Abstract
The ubiquitous presence of SPFH (Stomatin, Prohibitin, Flotillin, HflK/HflC) proteins in all domains of life suggests that their function would be conserved. However, SPFH functions are diverse with organism-specific attributes. SPFH proteins play critical roles in physiological processes such as mechanosensation and respiration. Here, we characterize the stomatin ORF19.7296/SLP3 in the opportunistic human pathogen Candida albicans. Consistent with the localization of stomatin proteins, a Slp3p-Yfp fusion protein formed visible puncta along the plasma membrane. We also visualized Slp3p within the vacuolar lumen. Slp3p primary sequence analyses identified four putative S-palmitoylation sites, which may facilitate membrane localization and are conserved features of stomatins. Plasma membrane insertion sequences are present in mammalian and nematode SPFH proteins, but are absent in Slp3p. Strikingly, Slp3p was present in yeast cells, but was absent in hyphal cells, thus categorizing it as a yeast-phase specific protein. Slp3p membrane fluorescence significantly increased in response to cellular stress caused by plasma membrane, cell wall, oxidative, or osmotic perturbants, implicating SLP3 as a general stress-response gene. A slp3Δ/Δ homozygous null mutant had no detected phenotype when slp3Δ/Δ mutants were grown in the presence of a variety of stress agents. Also, we did not observe a defect in ion accumulation, filamentation, endocytosis, vacuolar structure and function, cell wall structure, or cytoskeletal structure. However, SLP3 over-expression triggered apoptotic-like death following prolonged exposure to oxidative stress or when cells were induced to form hyphae. Our findings reveal the cellular localization of Slp3p, and for the first time associate Slp3p function with the oxidative stress response.
Introduction
The SPFH protein superfamily is widely conserved throughout all domains of life, but the distribution of SPFH proteins varies in different organisms [1]. Although the structurally conserved SPFH domain is the defining feature, the N- and C- terminal regions are highly divergent [2]. SPFH proteins localize to the plasma membrane and organelle membranes, such as the endoplasmic reticulum, mitochondria, vacuole, and lysosome [2]. SPFH protein functions have been extensively investigated in mammals, nematodes, and several microbes. SPFH proteins have roles in mechanosensation, cell fusion, apoptosis, respiration, morphogenesis, storage, transport, and cell signaling [2–6]. These processes are essential to the pathogenicity of Candida albicans, the major fungal pathogen of humans and focus of this study.
Stomatin (STOM), the founding member of SPFH proteins, was first isolated from the plasma membrane of human erythrocytes [7, 8]. The absence of STOM causes a non-lethal hemolytic anemia known as hereditary stomatocytosis [9]. Mutations to the orthologous stomatins MEC-2 in the nematode Caenorhabditis elegans and STOML-3 in mice resulted in a loss of sensitivity to mechanical stimuli [10, 11]. Importantly, inhibitors that target STOML-3 may represent a new class of drugs to treat patients with severe nerve injury or diabetic neuropathy and ameliorate acute touch-sensitive pain [12].
Stomatins display both homo- and hetero- oligomerization. The SPFH domain is important for oligomerization, because mutations to SPFH domain sequences abolished homo-oligomerization and plasma membrane localization in mammalian stomatin orthologs [13]. Hetero-oligomeric complexes were observed with human stomatin and various proteins such as the glucose transporter GLUT1, band 3 ion transporter, and the water transporter aquaporin-1 [14]. Also, STOML-3 oligomers interact with Piezo mechanically-gated ion channels in mechanosensation [12]. The diverse functional roles, widespread conservation of SPFH proteins and potential clinical applications underscores the biological significance of the SPFH family.
