Abstract
Calcium plays an integral role to many cellular processes including contraction, energy metabolism, gene expression, and cell death. The inositol 1, 4, 5-trisphosphate receptor (IP3R) is a calcium channel expressed in cardiac tissue. There are three IP 3R isoforms encoded by separate genes. In the heart, the IP3R-2 isoform is reported to being most predominant with regards to expression levels and functional significance. The functional roles of IP3R-1 and IP3R-3 in the heart are essentially unexplored despite measureable expression levels. Here we show that all three IP3Rs isoforms are expressed in both neonatal and adult rat ventricular cardiomyocytes, and in human heart tissue. The three IP3R proteins are expressed throughout the cardiomyocyte sarcoplasmic reticulum. Using isoform specific siRNA, we found that expression of all three IP 3R isoforms are required for hypertrophic signaling downstream of endothelin-1 stimulation. Mechanistically, IP 3Rs specifically contribute to activation of the hypertrophic program by mediating the positive inotropic effects of endothelin-1 and leading to downstream activation of nuclear factor of activated T-cells. Our findings highlight previously unidentified functions for IP 3R isoforms in the heart with specific implications for hypertrophic signaling in animal models and in human disease.
Keywords: Inositol Trisphosphate Receptor (InsP3R); Inositol 1,4,5-Trisphosphate (IP3); Calcium; Cardiac Hypertrophy; Calcium Channel
1. Introduction
In the heart calcium is an essential modulator of a wide variety of cellular functions including cardiomyocyte excitation-contraction coupling (ECC) and gene expression. Cardiomyocyte function can be modulated by neuro-hormonal agonists to accommodate cardiac demand. One example is endothelin-1 (ET-1), which is a potent vasoconstrictor that plays an important role in modulating muscle contractility, vascular tone, cardiomyocyte growth, and survival [1, 2]. Plasma levels of ET-1 are also increased during pathological conditions such as chronic heart failure, myocardial infarction, cardiac hypertrophy and in hypertension [3, 4]. As such, ET-1 has been linked to pathological remodeling of the heart [1, 5]. ET-1 signaling is initiated by ET-1 binding to G-protein coupled receptors at the plasma membrane leading to the activation of phospholipase C (PLC). PLC catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) which leads to increased production of the second messengers inositol 1,4,5-trisphosphate (IP3) and diacylglycerol. IP3 then acts as a second messenger that binds inositol 1,4,5-trisphosphate receptors (IP 3Rs), activating IP3-induced calcium release (IICR).
IP3Rs are a family of calcium channels involved in a variety of cellular functions. There are three different IP3R isoforms encoded by separate genes. The three IP 3Rs share a high degree of sequence homology and are found in a variety of tissues including the heart [6, 7]. Cardiac IP3Rs are implicated in regulating the progression of cardiac hypertrophy [8, 9]. Within the cardiomyocyte, IP3Rs are known to localize in the dyadic cleft, sarcoplasmic reticulum and at the outer/inner nuclear membrane [1, 8–10]. Several lines of evidence have also implicated nuclear calcium transients as a significant contributor to cardiomyocyte hypertrophy. Nuclear or perinuclear IP 3Rs may promote nuclear-restricted calcium release events that initiate gene transcription [10]. Nuclear calcium transients are involved in the activation of transcription factors such as histone deacetylase 5 (HDAC5) [1, 11]. However the mechanism by which cardiomyocytes can discriminate between calcium signals from ECC and calcium signals that target gene transcription it still unclear, as calcium release events mediated by ECC are also efficiently transmitted to the nuclear matrix. Hypertrophic agents such as ET-1 also increase contractility [9, 12], which may afford a mechanism for decoding IP 3-dependent signals without requiring subcellular compartment-specific IP3R activation.
The IP3R-2 isoform is considered the predominant isoform in the heart [13, 14]. However, transgenic IP3R-2 rodent models have either supported [15, 16], or contradicted [17, 18] the role of IP3R-2 channel in cardiac hypertrophy. As such, it is still unclear whether IP 3R channels are significant contributors to cardiac physiology and pathologic remodeling such as hypertrophy [19]. It has been shown that all three IP3R isoforms, at least at the mRNA level, are expressed in the heart of humans and mice [8, 20]. This opens the question of whether IP 3R-1 and -3 are able to functionally compensate for IP3R-2 deficiencies in these models.
We now show that all three IP3R isoforms are expressed in cardiomyocytes and that they are essential for the progression of ventricular hypertrophy induced by ET-1. IP3R-dependent activation of the hypertrophic program was not dependent upon nuclear-specific calcium transients, but instead was mediated by increased contractility induced by ET-1. Lastly, these results were independent of increased IP3R expression both in vitro and in vivo.
2. Materials and Methods
2.1. Antibodies, expression constructs, and reagents
Rabbit polyclonal antibody against type-1 IP3R was developed in-house and is specific for the type-1 isoform [21]. The rabbit polyclonal antibody against type-2 IP3R have been described elsewhere [6] and was kindly provided by Dr. Richard Wojcikiewicz (SUNY Upstate). Mouse monoclonal anti-IP3R type-3 was purchased from BD Bioscience. Mouse monoclonal anti-α-actinin and anti-ryanodine receptor antibody was purchased from Sigma-Aldrich. Mouse anti-SERCA2 antibody was from Thermo Fisher and rabbit anti-ANP was purchased from Abcam. Anti-alpha-fodrin was purchased from EMD Millipore. Secondary antibodies conjugated to Alexa-488 and Alexa-555 were from Molecular Probes, and peroxidase-conjugated antibodies were from Jackson ImmunoResearch. Expression constructs 9X NFAT-apha-MHC-Luc was a gift from Jeffery Molkentin (Addgene plasmid # 51941), pGP-CMV-GCaMP6s was a gift from Douglas Kim ([22]; Addgene plasmid # 40753) and Tol2-elavl3-H2B-GCaMP6s was a gift from Misha Ahrens (Addgene plasmid # 59530). Endothelin-1 was purchased from Bachem. Silencer pre-design siRNAs were purchased from Ambion. Fura-2 AM was purchased from Molecular Probes, and the dual luciferase reporter assay kit was from Promega. All other reagents were purchased from Sigma -Aldrich.
2.2. Preparation of primary neonatal cardiomyocytes
Neonatal rat ventricular cardiomyocytes (NRVM) were obtained from 1 to 2 day old Sprague-Dawley rat hearts as previously described, with minor modifications [23]. Cardiomyocytes were plated into fibronectin-coated culture dishes and incubated at 37°C in 5% CO2 incubator. Two days after plating, media was replaced with 50% Ham’s F10 - 50% DMEM culture medium with β-D-arabinofuranoside (ARA-C; 1 μM) to inhibit growth of fibroblasts. NRVMs were transfected using Lipofectamine 3000 following manufacturer’s instructions. All experiments were carried out 48 hours after transfection unless otherwise stated. All vertebrate animal procedures were approved by the Animal Welfare Committee (AWC) at UTHealth.
