Abstract
Clinical translation of proteomic technologies is often hampered by technical limitations, including inter-laboratory inconsistencies of label-free derived relative quantification, time-consuming analytical approaches and the subsequent challenge of performing proteomic analyses on large cohorts of subjects. Here we introduce plasma QconCAT-based targeted proteomics, an approach that allows the simultaneous absolute quantitation down to the picogram level of hundreds of proteins in a single liquid chromatography-selected reaction monitoring mass spectrometry run. We demonstrate the robustness of the approach by analyzing apheresis platelet concentrate supernatants at storage day 1 and the end of the shelf life for this blood-derived therapeutic, day 5. The targeted approach was repeatable and robust revealing potential gender-specific signatures across a set of three male and female donors. This technical note represents a proof-of-principle of the application of QconCAT-based MRM strategies to transfusion-medicine relevant issues, such as storage and gender-dependent proteomic signatures in blood-derived therapeutics.
Biological significance
Gender differences in the proteome composition of apheresis platelet supernatants have always been postulated, and might underlie a higher risk of adverse reactions when transfusing apheresis products from female donors. Preliminary proteomic studies provided an overview of gender-dependent relative compositional differences in the proteome of apheresis platelet supernatants during routine storage in the blood bank. Here we apply a proteomics approach for absolute quantitation of approximately 100 proteins in apheresis platelet supernatants from male and female donors at storage days 1 and 5. Absolute quantitative proteomic analyses allowed us to confirm and expand on previous observations about gender and storage-dependency of platelet supernatant protein profiles.
Keywords: Targeted proteomics, Transfusion medicine, Donor variability, Storage
Platelet proteomics is an important research endeavor [1] that holds immediate translational potential in the fields of transfusion medicine [2–4], immunology [5], and clinical biology [6]. Transfusion of apheresis platelet concentrates represents a key component of supportive care for thrombocytopenic patients. Even though apheresis platelet concentrates can be stored up to 5–7 days in most countries, the quality of platelets is affected by storage duration, resulting in partially compromised cell morphology, hypotonic shock responses, altered cell volume, density, activation and coagulant activity [2–4]. Early studies suggested that platelet storability might be affected by donor gender as well [7]. Gender differences in donated apheresis platelets include a higher risk of anti-HLA or anti-neutrophil antibodies in plasma from parous women, which could result in adverse transfusion reactions such as transfusion-related acute lung injury (TRALI) [7]. Recent proteomic evidence has been provided about the apparent increase in the levels of pro-inflammatory factors and activation markers in the supernatants of apheresis platelets from female donors during routine storage in the blood bank. The accumulation of these factors in apheresis platelet supernatants might further increase the risk of TRALI associated with transfusion of platelet concentrates from female donors [8]. In this view, it is worth noting that, in the last few years, the supernatants of platelets concentrated for transfusion purposes have emerged as key indicators of storage quality [4,8–10]. Unfortunately, these key translational studies are compromised by the limited capacity to perform proteomic analyses on large cohorts of subjects. Indeed, these studies often rely upon time-consuming workflows characterized by a long series of time consuming steps; protein extraction, solubilization, digestion, and separation via long nano-high performance liquid chromatography–tandem mass spectrometry—HPLC–MS/MS runs. In addition, most proteomic studies performed to date are based upon relative quantitative approaches, which hamper inter-laboratory comparability of the results. To cope with these inconveniences, we propose the application of a targeted, micro-LC-selected reaction monitoring mass spectrometry (SRM) approach. This approach is based upon the absolute quantitation of trypsin-digested protein mixtures against spiked in 13C-labeled quantitative concatenamer (QconCAT) derived internal standards [11]. So far, the QconCAT method has been applied to a limited number of clinically relevant issues [12,13], basically owing to the technical difficulties associated with the selection of adequate proteotypic peptides and the generation of QconCAT constructs. Here we show its application to plasma supernatants from apheresis platelet concentrates for transfusion purposes, and demonstrate the potential of this technology to monitor storage and gender-dependency of proteomic changes in apheresis platelet supernatants during routine storage in the blood. The present results might inform policies regarding gender issues associated with apheresis platelet storability and safety.