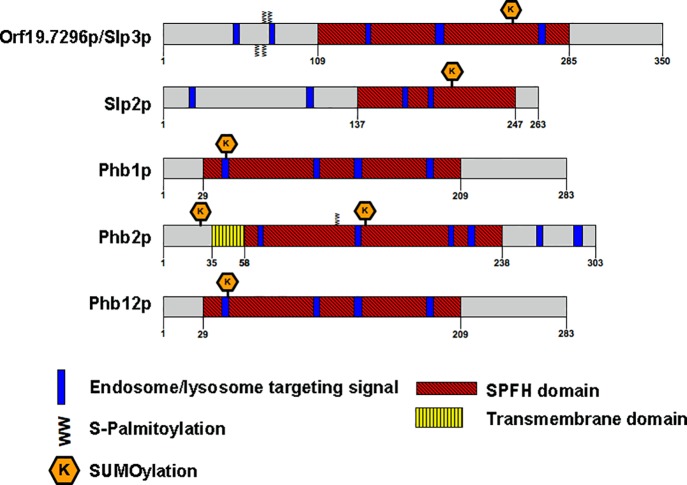
In contrast, SPFH functional information is limited in fungi. In the model yeast Saccharomyces cerevisiae the sole SPFH proteins, prohibitin 1 and prohibitin 2 (Phb1p/2p), interacts with Atp23p at the inner mitochondrial membrane to assist formation of ATP synthase [15]. The flotillin FloA, mediates formation of plasma membrane sterol-rich domains, and the stomatin StoA, is required for hyphal polarized growth in the filamentous fungus Aspergillus nidulans [6]. The C. albicans genome includes 5 SPFH family members: PHB1, PHB2, PHB12, SLP2 (stomatin-like protein 2) and Open Reading Frame (ORF) 19.7296 (Fig 1). We previously demonstrated that ORF19.7296 is a major target of the High Osmolarity Glycerol (HOG) signaling pathway. ORF19.7296 transcription is significantly increased in cells exposed to acute cationic stress and requires the HOG pathway signaling components Hog1p and Sko1p for full expression [16]. Proteomic analysis implied that Orf19.7296p resides in the plasma membrane [17]. However, Orf19.7296p function remains uncharacterized. More importantly, there is no functional information available for any C. albicans SPFH proteins.
Fig 1. Structure of C. albicans SPFH proteins.
Schematic representation of Orf19.7296p/Slp3p compared to C. albicans SPFH family members. Images are drawn to scale.
Here, we utilized a genetic and cellular approach to characterize ORF19.7296. The Candida Genome Database (CGD) lists the gene name SLP99 as an alias for ORF19.7296 [18]. However, SLP99 is also used to describe an unrelated gene. Therefore, we designate ORF19.7296 as SLP3 (Stomatin Like Protein 3). We show that SLP3 is a yeast-phase specific general stress response gene. Further, our mutational analysis revealed that Slp3p over-production caused apoptotic-like death when cells are subjected specifically to oxidative stress or when induced to form hyphae.
Methods
Yeast strains and media
All C. albicans strains used in this study were derived from the wild-type (wt) reference strain BWP17 (ura3Δ::λimm434/ura3Δ::λimm434, arg4::hisG/arg4::hisG, his1::hisG/his1::hisG) or its derivative strains DAY185 and DAY286 [19, 20]. All strains used in experimental assays were isogenic, and genotypes are listed in Table 1. Cultures were prepared in YPD+uri (1% yeast extract, 2% peptone, 2% dextrose, 80 mg/L uridine) media at 30°C with shaking at 225 rpm unless otherwise noted.
Table 1. Yeast strains used in this study.
| Strain | Genotype |
|---|---|
| DAY185 | ura3Δ::λimm434/ura3Δ::λimm434, ARG::URA3::arg4::hisG/arg4::hisG, HIS1::his1::hisG/his1::hisG |
| ES11 | ura3Δ::λimm434/ura3Δ::λimm434, arg4::hisG/arg4::hisG, his1::hisG::pHIS1/his1::hisG, slp3::URA3/slp3::ARG4 |
| ES26 | ura3Δ::λimm434/ura3Δ::λimm434, ARG4::URA3::arg4::hisG/arg4::hisG, his1::hisG/his1::hisG, SLP3/SLP3-pYFP-HIS1 |
| JMR249 | ura3Δ::λimm434/ura3Δ::λimm434, ARG4::URA3::arg4::hisG/arg4::hisG, his1::hisG/his1::hisG, SLP3/PTDH3SLP3-pYFP-HIS1 |
| JMR255 | ura3Δ::λimm434/ura3Δ::λimm434, ARG4::URA3::arg4::hisG/arg4::hisG, HIS1::his1::hisG/his1::hisG, SLP3/PTDH3SLP3 |
Construction of yeast strains
The SLP3-yellow fluorescent protein (YFP) tagged strain, slp3Δ/Δ homozygous null mutant strain, PTDH3SLP3, and PTDH3SLP3-YFP over-expressing strains were constructed by PCR-mediated homologous recombination [19, 20]. Oligonucleotide sequences are listed in Table 2. Yeast transformations followed the lithium acetate protocol and were verified by colony PCR [21]. We used plasmid pMG1656 [22] as a template to produce an amplicon containing YFP-, HIS1-, and SLP3- specific sequences. The amplicon was transformed into C. albicans strain DAY286 and integrated directly following the final amino acid encoding codon to produce strain ES26.