2.3. Preparation of adult cardiomyocytes
Calcium-tolerant adult rat ventricular myocytes (ARVM) were isolated from hearts of wildtype Sprague-Dawley rats (300–320 g) as described by Louch et. al. [24]. Briefly, animals were anesthetized with chloralhydrate (400mg/kg b.w. i.p.) and heparinized (5,000 U/kg b.w.) via direct injection into the vena cava inferior. The hearts were aseptically removed and directly placed in ice-cold Krebs-Henseleit (KH) buffer (133.5 mM NaCl, 4 mM KCl, 1.2 mM NaH2PO4, 10 mM HEPES, 1.2 mM MgSO4, 10 mM BDM) containing glucose (5.5 mM) before being perfused with a Langendorff preparation. Perfusion (3 min) with KH buffer at 37°C lacking Ca2+ was followed by perfusion with recirculating KH buffer containing 2% BSA (wt/vol), 50 μM Ca2+ and type II collagenase. After 20 minutes of perfusion, hearts were minced, and undigested tissue was separated with a 230 μm mesh sieve. The cell suspension was allowed to settle with gravity within 5 to 7 min, and the cell pellet was re-suspended in KH containing 2% BSA (wt/vol), and calcium was slowly reintroduced to a final concentration of 1 mM. Cardiomyocytes were plated and culture under 5% carbon dioxide.
2.4. Immunofluorescence labeling of isolated cardiomyocytes
Adult and neonatal cardiomyocytes were plated at a density of 300 cells per mm2 on glass coverslips coated with fibronectin. On day 4 the medium was exchanged and the cells were treated with 100 nM ET-1 for 48 hrs. Adult cardiomyocytes cells were fixed with 4% paraformaldehyde in PBS. Neonatal cardiomyocytes were fixed with 100% cold MeOH. Briefly, cells were incubated with either rabbit polyclonal anti-IP3R-1 (1:250), rabbit polyclonal anti-IP3R-2 (1:250), mouse monoclonal anti-IP3R-3 (1:250), mouse monoclonal anti-α-actinin (1:250), mouse anti-SERCA2 (1:250), or mouse anti-ryanodine2 (1:250), overnight at 4°C. Followed by incubation with secondary antibodies conjugated to Alexa-488 and Alexa-555 for 1 hr.
2.5. Immunofluorescent labeling of human heart failure samples
Disease heart tissue was obtained from patients undergoing heart transplantation due to advanced heart failure. Immediately after explant the tissue is flash frozen with liquid nitrogen for future analyses. Control heart tissue was obtained from organs that were declined for transplantation due to non-cardiac reasons. Frozen left ventricular cardiac tissues were cryo-sectioned onto charged glass slides. The sections were fixed with 4% paraformaldehyde in PBS. Tissue was stained with anti-IP3R-1 (1:100), anti-IP3R-2 (1:100), anti-IP3R-3 (1:100), or anti-α-actinin (1:100), for 1hr at 37°C. Subsequently, slides were washed and incubated with secondary antibodies conjugated to Alexa-488 and Alexa-555 for 1 hr. Total corrected tissue fluorescence was quantified using ImageJ exactly as described previously [25]. All experiments on human samples were approved by the Institutional Review Board (IRB) at the Houston Methodist Institute for Academic Medicine and the McGovern Medical School at UTHealth.
2.6. Cell size determination
NRVM were plated on glass coverslips. Two days after plating cells were transfected with control siRNA or triple IP3R siRNA. Following transfection cells were treated with ET-1 for 48 hours. After treatment cells were fixed with 4% paraformaldehyde in PBS. Subsequently, coverslips were stained with anti-α-actinin and secondary antibodies conjugated to Alexa-555. For measurement of cell area, at least 30 fields randomly chosen were analyzed in each coverslip. Cardiomyocytes area was measured in captured images using ImageJ software.
2.7. Western blotting
Cells were harvested by gently scraping plates with a cell scraper and washing once with cold PBS. Cell lysis buffer (150 mM NaCl, 50 mM Tris-HCl at pH 7.8, 1% Triton X-100 and 1 mM EDTA) was added to the cell pellet. Samples were cleared of insoluble debris by centrifugation at 20,000g at 4°C. Cell lysates were quenched with SDS sample buffer. Samples were resolved by SDS–PAGE and analyzed by western blotting. Where indicated, cardiomyocytes were treated 100 nM ET-1 for 48 hours or six days.
2.8. Calcium imaging
NRVM were plated on fibronectin-coated glass coverslips and were transfected with triple IP 3R siRNA targeting rat IP3R-1, IP3R-2 and IP3R-3. The total amount of siRNA transfected was 12.5 pmol per 350,000 cells. Transfected cells were identified by co-transfection with cDNA for YFP (0.25pmol per 350,000 cells). Cells were imaged after 48 hrs. For imaging, cardiomyocytes were incubated with 5 μM Fura-2 AM in imaging solution (1% BSA, 107mM NaCl, 20 mM HEPES, 2.5 mM MgCl2, 7.5 mM KCl, 11.5mM glucose, and 1 mM CaCl2) for 30 min at RT. The solution was replaced with imaging solution without Fura-2 AM for an additional 20 min. Images were acquired using a Nikon TiS inverted microscope as previously described [26]. Responses to 100 nM ET-1 were recorded on YFP only positive cells. In order to specifically look at nuclear calcium transients we used GCaMP6s fused to a sequence encoding human histone H2b at the 5′ end[27]. We sub-cloned H2b-GCAMP6s into the mammalian expression vector pcDNA 3.1(+).NRVM were transfected with different siRNAs (IP 3R-1, IP3R-2 or IP3R-3) and with H2b-GCAMP6s two days after plating. Cells were loaded with Fura-2 AM in imaging solution 48 hrs after transfection. Responses to 100nM ET-1 were acquired at 1 Hz during continuous recording. Oscillation frequency was determined manually. An oscillation was counted when the Fura-2 ratio rose 10% above the baseline ratio. Similar to Fura-2, an oscillation was counted when GCaMP6s fluorescence rose above 5% from baseline fluorescence.
2.9. NFAT luciferase
For assessment of NFAT activation cells were co-transfected with 9xNFAT-TATA luciferase plasmid [28] and pRL-TK control vector (2:1). All experiments were performed 48 hours after transfection. Cells were harvested and cell extracts were assayed using dual luciferase reporter assay as specified by manufacturer’s protocol (Promega). All data are shown as mean ± SEM, with statistical significance determined at p< 0.01 vs control using an unpaired Student’s t-test.
