Abstract
The identification of transcriptional enhancers in the human genome is a prime goal in biology. Enhancers are typically predicted via chromatin marks, yet their function is primarily assessed with plasmid-based reporter assays. Here, we show that two previous observations relating to plasmid-transfection into human cells render such assays unreliable: (1) the function of the bacterial plasmid origin-of-replication (ORI) as conflicting core-promoter and (2) the activation of a type-I-interferon (IFN-I) response. These problems cause strongly confounding false-positives and -negatives in luciferase assays and STARR-seq screens. We overcome both problems by employing the ORI as core-promoter and by inhibiting two IFN-I-inducing kinases. This corrects luciferase assays and enables genome-wide STARR-seq screens in human cells. In HeLa-S3 cells, we uncover strong enhancers, IFN-I-induced enhancers, and enhancers endogenously silenced at the chromatin level. Our findings apply to all episomal enhancer activity assays in mammalian cells, and are key to the characterization of human enhancers.
Introduction
While promoters are located at the 5’end of genes and initiate transcription locally, enhancers can activate transcription from distal core-promoters1. This defining property is assessed in enhancer activity assays that test candidate DNA fragments in reporter plasmids, outside their endogenous contexts. Typically, candidates are placed downstream of a reporter gene or a barcode sequence (Figure 1A), ensuring the assessment of enhancer- rather than promoter activity. Importantly however, in human cells reporter transcripts from the widely used pGL3/4 system (for MPRAs based on this system see Supplementary Table 1 and Santiago-Algarra et al.2) initiate predominantly in the bacterial plasmid origin-of-replication (ORI) rather than the minimal core-promoter3 (Figure 1A; fly cells do not seem to be affected4). While the function of the ORI as a core-promoter is not unexpected given the presence of core-promoter elements3 (Supplementary Figure 1A) and the ORI’s propensity to remain nucleosome free5, it will likely impact enhancer-activity measurements: the undefined 5’ UTR, differences in reporter transcript stability or transcriptional interference between the two core-promoters can affect assays that measure reporter abundance at the protein or RNA level, as all sequencing-based massively parallel reporter assays (MPRAs) including STARR-seq do1,2.
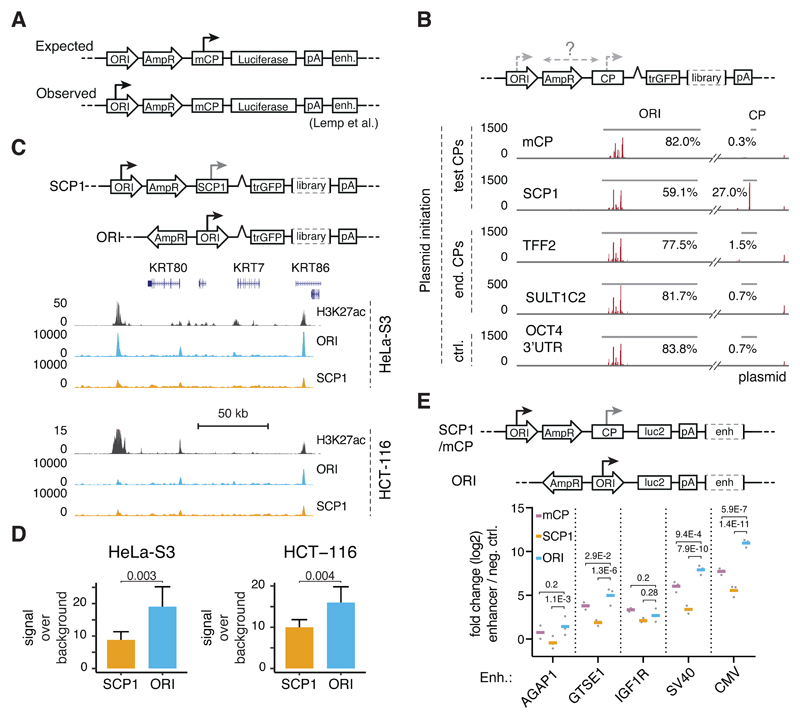
Figure 1. The ORI is an optimal core-promoter for STARR-seq and luciferase assays.
A, Typical layout of a reporter plasmid for enhancer-activity assays (e.g. pGL3/4) with the origin-of-replication (ORI), a resistance gene (AmpR), a minimal core-promoter (mCP), a reporter gene (Luciferase), a polyadenylation sequence (polyA) and an enhancer candidate (enh.). The major site of reporter-transcript initiation is indicated with an arrow (expected vs. observed according to Lemp et al.3). B, Reporter-transcript initiation on STARR-seq plasmids as measured by STAP-seq7 for setups with two synthetic core-promoters (mCP, SCP1) and two endogenous core-promoters (TTF2, SULT1C2) vs. a negative control (ctrl., OCT4 3’UTR). Red vertical lines indicate transcription initiation sites with the respective initiation frequencies according to STAP-seq. The percentages indicate the fraction of all initiation events in either the ORI or the respective core-promoter. C, Original and new setup of the STARR-seq plasmid (top) and STARR-seq profiles for screens using both setups in HeLa-S3 and HCT-116 cells (H3K27ac data from ENCODE and Rickels et al.47, see Supplementary Table 3) at a representative locus. D, STARR-seq signal-over-background between screens employing SCP1 or the ORI as a core-promoter over predicted enhancers for HeLa-S3 cells (n=39) or HCT-116 cells (n=27). Bars represent mean signal, error bars 75% confidence intervals, P-values as listed (two-sided paired t-test). See Supplementary Figure 1E,F,G for an equivalent analysis over luciferase validated regions in HeLa-S3 cells. E, Original and new setup of the luciferase plasmids (top) and average luciferase activity (bottom) for 3 cellular (AGAP1, GTSE1, IGF1R) and 2 viral (SV40, CMV) enhancers over a negative control (in log2 fold-change) in different reporter plasmid setups with the mCP (magenta), SCP1 (orange), and ORI (blue) as core-promoter. Bars represent mean signal across three independent transfections (grey dots), P-values as listed (two-sided Fisher’s LSD test).
Results
STARR-seq reporter transcripts initiate in the ORI rather than in core-promoters
Similar to single-candidate luciferase assays, STARR-seq tests candidates downstream of a core-promoter as comprehensive libraries with hundreds of millions of fragments6. To assess where reporter transcription initiates in STARR-seq, we mapped the initiation sites of reporter transcripts7 in HeLa-S3 cells, using STARR-seq libraries with two frequently used synthetic core-promoters, two endogenous core-promoters, and one non-core-promoter control. In line with the findings for luciferase reporters3, most reporter transcripts (≥77.5%) initiated within the ORI and almost no initiation occurred within the core-promoters (≤1.5%; Figure 1B) or other backbone positions (Supplementary Figure 1B). Even SCP18, which was the only exception giving rise to 27.0% of all reporter transcripts, presumably due to its high(er) basal activity, was less efficient than the ORI (59.1%).
ORI-derived STARR-seq reporter transcripts identify active enhancers
To test whether ORI-initiation affected the enhancer activity profiles, we processed the same samples using the STARR-seq protocol6 (Supplementary Figure 1C). All five setups show similar profiles that identify active enhancers, despite considerable background. When we separated reporters that initiated within the SCP1 or the ORI, we obtained highly similar enhancer-activity profiles (Supplementary Figure 1D), suggesting that the ORI functions as a highly inducible core-promoter that responds to human enhancers.
The ORI is an optimal core-promoter for STARR-seq and luciferase assays
To capitalize on the efficiency of the ORI as a core-promoter, we cloned STARR-seq libraries in which the ORI is used as core-promoter, placed immediately upstream of reporter gene and candidate-library. This should provide maximal enhancer-mediated activation, avoid the presence of two potentially conflicting core-promoters, and prevent transcription and splicing across the ~2kb intervening plasmid sequence3. Indeed, STARR-seq in HeLa-S3 cells with these constructs showed improved signal-over-background for putative enhancers predicted based on chromatin features (Figure 1C, D, Supplementary Figure 1E) and for luciferase-validated enhancers (Supplementary Figure 1F). Using the ORI as a core-promoter also improved STARR-seq signals in the colorectal cancer cell line HCT-116 (Figure 1C, D, Supplementary Figure 1G).
Importantly, these improvements also applied to single-candidate luciferase assays: reporter transcription was induced up to 10-fold more strongly by cellular and up to 40-fold more strongly by viral enhancers in the ORI-based setup compared to two core-promoter-based setups (Figure 1E). Together, these results suggest that employing the ORI as core-promoter improves the signal of single-candidate luciferase assays and MPRAs such as STARR-seq (see Supplementary Figure 1H for alternatives).
