodles.149 The SERS LOD of melamine was reported as 0.033 μg/mL compared to the LOD for HPLC of 1 μg/mL. Melamine was quantified using the characteristic vibrational band at 682 cm−1.149 Wen et al. used suspended gold NPs to detect melamine in five different materials such as sucrose, urea, arginine, histidine, and cell culture media.150 The LOD was calculated to be 10 ppb in a water/acetonitrile solvent but only 500 ppb in the raw materials.150
The insecticide, acephate, and its metabolite, methamidophos, were extracted from urine and detected using SERS.151 The major vibrational bands for acephate are associated with the P-O-C, P-S-C, and ketone features. To determine the spectral overlap between acephate and urea, urea was detected on the same electrochemically roughed substrates. It was also reported that acephate can be efficiently extracted from urine to remove the overwhelming urea interference to obtain ppb detection limits.151
Carrillo-Carrión et al. coupled capillary-LC and SERS detection through a microdispenser interface to separate and quantify the four pesticides: atrazine, terbuthylazine, chlortoluron, and diuron.152 A highly sensitive SERS-active Ag-Quantum dot substrate was fabricated for at line SERS detection. The calculated SERS LOD ranged between 0.1–0.2 μg/mL depending on the different pesticides. The LOD calculated for the LC-UV ranged from 1.0–1.5 μg/mL.152 The fungicide and animal repellent, thiram, was quantified by coupling capillary HPLC-SERS.153 The SERS substrate was fabricated at the inner wall of the capillary with Ag@Au NPs, achieving a LOD as low as 10−7 M.153
Metabolic Biomarkers
The neuroendocrine tumor marker, metanephrine, was identified and quantified using the NP sandwich technique.154 The LOD was calculated to be 50 μM. Many of the observed vibrational bands were associated with the catechol moiety especially the intense C-C stretching mode at 1279 cm−1. This is very similar to the epinephrine and dopamine SERS spectra. They reported that the C-N vibration was not enhanced in the SERS spectra compared to the normal Raman therefore it was concluded that metanephrine absorbed to the surface through the lone pair of the O10 atom resulting in a tilted orientation.154
Glycerophosphoinositol (GroPlns) is a metabolite associated with the oncogenic Ras transformation in epithelial cells. De Luca et al. compared the SERS spectra of GroPlns, myo-inositol, and glycerol when using traditional silver colloids with their lithographically designed gold fishnet planar substrates.155 The fishnet planar substrates allowed for a more reproducible spectrum. At λex= 532 nm, the detection and identification of the three biomarkers was achieved with a sensitivity of 200 nM. The phosphodioxy group in GroPlns showed a very strong vibrational band at 1080 cm−1, however, the C-C and C-O bonds also produced a complex array of vibrational bands. Glycerol was dominated by the C-C stretching and CH2 deformation modes. While myo-inositol showed spectral overlap with the GroPlns, it also showed an additional and intense C-C-O stretching band at 1005 cm−1.155
BACTERIA
Rapid detection and classification of microorganism is essential in the field of microbiology, pathology as well as medical diagnosis. Current technology for molecular identification of bacteria includes PCR, which can detect and amplify bacterial strains from the cultured bacterial cells samples using target primer sequences to facilitate specificity of bacterial DNA/RNA detection. Pathogenic bacteria have been major risk in clinical, environmental and biosecurity fields and there is a need for accurate and rapid identification of the bacteria. In many cases, especially when dealing with infectious diseases, the ability to obtain immediate result is essential to maximize the effectiveness of target antibiotics within their therapeutic window. For this reason, SERS has been proven to be a powerful analytical tool for highly sensitive and rapid bacteria detection down to molecular level without the need of lengthy cell culturing time.
Bodelon et al. used gold nanostructures porous substrates to investigate in vitro pyrocyanin production by P. aeruginosa by SERRS (λex= 785 nm).156 The LOD by wet state immersion was reported to be 10−14 M.156
Bell et al. employed SERS to detect dipicolinic acid (DPA), the metabolite responsible for the heat resistance of bacillus endospores and a biomarker for the bacterial spores.157 Different aggregating agents for silver colloids were tested and it was found that sulfate aggregation resulted in remarkable increase in SERS signals of DPA. The internal standard, CNS−, allowed for the quantitative analysis of DPA by plotting the intensity ratio between peak 1010 cm−1 (DPA) and the thiocyanate stretch at 2120 cm−1.157 However, Zhang et al. used silver film over nanospheres to detect Bacillus anthracis spores and calcium dipicolinate (CaDPA).158 They were able to detect the spores at concentrations below the infectious dose, reporting an LOD of 2.6×103 spores. The LOD for CaDPA was reported at 3.1 μM. Citrate reduced silver colloids were also used to detect DPA and glutaric acid.159 DPA was detected in both pure samples and when extracted from bacillus spores. Glutaric acid was used as an internal standard for quantification. The LOD for DPA was reported as 10.2 ppb. DPA has a strong vibrational band at 1006 cm−1 associated with the pyridine ring-breathing mode.159 Another SERS analysis of DPA used meso-droplets on super hydrophobic wires with hydrophilic tips to significantly decrease sample size.160 However, this technique increases sensitivity for DPA. It was reported that the LOD was 1×10−6 M which is equal to 3.34×10−5 μg in a 0.2 μL droplet.160
Bacteria Classification
Efrima and colleagues performed early experiments on SERS detection of bacteria by coating a silver metal colloidal layer on the bacterial cell wall.161–162 Intense SERS signal were obtained from four different bacteria: E. coli, A. calcoaceticus RAG-1, P. aeruginosa YS-7 and B. megaterium. However, under closer examination, they reported that the SERS spectra of these bacteria were quite similar despite having significant biological differences, such as Gram positive, Gram negative, and differing contents in the cell walls.162 Their control SERS experiments suggested that the bacteria spectra were dominated by the flavin adenine dinucleotide modes, which has a larger Raman cross-section compared to other elements of the cell wall, and complicating interpretation.
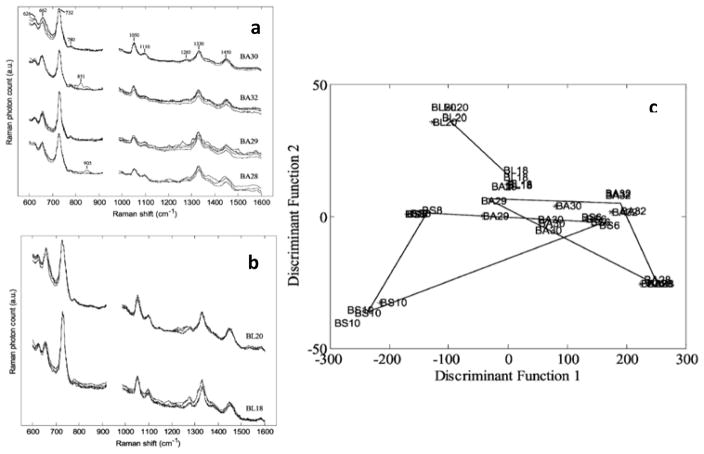
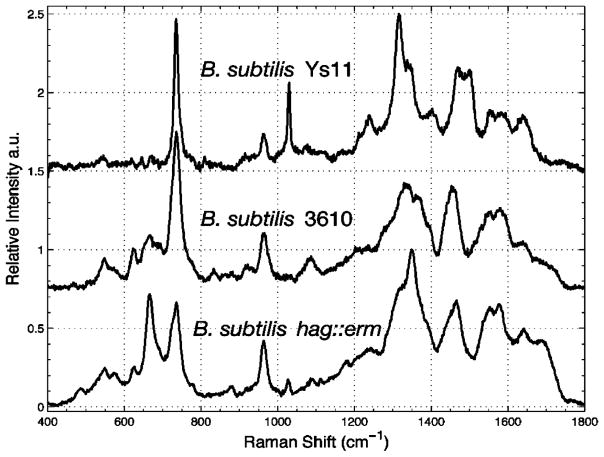
The first successful experiment to discriminate multiple bacteria strains was done by Goodacre et al. where they employed aggregated silver colloid substrate and multivariate statistical analysis to six bacterial species isolated from urinary tract infections.163 They observed SERS spectra of different bacteria species share the same main features, which are attributed to their similar constituents of the cell wall. For instance, they reported that the Gram-negative bacteria (two Klebsiella spp. and E. coli) expressed similar SERS spectra, such as the broad peaks at 651 cm−1 follow by small shoulder peaks at 720 cm−1, although some differences can still be observed within the region of 480 cm−1 to 800 cm−1. Interestingly, the Gram-negative Proteus and Gram-positive Enterococcus both show an intense peak at approximately 730 cm−1, which is assigned to vibrational modes of glycosidic ring of N-acetyl-D-glucosamine and N-acetylmuramic acid abundant in the cell walls. In a further study also done by this group, three related species of Bacillus bacteria (B. amyloliquefaciens, B. licheniformis, and B. subtilis) were successfully analyzed and discriminated by SERS.164 The peak at 626 cm−1 was observed in the SERS spectra of BS8 but not in the BS6 and BS10. Also, there were clear differences in the relative intensities of the peaks at 732 cm−1 and 662 cm−1 across the SERS spectra of the three B. subtilis isolates. Using cluster analysis and PC-DFA ordination plot as shown in Figure 10, different groups of bacteria can be discriminated based on their SERS spectral fingerprints. Their results indicate it is possible for SERS spectra to identify not only different bacterial species but also different strains/isolates within the same species. In addition to species differentiation, the SERS fingerprints was able to specify mutations in bacterial strains as demonstrated in Figure 11.165 In most cases, the strong peaks near 732 cm−1 can be attributed to the adenosine nucleic acid base of bacterial cell components58, 165 while the vibrational bands within 930 cm−1 to 1130 cm−1 have been previously assigned to phospholipids.163–165 Amide I, II, III vibrations and the carboxylic stretches that associated with the protein backbones are reported within the range of 1220 cm−1 to 1660 cm−1.164–166
Figure 10.