3. Results
3.1 Expression of IP3R protein in neonatal and adult ventricular cardiomyocytes
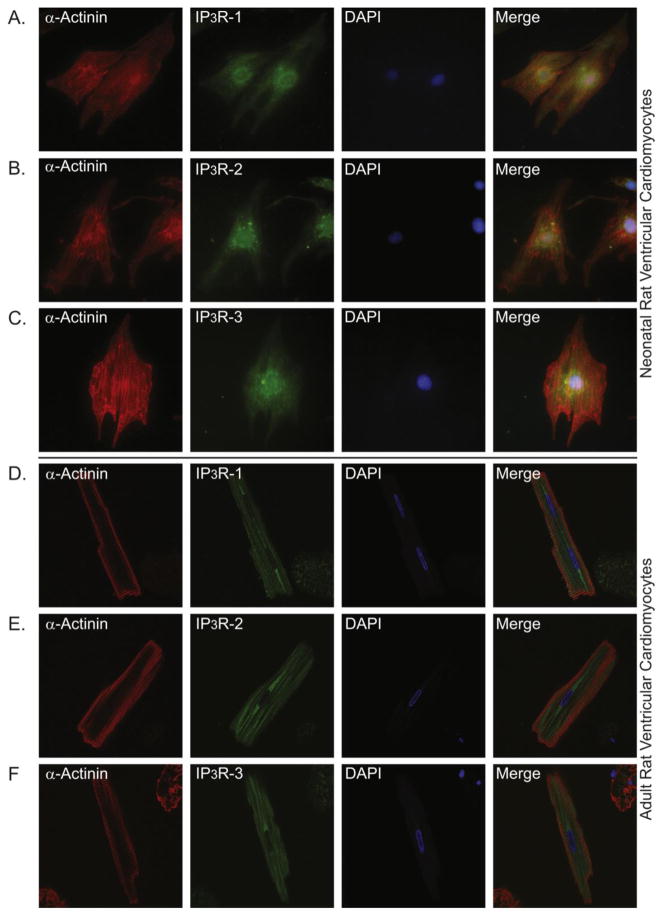
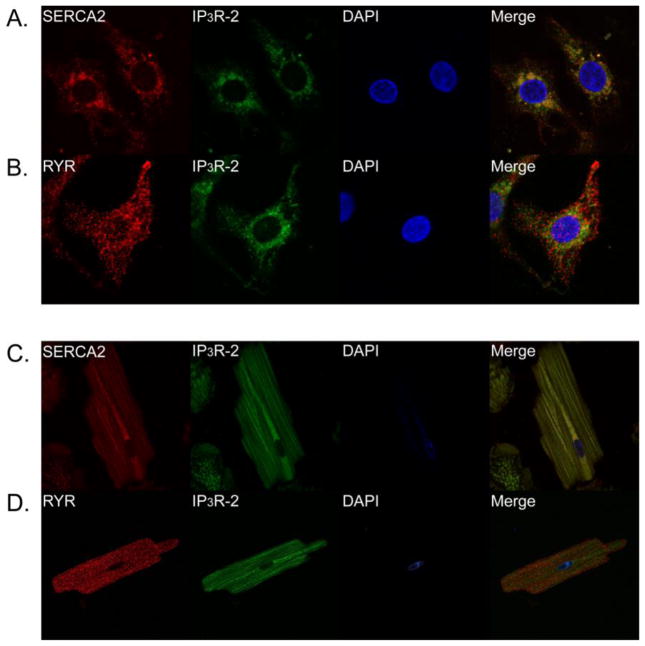
Previous studies have indicated that IP 3R-2 mRNA is predominant compared to other IP 3R isoforms in ventricular cardiomyocytes [13, 14]. Using highly specific antibodies we analyzed the protein expression and localization of the three IP 3R isoforms in primary neonatal and adult rat cardiomyocytes. In neonatal ventricular cardiomyocytes all three IP 3R isoforms were expressed throughout the cell (Fig. 1A–C, second column; Supplemental Fig. 1). Expression was more prominent in the perinuclear region consistent with previous reports [1]. Similarly, all three IP3R isoforms are expressed throughout the cell in adult rat cardiomyocytes (Fig. 1D–F, second column). In contrast to neonatal ventricular cardiomyocytes, IP3R expression in adult cardiomyocytes was more equally distributed throughout the cell with no obvious concentration in the perinuclear region. This expression pattern overlaps with SERCA2 localization indicating is it present throughout the sarcoplasmic reticulum in both neonatal and adult cardiomyocytes (Fig. 2A, C). As expected, IP3R localization was distinct from the localization with RYR ([1]; Fig. 2B, D; Supplemental Fig. 2). These results indicate that in addition to IP3R-2, IP3R isoforms type 1 and 3 are expressed in rat neonatal and adult ventricular cardiomyocytes.
Fig. 1. Expression and distribution IP3R isoforms in primary neonatal and adult ventricular cardiomyocytes.
Immunofluorescence staining of neonatal (rows A–C) and adult (rows D–F) ventricular cardiomyocytes. Column 1 is stained with α-actinin to label sarcomeres. Column 2 is stained with indicated IP3R antibodies. Column 3 is DAPI staining of the nucleus. Column 4 is the merged images of each row. See also Supplemental Fig. 1 for larger versions of the merged images.
Fig. 2. Expression and distribution of IP3R, SERCA2, and RYR in primary ventricular cardiomyocytes.
Immunofluorescence staining of neonatal ventricular cardiomyocytes stained with SERCA2 and IP3R-2 (A); RYR2 and IP3R-2 (B). Immunofluorescence staining of adult ventricular cardiomyocytes with SERCA2 and IP3R-2 (C); RYR2 and IP3R-2 (D). See also Supplemental Fig. 2 for larger versions of the merged images.