Table 2. Oligonucleotide sequences.
| Primer Name | Oligonucleotide Sequence 5' - 3' |
|---|---|
| SLP3FWDPR | AATATTTGAATTTGTGGGGTTTTGATCTGTTGTAGATGCTCATAACTATCAATCAATTATTGCATTGCCTCAATTCGTGTTTTAATTTGTGTAGATACTGTTTCCCAGTCACGACGTT |
| SLP3REVPR | AGCTATCTGTAGTGCCTAAGAATGCAGACACGTTAACCTTCTCCCAACCTCCATCACCACTTCGAGTACTATTTAAATTGGGTCAAGAGTATCACAAATTGTGGAATTGTGAGCGGATA |
| SLP3FWDdet | CCGCTCCCAGCTATGTACAA |
| SLP3REVdet | TCATACGAGCAAATGAACAA |
| SLP3FWDcomp | TTGA TCAGCCATTT ATTTTCGTT |
| SLP3REVcomp | TCATACGAGCAAATGAACAA |
| SLP3/YFPFWDPR | AAGGATTGGGTAGGTACTCCACAAAGTATTGGTATTAACAATAATAGGCATATCAGTGAGACAGTTGCATTGCAAGAAGCCATGAGAGTAGGTGGTGGTTCTAAAGGTGAAGAATTATT |
| SLP3/YFPRVPR | GGGGACAGGGGTCCACAAAAGATCAAATATCCAACCCAGTTGGAGATGGGGATAACAACTAAGAAACCATTAATA GAATTCCGGAATATTTATGAGAAAC |
| SLP3OEFWD | ACGTAATTGATCAGCCATTTATTTTCGTTTAAAACATTACCTTTGTGGTACGTGGTATCTCCGAGCTGTATACTGTCAATATCGATAGCAAAGAAGAAAAATCAAGCTTGCCTCGTCCCC |
| SLP3OERV | TCTCTGGTTTGTGAGTTGGTTGATCGGTAATTGCTGCTTGGGACTTTTTATAAGTATCAGGATCAAAAGAATCAGTGGAATGATTTGATTGAGCTTGCATATTTGAATTCAATTGTGATG |
| SLP3/OEFwDetPr1 | ACGTGGTATCTCCGAGCTGT |
| SLP3/OERvDetPr2 | TCAATGGTGGATCAACTGGA |
To construct the slp3Δ/Δ homozygous null mutant strain, ES11, both SLP3 alleles were replaced with URA3 and ARG4 via PCR-mediated gene deletion and brought to HIS prototrophy through transformation with NruI-digested plasmid pDDB78. To construct the PTDH3SLP3-YFP and PTDH3SLP3 over-expressing strains, JMR249 and JMR255, we used plasmid pCJN542 as a template to produce an amplicon containing the glyceraldehyde-3-phosphate dehydrogenase (TDH3) promoter sequences, nourseothricin resistance gene (NAT), and SLP3-specific sequences. We chose the TDH3 promoter for over-expression analysis, because it was shown to be constitutively active under C. albicans invasive conditions [23]. The amplicon was transformed into the C. albicans SLP3-YFP strain (ES26) and wt strain (DAY 185) and integrated directly upstream of the SLP3 start codon.