Our results are consistent with recent untargeted label-free proteomics analyses of plasma [14] and platelets [8] from our lab. At the same time, the reported results expand upon recent literature by providing absolute quantitation values for each detected protein. Though the technology is not novel, here we show for the first time its application to a transfusion medicine-relevant issue and the potential for its implementation in routine proteomics and possibly clinical analysis of plasma-related matrices (such as supernatants of blood-derived therapeutics).
Briefly, 6 healthy donors (3 females and 3males) donated one unit of apheresis platelets using a Cobe Trima apparatus with appropriate leukoreduction (<5 × 106/unit, consistent with AABB guidelines and the literature [8]). Samples were collected on storage days 1 and 5. Platelet supernatants were obtained through serial centrifugation, aliquoted and stored at −80 °C [8]. Before quantitative proteomics analysis, depletion of the top 2 most abundant plasma proteins (Serum Albumin & IgG) was performed by Multiple Affinity Removal System™columns (4.6 × 100 mm, Agilent—Palo Alto, CA), as previously reported [8].
Four QconCAT constructs (Supplementary table 1) were designed to quantify plasma and platelet specific proteins previously found in healthy individuals, and various trauma patients. Proteotypic peptides were selected from previous experimental data (non-targeted plasma proteomics [14]) and publicly accessible databases, including PeptideAtlas [15] SRM Atlas [16] and Global Proteome Database [17]. Criteria for proteotypic peptide selection and optimal QconCAT designing were consistent with the literature [18]. The resulting list of 139 unique peptides is provided in Supplementary table 1. QconCAT DNA constructs were synthesized de novo by Genscript (Piscataway, NJ) and cloned into pET21b. Escherichia coli auxotrophic strain BL21(l)DE3-LysA ArgA [19] was transformed with the plasmid and cultured in minimal medium supplemented either with unlabeled or 13C6 arginine and 13C6 lysine at 0.1 mg/ml (Sigma Aldrich). The cells were grown to mid-log phase (A600 0.6–0.8), at which point expression was induced by adding 1 mM isopropyl-D-1-thiogalactopyranoside. After 4 h of growth at 37 °C the cells were harvested by centrifugation and processed as previously described with minor modifications [11]. Briefly, the cells were lysed with the BugBuster Protein Extraction Reagent (EMD Millipore). Inclusion bodies were suspended in 20 mM phosphate buffer, 6 M guanidinium chloride, 0.5 M NaCl, 20 mM imidazole, pH 7.4. QconCAT proteins were purified by affinity chromatography using a nickel-based resin. The purified QconCAT was desalted by three rounds of dialysis against 100 volumes of 10 mM ammonium