Example of SERS spectra with four replicates for each strain taken from (a) B. amyloliquefaciens, and (b) B. licheniformis. A cross validated PCA-DFA (c) of all 9 strains of Bacillus with 7 PCs and discriminant function cluster algorithm (DFA) shows successful isolation between the bacterial strains with good experimental reproducibility. Adapted from Ref.164 with permission of The Royal Society of Chemistry.
Figure 11.
SERS fingerprints show evidence of mutants specificity in wild-type B. subtilis 3610 cells, wild-type B. subtilis YS11 cells, and B. subtilis 3610 congenic mutant cells lacking flagella with the hag gene is deleted (hag::erm). Reprinted with permission from Ref. 165. Copyright 2005 American Chemical Society.
Bacterial Biomarkers
Although SERS has shown excellent ability to differentiate strains of bacterial species, in many cases it is the bacterial spores that serves as markers for biohazards, contaminations, and biological warfare agents. There were multiple studies on SERS detection of dipicolinic acid (DPA), which is found in anthrax bacterial spores.157, 159–160, 164 The SERS spectra of DPA discussed in the above sections are evidence for SERS detection of the bacterial spores. Many approaches were developed to quantify DPA isolated from Bacillus spores such as adding an internal standard (thiocyanate157 or glutaric acid159), multivariate analysis to quantify the concentration of DPA and spore counts159–160, 164, or using SERS of meso-droplets on superhydrophobic wires for significant reduction in sample size160. Appropriate limit of detections for DPA by SERS were obtained by multiple methods with total sample mass ranging from 2 × 10−3 μg (1100 pores) – 3.34 × 10−5 μg (18.3 pores), which were considerably below the infective dose of anthrax. Recently, SERS has been applied widely into medical diagnosis for detection of viruses166–167, food borne pathogenic bacteria168, respiratory tract pathogens collected from true clinical throat swab samples169–170, and it was able to provide comparable results to that determined by qPCR.
It is very important to note that the peaks assignments in the above studies are tentative and a molecular level interpretation of these bacterial SERS spectra has been very challenging in the early SERS studies of bacteria. Development of a SERS spectral library for bacteria facilitates identifying different bacterial strains and species with high certainty. Ziegler et al. successfully employed cluster analysis method on SERS data to create a ‘barcoding’ library for bacterial species.171 Binary barcodes generated with PCA algorithms were applied onto the normalized second derivative SERS spectrum to differentiate spectra. The use of second derivative SERS spectra can reduce background and signal fluctuations that usually obstruct SERS reproducibility.171 Moreover, the bacterial ‘barcode’ was expected to be unique for each species and can be utilized into a priori reference library of known barcodes to facilitate rapid bacterial identification by SERS. In an expanded work, this barcoding bacterial SERS method was employed to successfully identify 17 isolates and microorganism from whole blood and blinded blood samples spiked with E. coli and S. aureus with high sensitivity and specificity.172
Multiple techniques have been developed to achieve high-throughput and highly reproducible SERS experiments in order to build a reliable reference spectral library for bacterial strains and species. Cheng et al. reported the development of a microfluidic platform for concentrating pathogens from blood onto the SERS-active roughen electrode while sorting out other macromolecules by hybrid electrokinetic or dielectrophoresis.173 They were successfully identify three bacteria (S. aureaus, E. coli and P. aeruginosa) from a mixture of whole red blood cells without the need to use any antibody. Knauer et al. performed a series of studies to develop a microarray readout chip based on SERS with immobilized antibodies and Ag colloid agglomeration for microorganism detections.174–176 This SERS based immunoassay was able to nondestructively detect and quantify two different bacteria in an aqueous environment. They reported the differences in their SERS spectra were associated with differences in cell wall constituents such as the Raman shift originated from amide I at 1640–1680 cm−1, deformation CH2 of saturated lipids at 1440–1460 cm−1 or the Phe skeletal bands at 620 cm−1. The strong intensity of both COO− and NH2 vibrational modes indicate that the silver colloids bind with the bacterial cells through amine and carboxylate linkages.174–175 In a later work, they were able to embed the antibodies immobilized chip into a flow cell where it is then flushed with bacterial samples.176 Quantification was performed by SERS mapping with LOD to be 4,485 cells/mL.
Recent work done by the Ziegler group combined SERS with spectral fitting and mass spectrometry for metabolomics profiling purposes with bacteria.177 The SERS capacity for bacterial identification as well as the biochemical origins of the vibrational fingerprints on SERS spectra was fully exploit. In general, SERS spectra of bacteria were obtained with Ag-coated SiO2 SERS chip substrate at 785 nm, which can eliminate strong SERS signals contributed from bacterial cell wall (lipids, polysaccharides, peptidoglycan, and proteins). Analysis of SERS spectra together with isotopic labeling and MS resulted in dominating features of purine degradation products: adenine, hypoxanthine, xanthine, guanine, uric acid, and AMP as the bacterial cell went through the starvation process.177 The results provided a fundamental point for the development of SERS as rapid bacterial detection as well as a mean to monitor cellular activity and the kinetic of metabolic pathway.
Summary and Future Outlook
The results reported above clearly demonstrate that SERS can both identify and quantify diverse biomolecules on the basis of the intrinsic vibrations within a molecule. While SERS-tags have evolved into regulatory approved clinical assays,178 the de novo identification of biomolecules is quickly catching up. De novo SERS detection described above indicates that a number of factors are important for SERS identification and quantification.
How the biomolecules interact with the nanostructure is key. The SERS spectrum is often different from the spontaneous Raman spectrum. For the different classes of molecules above, there are consistent trends in what is observed in the SERS spectrum.
First, the functional groups on the molecule that interact with the nanostructure are preferentially enhanced. The enhancement is dependent upon the orientation of molecule. In nucleotides, the 736 cm−1 band of adenine was observed to overwhelm other signals when oriented in an extended geometry from the surface but diminish in intensity when DNA oriented parallel to the surface.44 The interactions between the carbonyls and the nanostructures resulted in frequency shifts suggesting an adsorption geometry.42 Similarly for amino acids, red shifted C-S peaks suggest binding through thiols,67 while the observation of specific residues suggest how amino acids interact with the nanostructures. By changing the surface chemistry, such as coating the nanostructures with oxides,67 new molecules can bind to the nanostructures to be identified by SERS.
Aromatic functional groups are often observed in SERS spectra. This is well documented above, particularly for proteins. While a large number of different proteins have been detected by SERS, the aromatic amino acids are consistently reported in the spectra. The heavy influence of aromatic groups on the observed spectra may cause concern about the ability to identify non-aromatic molecules; however, examples above such as the differentiation of the 20 amino acids indicates that molecules still provide a unique pattern in the SERS spectrum,69 albeit often different from the spontaneous Raman spectrum.
The reproducibility of SERS spectra has long been attributed with nanostructures used for enhancement. This has perhaps resulted in a perception of irreproducibility with SERS data. Indeed results above suggest that changes in aggregation state can affect the observed signals;65 however, strong similarities were observed between SERS spectra of a protein mixed with aggregated NPs and TERS spectra from NPs interacting with the same protein on surfaces that suggest the nature of the molecules interaction with the nanostructure is most important.109–111 This hypothesis would not rule out signal changes observed with different metals comprising the nanostructures. Because the enhancement observed is also dependent upon the nature of the interaction with the nanostructures, varied signal intensities have also been observed. Here the use of internal standards has shown that the enhancements can be corrected and provide quantitative analysis.55–56, 120
Because the interaction with the surface is critical to the observed signals, there are consequences for SERS analysis in mixtures. While the narrow lines associated with Raman spectra have increased multiplexing capability with SERS tags,28 de novo SERS detection will always be sensitive to the molecule that interacts most strongly with the surface. Technological advances combining SERS with separation techniques, such as LC and CE,68, 120, 179–180 illustrates the potential of SERS for general-purpose molecular identification. Perhaps most important for the advance of SERS based molecular identification is the development of libraries for SERS spectra. Since Ziegler and coworkers provided proof-of-concept data for this idea,171 the number of spectra available for spectral matching remains low. Similar to the utility of GC-MS databases, SERS could also be used for library-matched identification. The advantage of SERS is the significantly reduced cost of Raman instrumentation relative to mass spectrometers.181–182
Overall, the rapid progress has poised SERS to play a key role for biomolecular identification and quantification, which may enable next generation diagnostics.