3.2 Altered contractility induced by ET-1 is attenuated by decreasing IP3R expression
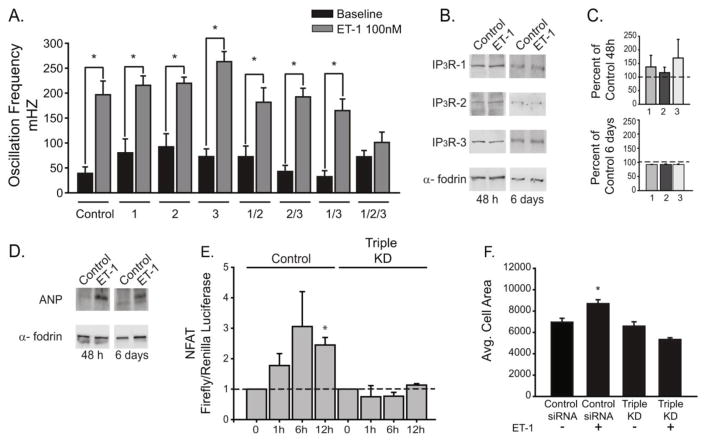
ET-1 is known act as a positive inotropic agent in cardiomyocytes [9, 12]. We monitored intracellular calcium in spontaneously contracting neonatal ventricular cardiomyocytes treated with ET-1. As shown in Fig. 3A (control bars) there is an increase in the calcium oscillation frequency in neonatal cardiomyocytes stimulated with 100 nM ET-1. Previous studies using overexpression of an IP3 5′-phosphatase to inhibit IP3 signaling suggested that IP 3R channels do not play a role in this response to ET-1 [1]. To evaluate the role of IP 3R channels in the process more specifically, we used siRNA knockdown of each individual IP3R isoform. Western blot analysis confirmed efficient and specific knockdown of each of the three IP3R isoforms (Supplemental Fig. S3). Knockdown of individual IP 3R isoforms did not have significant effects on the increased oscillation frequency induced by ET-1 stimulation (Fig. 3A). Next we used double knockdowns (IP3R 1/2, 2/3 and 1/3) and determined their effect on the response to ET-1. Similar to single knockdowns, in double knockdowns there is no significant effects on the increased oscillation frequency induced by ET-1 stimulation. Triple knockdown of all three IP3R isoforms completely suppressed the response of cardiomyocytes to ET-1 (Fig. 3A). Thus, the increased contractility in response to ET-1 requires IP3R activity, and furthermore all three IP 3Rs are expressed and play a functionally redundant role in the response to ET-1 stimulation.
Fig. 3. All three IP3R isoforms contribute to hypertrophic signaling by ET-1.
(A) Neonatal cardiomyocytes were transfected with control or IP 3R siRNA as indicated. Cells were treated with ET-1 and oscillation frequency was quantified from at least 4 separate experiments (control n=6, 1 n=6, 2 n=7, 3 n=6, 1/2 n=6, 2/3 n=5, 1/3 n=5 and 1/2/3 n=4). (B). Western blot of indicated proteins after 48 hours or 6 days treatment with 100 nM ET-1. (C). Quantification of IP 3R levels after ET-1 treatment for 48 hours or 6 days expressed as percent of control. (D) Atrial natriuretic peptide (ANP) levels after 48 hours and 6 days treatment with ET-1. Blotting with alpha-fodrin was used as control. (E) NFAT activity using luciferase reporter construct after ET-1 treatment for the indicated times in control siRNA and triple IP3R siRNA transfected cardiomyocytes. (F) Average cell area before and after 48 hours treatment with ET-1. Neonatal cardiomyocytes were transfected with control siRNA and triple IP 3R siRNA. All data are shown as mean ± SEM, *P < 0.01 vs control.
3.3 Attenuating IP3R expression inhibits ET-1 induced NFAT activation and hypertrophy
At the cellular level cardiac hypertrophy is characterized by activation of the hypertrophic transcriptional program by NFAT ultimately leading to increased cardiomyocyte cell size. ET-1 is one of the best described neuro-hormonal factors known to induce cardiomyocyte hypertrophy [10, 16]. ET-1 also induces hypertrophic remodeling of neonatal rat ventricular cardiomyocytes [1, 29], and thus we used this well-characterized model to assess the role of IP 3Rs in hypertrophic signaling. As mentioned previously, IP3R mRNA levels are altered during hypertrophy [7, 13], however the effect on IP 3R protein levels is not clear. In order to assess whether IP 3R protein expression is altered during ET-1 induced hypertrophy, we treated neonatal cardiomyocytes with ET-1 for 48 hours and 6 days. Using this paradigm, protein expression of all three IP 3R isoforms is unchanged compared to non-treated controls (Fig. 3B–C) despite robust induction of the hypertrophic marker protein atrial natriuretic peptide (ANP; Fig. 3D).
Next we determined whether IP 3R expression is essential for activation of the hypertrophic program downstream of ET-1 stimulation. Treatment of cells with ET-1 led to significant NFAT activation within 12 hours as determined by a luciferase reporter assay (Fig. 3E). Triple knockdown of all three IP3R isoforms completely abrogated NFAT activation in response to ET-1 stimulation (Fig. 3E), a finding consistent with our observation that triple IP 3R knockout suppresses the increased calcium oscillation frequency in response to ET-1 (Fig. 3A). Treatment of cardiomyocytes with ET-1 leads to an increase in cell size, the cardinal feature of hypertrophy ([1, 5]; Fig. 3F). Triple knockdown of all three IP3R isoforms completely abrogates the increased cell size induced by to ET-1 treatment as shown in Fig. 3F and Supplemental Fig. S5. Thus, all three IP3Rs contribute to signaling downstream of ET-1 in cardiomyocytes and are essential for activation of the hypertrophic program induced by ET-1 in vitro.
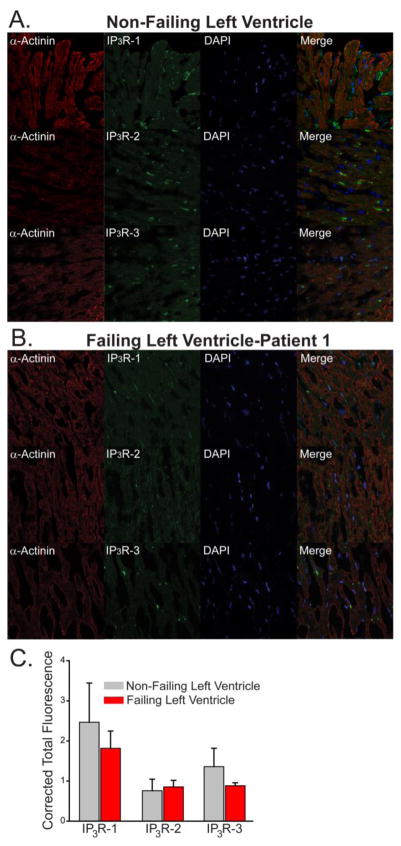
It has also been shown that IP3R mRNA levels are altered in both animal models [9, 17] and in the human failing heart [8]. To determine whether IP3R expression levels are altered in vivo, we compared IP3R expression levels in heart tissue samples from three patients diagnosed with dilated cardiomyopathy to biopsies of left ventricular heart tissue obtained from three healthy human subjects (non-failing hearts). Patients diagnosed with dilated cardiomyopathy were male, 60±3.5 years old, with a reduced ejection fraction of 20%, and with an increased left ventricular end diastolic diameter (LVEDD) and left ventricular pressure (LVP, 1.10±0.44) as assessed by transthoracic echocardiography (Supplemental Table 1). Protein expression of all three IP 3R isoforms could be readily detected in the three human heart samples. We found no significant differences in the protein expression level of IP3R isoforms in human failing hearts relative to control (Fig. 4 and Supplementary Fig. S4).