bicarbonate, pH 8.5. Known concentrations of recombinant isotopically labeled QconCAT proteins were mixed with the depleted platelet supernatants. Trypsinization (FASP protocol-sequencing grade modified trypsin — Promega, through a 10 kDa molecular weight cutoff filter and centrifugation at 14,000 g for 15 min) was performed upon reduction (10 mM DTT in 8 M urea in 0.1 M Tris–HCl, pH 8.5, incubation for 30 min at RT) and alkylation of the samples (55 mM iodoacetamide in 8 M urea in 0.1 M Tris–HCl, pH 8.5, incubated for 30 min at RT). Six washes were then performed, with 8 M urea in 0.1 M Tris–HCl, pH 8.5 solution followed by 50 mM ammonium bicarbonate buffer. Overnight trypsin digestion at 37 °C was performed before recovering peptides from the filter using 30% ACN. Samples were then dry vacuumed to ~2 µL and reconstituted to 50 µL with 0.1% formic acid. The resultant peptide mixture was analyzed in duplicate by LC-SRM. Briefly, a targeted SRM approach was performed using the QTRAP® 5500 interfaced with a capillary HPLC system (Agilent 1200, Palo Alto, Calif). Ten µL of each sample was injected and directly loaded onto an Agilent C18 column (Zorbax SB-C18, 5 µm 150 × 0.5 mm) with 5% acetonitrile (ACN), 0.1% FA at 30 µl/min for 3 min. Chromatography was performed with mobile phase A (Milli-Q water with 0.1% formic acid) and Solvent B (acetonitrile with 0.1% formic acid). A gradient of 5–28% ACN was run for 61 min to differentially elute QconCAT peptides. The mass spectrometer was run in positive ion mode with the following settings: a source temperature of 200 °C, spray voltage of 5300 V, curtain gas of 20 psi, and a source gas of 35 psi (nitrogen gas). Multiple SRM transitions were monitored with both Q1 and Q3 quadrupoles operating at unit mass resolution to maximize selectivity and ensure specificity. SRM assay optimization was performed with the aid of Skyline v2.2 software. Collision energies (CE) and declustering potential (DP) were optimized for each transition. The energy was ramped around the predicted value in 5 steps on both sides with 1 V increments. Method building and acquisition were performed using the instrument supplied Analyst® Software (Version 1.5.2). Raw SRM data files were imported to Skyline v2.2 software for data processing (monitored transitions are detailed in the Skyline file—Supplementary file 1). Quantification was based on the ratio of corresponding light and heavy peak areas. The method is linear (across four to five logs range of concentrations) and sensitive (down to picogram level – fmol per microgram of injected proteins), selective (up to four transitions were monitored for each peptide — Supplementary File 1) and robust (chromatographic repeatability is within a 12 ± 6 s SD time window for each peptide).