DNA transfection into human cells mounts an interferon response
As cytoplasmic DNA is prevalent during plasmid DNA transfection9, we hypothesized that enhancer activity assays in human cells might suffer from an innate immune response: most mammalian cell types and many immortalized cell lines sense cytoplasmic DNA and induce type-I-interferon (IFN-I) expression via cGAS, STING, TBK1, and IRF transcription factors10,11. This substantially changes gene expression, suggesting that the corresponding enhancer activities are also altered.
For example, expression of the interferon pathway genes cGAS, STING, TBK1, and PKR groups 19 ENCODE cell lines into three main clusters (Supplementary Figure 2A): four have low levels of STING mRNA, and four - including HCT-116 - have low cGAS mRNA levels, suggesting that their ability to induce interferon stimulated genes (ISGs) in response to cytosolic DNA might be compromised. This has indeed been observed for five of the eight cell lines (Supplementary Figure 2A and references therein), and we confirm the lack of ISG induction after transfection of a STARR-seq library by electroporation for HCT-116, K562, and SK-N-SH cells (Supplementary Figure 2B).
In contrast, the majority of all cell lines (10 out of 18), including the widely-used HeLa-S3 cells, show high expression of interferon pathway genes including cGAS and STING. Induction of ISGs is known for three of these (Supplementary Figure 2A and references therein) and we confirm ISG-induction for HeLa-S3 and GM12878 cells, as well as two additional widely used cell lines (U937 and THP1; Supplementary Figure 2C). ISG-induction was not specific to STARR-seq-library transfection, but also occurred for luciferase-reporter plasmids, pBluescript (see also Bridge et al.12), a plasmid without any CpG dinucleotides (Supplementary Figure 2D), and for chemical transfection (Supplementary Figure 2E). These findings together with similar reports in the literature (see references in Supplementary Figure 2A) demonstrate that IFN-I-related gene induction by plasmid transfection is a widespread phenomenon that affects many widely used cell lines.
Enhancer activity assays in HeLa-S3 cells are dominated by IFN-I-signaling
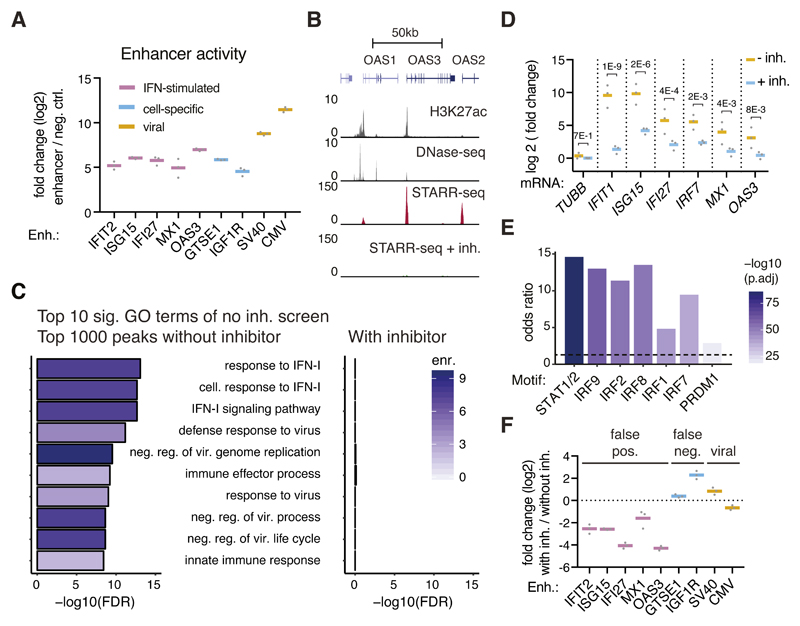
The IFN-I-related gene induction suggests that the corresponding enhancer and promoter activities are also changed. Indeed in HeLa-S3 cells, putative enhancers proximal to canonical ISGs were highly active in luciferase assays (Figure 2A), and a genome-wide STARR-seq screen was dominated by enhancers related to IFN-I signaling: genes proximal to the top 1000 enhancers were strongly enriched for GO-terms relating to cellular immunity and IFN-I signaling (Figure 2B, C), similar to a focused STARR-seq screen testing ~21,000 promoter regions for enhancer activity13. Additionally, disease-ontology analyses pointed towards viral infection rather than cervical cancer as one would expect for HeLa cells (Supplementary Figure 3A), in which genes related to IFN-I signaling are not particularly highly expressed14. Together with the fact that ISG-induction is independent of plasmid type and -delivery (Supplementary Figure 2D-E), these results argue that all plasmid-based luciferase assays and MPRAs should suffer from plasmid-induced IFN-I signaling. Indeed, we find luciferase and STARR-seq assays to be affected (Figure 2A, B, C) and several studies using synthetic, barcode-based MPRAs or libraries enriched for selected candidates reported high activities of IFN-I-related enhancers and/or enrichments of IFN-I-related transcription factor binding sites indicative of IFN-I signaling13,15,16.
Figure 2. Genome-wide enhancer screens are dominated by false-positive signals.
A, Mean enhancer activity (luciferase mRNA levels relative to negative control; log2) across three independent transfections (grey dots), assessed by qPCR in reporter assays employing the indicated enhancers. B, Representative STARR-seq enhancer activity profiles over a canonical ISG locus (GRCh37 Refseq genes indicated above) for genome-wide HeLa-S3 STARR-seq screens without and with inhibitors against TBK1/IKK/PKR (H3K27ac and DHS data from ENCODE, Supplementary Table 3). C, The 10 most significantly enriched GO terms for genes proximal to the top 1000 peaks in a HeLa-S3 STARR-seq screen. Shown are log10 transformed FDR-adjusted P-values (Fisher’s exact test) and fold-enrichments (shades of purple). The same terms were assessed for the TSSs proximal to the top 1000 peaks from the TBK1/IKK/PKR-inhibitor-treated screen. D, qPCR-based assessment of ISG-mRNA induction after DNA transfection in TBK1/IKK/PKR-inhibitor-treated vs. non-treated HeLa-S3 cells. Bars represent mean fold change across three independent transfections (grey dots), P-values as stated (two-sided Fisher’s LSD test). E, Odds ratios (FDR-adjusted P-values < 10-5, Fisher’s exact test) of indicated transcription factor motifs in STARR-seq enhancers 5-fold downregulated upon TBK1/IKK/PKR-treatment (FDR-adjusted P-value < 0.001, n=400) vs. unchanged enhancers (within +/- 1.5-fold change upon treatment, n=2245). F, Mean luciferase activity fold change across three independent transfections (grey dots) of luciferase mRNA expression in reporter assays employing the indicated enhancers in cells treated without PKR/TBK1 inhibitors over with inhibitors (log2).
TBK1/IKKε and PKR inhibition prevents dominant false-positive IFN-1-related enhancer signals
Cytoplasmic DNA leads to IFN-I induction via the key signaling kinases TANK-binding-kinase-1 (TBK1) and IκB-kinase-ε (IKKε), which activate IRF317. Similarly, double stranded RNA (dsRNA) can arise at plasmids and activate protein-kinase-R (PKR), affecting transgene expression18–20. The inhibition of these kinases (or other pathway components) should ameliorate the changes of gene expression and enhancer activities described above. Indeed, treating HeLa-S3 cells during plasmid transfection with the TBK1/IKKe inhibitor BX-79521 and the PKR inhibitor C1622 prevents the strong induction of ISGs observed after plasmid transfection in HeLa-S3 cells (Figure 2D).
Moreover, enhancers proximal to canonical ISGs that were previously among the strongest STARR-seq signals genome-wide, were only detected at background levels in an inhibitor-treated STARR-seq screen (Figure 2B). Genes near the top 1000 peaks were no longer enriched in interferon-signaling-related GO categories (Figure 2C, right panel, Supplementary Figure 3B), indicating that TBK1/IKK/PKR inhibition removes these dominant false-positive signals. Consistently, STARR-seq peaks that lost activity upon TBK1/IKK/PKR inhibition (≥5-fold down-regulation, P-value <0.001) were next to interferon-signaling-related genes (Supplementary Figure 3C, D) and were highly enriched in motifs of transcription factors involved in ISG-induction (Figure 2E). Finally, luciferase assays confirmed that enhancers proximal to ISGs showed strongly reduced activity in cells treated with TBK1/IKK/PKR inhibitors during plasmid transfection (Figure 2F). This demonstrates that plasmid-induced IFN-I-related false positive enhancer activities can be prevented by TBK1/IKK/PKR inhibition. The importance of TBK1/IKK/PKR inhibition for accurate enhancer activity assessment is particularly evident when analyzing disease ontology terms near the top STARR-seq peaks23: without inhibitors, the top terms relate to viral infection (likely due to strong interferon-signaling), while inhibitor-treatment reveals terms relating to female reproductive cancer, consistent with HeLa-S3 cell biology (Supplementary Figure 3A).