Fig. 4. Expression of IP3R isoforms in non-failing and end stage heart failure samples.
Immunofluorescence staining of non-failing human left ventricle (A). Left ventricular heart failure patient 1(B). Column 1 is stained with α-actinin to label sarcomeres. Column 2 is stained with indicated IP 3R antibodies. Column 3 is DAPI staining of the nucleus. Column 4 is the merged images of each row. (C) Quantified immunofluorescence from three control and three failing heart samples (see Supplemental Fig. S4 for additional staining).
3.4 Calcium release induced by ET-1 is not restricted to the nuclear compartment
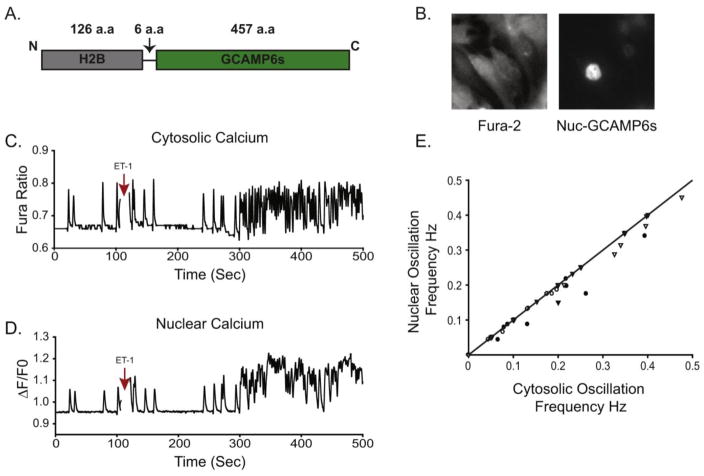
Rhythmic contraction of the heart is mediated by intracellular RyR calcium channels in a process termed calcium-induced calcium release (CICR). An important question is how IP3R-mediated calcium transients could be “decoded” in the constant background of CICR in the beating heart. One prevalent model is that nuclear-compartment specific IP 3R calcium transients mediate gene expression in response to hypertrophic agents [10]. However, as CICR calcium transients also diffuse into the nucleus the specific mechanism is not clear. Furthermore, NFAT is a cytosolic protein in the inactive state, and translocates to the nucleus in response to elevations in cytosolic calcium [5]. To resolve these discrepancies, we used the genetically encoded calcium indicator GCaMP6s targeted to the nucleus in order to measure nuclear calcium transients unambiguously (Fig. 5A–B). Using the spectrally separated indicator Fura-2 to measure cytosolic calcium, we were able to quantify calcium release in both nuclear and cytosolic compartments with high specificity. As shown in Fig. 5C–D, calcium release was detectable in both compartments of spontaneously beating cardiomyocytes. After ET-1 stimulation there is an increase in the oscillation frequency consistent with the positive inotropic effects of ET-1 (Fig. 5C–D), which we showed is absolutely dependent upon IP 3R expression (Fig. 3A). Visual observation of Fura-2 versus H2b-GCaMP6s traces revealed essentially overlapping plots (Fig. 5C–D), however the H2b-GCaMP6s signal had lower resolving power at higher oscillation frequencies. Expression of cytosolic GCaMP6s revealed that this was due to a buffering effect of the indicator, not an intrinsic property of the nuclear matrix (Fig. S6). In order to compare the cytosolic versus nuclear oscillations more quantitatively, we calculated the oscillation frequency over the course of the experiment in 20 second bins in both the cytosolic and nuclear compartments (Fig. 5E). Data points above the line in Fig. 5E would indicate a higher frequency in the nucleus (thus highlighting nuclear only events). Conversely, data points below the line are representative of events that occur in the cytosol but were not detectable in the nucleus. Presented in this way, we were unable to visualize any “nuclear only” release event either before or after ET-1 stimulation. We did, however, note a few “cytosol only” events at various frequencies. This may be due to either a cytosol restricted transient or insufficient sensitivity of the GCaMP6s sensor. Regardless, if there are nuclear restricted calcium transients they are either rare or low amplitude events. The results presented herein are also entirely consistent with the large body of evidence indicating that NFAT is activated by changes in calcium release frequency, not amplitude [30–32].
Fig. 5. Nuclear and cytosolic calcium in response to ET-1 stimulation.
(A) Schematic cartoon of H2B-GCaMP6s. (B) Representative image of neonatal cardiomyocytes expressing H2B-GCaMP6s loaded with Fura-2 AM. (C) Single cell imaging of a neonatal cardiomyocyte treated with 100 nM ET-1 at indicated time. (D) H2b-GCaMP6s signal in the same cell as in C. (E) Plot of cytosolic vs. nuclear oscillation frequency. Each symbol represents a single coverslip averaging 5–10 cells for a total of four separate coverslips. Frequency data was quantified from 20 seconds bins.
4. Discussion
IP3Rs likely play a key role in the progression of hypertrophy [1, 9, 16]. However, the majority of the studies on the role of IP 3Rs in hypertrophy have focused solely on IP3R-2, despite the fact that the other two isoforms are also expressed in the heart [8, 17, 20]. We now show that all three IP3R channels are expressed at readily detectable levels in cardiomyocytes in both rodent and human heart. Furthermore, all three channels contribute to calcium release and activation of the hypertrophic program in a functionally redundant manner. Lastly, our findings indicate that IP3Rs contribute to activation of the hypertrophic program after ET-1 stimulation primarily by mediating the increase in calcium release frequency/contractility. We do not find evidence, at least in this model, for nuclear-specific calcium events.