The peak integration was done automatically by the software, using Savitzky–Golay smoothing, and all the data were manually inspected to confirm correct peak detection. 12C/13C ratios were normalized to standard Saccharomyces cerevisiae alcohol dehydrogenase peptides to control for technical variability during sample preparation and then back calculated to fmol/ug of protein based on 100 fmol 13C QconCAT load. Alcohol dehydrogenase peptides control for minor errors in quantity of QconCAT spiked into the samples and tests for digestion efficiency and peptide recovery post-digestion; the normalization applied is typically between 2 and 10%. Quantitative results from Skyline were exported into .xls files (Supplementary table 1) and loaded into GENE-E (v. 3.0.200 — Broad Institute, Inc.) and Multibase (http://www.numericaldynamics.com/) to plot heat maps, perform hierarchical clustering analyses (one minus Pearson correlation) and partial least-square discriminant analysis (PLS-DA).
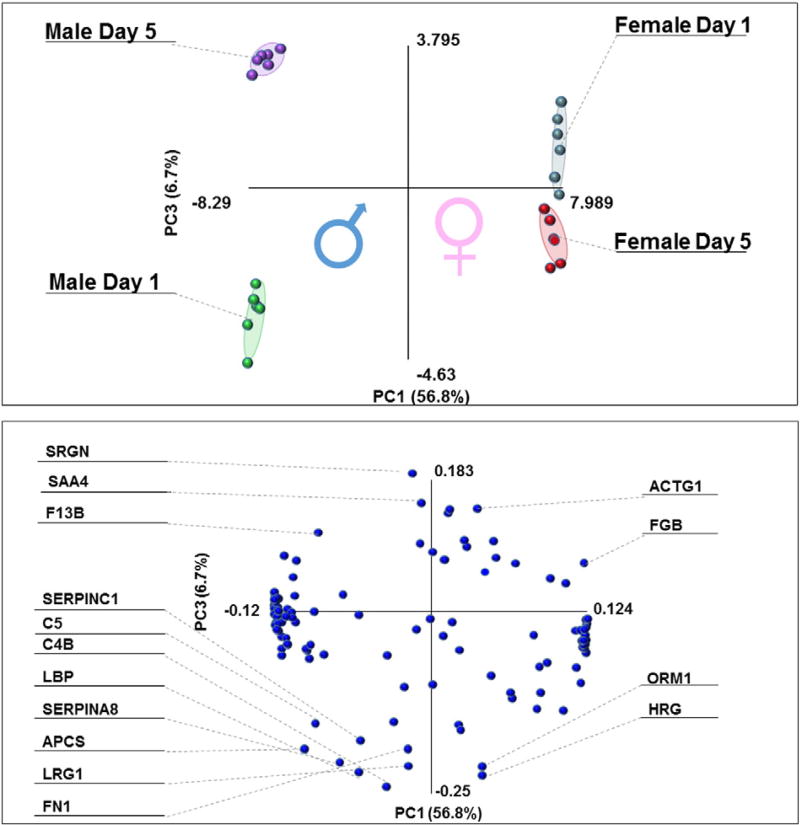
A total of 139 distinct peptides were monitored, out of which 109 were actually detected in the platelet supernatants from the 4 sample groups: female or male at storage day 1 or 5. Absolute quantitation results are provided in Supplementary table 1, while PLS-DA elaboration is reported in Fig. 1. Hierarchical clustering patterns highlighted storage (across PC1) and gender (PC2) related distributions, mostly attributable to complement proteins (C4B, C5), coagulation proteins and structural proteins (coagulation factor 13B, actin G1, fibronectin, fibrinogen beta chain). Quantitative changes in a storage and gender-dependent fashion were plotted as scatter plots and heat maps in Fig. 2. Heat maps indicate clear signatures of proteins increasing in a storage-dependent fashion or being constitutively higher in apheresis platelet supernatants from female or male donors (detailed names and hierarchical clustering – 1-Pearson correlations – are provided in Supplementary Fig. 1). From scatter plots it emerges that there is a higher susceptibility to storage lesions, as most of the proteins tended to increase at storage day 5 (Fig. 2 middle panel, upper right quadrant).
Fig. 1.
Partial least square discriminant analysis (PLS-DA) of quantitative targeted proteomics data on plasma from apheresis platelet supernatants from three male and female donors at storage days 1 and 5 (end of the shelf life) run in technical duplicates. Storage and gender dependent quantitative changes are observed across principal components 1 and 2, respectively (top panel). Correspondingly, protein variables (uniprot names) informing PLS-DA clusterization patterns are provided in the bottom panel.
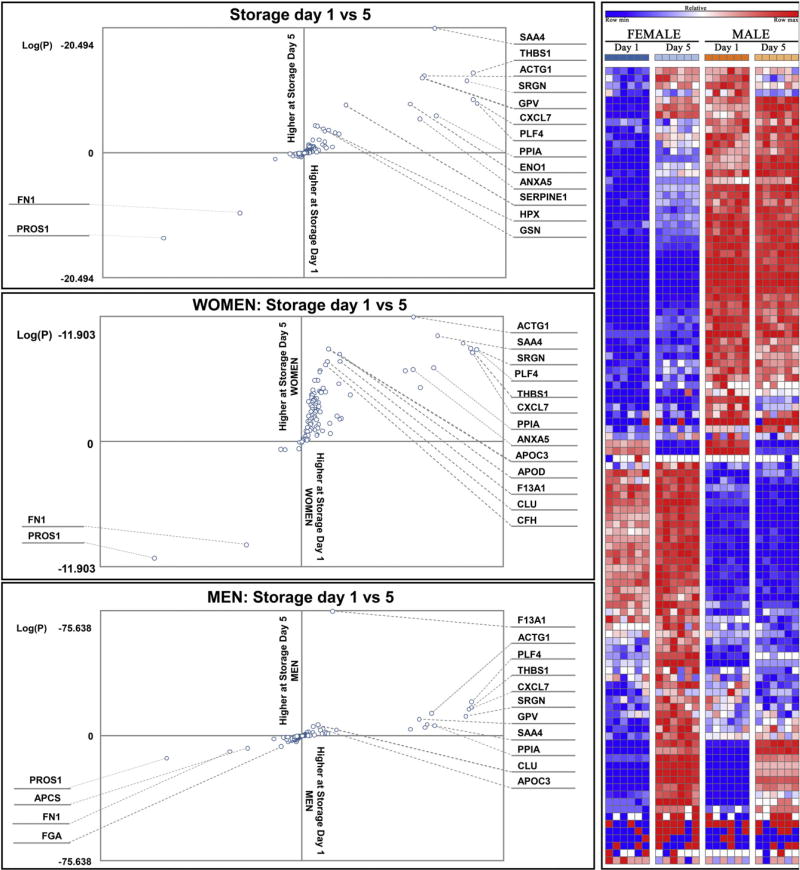
Fig. 2.