A genome-wide set of IFN-I related enhancers
The comparison of the genome-wide STARR-seq screens with and without TBK1/IKK/PKR inhibitors also defines a genome-wide set of IFN-I related enhancers and their respective induction strengths, some of which were more than 100-fold (Supplementary Figure 3E, Supplementary Table 2). Interestingly, many of these predominantly promoter-proximal enhancers were pre-marked by H3K27ac in unperturbed HeLa-S3 cells (Supplementary Figure 3E), which might be a general feature of rapidly inducible enhancers24. This genome-wide set of IFN-I-related enhancers (Supplementary Table 2) is a valuable resource for the study of IFN-I-mediated transcriptional regulation.
TBK1/IKK/PKR inhibition prevents false-negative signals and improves signal over background
Of the enhancers we tested individually in luciferase assays, the activity of two endogenous HeLa-S3 cell enhancers and one viral enhancer was increased after TBK1/IKK/PKR inhibition (Figure 2F). This suggests that IFN-I-signaling might also repress bona-fide enhancers, leading to an underestimation of their activities or false negatives. Indeed, the STARR-seq signal increased substantially in inhibitor-treated HeLa-S3 cells (Supplementary Figure 3F), but not in HCT-116 cells, which do not induce ISGs after DNA transfection (Supplementary Figure 3G). The signal-increase is therefore specific and inhibitor treatment should not otherwise impact enhancer activities.
STARR-seq enhancers are mostly intergenic or intronic and are enriched in enhancer-associated chromatin states
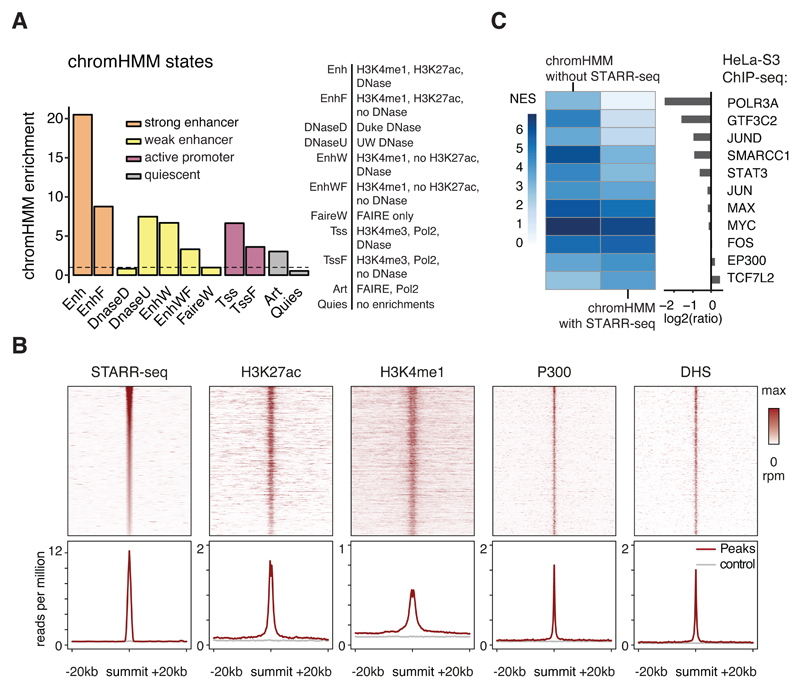
The genome-wide STARR-seq screen using the new screening setup and TBK1/IKK/PKR-inhibitors yielded 9,613 peaks, of which 2,508 have a corrected enrichment of ≥10-fold and 209 of ≥50-fold. The enhancer activity profiles are highly similar between replicates (PCC=0.98). Almost half of all peaks are in intergenic regions (48.3%) and 43.2% are within introns (Supplementary Figure 4A). The peaks significantly overlap with regions that exhibit enhancer- or promoter-associated chromatin states according to chromHMM25: all 9,613 STARR-seq peaks are enriched 20.5-fold in the strong enhancer state Enh, 6.7-fold in the weak enhancer state EnhW and 6.6-fold in the active promoter state TSS, compared to these states’ genomic abundance (Figure 3A). The enrichment for the Enh state was 40.3-fold for the top 500 peaks and still ~ 9-fold for the weakest peaks, suggesting that even sub-threshold peaks might be functional in vivo, albeit with weak effects on transcription (Supplementary Figure 4B, Supplementary Table 2). Furthermore, peaks that are accessible in HeLa-S3 cells according to ENCODE DNase-seq (42.3%, Supplementary Table 3) align precisely with characteristic enhancer features (Figure 3B).
Figure 3. STARR-seq enhancers are enriched in chromHMM enhancer-related states.
A, Enrichment of enhancer relevant ChromHMM states25 within STARR-seq enhancers (dotted line indicates no enrichment (=1)). B, Coverage heatmaps (top) and average coverage (bottom) of STARR-seq, H3K27ac, H3K4me1, P300 and DHS signal for STARR-seq enhancers accessible in HeLa-S3 cells (rpm: reads per million; grey: random control regions). C, Normalized enrichment scores for different HeLa-S3 ChIP-seq datasets (NES, i-cisTarget26) for chromHMM strong enhancers (‘Enh’) with or without STARR-seq support and the respective fold-differences (right, log2).
STARR-seq-negative enhancer-candidates are associated with Pol III transcription
ChromHMM Enh regions that do not show activity in enhancer activity assays such as STARR-seq would generally be considered non-functional1, yet 1,323 such regions (8.9%) are bound by RNA polymerase III (Pol III) or its general transcription factor 3C, yielding a strong enrichment according to i-cisTarget26 (Figure 3C). Pol III typically transcribes non-coding genes from promoters that are independent of enhancer-like upstream regions27, yet can bear chromatin marks reminiscent of Pol-II-enhancers28. The transcription factors JUN, MAX, MYC, or FOS were enriched to similar extents in ChromHMM regions with or without STARR-seq signals (EP300 and TCF7L2 were slightly more enriched in STARR-seq-positive regions, Figure 3C). Open STARR-seq enhancers that do not overlap chromHMM Enh regions show similar enrichments for different TFs and an even slightly higher enrichment for the TFs JUN and JUND and for EP300, as expected for bona-fide enhancers (Supplementary Figure 4C). Interestingly, TSS-distal and TSS-proximal enhancers differed in motif content (Supplementary Figure 4D), transcription factor binding (Supplementary Figure 4E) and histone modifications (Supplementary Figure 4F), suggesting that they might be regulated differently.
A substantial fraction of enhancers is silenced at the chromatin level
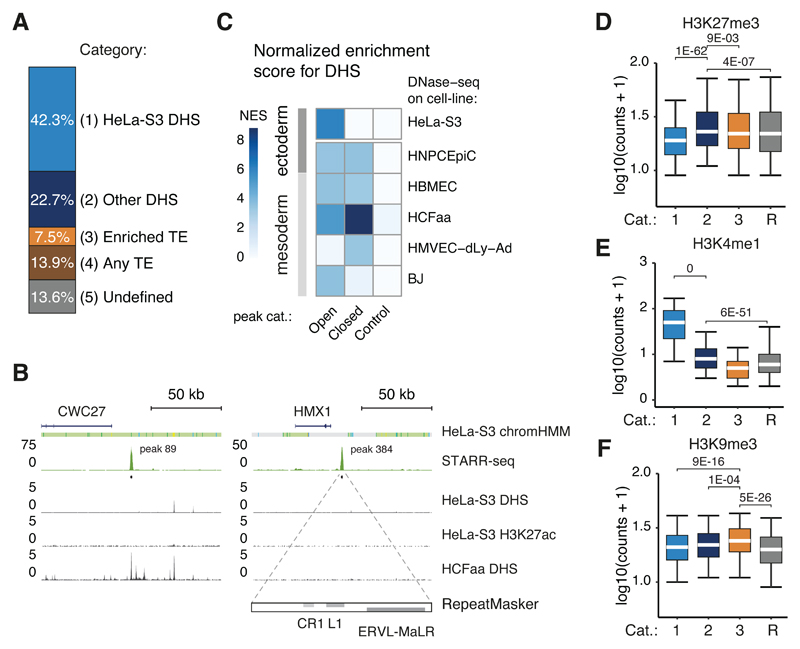
One of the advantages of ectopic assays such as STARR-seq is their ability to assess the enhancer activities of DNA sequences that are able to strongly activate transcription yet are silenced endogenously at the chromatin level6. While a large fraction of the STARR-seq enhancers were accessible in their endogenous genomic locations according to ENCODE DNase-seq (56.2% of the top 500 and 42.3% of all 9,613 enhancers, Supplementary Table 3), 57.7% of STARR-seq enhancers were not accessible, i.e. closed, and thus likely actively silenced (Figure 4A). This included DNA sequences that can function as very strong enhancers in HeLa-S3 cells, such as peak 89 in the CWC27 locus or peak 384 in the HMX1 locus (Figure 4B).