Our finding that IP3Rs play functionally redundant roles in ventricular cardiomyocytes may resolve some controversies regarding transgenic models of IP 3R function in the heart. Previously, the IP3R-2 was thought to be the predominant isoform expressed in the heart. Consequently, most of the studies that focus on the role of IP 3R in the heart have focused solely on IP3R-2 function [13, 33]. However, global genetic knockout of the IP3R-2 in mice does not cause any significant difference in the hypertrophic response in pressure overload or dilated cardiomyopathy mouse models [17]. However, the potential involvement of IP3R-1 and IP3R-3 activity has not been investigated further. Another group has shown that global knockout of IP3R-2 eliminates the positive inotropic effects of endothelin-1 (ET-1) in the atria and protects against arrhythmias [15]. Overexpression of the ligand binding domain of the IP 3R inhibits signaling through the channel by buffering IP 3 levels (so called “IP3 sponge” [34]). Targeted inducible overexpression of the IP 3 sponge in the heart inhibited hypertrophy in response to isoproterenol and angiotensin-II [16]. As the IP3 sponge would inhibit signaling through all three IP3R channels, these findings are consistent with our results suggesting that all three IP 3Rs can contribute to hypertrophic signaling in cardiomyocytes. Similarly, another study using overexpression of an IP3 5′-phosphatase (presumably INPP5A) demonstrated that IP3 signaling is required for the hypertrophic effects of ET-1 in neonatal rat ventricular myocytes [1]. Interestingly, overexpression of this enzyme did not inhibit the increased beating frequency in response to ET-1 treatment [1]. One possible explanation for this discrepancy with our results in Fig. 3 is that this enzyme is effective in buffering long-term elevation of IP3 levels (such as during chronic exposure to ET-1) but is not as effective at metabolizing IP3 after acute exposure to agonist. This explanation is consistent with other studies indicating that IP3 5′-phosphatases becomes the dominant mediator of IP 3 metabolism only at high IP3 concentrations [35, 36].
The subcellular localization and precise aspects of how IP 3Rs regulate spatio-temporal aspects of calcium release in the contracting myocyte is an area of debate. In particular, how IP 3R signals are decoded to regulate processes such as transcriptional activation in the beating cardiomyocyte is unclear. IP3Rs are known to be primarily localized at the ER/SR membranes in most cells, however in the heart it is thought to be concentrated at the nuclear and perinuclear membranes, where it is thought to play a key role in gene transcription [1, 10, 11, 37]. Nuclear localized IP 3R would thus facilitate spatially restricted calcium transients to the nuclear matrix. It has been shown that IP 3 and ET-1 can trigger nuclear calcium sparks and nuclear localized calcium transients [10, 37–39], however it is unclear whether these transients originate from the cytosol/perinuclear area. We show now that all three IP3Rs are expressed throughout the SR of the cardiomyocyte in both isolated rat cardiomyocytes and human tissue. Using nuclear-localized GCaMP6s to unambiguously monitor nuclear calcium, our results simultaneously imaging nuclear and cytosolic calcium indicate that nuclear-only calcium transients, if present, are an exceedingly rare events. This is consistent with a study by Nakao et. al. using chelators and targeted parvalbumin which concluded that nuclear calcium signals in cardiomyocytes after electrical stimulation originate in the cytoplasm [29]. This report also compared cytosolic and nuclear release events induced by the IP3-coupled agonist IGF-1. Nuclear release events were of higher amplitude compared to cytosolic events as measured with the genetically encoded calcium sensor GECO1 [29]. The ability of nuclear and cytosolic calcium chelators to block these events were not investigated, so it is unclear whether IGF-1 can specifically stimulate nuclear or perinuclear calcium release via the IP3R. Similar to our results Nakao et. al. also observed that the IP3R is mainly localized at the perinuclear membrane in NRVMs where it interacts with NCS-1 [29]. Thus, a possible explanation for the difference in calcium peak amplitude observed in the nucleus after IGF-1-induced calcium release is that perinuclear localization of the IP3R may allow calcium to diffuse rapidly into the nucleus. Consistent with the results of Nakao et. al. we have also observed that agonist-induced calcium release is able to induce cytosolic calcium oscillations (Fig. 5) which is relevant to the activation of different key transcription factors. Further work will be necessary to evaluate the possibility that IGF-1, ET-1, or other IP3-coupled agonists can activate a pool of nuclear or perinuclear receptors to elicit nuclear-specific calcium transients.
GCaMP6s has been shown to be more sensitive to calcium than other genetically encoded calcium indicators and synthetic calcium dyes such as Fura-2 [22, 40]. One limitation of GCaMP6s is the relatively slow kinetics which could preclude the possibility of imaging fast calcium release events [40]. However, using cytosolic GCaMP6s we can detect cytosolic events not detected by Fura-2 (Fig. S6). The superior sensitivity of GCaMP6s have been used to detect cytosolic and nuclear restricted calcium transients [27]. However, it remains entirely possible that there are low amplitude calcium release events in the nucleus which are below the detection capability of GCaMP6s. As it is highly likely that more sensitive genetically encoded calcium indicators will be developed, this possibility can be tested in future studies. Another limitation of our study is the use of neonatal rat ventricular cardiomyocytes (NRVMs) as a model of hypertrophy. Although they display many characteristics of hypertrophy in vitro, they do not precisely reflect the response of adult cardiomyocytes in vitro or in vivo. Neonatal and adult cardiomyocytes are well known to have differences in calcium-handling properties due to obvious differences in T-tubule/SR morphology and physiology. Interestingly we found that the perinuclear localization of the IP3R was much less prominent in adult cardiomyocytes when compared to NRVMs. This is strongly suggestive that the spatio-temporal pattern of calcium release in adult cardiomyocytes would be very different compared to NRVMs. Indeed, this has been intensively investigated (reviewed in [41]), however the remodeling of IP3R signaling in the developing myocyte is still at an early stage of understanding [42]. Thus it will be important in future studies to compare the nuclear versus cytosolic release events due to IP3R activation in adult cardiomyocytes. It would also be interesting to examine in vivo models such as cardiac-specific knockout of each of the three IP3Rs in rodent models of heart failure. Finally, it has also become increasingly apparent that epigenetics plays a major role in cardiac hypertrophy, and this likely contributes to widespread transcriptional changes leading to fetal reprogramming [43–45]. It would also be of interest in future studies to examine how calcium/IP3 signaling pathways are regulated by epigenetic mechanism
Supplementary Material
Highlights.
All three IP3R isoforms are expressed in rat ventricular cardiomyocytes and in human heart tissue.
All three IP3R isoforms are required for ET-1 induced hypertrophic signaling.
Calcium release induced by ET-1 is not restricted to the nuclear compartment in rat ventricular cardiomyocytes.
Acknowledgments
We would like to thank Dr. Richard Wojcikiewicz (SUNY Upstate) for his generous gift of IP3R-2 antisera. We would also like to thank Dr. Shane Cunha (McGovern Medical School at UTHealth) for advice regarding primary cardiomyocyte preparation. Lastly, we would like to thank Ann-Bin Shyu (McGovern Medical School at UTHealth) for assistance with the luciferase assays. This work was support by NIH grants R01GM081685 (DB) a Research Supplement to Promote Diversity in Health-Related Research on the same grant (to MIG), R01HL61483 (HT), Friede Springer Herz Stiftung (AK), and the Roderick MacDonald Research Fund 15RDM005 (AK). This work was also supported by startup funds provided by the McGovern Medical School at UTHealth (DB).