Log scale quantitative changes of proteins in storage day 1 vs 5 (top panel), either from women (middle) and men (bottom panel) donors are provided. In the right panel, quantitative trends of plasma proteins are represented as a heat map, whereby color codes are representative of changes during storage duration (blue = low, red = high). Apheresis platelets plasma proteomes from women are shifted towards progressive increase, while men donor counterparts are apparently more stable.
Consistent with previous label free proteomic results [8], storage affects platelet supernatants resulting in the accumulation of activation-associated structural proteins (actin G1), proteins involved in wound responses, and components of the complement system, especially in women (Fig. 2). These results are relevant in that they strengthen the case and provide molecular insight for the exclusion of women as apheresis platelet donors, which is already routine practice in certain countries (e.g. France and United Kingdom) due to an increased likelihood of TRALI.
Here we describe the feasibility of a clinical proteomic approach to obtain absolute protein quantitation of plasma proteins in a complex biological matrix of relevance in the field of transfusion medicine. The main advantage of the described approach is the capacity to provide absolute quantitation of hundreds of proteins in a 90 minute assay. Previous approaches in the field either relied upon 2DE [10], combinatorial hexapeptide-ligand libraries-2DE-nanoLC-MS [20], GeLC-MS strategies for preliminary pre-fractionation [8], or extended chromatographic gradients (up to 240 min) coupled with label free quantitation of detected proteins [21]. While labeling proteomics approaches like ICAT or iTRAQ [22] have been applied in the field of platelet proteomics for transfusion medicine-relevant issues or to expand our understanding of the murine platelet proteome, these approaches have resulted only in relative quantitation. Alternatively, indirect-determination of absolute protein levels has been reported for 4000 proteins, although concentrations were determined by relative comparison against thirteen platelet proteins by SILAC-Protein Epitope Signature Tags (PrESTs) [23] and not through internal spiked-in labeled proteotypic peptides for each protein of interest. The approach we describe here will be further optimized in the next few years as to expand the QconCAT protein library to include more reporter “proteotypic” peptides per target protein and to cover additional proteins of interest. Faster analytical workflows and improved sensitivity will result from the implementation of latest generation UPLCs and faster scanning MS instruments.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2015.02.010.
Supplementary Material
Acknowledgments
The authors are grateful to Drs. Christopher C. Silliman and Marguerite R. Kelher (Bonfils Blood Center, Denver, CO, USA) for providing the samples assayed in this study.
Financial support
This work was supported in part by grants from the National Institutes of Health, National Institute of General Medical Sciences grants: T32-GM008315 and P50-GM049222, and by NIH/NCATS Colorado CTSA (UL1 TR001082).
Footnotes
Conflict of interest
The authors disclose no conflict of interest.
Transparency Document
The Transparency document associated with this article can be found, in the online version.