Acknowledgments
The authors acknowledge support from the National Science Foundation award DBI-1455445, and the National Cancer Institute, part of the United States National Institutes of Health, award R33 CA206922.
References
- 1.Abalde-Cela S, Aldeanueva-Potel P, Mateo-Mateo C, Rodriguez-Lorenzo L, Alvarez-Puebla RA, Liz-Marzan LM. Surface-enhanced Raman scattering biomedical applications of plasmonic colloidal particles. J R Soc Interface. 2010;7(Suppl 4):S435–50. doi: 10.1098/rsif.2010.0125.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harper MM, McKeating KS, Faulds K. Recent developments and future directions in SERS for bioanalysis. Phys Chem Chem Phys. 2013;15(15):5312–28. doi: 10.1039/c2cp43859c. [DOI] [PubMed] [Google Scholar]
- 3.Hering K, Cialla D, Ackermann K, Dorfer T, Moller R, Schneidewind H, Mattheis R, Fritzsche W, Rosch P, Popp J. SERS: a versatile tool in chemical and biochemical diagnostics. Anal Bioanal Chem. 2008;390(1):113–24. doi: 10.1007/s00216-007-1667-3. [DOI] [PubMed] [Google Scholar]
- 4.Clarke CJ, Haselden JN. Metabolic Profiling as a Tool for Understanding Mechanisms of Toxicity. Toxicologic Pathology. 2008;36:140–147. doi: 10.1177/0192623307310947. [DOI] [PubMed] [Google Scholar]
- 5.Fan X, White IM, Shopova SI, Zhu H, Suter JD, Sun Y. Sensitive optical biosensors for unlabeled targets: a review. Anal Chim Acta. 2008;620(1–2):8–26. doi: 10.1016/j.aca.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson CH, Gonzalez FJ. Challenges and opportunities of metabolomics. Journal of cellular physiology. 2012;227:2975–81. doi: 10.1002/jcp.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 8.Wolfbeis OS. Fluorescence spectroscopy: new methods and applications. Springer Science & Business Media; 2012. [Google Scholar]
- 9.Hubert J, Nuzillard JM, Purson S, Hamzaoui M, Borie N, Reynaud R, Renault JH. Identification of natural metabolites in mixture: a pattern recognition strategy based on (13)C NMR. Anal Chem. 2014;86(6):2955–62. doi: 10.1021/ac403223f. [DOI] [PubMed] [Google Scholar]
- 10.Lenz EM, Wilson ID. Analytical strategies in metabonomics. Journal of proteome research. 2007;6:443–58. doi: 10.1021/pr0605217. [DOI] [PubMed] [Google Scholar]
- 11.Pasikanti KK, Ho PC, Chan ECY. Gas chromatography/mass spectrometry in metabolic profiling of biological fluids. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2008;871:202–11. doi: 10.1016/j.jchromb.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 12.Ramautar R, Nevedomskaya E, Mayboroda OA, Deelder AM, Wilson ID, Gika HG, Theodoridis GA, Somsen GW, de Jong GJ. Metabolic profiling of human urine by CE-MS using a positively charged capillary coating and comparison with UPLC-MS. Molecular bioSystems. 2011;7:194–9. doi: 10.1039/c0mb00032a. [DOI] [PubMed] [Google Scholar]
- 13.Theodoridis G, Gika HG, Wilson ID. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. TrAC Trends in Analytical Chemistry. 2008;27:251–260. [Google Scholar]
- 14.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proceedings of the National Academy of Sciences. 2015;112:201516878. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barth A. Infrared spectroscopy of proteins. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2007;1767(9):1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Baker MJ, Trevisan J, Bassan P, Bhargava R, Butler HJ, Dorling KM, Fielden PR, Fogarty SW, Fullwood NJ, Heys KA, Hughes C, Lasch P, Martin-Hirsch PL, Obinaju B, Sockalingum GD, Sulé-Suso J, Strong RJ, Walsh MJ, Wood BR, Gardner P, Martin FL. Using Fourier transform IR spectroscopy to analyze biological materials. Nat Protocols. 2014;9(8):1771–1791. doi: 10.1038/nprot.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonio KA, Schultz ZD. Advances in Biomedical Raman Microscopy. Anal Chem. 2014;86(1):30–46. doi: 10.1021/ac403640f. [DOI] [PubMed] [Google Scholar]
- 18.Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. Surface-enhanced Raman spectroscopy. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:601–26. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 19.Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chemical Physics Letters. 1974;26(2):163–166. [Google Scholar]
- 20.Jeanmaire DL, Duyne RPV. Surface Raman Spectroelectrochemistry Part 1. Heterocyclic, Aromatic, and Aliphatic Amine Adsorbed on the Anodized Silver Electrode. J Electroanal Chem. 1977;84(1):1–20. [Google Scholar]
- 21.Asiala SM, Schultz ZD. Surface enhanced Raman correlation spectroscopy of particles in solution. Anal Chem. 2014;86(5):2625–32. doi: 10.1021/ac403882h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camden JP, Dieringer JA, Zhao J, Van Duyne RP. Controlled plasmonic nanostructures for surface-enhanced spectroscopy and sensing. Acc Chem Res. 2008;41(12):1653–61. doi: 10.1021/ar800041s. [DOI] [PubMed] [Google Scholar]
- 23.Hakonen A, Svedendahl M, Ogier R, Yang ZJ, Lodewijks K, Verre R, Shegai T, Andersson PO, Kall M. Dimer-on-mirror SERS substrates with attogram sensitivity fabricated by colloidal lithography. Nanoscale. 2015;7(21):9405–10. doi: 10.1039/c5nr01654a. [DOI] [PubMed] [Google Scholar]
- 24.McNay G, Eustace D, Smith WE, Faulds K, Graham D. Surface-Enhanced Raman Scattering (SERS) and Surface-Enhanced Resonance Raman Scattering (SERRS): A Review of Applications. Applied Spectroscopy. 2011;65(8):825–837. doi: 10.1366/11-06365. [DOI] [PubMed] [Google Scholar]
- 25.Tolaieb B, Constantino CJL, Aroca RF. Surface-enhanced resonance Raman scattering as an analytical tool for single molecule detection. Analyst. 2004;129(4):337–341. [Google Scholar]
- 26.Dieringer JA, Wustholz KL, Masiello DJ, Camden JP, Kleinman SL, Schatz GC, Van Duyne RP. Surface-Enhanced Raman Excitation Spectroscopy of a Single Rhodamine 6G Molecule. Journal of the American Chemical Society. 2009;131(2):849–854. doi: 10.1021/ja8080154. [DOI] [PubMed] [Google Scholar]
- 27.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS) Physical Review Letters. 1997;78(9):1667–1670. [Google Scholar]
- 28.Laing S, Gracie K, Faulds K. Multiplex in vitro detection using SERS. Chem Soc Rev. 2016;45(7):1901–18. doi: 10.1039/c5cs00644a. [DOI] [PubMed] [Google Scholar]
- 29.Nolan JP, Duggan E, Liu E, Condello D, Dave I, Stoner SA. Single cell analysis using surface enhanced Raman scattering (SERS) tags. Methods. 2012;57(3):272–9. doi: 10.1016/j.ymeth.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nolan JP, Sebba DS. Surface-enhanced Raman scattering (SERS) cytometry. Methods Cell Biol. 2011;102:515–32. doi: 10.1016/B978-0-12-374912-3.00020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guven B, Basaran-Akgul N, Temur E, Tamer U, Boyaci IH. SERS-based sandwich immunoassay using antibody coated magnetic nanoparticles for Escherichia coli enumeration. Analyst. 2011;136(4):740–8. doi: 10.1039/c0an00473a. [DOI] [PubMed] [Google Scholar]
- 32.Hwang J, Lee S, Choo J. Application of a SERS-based lateral flow immunoassay strip for the rapid and sensitive detection of staphylococcal enterotoxin B. Nanoscale. 2016;8(22):11418–25. doi: 10.1039/c5nr07243c. [DOI] [PubMed] [Google Scholar]
- 33.Rong Z, Wang C, Wang J, Wang D, Xiao R, Wang S. Magnetic immunoassay for cancer biomarker detection based on surface-enhanced resonance Raman scattering from coupled plasmonic nanostructures. Biosens Bioelectron. 2016;84:15–21. doi: 10.1016/j.bios.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Xia X, Li W, Zhang Y, Xia Y. Silica-coated dimers of silver nanospheres as surface-enhanced Raman scattering tags for imaging cancer cells. Interface Focus. 2013;3(3):20120092. doi: 10.1098/rsfs.2012.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu L, Yan W, Ma W, Kuang H, Wu X, Liu L, Zhao Y, Wang L, Xu C. SERS encoded silver pyramids for attomolar detection of multiplexed disease biomarkers. Adv Mater. 2015;27(10):1706–11. doi: 10.1002/adma.201402244. [DOI] [PubMed] [Google Scholar]