Footnotes
Disclosures
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Higazi DR, Fearnley CJ, Drawnel FM, Talasila A, Corps EM, Ritter O, McDonald F, Mikoshiba K, Bootman MD, Roderick HL. Endothelin-1-stimulated InsP3-induced Ca2+ release is a nexus for hypertrophic signaling in cardiac myocytes. Mol Cell. 2009;33(4):472–82. doi: 10.1016/j.molcel.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Shubeita HE, McDonough PM, Harris AN, Knowlton KU, Glembotski CC, Brown JH, Chien KR. Endothelin induction of inositol phospholipid hydrolysis, sarcomere assembly, and cardiac gene expression in ventricular myocytes. A paracrine mechanism for myocardial cell hypertrophy. J Biol Chem. 1990;265(33):20555–62. [PubMed] [Google Scholar]
- 3.Stewart DJ, Kubac G, Costello KB, Cernacek P. Increased plasma endothelin-1 in the early hours of acute myocardial infarction. J Am Coll Cardiol. 1991;18(1):38–43. doi: 10.1016/s0735-1097(10)80214-1. [DOI] [PubMed] [Google Scholar]
- 4.Yorikane R, Sakai S, Miyauchi T, Sakurai T, Sugishita Y, Goto K. Increased production of endothelin-1 in the hypertrophied rat heart due to pressure overload. FEBS Lett. 1993;332(1–2):31–4. doi: 10.1016/0014-5793(93)80476-b. [DOI] [PubMed] [Google Scholar]
- 5.Colella M, Grisan F, Robert V, Turner JD, Thomas AP, Pozzan T. Ca2+ oscillation frequency decoding in cardiac cell hypertrophy: role of calcineurin/NFAT as Ca2+ signal integrators. Proc Natl Acad Sci U S A. 2008;105(8):2859–64. doi: 10.1073/pnas.0712316105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wojcikiewicz RJ. Type I, II, and III inositol 1,4,5-trisphosphate receptors are unequally susceptible to down-regulation and are expressed in markedly different proportions in different cell types. J Biol Chem. 1995;270(19):11678–83. doi: 10.1074/jbc.270.19.11678. [DOI] [PubMed] [Google Scholar]
- 7.Perez PJ, Ramos-Franco J, Fill M, Mignery GA. Identification and functional reconstitution of the type 2 inositol 1,4,5-trisphosphate receptor from ventricular cardiac myocytes. J Biol Chem. 1997;272(38):23961–9. doi: 10.1074/jbc.272.38.23961. [DOI] [PubMed] [Google Scholar]
- 8.Signore S, Sorrentino A, Ferreira-Martins J, Kannappan R, Shafaie M, Del Ben F, Isobe K, Arranto C, Wybieralska E, Webster A, Sanada F, Ogórek B, Zheng H, Liu X, Del Monte F, D’Alessandro DA, Wunimenghe O, Michler RE, Hosoda T, Goichberg P, Leri A, Kajstura J, Anversa P, Rota M. Inositol 1, 4, 5-trisphosphate receptors and human left ventricular myocytes. Circulation. 2013;128(12):1286–97. doi: 10.1161/CIRCULATIONAHA.113.002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harzheim D, Movassagh M, Foo RS, Ritter O, Tashfeen A, Conway SJ, Bootman MD, Roderick HL. Increased InsP3Rs in the junctional sarcoplasmic reticulum augment Ca2+ transients and arrhythmias associated with cardiac hypertrophy. Proc Natl Acad Sci U S A. 2009;106(27):11406–11. doi: 10.1073/pnas.0905485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arantes LA, Aguiar CJ, Amaya MJ, Figueiró NC, Andrade LM, Rocha-Resende C, Resende RR, Franchini KG, Guatimosim S, Leite MF. Nuclear inositol 1,4,5-trisphosphate is a necessary and conserved signal for the induction of both pathological and physiological cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2012;53(4):475–86. doi: 10.1016/j.yjmcc.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Zhang T, Bossuyt J, Li X, McKinsey TA, Dedman JR, Olson EN, Chen J, Brown JH, Bers DM. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J Clin Invest. 2006;116(3):675–82. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proven A, Roderick HL, Conway SJ, Berridge MJ, Horton JK, Capper SJ, Bootman MD. Inositol 1,4,5-trisphosphate supports the arrhythmogenic action of endothelin-1 on ventricular cardiac myocytes. J Cell Sci. 2006;119(Pt 16):3363–75. doi: 10.1242/jcs.03073. [DOI] [PubMed] [Google Scholar]
- 13.Lipp P, Laine M, Tovey SC, Burrell KM, Berridge MJ, Li W, Bootman MD. Functional InsP3 receptors that may modulate excitation-contraction coupling in the heart. Curr Biol. 2000;10(15):939–42. doi: 10.1016/s0960-9822(00)00624-2. [DOI] [PubMed] [Google Scholar]
- 14.Anger M, Lompré AM, Vallot O, Marotte F, Rappaport L, Samuel JL. Cellular distribution of Ca2+ pumps and Ca2+ release channels in rat cardiac hypertrophy induced by aortic stenosis. Circulation. 1998;98(22):2477–86. doi: 10.1161/01.cir.98.22.2477. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Zima AV, Sheikh F, Blatter LA, Chen J. Endothelin-1-induced arrhythmogenic Ca2+ signaling is abolished in atrial myocytes of inositol-1,4,5-trisphosphate(IP3)-receptor type 2-deficient mice. Circ Res. 2005;96(12):1274–81. doi: 10.1161/01.RES.0000172556.05576.4c. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama H, Bodi I, Maillet M, DeSantiago J, Domeier TL, Mikoshiba K, Lorenz JN, Blatter LA, Bers DM, Molkentin JD. The IP3 receptor regulates cardiac hypertrophy in response to select stimuli. Circ Res. 2010;107(5):659–66. doi: 10.1161/CIRCRESAHA.110.220038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooley N, Ouyang K, McMullen JR, Kiriazis H, Sheikh F, Wu W, Mu Y, Du XJ, Chen J, Woodcock EA. No contribution of IP3-R(2) to disease phenotype in models of dilated cardiomyopathy or pressure overload hypertrophy. Circ Heart Fail. 2013;6(2):318–25. doi: 10.1161/CIRCHEARTFAILURE.112.972158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santulli G, Xie W, Reiken SR, Marks AR. Mitochondrial calcium overload is a key determinant in heart failure. Proc Natl Acad Sci U S A. 2015;112(36):11389–94. doi: 10.1073/pnas.1513047112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodcock EA, Matkovich SJ. Ins(1,4,5)P3 receptors and inositol phosphates in the heart-evolutionary artefacts or active signal transducers? Pharmacol Ther. 2005;107(2):240–51. doi: 10.1016/j.pharmthera.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Garcia MI, Boehning D. Cardiac inositol 1,4,5-trisphosphate receptors. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbamcr.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehning D, Patterson RL, Sedaghat L, Glebova NO, Kurosaki T, Snyder SH. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat Cell Biol. 2003;5(12):1051–61. doi: 10.1038/ncb1063. [DOI] [PubMed] [Google Scholar]