Microscopy and flow cytometry
Approximately 5 μL of cell suspension was spotted onto poly-lysine coated slides for microscopic analysis. Samples were observed at a total magnification of 1000X with an EVOS compound light microscope (Thermofisher) equipped with the appropriate light cubes for fluorescent visualization. All photographs were processed with ImageJ software. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times unless otherwise noted, and data presented represents one representative experiment.
Fluorescent cells were quantified using an Attune NxT Flow Cytometer (Life Technologies) equipped with a 50 mW, 488 nm LED laser and 530/30 nm emissions filters. Data was analyzed using Attune NxT Software v2.2. Acquisition settings were set to wt untagged control cells by adjusting the voltage to the third logarithmic decade of the fluorescence channel on a histogram plot. We gated out all events with fluorescence below the maximum value for wt untagged control cells to account for potential yeast cell auto-fluorescence, unless otherwise noted. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Ten thousand events were collected for each sample, and the Median Fluorescent Intensity (MFI) was recorded. The average MFI was determined, and statistical differences between samples were analyzed in paired t-tests.
Slp3p localization assays
C. albicans cultures were grown overnight for 16 hours in 5 mL YPD+uri. Approximately 5.0 x 107 cells were introduced to fresh YPD+uri and incubated for 3–4 hours. Approximately 3.0 x 107 cells were harvested, and resuspended in 100 μL of YPD+uri. To visualize Slp3p within the vacuole, samples were incubated with 160 μM of the lipophilic styryl dye FM 4–64 (Life Technologies) and incubated in the dark at room temperature for 40 minutes (pulse). Cells were harvested, resuspended in 1 mL of YPD+uri and incubated for 2.5 hours at 30°C (chase). Since FM 4–64 can be used to visualize endosome to vacuolar membrane transport in a time-dependent manner, chase was performed at 15 and 30 minutes to monitor endocytosis and 2.5 hours to ensure only visualization of the vacuole [24]. Cells were washed three times with 1X Phosphate-buffered saline (PBS) solution pH 7.0, resuspended in 1X PBS, and examined under the microscope.
To visualize Slp3p within the plasma membrane, overnight cultures were diluted to an OD600nm of 0.2 in 5 mL of YPD+uri and incubated until an OD600nm of ~0.8–1.0 was obtained. Cells were harvested, washed once with 1X PBS, and 1 x 105 cells were resuspended in 1 mL of 1X PBS. Samples were incubated with 200 μg/mL Filipin III (Sigma) for 5 minutes at room temperature in the dark and examined under the microscope.
To examine Slp3p localization kinetics, overnight cultures were inoculated to an OD600nm of 0.2 in 5 mL of YPD+uri and incubated for the indicated culture time periods. The time points of inoculation were staggered to achieve the indicated culture times and allow for analysis of the samples at the same time. Once all indicated culture time periods were achieved, samples were washed with 1X PBS, diluted to an OD600nm of 1.0 in 1 mL of 1X PBS, and visualized under the microscope. Fluorescent cells were quantified with flow cytometry.
To examine Slp3p localization under environmental stress, cells from overnight cultures were diluted to an OD600nm of 0.2 in 5 mL of YPD+uri and incubated until an OD600nm of ~0.8–1.0 was obtained. Next, cells were treated with the indicated stress agent (or dH2O as a control) and incubated for 45 minutes at 30°C. Following treatment, samples were immediately examined under the microscope, and fluorescence was quantified with flow cytometry.
To examine Slp3p in the yeast-to-hyphae transition, yeast cells from overnight cultures were harvested and washed three times with 1X PBS buffer. Samples were diluted to an OD600nm of 0.2 in 5 mL of YPD+uri supplemented with 10% Fetal Bovine Serum (FBS) or in Spider medium (1% nutrient broth, 1% mannitol, 11.5 M potassium phosphate, pH 7.2). The cultures were grown for approximately 16–24 hours at 37°C, washed three times with 1X PBS, resuspended in 1 mL of PBS, and examined under the microscope. To examine stress-induced localization of Slp3p in hyphal cells, samples were prepared as described above; however, hyphal cells were resuspended in 5 mL of YPD+uri supplemented with 10% FBS and the indicated additive (or dH20 for control). Cultures were incubated at 37°C with shaking for 45 minutes followed by microscopic examination.