- 36.Wang HN, Vo-Dinh T. Multiplex detection of breast cancer biomarkers using plasmonic molecular sentinel nanoprobes. Nanotechnology. 2009;20(6):065101. doi: 10.1088/0957-4484/20/6/065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham D, Thompson DG, Smith WE, Faulds K. Control of enhanced Raman scattering using a DNA-based assembly process of dye-coded nanoparticles. Nat Nanotechnol. 2008;3(9):548–51. doi: 10.1038/nnano.2008.189. [DOI] [PubMed] [Google Scholar]
- 38.Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Femtomolar detection of prostate-specific antigen: an immunoassay based on surface-enhanced Raman scattering and immunogold labels. Anal Chem. 2003;75(21):5936–43. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 39.Li M, Cushing SK, Zhang J, Suri S, Evans R, Petros WP, Gibson LF, Ma D, Liu Y, Wu N. Three-dimensional hierarchical plasmonic nano-architecture enhanced surface-enhanced Raman scattering immunosensor for cancer biomarker detection in blood plasma. ACS Nano. 2013;7(6):4967–76. doi: 10.1021/nn4018284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou L, Ding F, Chen H, Ding W, Zhang W, Chou SY. Enhancement of immunoassay’s fluorescence and detection sensitivity using three-dimensional plasmonic nano-antenna-dots array. Anal Chem. 2012;84(10):4489–95. doi: 10.1021/ac3003215. [DOI] [PubMed] [Google Scholar]
- 41.Bell SEJ, Sirimuthu NMS. Quantitative surface-enhanced Raman spectroscopy. Chemical Society reviews. 2008;37:1012–24. doi: 10.1039/b705965p. [DOI] [PubMed] [Google Scholar]
- 42.Otto C, Tweel TJJvd, Mul FFMd, Greve J. Surface-enhanced Raman spectroscopy of DNA bases Journal of Raman Spectroscopy. [accessed 01];Journal of Raman Spectroscopy [Online] 1986 17(3):289–298. http://onlinelibrary.wiley.com/doi/10.1002/jrs.1250170311/abstract. [Google Scholar]
- 43.Kneipp K, Kneipp H, Kartha VB, Manoharan R, Deinum G, Itzkan I, Dasari RR, Feld MS. Detection and identification of a single DNA base molecule using surface-enhanced Raman scattering (SERS) Physical Review E. 1998;57(6):R6281–R6284. [Google Scholar]
- 44.Bell SE, Sirimuthu NM. Surface-enhanced Raman spectroscopy (SERS) for sub-micromolar detection of DNA/RNA mononucleotides. J Am Chem Soc. 2006;128(49):15580–1. doi: 10.1021/ja066263w. [DOI] [PubMed] [Google Scholar]
- 45.Lo HC, Hsiung HI, Chattopadhyay S, Han HC, Chen CF, Leu JP, Chen KH, Chen LC. Label free sub-picomole level DNA detection with Ag nanoparticle decorated Au nanotip arrays as surface enhanced Raman spectroscopy platform. Biosens Bioelectron. 2011;26(5):2413–8. doi: 10.1016/j.bios.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 46.Barhoumi A, Halas NJ. Label-free detection of DNA hybridization using surface enhanced Raman spectroscopy. J Am Chem Soc. 2010;132(37):12792–3. doi: 10.1021/ja105678z. [DOI] [PubMed] [Google Scholar]
- 47.Barhoumi A, Halas NJ. Detecting Chemically Modified DNA Bases Using Surface Enhanced Raman Spectroscopy. J Phys Chem Lett. 2011;2(24):3118–3123. doi: 10.1021/jz201423b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barhoumi A, Zhang D, Tam F, Halas NJ. Surface-enhanced Raman spectroscopy of DNA. J Am Chem Soc. 2008;130(16):5523–9. doi: 10.1021/ja800023j. [DOI] [PubMed] [Google Scholar]
- 49.Papadopoulou E, Bell SE. DNA reorientation on Au nanoparticles: label-free detection of hybridization by surface enhanced Raman spectroscopy. Chem Commun (Camb) 2011;47(39):10966–8. doi: 10.1039/c1cc13705k. [DOI] [PubMed] [Google Scholar]
- 50.Torres-Nunez A, Faulds K, Graham D, Alvarez-Puebla RA, Guerrini L. Silver colloids as plasmonic substrates for direct label-free surface-enhanced Raman scattering analysis of DNA. Analyst. 2016;141(17):5170–80. doi: 10.1039/c6an00911e. [DOI] [PubMed] [Google Scholar]
- 51.Abell JL, Garren JM, Driskell JD, Tripp RA, Zhao Y. Label-free detection of micro-RNA hybridization using surface-enhanced Raman spectroscopy and least-squares analysis. J Am Chem Soc. 2012;134(31):12889–92. doi: 10.1021/ja3043432. [DOI] [PubMed] [Google Scholar]
- 52.Driskell JD, Primera-Pedrozo OM, Dluhy RA, Zhao Y, Tripp RA. Quantitative surface-enhanced Raman spectroscopy based analysis of microRNA mixtures. Appl Spectrosc. 2009;63(10):1107–14. doi: 10.1366/000370209789553183. [DOI] [PubMed] [Google Scholar]
- 53.Prado E, Daugey N, Plumet S, Servant L, Lecomte S. Quantitative label-free RNA detection using surface-enhanced Raman spectroscopy. Chem Commun (Camb) 2011;47(26):7425–7. doi: 10.1039/c1cc11925g. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Joshi P, Zhou Y, Ding R, Zhang P. Quantitative SERS-based DNA detection assisted by magnetic microspheres. Chem Commun (Camb) 2015;51(83):15284–6. doi: 10.1039/c5cc06571b. [DOI] [PubMed] [Google Scholar]
- 55.Morla-Folch J, Alvarez-Puebla RA, Guerrini L. Direct Quantification of DNA Base Composition by Surface-Enhanced Raman Scattering Spectroscopy. J Phys Chem Lett. 2016;7(15):3037–41. doi: 10.1021/acs.jpclett.6b01424. [DOI] [PubMed] [Google Scholar]
- 56.Xu LJ, Lei ZC, Li J, Zong C, Yang CJ, Ren B. Label-free surface-enhanced Raman spectroscopy detection of DNA with single-base sensitivity. J Am Chem Soc. 2015;137(15):5149–54. doi: 10.1021/jacs.5b01426. [DOI] [PubMed] [Google Scholar]
- 57.Papadopoulou E, Bell SE. Label-free detection of single-base mismatches in DNA by surface-enhanced Raman spectroscopy. Angew Chem Int Ed Engl. 2011;50(39):9058–61. doi: 10.1002/anie.201102776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papadopoulou E, Bell SE. Label-free detection of nanomolar unmodified single- and double-stranded DNA by using surface-enhanced Raman spectroscopy on Ag and Au colloids. Chemistry. 2012;18(17):5394–400. doi: 10.1002/chem.201103520. [DOI] [PubMed] [Google Scholar]
- 59.Guerrini L, Krpetic Z, van Lierop D, Alvarez-Puebla RA, Graham D. Direct surface-enhanced Raman scattering analysis of DNA duplexes. Angew Chem Int Ed Engl. 2015;54(4):1144–8. doi: 10.1002/anie.201408558. [DOI] [PubMed] [Google Scholar]
- 60.Morla-Folch J, Xie HN, Gisbert-Quilis P, Gomez-de Pedro S, Pazos-Perez N, Alvarez-Puebla RA, Guerrini L. Ultrasensitive Direct Quantification of Nucleobase Modifications in DNA by Surface-Enhanced Raman Scattering: The Case of Cytosine. Angew Chem Int Ed Engl. 2015;54(46):13650–4. doi: 10.1002/anie.201507682. [DOI] [PubMed] [Google Scholar]
- 61.Morla-Folch J, Xie HN, Alvarez-Puebla RA, Guerrini L. Fast Optical Chemical and Structural Classification of RNA. ACS Nano. 2016;10(2):2834–42. doi: 10.1021/acsnano.5b07966. [DOI] [PubMed] [Google Scholar]
- 62.Chen Y, Chen G, Feng S, Pan J, Zheng X, Su Y, Chen Y, Huang Z, Lin X, Lan F, Chen R, Zeng H. Label-free serum ribonucleic acid analysis for colorectal cancer detection by surface-enhanced Raman spectroscopy and multivariate analysis. J Biomed Opt. 2012;17(6):067003. doi: 10.1117/1.JBO.17.6.067003. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H, Liu Y, Gao J, Zhen J. A sensitive SERS detection of miRNA using a label-free multifunctional probe. Chem Commun (Camb) 2015;51(94):16836–9. doi: 10.1039/c5cc06225j. [DOI] [PubMed] [Google Scholar]
- 64.O’Neal PD, Motamedi M, Lin WC, Chen J, Cote GL. Feasibility study using surface-enhanced Raman spectroscopy for the quantitative detection of excitatory amino acids. J Biomed Opt. 2003;8(1):33–9. doi: 10.1117/1.1528208. [DOI] [PubMed] [Google Scholar]
- 65.Nascimento FC, Carneiro CE, de Santana H, Zaia DA. The effect of artificial seawater on SERS spectra of amino acids-Ag colloids: an experiment of prebiotic chemistry. Spectrochim Acta A Mol Biomol Spectrosc. 2014;118:251–9. doi: 10.1016/j.saa.2013.08.113. [DOI] [PubMed] [Google Scholar]