Figure 4. STARR-seq identifies enhancers silenced endogenously.
A, Percentages of STARR-seq enhancers that have significant DNase-seq signal in HeLa-S3 cells (P-value < 0.05, one-tailed binomial test), are accessible in other enriched cell types, contain repetitive elements from three enriched repeat families (see Figure 5G), contain other repetitive elements, or none of the above (undefined). B, Enhancer activity profiles over two gene loci (indicated above; DHS and H3K27ac data from ENCODE, Supplementary Table 3), representative of category 2 (CWC27, left panel) and 3 (HMX1, right panel). The right panel includes the RepeatMasker track, displaying elements of the indicated repeat families within the STARR-seq peak above. C, Normalized enrichment scores (NES, i-cisTarget26) for ENCODE DNase-seq datasets within STARR-seq enhancers that are open or closed in HeLa-S3 cells (P-value < 0.05, one-tailed binomial test). NES scores for random regions are shown as control. D, E, F, Boxplots of H3K27me3 (D), H3K4me1 (E) and H3K9me3 (F) read coverage in log10 (counts + 1) for STARR-seq enhancers of the categories defined in (A, N= 4071 (1), 2180 (2), 721 (3)) and random regions (R, N= 9613). Lower whisker: 5th percentile, lower hinge: 25th percentile, median, upper hinge: 75th percentile, upper whisker: 95th percentile. P-values as stated (one-sided Wilcoxon rank sum test).
Closed STARR-seq enhancers are open in other cell types and H3K27me3-marked
Interestingly, the closed enhancers are strongly enriched in DNase-I-hypersensitive regions of five ENCODE cell lines according to i-cisTarget26 (e.g. HMVEC or HCFaa cells; Figure 4C). Together, DNase-I hypersensitivity in these five non-HeLa cell types accounts for 39.3% of the closed HeLa-S3 enhancers (Figure 4A, see Supplementary Table 3) and almost all HeLa-S3 STARR-seq enhancers appear accessible in at least one of 13 ENCODE cell lines (Supplementary Figure 5A; though an increasing fraction cannot be discerned from chance-expectation given the increasing number of DNase-I-hypersensitive sites).
Closed HeLa-S3 enhancers were enriched for H3K27me3 (Figure 4D), consistent with Polycomb-mediated repression29 and marked by H3K4me1 (albeit less than accessible enhancers; Figure 4E). This suggests that they might be recognized as enhancers (as in flies6) and is consistent with previous reports that H3K4me1 labels enhancers independently of their activity30–32.
Closed STARR-seq peaks are enriched for TEs and are H3K9me3-marked
Many (39.8%) of the remaining closed peaks (24.1% of all closed peaks) contained repetitive elements annotated by RepeatMasker33. While long interspersed nuclear elements (LINEs) were depleted overall within closed peaks, elements from 3 families of endogenous retroviruses (ERVs) were highly enriched and overlapped with 13.0% of all closed peaks (Supplementary Figure 5B). These peaks exhibited a significantly higher signal of H3K9me3 than other regions (Figure 4F), suggesting that they constitute active regulatory regions that are repressed at the chromatin level34.
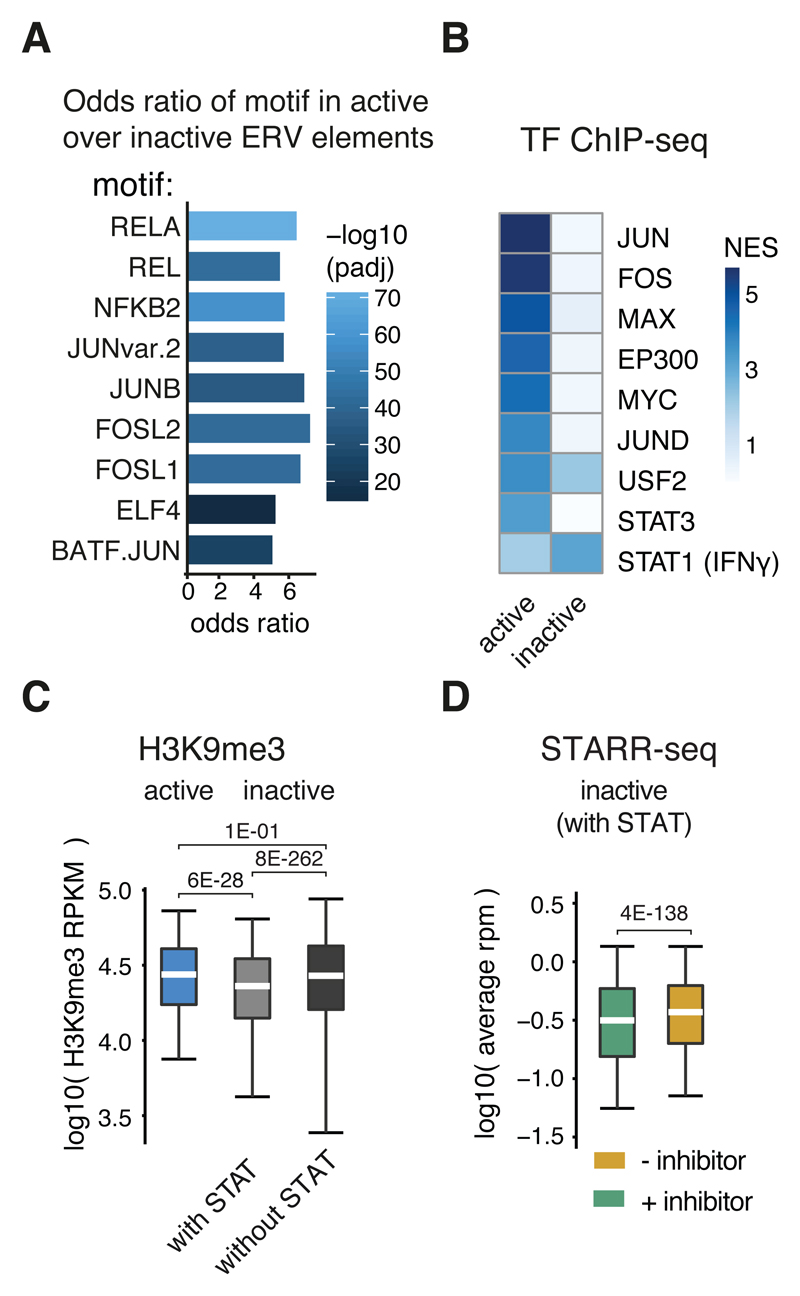
Consistent with their function as bona-fide enhancers, ERVs that are active according to STARR-seq are enriched in binding motifs of various transcription factors (e.g. FOS and JUN) compared to inactive ERVs (Figure 5A). Consistently, active ERVs were enriched for ChIP-seq-determined binding of several TFs according to i-cisTarget (e.g. FOS and JUN; Figure 5B), suggesting that such elements can escape silencing and/or might be co-opted for transcriptional regulation35,36.
Figure 5. ERV elements are co-opted for IFN-I signaling.