References
- 1.Burkhart JM, Gambaryan S, Watson SP, Jurk K, Walter U, Sickmann A, et al. What can proteomics tell us about platelets? Circ Res. 2014;114:1204–19. doi: 10.1161/CIRCRESAHA.114.301598. http://dx.doi.org/10.1161/CIRCRESAHA.114.301598. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Schubert P, Culibrk B, Devine DV. p38MAPK is involved in apoptosis development in apheresis platelet concentrates after riboflavin and ultraviolet light treatment. Transfusion (Paris) 2014 doi: 10.1111/trf.12905. http://dx.doi.org/10.1111/trf.12905. [DOI] [PubMed]
- 3.Prudent M, D’Alessandro A, Cazenave J-P, Devine DV, Gachet C, Greinacher A, et al. Proteome changes in platelets after pathogen inactivation—an interlaboratory consensus. Transfus Med Rev. 2014;28:72–83. doi: 10.1016/j.tmrv.2014.02.002. http://dx.doi.org/10.1016/j.tmrv.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Thiele T, Iuga C, Janetzky S, Schwertz H, Gesell Salazar M, Fürll B, et al. Early storage lesions in apheresis platelets are induced by the activation of the integrin αIIbβ3 and focal adhesion signaling pathways. J Proteomics. 2012;76:297–315. doi: 10.1016/j.jprot.2012.04.057. http://dx.doi.org/10.1016/j.jprot.2012.04.057 [Spec] [DOI] [PubMed] [Google Scholar]
- 5.Klockenbusch C, Walsh GM, Brown LM, Hoffman MD, Ignatchenko V, Kislinger T, et al. Global proteome analysis identifies active immunoproteasome subunits in human platelets. Mol Cell Proteomics MCP. 2014 doi: 10.1074/mcp.M113.031757. http://dx.doi.org/10.1074/mcp.M113.031757. [DOI] [PMC free article] [PubMed]
- 6.Van Holten TC, Bleijerveld OB, Wijten P, de Groot PG, Heck AJR, Barendrecht AD, et al. Quantitative proteomics analysis reveals similar release profiles following specific PAR-1 or PAR-4 stimulation of platelets. Cardiovasc Res. 2014;103:140–6. doi: 10.1093/cvr/cvu113. http://dx.doi.org/10.1093/cvr/cvu113. [DOI] [PubMed] [Google Scholar]
- 7.Gajic O, Yilmaz M, Iscimen R, Kor DJ, Winters JL, Moore SB, et al. Transfusion from male-only versus female donors in critically ill recipients of high plasma volume components. Crit Care Med. 2007;35:1645–8. doi: 10.1097/01.CCM.0000269036.16398.0D. http://dx.doi.org/10.1097/01.CCM.0000269036.16398.0D. [DOI] [PubMed] [Google Scholar]
- 8.Dzieciatkowska M, D’Alessandro A, Burke TA, Kelher MR, Moore EE, Banerjee A, et al. Proteomics of apheresis platelet supernatants during routine storage: gender-related differences. J Proteomics. 2014;112C:190–209. doi: 10.1016/j.jprot.2014.08.016. http://dx.doi.org/10.1016/j.jprot.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schubert P, Culibrk B, Karwal S, Slichter SJ, Devine DV. Optimization of platelet concentrate quality: application of proteomic technologies to donor management. J Proteomics. 2012;76:329–36. doi: 10.1016/j.jprot.2012.06.023. http://dx.doi.org/10.1016/j.jprot.2012.06.023 [Spec] [DOI] [PubMed] [Google Scholar]
- 10.Glenister KM, Payne KA, Sparrow RL. Proteomic analysis of supernatant from pooled buffy-coat platelet concentrates throughout 7-day storage. Transfusion (Paris) 2008;48:99–107. doi: 10.1111/j.1537-2995.2007.01487.x. http://dx.doi.org/10.1111/j.1537-2995.2007.01487.x. [DOI] [PubMed] [Google Scholar]