- 66.López-Ramírez MR, Arenas JF, Otero JC, Castro JL. Surface-enhanced Raman scattering of D-penicillamine on silver colloids Journal of Raman Spectroscopy. [accessed 01];Journal of Raman Spectroscopy [Online] 2004 35(5):390–394. http://onlinelibrary.wiley.com/doi/10.1002/jrs.1138/abstract. [Google Scholar]
- 67.Du P, Ma L, Cao Y, Li D, Liu Z, Wang Z, Sun Z. Stable Ag@oxides nanoplates for surface-enhanced Raman spectroscopy of amino acids. ACS Appl Mater Interfaces. 2014;6(11):8853–8. doi: 10.1021/am501637d. [DOI] [PubMed] [Google Scholar]
- 68.He L, Natan MJ, Keating CD. Surface-enhanced Raman scattering: a structure-specific detection method for capillary electrophoresis. Anal Chem. 2000;72(21):5348–55. doi: 10.1021/ac000583v. [DOI] [PubMed] [Google Scholar]
- 69.Negri P, Schultz ZD. Online SERS detection of the 20 proteinogenic-amino acids separated by capillary zone electrophoresis. The Analyst. 2014;139:5989–5998. doi: 10.1039/c4an01177e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei F, Zhang D, Halas NJ, Hartgerink JD. Aromatic amino acids providing characteristic motifs in the Raman and SERS spectroscopy of peptides. J Phys Chem B. 2008;112(30):9158–64. doi: 10.1021/jp8025732. [DOI] [PubMed] [Google Scholar]
- 71.Negri P, Sarver Sa, Schiavone NM, Dovichi NJ, Schultz ZD. Online SERS detection and characterization of eight biologically-active peptides separated by capillary zone electrophoresis. The Analyst. 2015;140:1516–1522. doi: 10.1039/c4an01980f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blum C, Schmid T, Opilik L, Metanis N, Weidmann S, Zenobi R. Missing Amide I Mode in Gap-Mode Tip-Enhanced Raman Spectra of Proteins. The Journal of Physical Chemistry C. 2012;116(43):23061–23066. [Google Scholar]
- 73.Kurouski D, Postiglione T, Deckert-Gaudig T, Deckert V, Lednev IK. Amide I vibrational mode suppression in surface (SERS) and tip (TERS) enhanced Raman spectra of protein specimens. Analyst. 2013;138(6):1665–73. doi: 10.1039/c2an36478f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dou X, Yamaguchi Y, Yamamoto H, Doi S, Ozaki Y. NIR SERS detection of immune reaction on gold colloid particles without bound/free antigen separation. Journal of Raman Spectroscopy. 1998;29(8):739–742. [Google Scholar]
- 75.Xu H, Bjerneld EJ, Käll M, Börjesson L. Spectroscopy of Single Hemoglobin Molecules by Surface Enhanced Raman Scattering. Physical Review Letters. 1999;83(21):4357–4360. [Google Scholar]
- 76.Bizzarri AR, Cannistraro S. Surface-enhanced resonance Raman spectroscopy signals from single myoglobin molecules. Applied Spectroscopy. 2002;56(12):1531–1537. [Google Scholar]
- 77.Feng M, Tachikawa H. Surface-Enhanced Resonance Raman Spectroscopic Characterization of the Protein Native Structure. Journal of the American Chemical Society. 2008;130(23):7443–7448. doi: 10.1021/ja8006337. [DOI] [PubMed] [Google Scholar]
- 78.Habuchi S, Cotlet M, Gronheid R, Dirix G, Michiels J, Vanderleyden J, De Schryver FC, Hofkens J. Single-molecule surface enhanced resonance Raman spectroscopy of the enhanced green fluorescent protein. J Am Chem Soc. 2003;125(28):8446–7. doi: 10.1021/ja0353311. [DOI] [PubMed] [Google Scholar]
- 79.Delfino I, Bizzarri AR, Cannistraro S. Single-molecule detection of yeast cytochrome c by Surface-Enhanced Raman Spectroscopy. Biophysical Chemistry. 2005;113(1):41–51. doi: 10.1016/j.bpc.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 80.Zheng J, Zhou Q, Zhou Y, Lu T, Cotton TM, Chumanov G. Surface-enhanced resonance Raman spectroscopic study of yeast iso-1-cytochrome c and its mutant. Journal of Electroanalytical Chemistry. 2002;530(1–2):75–81. [Google Scholar]
- 81.Han XX, Kitahama Y, Itoh T, Wang CX, Zhao B, Ozaki Y. Protein-mediated sandwich strategy for surface-enhanced Raman scattering: application to versatile protein detection. Anal Chem. 2009;81(9):3350–5. doi: 10.1021/ac802553a. [DOI] [PubMed] [Google Scholar]
- 82.Etchegoin P, Liem H, Maher RC, Cohen LF, Brown RJC, Milton MJT, Gallop JC. Observation of dynamic oxygen release in hemoglobin using surface enhanced Raman scattering. Chemical Physics Letters. 2003;367(1–2):223–229. [Google Scholar]
- 83.Drachev VP, Thoreson MD, Khaliullin EN, Davisson VJ, Shalaev VM. Surface-Enhanced Raman Difference between Human Insulin and Insulin Lispro Detected with Adaptive Nanostructures. The Journal of Physical Chemistry B. 2004;108(46):18046–18052. [Google Scholar]
- 84.Wang P, Liang O, Zhang W, Schroeder T, Xie YH. Ultra-sensitive graphene-plasmonic hybrid platform for label-free detection. Adv Mater. 2013;25(35):4918–24. doi: 10.1002/adma.201300635. [DOI] [PubMed] [Google Scholar]
- 85.Chowdhury MH, Gant VA, Trache A, Baldwin A, Meininger GA, Cote GL. Use of surface-enhanced Raman spectroscopy for the detection of human integrins. J Biomed Opt. 2006;11(2):024004. doi: 10.1117/1.2187022. [DOI] [PubMed] [Google Scholar]
- 86.Yang X, Gu C, Qian F, Li Y, Zhang JZ. Highly sensitive detection of proteins and bacteria in aqueous solution using surface-enhanced Raman scattering and optical fibers. Anal Chem. 2011;83(15):5888–94. doi: 10.1021/ac200707t. [DOI] [PubMed] [Google Scholar]
- 87.Xu LJ, Zong C, Zheng XS, Hu P, Feng JM, Ren B. Label-free detection of native proteins by surface-enhanced Raman spectroscopy using iodide-modified nanoparticles. Anal Chem. 2014;86(4):2238–45. doi: 10.1021/ac403974n. [DOI] [PubMed] [Google Scholar]
- 88.Ivleva NP, Wagner M, Szkola A, Horn H, Niessner R, Haisch C. Label-free in situ SERS imaging of biofilms. J Phys Chem B. 2010;114(31):10184–94. doi: 10.1021/jp102466c. [DOI] [PubMed] [Google Scholar]
- 89.Avci E, Culha M. Influence of protein size on surface-enhanced Raman scattering (SERS) spectra in binary protein mixtures. Appl Spectrosc. 2014;68(8):890–9. doi: 10.1366/13-07445. [DOI] [PubMed] [Google Scholar]
- 90.Han XX, Huang GG, Zhao B, Ozaki Y. Label-Free Highly Sensitive Detection of Proteins in Aqueous Solutions Using Surface-Enhanced Raman Scattering. Analytical Chemistry. 2009;81(9):3329–3333. doi: 10.1021/ac900395x. [DOI] [PubMed] [Google Scholar]
- 91.Combs ZA, Chang S, Clark T, Singamaneni S, Anderson KD, Tsukruk VV. Label-free Raman mapping of surface distribution of protein a and IgG biomolecules. Langmuir. 2011;27(6):3198–205. doi: 10.1021/la104787w. [DOI] [PubMed] [Google Scholar]
- 92.Kralova ZO, Orinak A, Orinakova R, Skantarova L, Radonak J. Enhanced detection of human plasma proteins on nanostructured silver surfaces. Nanomater nanotechnol. 2013;3:1–8. [Google Scholar]
- 93.Hughes J, Izake EL, Lott WB, Ayoko GA, Sillence M. Ultra sensitive label free surface enhanced Raman spectroscopy method for the detection of biomolecules. Talanta. 2014;130:20–25. doi: 10.1016/j.talanta.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 94.Buividas R, Dzingelevicius N, Kubiliute R, Stoddart PR, Khanh Truong V, Ivanova EP, Juodkazis S. Statistically quantified measurement of an Alzheimer’s marker by surface-enhanced Raman scattering. J Biophotonics. 2015;8(7):567–74. doi: 10.1002/jbio.201400017. [DOI] [PubMed] [Google Scholar]
- 95.Kahraman M, Sur I, Culha M. Label-free detection of proteins from self-assembled protein-silver nanoparticle structures using surface-enhanced Raman scattering. Anal Chem. 2010;82(18):7596–602. doi: 10.1021/ac101720s. [DOI] [PubMed] [Google Scholar]
- 96.Keskin S, Kahraman M, Culha M. Differential separation of protein mixtures using convective assembly and label-free detection with surface enhanced Raman scattering. Chem Commun (Camb) 2011;47(12):3424–6. doi: 10.1039/c0cc05275b. [DOI] [PubMed] [Google Scholar]