A, Odds ratios (FDR-adjusted P-value < 0.05, two-sided Fisher’s exact test) of transcription factor motifs in active over inactive (based on STARR-seq signal) ERV elements from the three enriched ERV families (see panel G). B, i-cisTarget normalized enrichments scores for ENCODE ChIP-seq datasets within active or inactive ERV elements. C, Boxplot of H3K9me3 read coverage per kb (RPKM) in log10 over active (n=1783) or inactive ERV elements with (n=26809) or without (n=491157) STAT motifs. D, STARR-seq read coverage in log10 for STARR-seq screens with (green) or without (red) TBK1/IKK/PKR inhibition over inactive ERV elements with STAT motifs (n=26809). C,D: Lower whisker: 5th percentile, lower hinge: 25th percentile, median, upper hinge: 75th percentile, upper whisker: 95th percentile. P-values as stated (one-sided Wilcoxon rank sum test).
The only enrichment specific to inactive ERV elements was for an ENCODE dataset that assessed STAT1 binding after IFNγ treatment (Supplementary Table 3). This suggests that some ERVs are activated by IFNγ, either because they evolved interferon-responsiveness or because such elements were co-opted as cellular interferon-responsive enhancers35. Indeed, inactive ERVs with STAT transcription factor binding motifs were less enriched for the repressive histone mark H3K9me3 compared to both, active elements or inactive elements without STAT binding motifs (Figure 5C). Additionally, ERVs with STAT motifs are induced during interferon signaling, i.e. appear active in STARR-seq screens without inhibitor treatment (Figure 5D).
Discussion
The past years have seen tremendous progress in predicting human enhancers based on chromatin properties and non-coding transcription that correlate with enhancer activities1. The direct functional assessment of enhancers by reporter assays has therefore become increasingly important and is a major aim of large consortia efforts37.
Here, we show that two previously reported effects – the core-promoter function of a bacterial ORI and the IFN-I response triggered by cytoplasmic DNA – confound enhancer activity assays in mammalian cells. Our results indicate that previous approaches might have substantially underestimated enhancer activities (Figure 1C-E), potentially missing up to 75% of all enhancers (Supplementary Figure 6A, B); and that enhancer-activity screens in cells with intact innate-immune signaling are dominated by false-positives (Figure 2C), grossly altering the cells’ enhancer landscapes (Supplementary Figure 3A, E).
Importantly, all published MPRAs are based on the same pGL3/4-derived backbone (Supplementary Table 1) and reporter-transcript initiation sites have typically not been mapped. It is therefore possible that the reporter transcripts do not initiate within the core-promoters but in the ORI (here and Lemp et al.3) or – for candidates upstream of reporter-gene or barcode – within the enhancer-candidates (consistent with eRNA-transcription in vivo38 and equivalent to promoter-activity assays39) or elsewhere on the plasmid-backbone. In addition, all plasmid-based enhancer activity assays should be affected by the IFN-I response, which is independent of plasmid type (Supplementary Figure 2D). Indeed, we found evidence of an active IFN-response, such as ISG-enhancer activities and STAT1/2- or IRF-motif enrichments, in publications covering ISG-enhancer-regions with different MPRAs, two with synthetic, barcode-based reporters15,16 and three that used enriched candidate libraries13,40,41.
The strategy to specifically use the ORI as a core-promoter is also applicable to barcode-based MPRAs that rely on different cloning strategies (Supplementary Figure 7A, B). Alternatively, approaches to suppress ORI-derived transcripts (Supplementary Figure 1H) or the use of other low(er)-copy ORIs, ORI-less constructs42,43, or reporter-delivery using AAV-44 or lentiviral vectors45,46 should allow for screens with different core-promoters. We note that retroviral integration preferences and promoter/enhancer elements on viral backbones (e.g. LTRs) should be carefully considered as they might confound enhancer-activity assays. Lastly, other means to inactivate key signaling factors of innate immune signaling (e.g. knockout cell lines or cGAS inhibition) are alternatives to the inhibitors we propose.
The methods presented here overcome key problems associated with plasmid-based enhancer-activity assays, correcting individual candidate luciferase assays and enabling genome-wide STARR-seq screens. In HeLa-S3 cells, such screens revealed thousands of human enhancer sequences with activities approaching the strengths of strong viral enhancers. Similar to previous results in flies6, many enhancers are silenced in their endogenous loci at the chromatin level. These enhancers are invisible to predictions based on chromatin features, yet constitute attractive examples to study mechanisms of chromatin-mediated repression. Given these results, we recommend that cellular enhancer function is assessed by combining enhancer activity assays with DNase I hypersensitivity assays or similar.
The tools and protocols presented here are applicable to all episomal reporter assays in mammalian cells and should become a central component of our efforts to identify all gene regulatory elements of the human genome and understand how their sequences encode cell type-specific gene expression.
Online Methods
The methods section is accompanied by laboratory protocols for STARR-seq library cloning (Supplementary Protocol 1) and screening (Supplementary Protocol 2). We further provide protocols for qPCR testing of IFN-I-response (Supplementary Protocol 3) and qPCR reporter assays (Supplementary Protocol 4). All protocols are also available from ‘Protocols Exchange’. All files are available at http://starklab.org/data/muerdter_boryn_2017/. Plasmids are available from Addgene. Details on experimental design can be found in the “Life Sciences Reporting Summary” at the end of this manuscript.
Experimental Methods
Cell lines
We purchased HeLa-S3, HCT-116, SK-N-SH cells from ATCC (cat# CCL-2.2, CCL-247, CCL-243, HTB-11) and GM12878 cells from Coriell Institute. K562 were a gift from the Zuber lab (IMP) and THP-1 and U937 cells were a gift from the Decker and Versteeg labs (MPFL). Cells were cultured in DMEM (HeLa-S3, HC-116, SK-N-SH; Gibco; cat# 52100-047) or RMPI-1640 (GM12878, K562, U937, THP1; Gibco; cat# 13018-031), supplemented with 10% heat-inactivated FBS (Sigma; cat# F7524, 15% for GM12878) and 2 mM L-glutamine (Sigma; cat# G7513) at 37°C in a 5% CO2 enriched atmosphere.
Electroporation
Cells were electroporated using the MaxCyte-STX system at a density of 1x107 cells per 100 µl and 20 µg of DNA using the pre-set protocols (except for opt-program 4 for GM12878 cells).
Chemical transfection
8x105 HeLa-S3 cells were plated 24 h prior to being transfected with 8.8 µg of STARR-seq library using FuGENE-HD reagent (3.5:1 reagent-to-DNA ratio, Promega; cat# E2312).
Plasmid purification
Plasmids for electroporation were purified with Qiagen Plasmid-Plus kits (cat# 12965, 12991) and resuspended in H2O. For endotoxin free preps, Qiagen EndoFree Plasmid maxi kits were used (cat# 12362).
Inhibitor treatment
We used PKR (C16, Sigma; cat# I9785-5MG) and TBK1/IKK inhibitors (BX-795, Sigma; cat# SML0694-5MG) as recommended in the literature21,22, adding them to the electroporated cells directly after resuspension at a final concentration of 1 µM per inhibitor.
Mapping of STARR-seq transcript initiation sites by STAP-seq
STAP-seq was performed as described7. Briefly, 50 µg of DNaseI-treated mRNA from STARR-seq library electroporated cells were treated with 25 µl of CIP (NEB; cat# M0290L) for 1.5h at 37°C. The CIP-treated RNA was purified with Qiagen RNeasy MinElute kits (cat# 74204), with beta-Mercaptoethanol (Sigma-Aldrich; cat# 63689) supplemented RLT buffer. The CIP-treated RNA was treated with 0.05 μl Tobacco Alkaline Phosphatase (Epicentre; discontinued) per 1 μg RNA and purified with Agencourt RNAClean XP beads (Beckman Coulter; cat# A66514, beads-to-RNA ratio 1.8). We ligated 10 µM RNA adapter (STAP_adapter, Supplementary Table 4) to the 5’ ends of each 1 µg TAP-treated mRNA using 0.2 µl T4 RNA Ligase 1 (NEB; cat# M0204L) for 16h at 16°C. The RNA was purified with Agencourt RNAClean XP beads (beads-to-RNA ratio 1.0).
First-strand cDNA synthesis was performed on the total amount of adapter-ligated RNA. Per reaction 2.5–5 µg adapter-ligated RNA was reverse-transcribed with 1 μl of Invitrogen's Superscript III (50 °C for 60 min, 70 °C for 15 min; cat# 18080085) and a reporter-RNA-specific primer (STAP_GSP, Supplementary Table 4). Five reactions were pooled and 1 μl of 10 mg/ml RNaseA was added (37 °C for 1 h) followed by Agencourt AMPureXP DNA bead purification (Beckman-Coulter; cat# A63881, ratio beads/RT reaction 1.8). We amplified the total amount of reporter cDNA for Illumina sequencing. For focused libraries, we performed two PCR reactions using the KAPA real-time library amplification kit (KAPA Biosystems, cat# KK2702), with STAP_fwd (Supplementary Table 4) forward primer and one of NEBNext Multiplex Oligos for Illumina (NEB; cat# E7335 or E7500) reverse primers. PCR products were purified with AMPureXP DNA beads (ratio beads/PCR 1.25).