- 11.Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat Protoc. 2006;1:1029–43. doi: 10.1038/nprot.2006.129. http://dx.doi.org/10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- 12.Achour B, Russell MR, Barber J, Rostami-Hodjegan A. Simultaneous quantification of the abundance of several cytochrome P450 and uridine 5′-diphospho-glucuronosyltransferase enzymes in human liver microsomes using multiplexed targeted proteomics. Drug Metab Dispos. 2014;42:500–10. doi: 10.1124/dmd.113.055632. http://dx.doi.org/10.1124/dmd.113.055632. [DOI] [PubMed] [Google Scholar]
- 13.Russell MR, Achour B, Mckenzie EA, Lopez R, Harwood MD, Rostami-Hodjegan A, et al. Alternative fusion protein strategies to express recalcitrant QconCAT proteins for quantitative proteomics of human drug metabolizing enzymes and transporters. J Proteome Res. 2013;12:5934–42. doi: 10.1021/pr400279u. http://dx.doi.org/10.1021/pr400279u. [DOI] [PubMed] [Google Scholar]
- 14.Silliman CC, Dzieciatkowska M, Moore EE, Kelher MR, Banerjee A, Liang X, et al. Proteomic analyses of human plasma: Venus versus Mars. Transfusion (Paris) 2012;52:417–24. doi: 10.1111/j.1537-2995.2011.03316.x. http://dx.doi.org/10.1111/j.1537-2995.2011.03316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–34. doi: 10.1038/embor.2008.56. http://dx.doi.org/10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picotti P, Lam H, Campbell D, Deutsch E, Mirzaei H, Ranish J, et al. A database of validated assays for the targeted mass spectrometric analysis of the S. cerevisiae proteome. Nat Methods. 2008;5:913–4. doi: 10.1038/nmeth1108-913. http://dx.doi.org/10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–42. doi: 10.1021/pr049882h. http://dx.doi.org/10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 18.Brownridge P, Holman SW, Gaskell SJ, Grant CM, Harman VM, Hubbard SJ, et al. Global absolute quantification of a proteome: challenges in the deployment of a QconCAT strategy. Proteomics. 2011;11:2957–70. doi: 10.1002/pmic.201100039. http://dx.doi.org/10.1002/pmic.201100039. [DOI] [PubMed] [Google Scholar]
- 19.Matic I, Jaffray EG, Oxenham SK, Groves MJ, Barratt CLR, Tauro S, et al. Absolute SILAC-compatible expression strain allows Sumo-2 copy number determination in clinical samples. J Proteome Res. 2011;10:4869–75. doi: 10.1021/pr2004715. http://dx.doi.org/10.1021/pr2004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egidi MG, Rinalducci S, Marrocco C, Vaglio S, Zolla L. Proteomic analysis of plasma derived from platelet buffy coats during storage at room temperature. an application of ProteoMiner™ technology. Platelets. 2011;22:252–69. doi: 10.3109/09537104.2010.550348. http://dx.doi.org/10.3109/09537104.2010.550348. [DOI] [PubMed] [Google Scholar]
- 21.Smalley DM, Root KE, Cho H, Ross MM, Ley K. Proteomic discovery of 21 proteins expressed in human plasma-derived but not platelet-derived microparticles. Thromb Haemost. 2007;97:67–80. [PubMed] [Google Scholar]
- 22.Thon JN, Schubert P, Duguay M, Serrano K, Lin S, Kast J, et al. Comprehensive proteomic analysis of protein changes during platelet storage requires complementary proteomic approaches. Transfusion (Paris) 2008;48:425–35. doi: 10.1111/j.1537-2995.2007.01546.x. http://dx.doi.org/10.1111/j.1537-2995.2007.01546.x. [DOI] [PubMed] [Google Scholar]
- 23.Frese CK, Altelaar AFM, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, et al. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J Proteome Res. 2011;10:2377–88. doi: 10.1021/pr1011729. http://dx.doi.org/10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.