- 97.Wong-Ek K, Chailapakul O, Eiamchai P, Horpratum M, Limnonthakul P, Patthanasettakul V, Sutapan B, Tuantranont A, Chindaudom P, Nuntawong N. Surface-enhanced Raman scattering using silver nanocluster on anodic aluminum oxide template sensor toward protein detection. Biomed Tech (Berl) 2011;56(4):235–40. doi: 10.1515/BMT.2011.103. [DOI] [PubMed] [Google Scholar]
- 98.Keskin S, Culha M. Label-free detection of proteins from dried-suspended droplets using surface enhanced Raman scattering. Analyst. 2012;137(11):2651–7. doi: 10.1039/c2an16296b. [DOI] [PubMed] [Google Scholar]
- 99.Kahraman M, Balz BN, Wachsmann-Hogiu S. Hydrophobicity-driven self-assembly of protein and silver nanoparticles for protein detection using surface-enhanced Raman scattering. Analyst. 2013;138(10):2906–13. doi: 10.1039/c3an00025g. [DOI] [PubMed] [Google Scholar]
- 100.Kahraman M, Wachsmann-Hogiu S. Label-free and direct protein detection on 3D plasmonic nanovoid structures using surface-enhanced Raman scattering. Anal Chim Acta. 2015;856:74–81. doi: 10.1016/j.aca.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 101.Chou IH, Benford M, Beier HT, Cote GL, Wang M, Jing N, Kameoka J, Good TA. Nanofluidic biosensing for beta-amyloid detection using surface enhanced Raman spectroscopy. Nano Lett. 2008;8(6):1729–35. doi: 10.1021/nl0808132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou J, Ren K, Zhao Y, Dai W, Wu H. Convenient formation of nanoparticle aggregates on microfluidic chips for highly sensitive SERS detection of biomolecules. Analytical and Bioanalytical Chemistry. 2012;402(4):1601–1609. doi: 10.1007/s00216-011-5585-z. [DOI] [PubMed] [Google Scholar]
- 103.Pavel I, McCarney E, Elkhaled A, Morrill A, Plaxco K, Moskovits M. Label-Free SERS Detection of Small Proteins Modified to Act as Bifunctional Linkers. The journal of physical chemistry. C, Nanomaterials and interfaces. 2008;112(13):4880–4883. doi: 10.1021/jp710261y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Das G, Mecarini F, Gentile F, De Angelis F, Mohan Kumar H, Candeloro P, Liberale C, Cuda G, Di Fabrizio E. Nano-patterned SERS substrate: application for protein analysis vs. temperature. Biosens Bioelectron. 2009;24(6):1693–9. doi: 10.1016/j.bios.2008.08.050. [DOI] [PubMed] [Google Scholar]
- 105.Keskin S, Efeoglu E, Kececi K, Culha M. Label-free detection of proteins in ternary mixtures using surface-enhanced Raman scattering and protein melting profiles. J Biomed Opt. 2013;18(3):037007. doi: 10.1117/1.JBO.18.3.037007. [DOI] [PubMed] [Google Scholar]
- 106.Li YT, Li DW, Cao Y, Long YT. Label-free in-situ monitoring of protein tyrosine nitration in blood by surface-enhanced Raman spectroscopy. Biosens Bioelectron. 2015;69:1–7. doi: 10.1016/j.bios.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 107.Li X, Martin SJ, Chinoy ZS, Liu L, Rittgers B, Dluhy RA, Boons GJ. Label-Free Detection of Glycan-Protein Interactions for Array Development by Surface-Enhanced Raman Spectroscopy (SERS) Chemistry. 2016;22(32):11180–5. doi: 10.1002/chem.201602706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang H, Carrier SL, Park S, Schultz ZD. Selective TERS detection and imaging through controlled plasmonics. Faraday Discussions. 2015;178(0):221–235. doi: 10.1039/c4fd00190g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang H, Schultz ZD. The chemical origin of enhanced signals from tip-enhanced Raman detection of functionalized nanoparticles. Analyst. 2013;138(11):3150–3157. doi: 10.1039/c3an36898j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang H, Schultz ZD. TERS Detection of αVβ3 Integrins in Intact Cell Membranes. ChemPhysChem. 2014;15(18):3944–3949. doi: 10.1002/cphc.201402466. [DOI] [PubMed] [Google Scholar]
- 111.Xiao L, Wang H, Schultz ZD. Selective Detection of RGD-Integrin Binding in Cancer Cells Using Tip Enhanced Raman Scattering Microscopy. Analytical Chemistry. 2016;88(12):6547–6553. doi: 10.1021/acs.analchem.6b01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han XX, Jia HY, Wang YF, Lu ZC, Wang CX, Xu WQ, Zhao B, Ozaki Y. Analytical technique for label-free multi-protein detection based on Western blot and surface-enhanced Raman scattering. Anal Chem. 2008;80(8):2799–804. doi: 10.1021/ac702390u. [DOI] [PubMed] [Google Scholar]
- 113.Wang J, Lin D, Lin J, Yu Y, Huang Z, Chen Y, Lin J, Feng S, Li B, Liu N, Chen R. Label-free detection of serum proteins using surface-enhanced Raman spectroscopy for colorectal cancer screening. J Biomed Opt. 2014;19(8):087003. doi: 10.1117/1.JBO.19.8.087003. [DOI] [PubMed] [Google Scholar]
- 114.Bailey MR, Pentecost AM, Selimovic A, Martin RS, Schultz ZD. Sheath-flow microfluidic approach for combined surface enhanced Raman scattering and electrochemical detection. Anal Chem. 2015;87(8):4347–55. doi: 10.1021/acs.analchem.5b00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stokes RJ, McBride E, Wilson CG, Girkin JM, Smith WE, Graham D. Surface-enhanced Raman scattering spectroscopy as a sensitive and selective technique for the detection of folic acid in water and human serum. Appl Spectrosc. 2008;62(4):371–6. doi: 10.1366/000370208784046812. [DOI] [PubMed] [Google Scholar]
- 116.Stevenson R, Stokes RJ, MacMillan D, Armstrong D, Faulds K, Wadsworth R, Kunuthur S, Suckling CJ, Graham D. In situ detection of pterins by SERS. Analyst. 2009;134(8):1561–4. doi: 10.1039/b905562b. [DOI] [PubMed] [Google Scholar]
- 117.Yang J, Tan X, Shih W-C, Cheng MM-C. A sandwich substrate for ultrasensitive and label-free SERS spectroscopic detection of folic acid/methotrexate. Biomedical Microdevices. 2014;16(5):673–679. doi: 10.1007/s10544-014-9871-3. [DOI] [PubMed] [Google Scholar]
- 118.Casella M, Lucotti A, Tommasini M, Bedoni M, Forvi E, Gramatica F, Zerbi G. Raman and SERS recognition of beta-carotene and haemoglobin fingerprints in human whole blood. Spectrochim Acta A Mol Biomol Spectrosc. 2011;79(5):915–9. doi: 10.1016/j.saa.2011.03.048. [DOI] [PubMed] [Google Scholar]
- 119.DeVault GL, Sepaniak MJ. Spatially focused deposition of capillary electrophoresis effluent onto surface-enhanced Raman-active substrates for off-column spectroscopy. Electrophoresis. 2001;22(11):2303–11. doi: 10.1002/1522-2683(20017)22:11<2303::AID-ELPS2303>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 120.Nguyen A, Schultz ZD. Quantitative online sheath-flow surface enhanced Raman spectroscopy detection for liquid chromatography. Analyst. 2016;141(12):3630–3635. doi: 10.1039/c6an00155f. [DOI] [PubMed] [Google Scholar]
- 121.Wang P, Xia M, Liang O, Sun K, Cipriano AF, Schroeder T, Liu H, Xie YH. Label-Free SERS Selective Detection of Dopamine and Serotonin Using Graphene-Au Nanopyramid Heterostructure. Anal Chem. 2015;87(20):10255–61. doi: 10.1021/acs.analchem.5b01560. [DOI] [PubMed] [Google Scholar]
- 122.Bailey MR, Martin RS, Schultz ZD. Role of Surface Adsorption in the Surface-Enhanced Raman Scattering and Electrochemical Detection of Neurotransmitters. The Journal of Physical Chemistry C. 2016;120(37):20624–20633. doi: 10.1021/acs.jpcc.6b01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li M, Zhao F, Zeng J, Qi J, Lu J, Shih W-C. Microfluidic surface-enhanced Raman scattering sensor with monolithically integrated nanoporous gold disk arrays for rapid and label-free biomolecular detection. Journal of Biomedical Optics. 2014;19(11):111611–111611. doi: 10.1117/1.JBO.19.11.111611. [DOI] [PubMed] [Google Scholar]
- 124.Stosch R, Henrion A, Schiel D, Guttler B. Surface-enhanced Raman scattering based approach for quantitative determination of creatinine in human serum. Anal Chem. 2005;77(22):7386–92. doi: 10.1021/ac0511647. [DOI] [PubMed] [Google Scholar]
- 125.Premasiri WR, Clarke RH, Womble ME. Urine analysis by laser Raman spectroscopy. Lasers Surg Med. 2001;28(4):330–4. doi: 10.1002/lsm.1058. [DOI] [PubMed] [Google Scholar]