STARR-seq
Screening Vectors
All STARR-seq vectors are based on the original human STARR-seq vector6 with the following changes: The GFP coding sequence is truncated, the synthetic intron is replaced with a chimeric intron (Promega, technical bulletin #TB206), the core-promoter is replaced with a panel of different minimal promoters or the ORI (Supplementary Table 4).
To enable using a core-promoter in the presence of the ORI, we generated four vectors containing SCP1 and the following changes (see Supplementary Figure 1H): Blocking variant 1 contains RBGpA polyA-signal48 and the bGHpA polyA-signal49 in the SpeI site. Blocking Variant 2 additionally contains bGHpA in the PciI site. Variant 3 contains the bcl-2 splice acceptor and four SV40 polyA-signals at the SpeI site49. Variant 4 additionally contains bGHpA at the PciI site.
Cloning of STARR-seq plasmid libraries
Focused STARR-seq libraries were generated from Bacterial Artificial Chromosome (BAC) DNA (see Supplementary Table 4, BAC mixes). BAC insert 1 was cloned into STARR-seq vectors that harbor different core promoters. BAC insert 2 was used to clone the libraries for all other screens. The library inserts (1000-1500bp) were generated as described previously6 with the following changes: We used a 45s extension during the final PCR amplification and 10 instead of 8 PCR reactions (primers in-fusion_fwd & in-fusion_rev, Supplementary Table 4). To generate genome-wide STARR-seq libraries we followed the same protocol with changes: We used 15 µg of size-selected genomic DNA (1000bp-1500bp) for library insert generation, 30 PCR reactions for library insert amplification, 20 In-Fusion HD reactions for library cloning and transformation of 25 aliquots of Invitrogen MegaX-DH10B Electrocompetent Cells. The bacteria culture was grown in 24 liters of LB.
STARR-seq screening
For focused BAC screens, we electroporated 8x107 cells per screen, for genome-wide STARR-seq libraries we electroporated 8x108 cells per screen. Genome-wide STARR-seq screens in HeLa-S3 cells were done in duplicate (inhibitor screens) or quadruplicate (non-inhibitor screens). Screens were performed as described previously6 with the following changes: We harvested total RNA 6h after electroporation, reverse transcription was performed with primer STARR_GSP (Supplementary Table 4) in a total of 10-20 reactions for focused and 40-60 reactions for genome-wide STARR-seq screens.
We purified cDNA using AMPureXP DNA beads (beads-to-cDNA ratio of 1.8) and amplified it using human STARR-seq specific primers (junction_fwd, junction_rev, Supplementary Table 4, 98°C for 45s; followed by 15 cycles of 98°C for 15s, 65°C for 30s, 72°C for 70s). We also increased the extension to 45s for the second PCR step (primers PE1.0 as forward primer and MP2.0 or TruSeq idx primers as reverse primer, Supplementary Table 4). We set up 2 reactions for focused and 10 reactions for genome-wide screens. PCR products were purified using SPRIselect beads (Becker Coulter; cat# B23318, 0.5 beads-to-PCR ratio). To sequence the un-transfected STARR-seq plasmid library, twenty PCR reactions with 100 ng STARR-seq library were performed (98°C for 45s; followed by 6-15 cycles of 98°C for 15s, 65°C for 30s, 72°C for 45s) with the KAPA Hifi Hot Start Ready Mix and NEBNext Multiplex Oligos for Illumina (cat# E7335L).
Next-generation sequencing
Next-generation sequencing was performed at the VBCF NGS Unit on an Illumina HiSeq2500. Genome-wide STARR-seq screens were sequenced as paired-end 50 cycle runs, focused BAC screens for signal-to-noise analysis as single-end 50 cycle runs, using the standard Illumina primer mix. STAP-seq screens were sequenced as paired-end 50 cycle runs with TruSeq RP1 primers as read 1 primer.
Measuring ISG expression levels
5x106 cells were electroporated with a focused STARR-seq library in three independent transfections with mock electroporations as negative controls. 6h after transfection, cells were lysed using Qiashredder columns (Qiagen; cat# 79654) and total RNA was extracted using the RNeasy mini prep kit (Qiagen; cat# 74104), with beta-Mercaptoethanol supplemented RLT buffer. 1 µg of total RNA was treated with recombinant DNaseI (rDNaseI; Ambion, cat# AM1906) for 30 min at 37ºC followed by the removal of rDNaseI using a DNase inactivation reagent (Ambion; cat.no. AM1906). The DNaseI treated RNA was reverse transcribed using Invitrogen's Superscript III and Oligo-dT20 primers (Invitrogen; cat# 18418020) (25º for 5 min, 50 °C for 50 min, 70 °C for 15 min), followed by qPCR on 2 µl diluted (1:5) cDNA using Go Tag SYBR Green qPCR Master Mix (Promega; cat# A6001) in a total volume of 20 µl with 0.5 µM gene-specific qPCR primers (95ºC, 2 min; 95ºC, 3s; 60ºC, 30s; 40 cycles total, see Supplementary Table 4 for primers).
qPCR based reporter assay
5x106 HeLa-S3 cells were electroporated with 18 µg of a firefly luciferase reporter plasmid and 2 µg of a Renilla luciferase control plasmid (pRL-CMV, Promega; cat# E2261). After 30 min, C16 and BX795 inhibitors were added to the cells at 1 µM concentration. qPCR to quantify firefly luciferase transcripts normalized to Renilla luciferase transcripts was performed as described above, with the exception that we used Turbo DNase (Ambion; cat# AM1907) for cells transfected with reporter plasmids (see Supplementary Table 4 for primer sequences).
Computational Methods
TSS determination of STARR-seq reporter transcripts
All sequenced read pairs were subsampled to 500,000 fragments using reservoir sampling50. Mate 1 was mapped to the STARR-seq plasmid backbone using bowtie51. The 5’ 8nt random barcode introduced during the RNA adapter ligation7 was removed and the following 15nt were mapped uniquely allowing for 3 mismatches (bowtie option –v 3 -m 1 --best –strata). The resulting mapping locations were collapsed based on the random 8nt barcode, allowing for molecular counting of all reporter mRNA 5’ ends.
Stratification of STARR-seq signal
To stratify genomic fragments based on their origin within the plasmid (e.g. ORI vs. CP), mate 1 was used to define the origin, followed by mate 2 mapping to the human genome (bowtie options -v 3 -m 1 --best --strata).
STARR-seq data processing
STARR-seq single- and paired-end sequencing reads were mapped in fasta format to the human genome (hg19), only considering the regular chromosomes 1-22, and X using bowtie version 0.12.9 (bowtie options -v 3 -m 1 --best –strata –X 2000)51. Genome-wide screens without inhibitors and focused screens for signal-to-noise analysis were mapped as 50-mers. Genome-wide screens with inhibitors were mapped as 36-mers due to poor base calls at the 3’ end of the read. Genome coverage bigwig files were generated using all reads with bedtools genomecov version 2.19.152 and normalized to reads per million (r.p.m.). For both genome-wide STARR-seq and input libraries, reads from biological replicates were combined.
Peak calling
Enriched regions were shortlisted using all reads from the combined STARR-seq replicates versus input as described6 with a P-value cutoff of 1x10-5 and an enrichment cutoff ≥ 3 (enrichment over input, where input coverage was evaluated locally at the peak summit or over a fixed input window of 20 kb surrounding the summit, whichever was higher) and peaks were then called with a corrected enrichment ≥4 (correction as in Stark et al.53, z=3). We discarded peaks for which a single fragment accounted for more than 50% of all fragments overlapping with the peak and peaks overlapping ENCODE blacklisted regions. Peaks were annotated as open if they had significant enrichments in DHS datasets (binomial P-value < 0.05). This P-value was calculated over the entire STARR-seq peak using the maximum DHS coverage (r.p.m.) over the median coverage in the input, yielding a FDR of 13.6% when applied to random regions. Peaks were annotated with an Ensembl gene ID based on the transcript TSS nearest to the peak’s edges using the R package GenomicRanges (version 1.20.8) and its function distanceToNearest. Gene ID annotations were obtained from Ensembl version 75 IDs54.