- 22.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma S, Guthrie PH, Chan SS, Haq S, Taegtmeyer H. Glucose phosphorylation is required for insulin-dependent mTOR signalling in the heart. Cardiovasc Res. 2007;76(1):71–80. doi: 10.1016/j.cardiores.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louch WE, Sheehan KA, Wolska BM. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol. 2011;51(3):288–98. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13(9):1400–12. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedgepeth SC, Garcia MI, Wagner LE, Rodriguez AM, Chintapalli SV, Snyder RR, Hankins GD, Henderson BR, Brodie KM, Yule DI, van Rossum DB, Boehning D. The BRCA1 tumor suppressor binds to inositol 1,4,5-trisphosphate receptors to stimulate apoptotic calcium release. J Biol Chem. 2015;290(11):7304–13. doi: 10.1074/jbc.M114.611186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeman J, Vladimirov N, Kawashima T, Mu Y, Sofroniew NJ, Bennett DV, Rosen J, Yang CT, Looger LL, Ahrens MB. Mapping brain activity at scale with cluster computing. Nature methods. 2014;11(9):941–50. doi: 10.1038/nmeth.3041. [DOI] [PubMed] [Google Scholar]
- 28.Wilkins BJ, Dai YS, Bueno OF, Parsons SA, Xu J, Plank DM, Jones F, Kimball TR, Molkentin JD. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ Res. 2004;94(1):110–8. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- 29.Nakao S, Wakabayashi S, Nakamura TY. Stimulus-dependent regulation of nuclear Ca2+ signaling in cardiomyocytes: a role of neuronal calcium sensor-1. PLoS One. 2015;10(4):e0125050. doi: 10.1371/journal.pone.0125050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392(6679):933–6. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 31.Li W, Llopis J, Whitney M, Zlokarnik G, Tsien RY. Cell-permeant caged InsP3 ester shows that Ca2+ spike frequency can optimize gene expression. Nature. 1998;392(6679):936–41. doi: 10.1038/31965. [DOI] [PubMed] [Google Scholar]
- 32.Tomida T, Hirose K, Takizawa A, Shibasaki F, Iino M. NFAT functions as a working memory of Ca2+ signals in decoding Ca2+ oscillation. EMBO J. 2003;22(15):3825–32. doi: 10.1093/emboj/cdg381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drawnel FM, Archer CR, Roderick HL. The role of the paracrine/autocrine mediator endothelin-1 in regulation of cardiac contractility and growth. Br J Pharmacol. 2013;168(2):296–317. doi: 10.1111/j.1476-5381.2012.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uchiyama T, Yoshikawa F, Hishida A, Furuichi T, Mikoshiba K. A novel recombinant hyperaffinity inositol 1,4,5-trisphosphate (IP(3)) absorbent traps IP(3), resulting in specific inhibition of IP(3)-mediated calcium signaling. J Biol Chem. 2002;277(10):8106–13. doi: 10.1074/jbc.M108337200. [DOI] [PubMed] [Google Scholar]
- 35.Sims CE, Allbritton NL. Metabolism of inositol 1,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate by the oocytes of Xenopus laevis. J Biol Chem. 1998;273(7):4052–8. doi: 10.1074/jbc.273.7.4052. [DOI] [PubMed] [Google Scholar]
- 36.Kasumu AW, Liang X, Egorova P, Vorontsova D, Bezprozvanny I. Chronic suppression of inositol 1,4,5-triphosphate receptor-mediated calcium signaling in cerebellar purkinje cells alleviates pathological phenotype in spinocerebellar ataxia 2 mice. J Neurosci. 2012;32(37):12786–96. doi: 10.1523/JNEUROSCI.1643-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo D, Yang D, Lan X, Li K, Li X, Chen J, Zhang Y, Xiao RP, Han Q, Cheng H. Nuclear Ca2+ sparks and waves mediated by inositol 1,4,5-trisphosphate receptors in neonatal rat cardiomyocytes. Cell Calcium. 2008;43(2):165–74. doi: 10.1016/j.ceca.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zima AV, Bare DJ, Mignery GA, Blatter LA. IP3-dependent nuclear Ca2+ signalling in the mammalian heart. J Physiol. 2007;584(Pt 2):601–11. doi: 10.1113/jphysiol.2007.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ibarra C, Vicencio JM, Estrada M, Lin Y, Rocco P, Rebellato P, Munoz JP, Garcia-Prieto J, Quest AF, Chiong M, Davidson SM, Bulatovic I, Grinnemo KH, Larsson O, Szabadkai G, Uhlén P, Jaimovich E, Lavandero S. Local control of nuclear calcium signaling in cardiac myocytes by perinuclear microdomains of sarcolemmal insulin-like growth factor 1 receptors. Circ Res. 2013;112(2):236–45. doi: 10.1161/CIRCRESAHA.112.273839. [DOI] [PubMed] [Google Scholar]
- 40.Garcia MI, Chen JJ, Boehning D. Genetically encoded calcium indicators for studying long-term calcium dynamics during apoptosis. Cell Calcium. 2017;61:44–49. doi: 10.1016/j.ceca.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louch WE, Koivumaki JT, Tavi P. Calcium signalling in developing cardiomyocytes: implications for model systems and disease. J Physiol. 2015;593(5):1047–63. doi: 10.1113/jphysiol.2014.274712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kockskamper J, Zima AV, Roderick HL, Pieske B, Blatter LA, Bootman MD. Emerging roles of inositol 1,4,5-trisphosphate signaling in cardiac myocytes. J Mol Cell Cardiol. 2008;45(2):128–47. doi: 10.1016/j.yjmcc.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahmoud SA, Poizat C. Epigenetics and chromatin remodeling in adult cardiomyopathy. J Pathol. 2013;231(2):147–57. doi: 10.1002/path.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaturvedi P, Tyagi SC. Epigenetic mechanisms underlying cardiac degeneration and regeneration. International journal of cardiology. 2014;173(1):1–11. doi: 10.1016/j.ijcard.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thienpont B, Aronsen JM, Robinson EL, Okkenhaug H, Loche E, Ferrini A, Brien P, Alkass K, Tomasso A, Agrawal A, Bergmann O, Sjaastad I, Reik W, Roderick HL. The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy. J Clin Invest. 2017;127(1):335–348. doi: 10.1172/JCI88353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.