- 126.Li M, Du Y, Zhao F, Zeng J, Mohan C, Shih WC. Reagent- and separation-free measurements of urine creatinine concentration using stamping surface enhanced Raman scattering (S-SERS) Biomed Opt Express. 2015;6(3):849–58. doi: 10.1364/BOE.6.000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lussier F, Brule T, Vishwakarma M, Das T, Spatz JP, Masson JF. Dynamic-SERS Optophysiology: A Nanosensor for Monitoring Cell Secretion Events. Nano Lett. 2016;16(6):3866–71. doi: 10.1021/acs.nanolett.6b01371. [DOI] [PubMed] [Google Scholar]
- 128.Yonzon CR, Haynes CL, Zhang X, Walsh JT, Van Duyne RP. A Glucose Biosensor Based on Surface-Enhanced Raman Scattering: Improved Partition Layer, Temporal Stability, Reversibility, and Resistance to Serum Protein Interference. Analytical Chemistry. 2004;76(1):78–85. doi: 10.1021/ac035134k. [DOI] [PubMed] [Google Scholar]
- 129.Westley C, Xu Y, Carnell AJ, Turner NJ, Goodacre R. Label-Free Surface Enhanced Raman Scattering Approach for High-Throughput Screening of Biocatalysts. Anal Chem. 2016;88(11):5898–903. doi: 10.1021/acs.analchem.6b00813. [DOI] [PubMed] [Google Scholar]
- 130.Farquharson S, Gift A, Shende C, Inscore F, Ordway B, Farquharson C, Murren J. Surface-enhanced Raman spectral measurements of 5-fluorouracil in saliva. Molecules. 2008;13(10):2608–27. doi: 10.3390/molecules13102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Han G, Liu R, Han M-Y, Jiang C, Wang J, Du S, Liu B, Zhang Z. Label-Free Surface-Enhanced Raman Scattering Imaging to Monitor the Metabolism of Antitumor Drug 6-Mercaptopurine in Living Cells. Analytical Chemistry. 2014;86(23):11503–11507. doi: 10.1021/ac503539w. [DOI] [PubMed] [Google Scholar]
- 132.Litti L, Amendola V, Toffoli G, Meneghetti M. Detection of low-quantity anticancer drugs by surface-enhanced Raman scattering. Analytical and Bioanalytical Chemistry. 2016;408(8):2123–2131. doi: 10.1007/s00216-016-9315-4. [DOI] [PubMed] [Google Scholar]
- 133.Yuen C, Zheng W, Huang Z. Low-level detection of anti-cancer drug in blood plasma using microwave-treated gold-polystyrene beads as surface-enhanced Raman scattering substrates. Biosens Bioelectron. 2010;26(2):580–4. doi: 10.1016/j.bios.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 134.Jaworska A, Wietecha-Posluszny R, Wozniakiewicz M, Koscielniak P, Malek K. Evaluation of the potential of surface enhancement Raman spectroscopy for detection of tricyclic psychotropic drugs. Case studies on imipramine and its metabolite. Analyst. 2011;136(22):4704–9. doi: 10.1039/c1an15598a. [DOI] [PubMed] [Google Scholar]
- 135.Choi CJ, Wu H-Y, George S, Weyhenmeyer J, Cunningham BT. Biochemical sensor tubing for point-of-care monitoring of intravenous drugs and metabolites. Lab on a Chip. 2012;12(3):574–581. doi: 10.1039/c2lc20586f. [DOI] [PubMed] [Google Scholar]
- 136.Levene C, Correa E, Blanch EW, Goodacre R. Enhancing Surface Enhanced Raman Scattering (SERS) Detection of Propranolol with Multiobjective Evolutionary Optimization. Analytical Chemistry. 2012;84(18):7899–7905. doi: 10.1021/ac301647a. [DOI] [PubMed] [Google Scholar]
- 137.Clarke SJ, Littleford RE, Smith WE, Goodacre R. Rapid monitoring of antibiotics using Raman and surface enhanced Raman spectroscopy. Analyst. 2005;130(7):1019–1026. doi: 10.1039/b502540k. [DOI] [PubMed] [Google Scholar]
- 138.Alharbi O, Xu Y, Goodacre R. Simultaneous multiplexed quantification of caffeine and its major metabolites theobromine and paraxanthine using surface-enhanced Raman scattering. Analytical and Bioanalytical Chemistry. 2015;407(27):8253–8261. doi: 10.1007/s00216-015-9004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mamian-Lopez MB, Poppi RJ. Standard addition method applied to the urinary quantification of nicotine in the presence of cotinine and anabasine using surface enhanced Raman spectroscopy and multivariate curve resolution. Anal Chim Acta. 2013;760:53–9. doi: 10.1016/j.aca.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 140.Alharbi O, Xu Y, Goodacre R. Simultaneous multiplexed quantification of nicotine and its metabolites using surface enhanced Raman scattering. The Analyst. 2014;139:4820. doi: 10.1039/c4an00879k. [DOI] [PubMed] [Google Scholar]
- 141.Yang T, Guo X, Wang H, Fu S, wen Y, Yang H. Magnetically optimized SERS assay for rapid detection of trace drug-related biomarkers in saliva and fingerprints. Biosensors and Bioelectronics. 2015;68:350–357. doi: 10.1016/j.bios.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 142.Sanles-Sobrido M, Rodriguez-Lorenzo L, Lorenzo-Abalde S, Gonzalez-Fernandez A, Correa-Duarte MA, Alvarez-Puebla RA, Liz-Marzan LM. Label-free SERS detection of relevant bioanalytes on silver-coated carbon nanotubes: The case of cocaine. Nanoscale. 2009;1(1):153–158. doi: 10.1039/b9nr00059c. [DOI] [PubMed] [Google Scholar]
- 143.Riordan CM, Jacobs KT, Negri P, Schultz ZD. Sheath flow SERS for chemical profiling in urine. Faraday Discussions. 2016;187(0):473–484. doi: 10.1039/c5fd00155b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sägmüller B, Schwarze B, Brehm G, Trachta G, Schneider S. Identification of illicit drugs by a combination of liquid chromatography and surface-enhanced Raman scattering spectroscopy. Journal of Molecular Structure. 2003;661–662:279–290. [Google Scholar]
- 145.Trachta G, Schwarze B, Sägmüller B, Brehm G, Schneider S. Combination of high-performance liquid chromatography and SERS detection applied to the analysis of drugs in human blood and urine. Journal of Molecular Structure. 2004;693:175–185. [Google Scholar]
- 146.Inscore F, Shende C, Sengupta A, Huang H, Farquharson S. Detection of drugs of abuse in saliva by surface-enhanced Raman spectroscopy (SERS) Appl Spectrosc. 2011;65(9):1004–8. doi: 10.1366/11-06310. [DOI] [PubMed] [Google Scholar]
- 147.Siddhanta S, Wrobel MS, Barman I. Integration of protein tethering in a rapid and label-free SERS screening platform for drugs of abuse. Chem Commun (Camb) 2016;52(58):9016–9. doi: 10.1039/c6cc00518g. [DOI] [PubMed] [Google Scholar]
- 148.Fabriciova G, Sanchez-Cortes S, Garcia-Ramos JV, Miskovsky P. Surface-enhanced Raman spectroscopy study of the interaction of the antitumoral drug emodin with human serum albumin. Biopolymers. 2004;74(1–2):125–130. doi: 10.1002/bip.20058. [DOI] [PubMed] [Google Scholar]
- 149.Lin M, He L, Awika J, Yang L, Ledoux DR, Li H, Mustapha A. Detection of Melamine in Gluten, Chicken Feed, and Processed Foods Using Surface Enhanced Raman Spectroscopy and HPLC. Journal of Food Science. 2008;73(8):T129–T134. doi: 10.1111/j.1750-3841.2008.00901.x. [DOI] [PubMed] [Google Scholar]
- 150.Wen ZQ, Li G, Ren D. Detection of trace melamine in raw materials used for protein pharmaceutical manufacturing using surface-enhanced Raman spectroscopy (SERS) with gold nanoparticles. Appl Spectrosc. 2011;65(5):514–21. doi: 10.1366/10-06089. [DOI] [PubMed] [Google Scholar]
- 151.Clauson SL, Sylvia JM, Arcury TA, Summers P, Spencer KM. Detection of Pesticides and Metabolites Using Surface-Enhanced Raman Spectroscopy (SERS): Acephate. Appl Spectrosc. 2015;69(7):785–93. doi: 10.1366/14-07594. [DOI] [PubMed] [Google Scholar]
- 152.Carrillo-Carrión C, Simonet BM, Valcárcel M, Lendl B. Determination of pesticides by capillary chromatography and SERS detection using a novel Silver-Quantum dots “sponge” nanocomposite. Journal of chromatography A. 2012;1225:55–61. doi: 10.1016/j.chroma.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 153.Wang W, Xu M, Guo Q, Yuan Y, Gu R, Yao J. Rapid separation and on-line detection by coupling high performance liquid chromatography with surface-enhanced Raman spectroscopy. RSC Adv. 2015;5:47640–47646. [Google Scholar]