ChIP-seq coverage
Counts for H3K27me3, H3K4me1, and H3K9me3 were calculated for the groups defined by DHS and enriched TEs using bedtools coverage (options –counts –F 0.1). For plotting, a pseudo-count of 1 was added to counts before log10 transformation.
Random control regions
We selected 9,613 random control regions from all fragments in the STARR-seq input library with a size distribution of 1000 to 1600bp by reservoir sampling (sample version 1.0.2 https://travis-ci.org/alexpreynolds/sample). To match the peak size, we extended each region by 641bp from the center of the fragment.
Comparison of signal-to-noise
To assess improvements in signal-to-noise we (1) evaluated STARR-seq signal-to-noise ratios on a small set of positive (n=4) and negative (n=6) regions according to luciferase assays and (2) defined a high-confidence subset of STARR-seq peaks in focused screens with support for endogenous enhancer activity based on DNase-seq and H3K27 acetylation (datasets from ENCODE, Supplementary Table 3). We called peaks as described above (P-value cutoff of 1x10-4, enrichment cutoff ≥ 3, corrected enrichment ≥ 4, z=1.67). A binomial P-value for DHS enrichment was calculated in a 20nt window around the peak summit for the median DHS coverage (r.p.m.) over the median coverage in the input of a window of 250. For H3K27ac the maximum coverage was identified in a 500 nt window around the peak summit. For this local maximum, a binomial P-value for H3K27ac enrichment was calculated as described for DHS. Peaks with an FDR adjusted P-value < 0.05 in both the DHS and H3K27ac were used as predicted positive regions. Background signal was assessed over BAC regions minus positive regions. The average coverage was calculated within each positive peak then averaged across all positive peak regions (merged between screens) and divided by the background to give the final signal-to-noise value. For the signal-over-background scatter plots, we used the average STARR-seq read coverage for each peak and divided it by the background as defined above for bar graphs. Correlations were calculated using the Pearson Correlation Coefficient using R’s cor function.
Differential peak analysis
Peak calls for PKR/TBK1/IKK inhibited and non-inhibited genome-wide STARR-seq screens were combined and collapsed into one region if peaks overlapped by at least 85%. Differential peaks were called using a hypergeometric test using the maximum count in each peak region for each dataset. P-values were adjusted to FDRs using R’s p.adjust. Peaks less than 2.5 kb away from a nearest TSS that belongs to the top 3 interferon enriched GO terms (response to type I interferon, type I interferon signaling pathway, and cellular response to type I interferon) were highlighted in the scatter plot.
Nearest TSS Gene Ontology
Gene ontology analysis of nearest TSSs was done using topGO version 2.20.055, which reports a Fisher’s exact test p-value. All protein coding genes were provided as a background to topGO using Ensembl version 75 IDs54. Enrichments were calculated by dividing the significant number of genes in each term by the number of genes that one would expect by chance reported in the topGO output. topGO results were reported for all terms, and P-values were adjusted to FDRs using R’s p.adjust.
Disease Ontology
Disease ontology analysis was done using GREAT version 3.0.023 with default settings for the top 500 accessible peaks (P-value < 0.05 for DHS) in genome wide screens with and without inhibitor treatment. Bars represent binomial FDR q-values and are colored by binomial fold enrichment.
Motif analysis
For repeat analysis, the sequence of the entire element was used, for distal vs. proximal STARR-seq peaks and differentially active STARR-seq peaks, we used a 700bp window around the peak summits. For differentially active STARR-seq peaks we only considered windows that were at least 50bp away from any transcript TSS (Ensembl version 7554). Motifs were called using MAST from the MEME suite version 4.8.156 using options -hit_list -mt 0.00001. A motif was only counted once within each peak. Odds ratios and P-values were derived using a one-sided Fisher’s exact test for comparisons defined in the text. P-values were adjusted with the Benjamini & Hochberg method (FDR) in R.
ChromHMM enrichments
The ChromHMM segmentations were obtained from UCSC25. The peak regions were overlapped with these segmentations using bedtools intersect version 2.19.1. The sum of lengths of overlaps for each annotation term was divided over the total length of all terms for either all peaks or all ranked regions in non-overlapping bins of 500. Enrichments were calculated by dividing these fractions by the fraction of the genome in each respective term either in the bin or the 9613 peaks. For binned plots, this was also done for the peak regions plus 50kb.
Genomic distributions
The genomic annotation hg19 was downloaded from UCSC. Upstream was defined as 2kb upstream of the first position of the first exon in a gene. Percentages were then calculated as described above for ChromHMM for all peaks.
STARR-seq centered heatmaps and average profile plots
The average coverage was calculated in 50bp non-overlapping windows for 40kb regions centered on STARR-seq peak summits using custom scripts in R and sorted by the total occupancy in a 2kb window around the peak summit. Peaks were separated by the presence of DHS over input with a binomial P-value < 0.05 for heatmaps and meta plots. Heatmaps were made using Java TreeView version 1.6.457. Average profile plots were constructed using the colMeans function in R.
STARR-seq distal vs proximal open peaks
Peaks were considered proximal (n=951) if the peak edges were within 10kb of the nearest annotated transcript TSS. Distal intronic peaks (n=460) were more than 50kb away from the nearest annotated TSS and had a summit overlapping an intron. Intergenic peaks (N=954) were more than 50kb away from the nearest TSS and had a summit overlapping intergenic regions. All three categories required a DHS P-value < 0.001. For these 3 categories motif analysis was done over the 9613 random regions. The average coverage for ENCODE ChIP-seq datasets (Supplementary Table 3) was calculated in 50bp non-overlapping windows for 10kb regions centered on STARR-seq peak summits using custom scripts in R. Average profile plots were constructed from coverage window averages using the colMeans function in R.
ENCODE RNA-seq processing
Fastq files were downloaded from the ENCODE RNA-seq dashboard. We only considered cell lines for which polyA-selected total RNA was paired-end sequenced in at least 2 replicates. Protein coding transcripts (Gencode release 2358) were quantified using kallisto 0.43.059 with sequence bias correction (--bias) and sample bootstrapping (-b 30). For each transcript, counts were normalized to sequencing depth (cpm), summarized to gene level and over replicates and then log2 transformed using EdgeR’s cpm function (prior.count = 2).
ENCODE RNA-seq based clustering
Log2 transformed, gene-level counts were clustered using pheatmap 1.0.860 (maximum distance matrix, complete linkage clustering).
Normalized enrichment scores from i-cisTarget
Normalized enrichment scores (NES) for DNase-seq and ChIP-seq datasets were obtained from i-cisTarget26 with default settings (minimum fraction of overlap of 0.4, ROC threshold of 0.005). We filtered all DHS datasets for ENCODE DNase-seq datasets only (NES cutoff > 3.5) and only considered HeLa-S3 ChIP-seq datasets (NES cutoff > 3.0). Only the maximum score for multiple scores from the same feature description was kept. The NES scores were visualized using pheatmap (v1.0.860).
Repeat enrichments
Peaks containing repeat elements were identified by coordinate intersection with the UCSC RepeatMasker track for release GRCh3733 using bedtools. (-a peak -b repeat -F 1.0, -f 0.1). Genomic background frequencies were calculated within 1x106 randomly sampled genomic regions with the same size and chromosomal distribution as STARR-seq input fragments. Odds ratios and P-values were calculated with a two-sided Fisher’s exact test using contingency tables of insertion frequencies. P-values were adjusted to FDRs using R’s p.adjust.
Active vs. inactive ERV elements
Active and inactive ERV elements were defined by intersection with STARR-seq peaks using bedtools intersect (-a peak -b repeat with option -F 1.0 –f 0.1). For i-cisTarget analysis, we repeatedly (n=5) sampled 1,783 inactive elements and averaged NES scores from the individual subsamples. For motif analysis, all inactive elements were considered. An inactive ERV element was annotated as STAT containing if either STAT1, STAT1:STAT2, or STAT3 motifs were present within the element (N=25,203). For each element in the three groups, H3K9me3 counts were calculated with bedtools coverage with options –counts –F 0.1 and then divided by the length multiplied by 106 and displayed in log10. For STARR-seq box plots, the average coverage was calculated in R and displayed as log10 values. For both analyses, negative infinite values resulting from log10 transforming were set to the minimal in each group (for plotting only). P-values were calculated with one-sided Wilcoxon-rank-sum tests in R on non-transformed values.
Core promoter motifs in ORI
We searched for known core promoter motifs in the ORI using position-weight matrices (PWMs) for 5 selected core promoter motifs61,62. PWMs for TATA-box, Initiator (INR), downstream promoter element (DPE) and E-box were obtained from Ohler et al.62 and PWM for TCT motif from Parry et al.63. At every position along the ORI the sequence was scored against the respective PWM and the score was converted to the percentage of the maximal possible PWM score (perfect motif match). Strong motif matches (x≥90%) were visualized along the beginning of the ORI sequence around the main initiation sites within the ORI.
qPCR analysis for ISG expression
Ct values for each target gene were normalized to ACTB using the delta-Ct method64. Delta-delta-Ct values were calculated between electroporations with and without DNA and displayed in log2.
qPCR analysis for reporter assay
Firefly luciferase Ct values for each candidate enhancer were normalized to Renilla firefly Ct values using the delta-Ct method64. Delta-delta-Ct values were calculated between enhancer candidates and a negative control and displayed in log2. In the case of enhancer activity changes upon inhibitor treatment (Figure 2F), delta-delta-Ct values were calculated between electroporations with and without inhibitor treatment.
Statistics and Reproducibility
Supplementary Table 5 contains all P-values and details for t and Wilcoxon tests. We performed two replicates per NGS experiment, except if equivalent experiments independently confirmed the results.
Supplementary Material
Acknowledgements
We thank Thomas Decker and Gijs Versteeg (Max F. Perutz Laboratories & University of Vienna), Stein Aerts (VIB-KU Leuven), Petr Svoboda (Institute of Molecular Genetics of the ASCR), Peter Andersen (IMBA) and Johannes Zuber (IMP) for helpful discussions and reagents. Deep sequencing was performed at the VBCF Next-Generation Sequencing Unit (http://vbcf.ac.at). F.M. was supported by an EMBO long-term fellowship (EMBO ALTF 491–2014). Research in the Stark group is supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 647320) and by the Austrian Science Fund (FWF, F4303-B09). Basic research at the IMP is supported by Boehringer Ingelheim GmbH and the Austrian Research Promotion Agency (FFG).
Footnotes
Author Contributions
F.M. and L.M.B. are shared first authors, A.R.W and C.N. are shared second authors. F.M., L.M.B., C.N., M.R., M.P., R.R.C., K.S. and C.D.A. performed experiments. L.M.B. designed and cloned all STARR-seq and luciferase vectors. A.R.W. and F.M. performed the computational analysis with the help of M.A.Z.; V.H. and T.K. analyzed the ORI sequence. F.M. and A.S. wrote the manuscript with help from all authors. A.S supervised the project.
Competing Financial Interests Statement
The authors declare no competing financial interests.
Code availability
Custom scripts for analysis are available upon request.
Data availability
All next-generation sequencing data is available at http://www.starklab.org/ and was deposited to GEO (GSE100432). All external datasets are listed in Supplementary Table 3.
References
- 1.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 2.Santiago-Algarra D, Dao LTM, Pradel L, España A, Spicuglia S. Recent advances in high-throughput approaches to dissect enhancer function. F1000Res. 2017;6:939. doi: 10.12688/f1000research.11581.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemp NA, Hiraoka K, Kasahara N, Logg CR. Cryptic transcripts from a ubiquitous plasmid origin of replication confound tests for cis-regulatory function. Nucleic Acids Res. 2012;40:7280–7290. doi: 10.1093/nar/gks451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zabidi MA, et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saragosti S, Moyne G, Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980;20:65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- 6.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 7.Arnold CD, et al. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat Biotechnol. 2017;35:136–144. doi: 10.1038/nbt.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods. 2006;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 9.Pine R, Levy DE, Reich N, Darnell JE., Jr Transcriptional stimulation by CaPO4-DNA precipitates. Nucleic Acids Res. 1988;16:1371–1378. doi: 10.1093/nar/16.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz A-L, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 13.Dao LTM, et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat Genet. 2017 doi: 10.1038/ng.3884. [DOI] [PubMed] [Google Scholar]
- 14.Landry JJM, et al. The genomic and transcriptomic landscape of a HeLa cell line. G3. 2013;3:1213–1224. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tewhey R, et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen TA, et al. High-throughput functional comparison of promoter and enhancer activities. Genome Res. 2016;26:1023–1033. doi: 10.1101/gr.204834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 18.Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nejepinska J, Malik R, Wagner S, Svoboda P. Reporters transiently transfected into mammalian cells are highly sensitive to translational repression induced by dsRNA expression. PLoS One. 2014;9:e87517. doi: 10.1371/journal.pone.0087517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nejepinska J, Malik R, Moravec M, Svoboda P. Deep sequencing reveals complex spurious transcription from transiently transfected plasmids. PLoS One. 2012;7:e43283. doi: 10.1371/journal.pone.0043283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J Biol Chem. 2009;284:14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jammi NV, Whitby LR, Beal PA. Small molecule inhibitors of the RNA-dependent protein kinase. Biochem Biophys Res Commun. 2003;308:50–57. doi: 10.1016/s0006-291x(03)01318-4. [DOI] [PubMed] [Google Scholar]
- 23.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guertin MJ, Lis JT. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Curr Opin Genet Dev. 2013;23:116–123. doi: 10.1016/j.gde.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imrichová H, Hulselmans G, Atak ZK, Potier D, Aerts S. i-cisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res. 2015;43:W57–64. doi: 10.1093/nar/gkv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet. 2011;12:459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 28.Oler AJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 31.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smit A, Hubley R, Green P. RepeatMasker Open-4.0. 2013-2015. 2014 http://repeatmasker.org.
- 34.Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol. 2015;31:429–451. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- 35.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 37.Stamatoyannopoulos JA. What does our genome encode? Genome Res. 2012;22:1602–1611. doi: 10.1101/gr.146506.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 39.van Arensbergen J, et al. Genome-wide mapping of autonomous promoter activity in human cells. Nat Biotechnol. 2017;35:145–153. doi: 10.1038/nbt.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barakat TS, et al. Functional dissection of the enhancer repertoire in human embryonic stem cells. bioRxiv. 2017 doi: 10.1101/146696. 146696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, et al. High-resolution genome-wide functional dissection of transcriptional regulatory regions in human. bioRxiv. 2017 doi: 10.1101/193136. 193136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nehlsen K, Broll S, Bode J. Replicating minicircles: Generation of nonviral episomes for the efficient modification of dividing cells. Gene Ther Mol Biol. 2006;10:233–244. [Google Scholar]
- 43.Walters AA, et al. Comparative analysis of enzymatically produced novel linear DNA constructs with plasmids for use as DNA vaccines. Gene Ther. 2014;21:645–652. doi: 10.1038/gt.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen SQ, et al. Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 2016;26:238–255. doi: 10.1101/gr.193789.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue F, et al. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 2017;27:38–52. doi: 10.1101/gr.212092.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maricque BB, Dougherty JD, Cohen BA. A genome-integrated massively parallel reporter assay reveals DNA sequence determinants of cis-regulatory activity in neural cells. Nucleic Acids Res. 2017;45:e16. doi: 10.1093/nar/gkw942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rickels R, et al. An Evolutionary Conserved Epigenetic Mark of Polycomb Response Elements Implemented by Trx/MLL/COMPASS. Mol Cell. 2016;63:318–328. doi: 10.1016/j.molcel.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanoix J, Acheson NH. A rabbit beta-globin polyadenylation signal directs efficient termination of transcription of polyomavirus DNA. EMBO J. 1988;7:2515–2522. doi: 10.1002/j.1460-2075.1988.tb03099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishida Y, Leder P. RET: a poly A-trap retrovirus vector for reversible disruption and expression monitoring of genes in living cells. Nucleic Acids Res. 1999;27:e35. doi: 10.1093/nar/27.24.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitter JS. Random sampling with a reservoir. ACM Trans Math Softw. 1985;11:37–57. [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aken BL, et al. Ensembl 2017. Nucleic Acids Res. 2017;45:D635–D642. doi: 10.1093/nar/gkw1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. 2016 [Google Scholar]
- 56.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 57.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 58.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 60.Kolde R. pheatmap: Pretty heatmaps [Software] 2015 [Google Scholar]
- 61.FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohler U, Liao G-C, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parry TJ, et al. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 2010;24:2013–2018. doi: 10.1101/gad.1951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.