- 154.Boca S, Farcau C, Baia M, Astilean S. Metanephrine neuroendocrine tumor marker detection by SERS using Au nanoparticle/Au film sandwich architecture. Biomed Microdevices. 2016;18(1):12. doi: 10.1007/s10544-016-0037-3. [DOI] [PubMed] [Google Scholar]
- 155.De Luca AC, Reader-Harris P, Mazilu M, Mariggiò S, Corda D, Di Falco A. Reproducible Surface-Enhanced Raman Quantification of Biomarkers in Multicomponent Mixtures. ACS Nano. 2014;8(3):2575–2583. doi: 10.1021/nn406200y. [DOI] [PubMed] [Google Scholar]
- 156.Bodelon G, Montes-Garcia V, Lopez-Puente V, Hill EH, Hamon C, Sanz-Ortiz MN, Rodal-Cedeira S, Costas C, Celiksoy S, Perez-Juste I, Scarabelli L, La Porta A, Perez-Juste J, Pastoriza-Santos I, Liz-Marzan LM. Detection and imaging of quorum sensing in Pseudomonas aeruginosa biofilm communities by surface-enhanced resonance Raman scattering. Nat Mater. 2016 doi: 10.1038/nmat4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bell SE, Mackle JN, Sirimuthu NM. Quantitative surface-enhanced Raman spectroscopy of dipicolinic acid--towards rapid anthrax endospore detection. Analyst. 2005;130(4):545–9. doi: 10.1039/b415290e. [DOI] [PubMed] [Google Scholar]
- 158.Zhang X, Young MA, Lyandres O, Van Duyne RP. Rapid Detection of an Anthrax Biomarker by Surface-Enhanced Raman Spectroscopy. Journal of the American Chemical Society. 2005;127(12):4484–4489. doi: 10.1021/ja043623b. [DOI] [PubMed] [Google Scholar]
- 159.Cowcher DP, Xu Y, Goodacre R. Portable, Quantitative Detection of Bacillus Bacterial Spores Using Surface-Enhanced Raman Scattering. Analytical Chemistry. 2013;85(6):3297–3302. doi: 10.1021/ac303657k. [DOI] [PubMed] [Google Scholar]
- 160.Cheung M, Lee WW, Cowcher DP, Goodacre R, Bell SE. SERS of meso-droplets supported on superhydrophobic wires allows exquisitely sensitive detection of dipicolinic acid, an anthrax biomarker, considerably below the infective dose. Chem Commun (Camb) 2016 doi: 10.1039/c6cc03521c. [DOI] [PubMed] [Google Scholar]
- 161.Efrima S, Bronk BV. Silver Colloids Impregnating or Coating Bacteria. The Journal of Physical Chemistry B. 1998;102(31):5947–5950. [Google Scholar]
- 162.Zeiri L, Bronk BV, Shabtai Y, Czégé J, Efrima S. Silver metal induced surface enhanced Raman of bacteria. Colloids and Surfaces A: Physicochemical and Engineering Aspects. 2002;208(1–3):357–362. [Google Scholar]
- 163.Jarvis RM, Goodacre R. Discrimination of Bacteria Using Surface-Enhanced Raman Spectroscopy. Analytical Chemistry. 2004;76(1):40–47. doi: 10.1021/ac034689c. [DOI] [PubMed] [Google Scholar]
- 164.Jarvis RM, Brooker A, Goodacre R. Surface-enhanced Raman scattering for the rapid discrimination of bacteria. Faraday Discuss. 2006;132:281–92. doi: 10.1039/b506413a. discussion 309–19. [DOI] [PubMed] [Google Scholar]
- 165.Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones G, 2nd, Ziegler LD. Characterization of the surface enhanced raman scattering (SERS) of bacteria. J Phys Chem B. 2005;109(1):312–20. doi: 10.1021/jp040442n. [DOI] [PubMed] [Google Scholar]
- 166.Shanmukh S, Jones L, Driskell J, Zhao Y, Dluhy R, Tripp RA. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 2006;6(11):2630–6. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- 167.Lim JY, Nam JS, Yang SE, Shin H, Jang YH, Bae GU, Kang T, Lim KI, Choi Y. Identification of Newly Emerging Influenza Viruses by Surface-Enhanced Raman Spectroscopy. Anal Chem. 2015;87(23):11652–9. doi: 10.1021/acs.analchem.5b02661. [DOI] [PubMed] [Google Scholar]
- 168.Chu H, Huang Y, Zhao Y. Silver Nanorod Arrays as a Surface-Enhanced Raman Scattering Substrate for Foodborne Pathogenic Bacteria Detection. Applied Spectroscopy. 2008;62(8):922–931. doi: 10.1366/000370208785284330. [DOI] [PubMed] [Google Scholar]
- 169.Henderson KC, Sheppard ES, Rivera-Betancourt OE, Choi JY, Dluhy RA, Thurman KA, Winchell JM, Krause DC. The multivariate detection limit for Mycoplasma pneumoniae as determined by nanorod array-surface enhanced Raman spectroscopy and comparison with limit of detection by qPCR. Analyst. 2014;139(24):6426–34. doi: 10.1039/c4an01141d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hennigan SL, Driskell JD, Dluhy RA, Zhao Y, Tripp RA, Waites KB, Krause DC. Detection of Mycoplasma pneumoniae in simulated and true clinical throat swab specimens by nanorod array-surface-enhanced Raman spectroscopy. PLoS One. 2010;5(10):e13633. doi: 10.1371/journal.pone.0013633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Patel IS, Premasiri WR, Moir DT, Ziegler LD. Barcoding bacterial cells: A SERS based methodology for pathogen identification. J Raman Spectrosc. 2008;39(11):1660–1672. doi: 10.1002/jrs.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Boardman AK, Wong WS, Premasiri WR, Ziegler LD, Lee JC, Miljkovic M, Klapperich CM, Sharon A, Sauer-Budge AF. Rapid Detection of Bacteria from Blood with Surface-Enhanced Raman Spectroscopy. Anal Chem. 2016;88(16):8026–35. doi: 10.1021/acs.analchem.6b01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cheng IF, Chang HC, Chen TY, Hu C, Yang FL. Rapid (<5 min) identification of pathogen in human blood by electrokinetic concentration and surface-enhanced Raman spectroscopy. Sci Rep. 2013;3:2365. doi: 10.1038/srep02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Knauer M, Ivleva NP, Liu X, Niessner R, Haisch C. Surface-enhanced Raman scattering-based label-free microarray readout for the detection of microorganisms. Anal Chem. 2010;82(7):2766–72. doi: 10.1021/ac902696y. [DOI] [PubMed] [Google Scholar]
- 175.Knauer M, Ivleva NP, Niessner R, Haisch C. Optimized surface-enhanced Raman scattering (SERS) colloids for the characterization of microorganisms. Anal Sci. 2010;26(7):761–6. doi: 10.2116/analsci.26.761. [DOI] [PubMed] [Google Scholar]
- 176.Knauer M, Ivleva NP, Niessner R, Haisch C. A flow-through microarray cell for the online SERS detection of antibody-captured E. coli bacteria. Anal Bioanal Chem. 2012;402(8):2663–7. doi: 10.1007/s00216-011-5398-0. [DOI] [PubMed] [Google Scholar]
- 177.Premasiri WR, Lee JC, Sauer-Budge A, Theberge R, Costello CE, Ziegler LD. The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS. Anal Bioanal Chem. 2016;408(17):4631–47. doi: 10.1007/s00216-016-9540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.White PL, Hibbitts SJ, Perry MD, Green J, Stirling E, Woodford L, McNay G, Stevenson R, Barnes RA. Evaluation of a Commercially Developed Semiautomated PCR–Surface-Enhanced Raman Scattering Assay for Diagnosis of Invasive Fungal Disease. Journal of Clinical Microbiology. 2014;52(10):3536–3543. doi: 10.1128/JCM.01135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Negri P, Flaherty RJ, Dada OO, Schultz ZD. Ultrasensitive online SERS detection of structural isomers separated by capillary zone electrophoresis. Chemical Communications. 2014;50(21):2707–2710. doi: 10.1039/c3cc49030k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cowcher DP, Jarvis R, Goodacre R. Quantitative online liquid chromatography-surface-enhanced Raman scattering of purine bases. Analytical chemistry. 2014;86:9977–84. doi: 10.1021/ac5029159. [DOI] [PubMed] [Google Scholar]
- 181.Hakonen A, Wang F, Andersson PO, Wingfors H, Rindzevicius T, Schmidt MS, Soma VR, Xu S, Li Y, Boisen A, Wu H. Hand-Held Femtogram Detection of Hazardous Picric Acid with Hydrophobic Ag Nanopillar SERS Substrates and Mechanism of Elasto-Capillarity. ACS Sensors. 2017;2(2):198–202. doi: 10.1021/acssensors.6b00749. [DOI] [PubMed] [Google Scholar]
- 182.Hakonen A, Rindzevicius T, Schmidt MS, Andersson PO, Juhlin L, Svedendahl M, Boisen A, Kall M. Detection of nerve gases using surface-enhanced Raman scattering substrates with high droplet adhesion. Nanoscale. 2016;8(3):1305–8. doi: 10.1039/c5nr06524k. [DOI] [PubMed] [Google Scholar]