Phenotype assays
To examine cytoskeleton structure, cells from overnight cultures were diluted to an OD600nm of 0.2 in YPD+uri and grown for 3 and 16 hours with shaking. Cultures were standardized to an OD600nm of 1.0 and were incubated with 6.6 μM phalloidin rhodamine (Invitrogen) to stain actin filaments as previously described [25]. To visualize tubulin, samples were stained with 150 nM Tubulin Tracker Green (Invitrogen) following manufacturer’s instructions. To visualize the cell wall, cells were stained with 100 μg/mL calcofluor white as previously described [25]. All samples were examined under a microscope as described above.
To examine vacuolar acidification, overnight cultures were standardized to 5.0 x 107 cells/mL in YPD+uri and incubated for 3 hours. Approximately 3.0 x 107 cells were stained with 10 μM 2′,7′-Bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester (BCECF AM; Invitrogen) for 40 minutes. Cells were washed three times with 1X PBS and examined under the microscope. To quantitatively examine vacuolar acidification in exponential and stationary phase cultures, the time points of inoculation and sample preparation were performed as described in the Slp3p kinetic localization assays. Samples were quantified using flow cytometry.
Growth assays on solid nutrient growth medium were performed as previously described [26]. Briefly, C. albicans overnight cultures were diluted to a starting OD600nm of 3.0. Samples were serially diluted, spotted onto designated plates, incubated at 30°C, and photographed after 1–3 days of growth.
To examine growth kinetics under oxidative stress, overnight cultures were standardized to an OD600nm of 0.2 in 200 μL YPD+uri and YPD+uri supplemented with 0.08% sodium dodecyl sulfate (SDS) or 0.17% hydrogen peroxide (H2O2) in a 96 well plate. Samples were grown for 24 hours with shaking, and the OD600nm was acquired every 30 minutes using a Synergy Mx plate reader (Biotek).
Inductively coupled plasma mass spectrometry was performed to assess intracellular ion levels. C. albicans cultures were prepared by the procedure outlined in Eide et al [27] with the following adjustments. Overnight cultures of C. albicans were grown for 16 hours and diluted to an OD600nm of 0.2 in 25 mL YPD+uri media. Samples were incubated with shaking at 30°C to an OD600nm of 1.0. To examine the effects of cation stress, cultures were split and one half was treated with distilled water, while the other was treated with 1.0 M NaCl. The cells were harvested and washed three times in 1 μM EDTA buffer, pH 7.0. Cell pellets were lyophilized overnight at 4°C and the mass recorded. Approximately 100 mg of each sample was acid-digested and analyzed by inductively coupled plasma mass spectrometry at the University of Georgia Center for Isotope Studies. Three biological replicates of each strain were used for each assay, and two independent experiments were performed.
Apoptosis assays
Overnight cultures were diluted to an OD600nm of 0.2 in 5 mL of YPD+uri supplemented with 0.08% SDS or 0.17% H2O2 and incubated. Cells were harvested after 3 and 16 hours, washed with 1X PBS, and approximately 1.0 x 106 cells were resuspended in 1 mL of 1X PBS. Samples were treated with 14 μM Propidium Iodide (PI, Thermofisher) and incubated in the dark for 20 minutes at room temperature. Cells were harvested, washed once with 1X PBS, and examined under the microscope. PI-labeled cells were quantified with flow cytometry. We used heat-killed wt cells to establish a profile of inviable cells and used 574/26 nm emissions filters. At least three independent experiments were performed.
To determine the mitochondrial membrane potential, cultures were grown, subjected to oxidative stress, and prepared as described above. Samples were treated with 1X JC-10 (Sigma) following the manufacturer’s protocol. Untreated samples were used as a negative control. We treated wt cells with 10 μM of the mitochondrial membrane perturbant carbonyl cyanide 4-(trifluoromethoxy) phenyl hydrazone (FCCP; ABCAM) as a positive control to profile cells with depolarized mitochondria. JC-10 fluorescence was monitored by flow cytometry using 530/30 nm and 574/26 nm emissions filters.
To determine ROS production, cultures were grown, subjected to oxidative stress, and prepared as described above. Samples were incubated with 5 μg/mL dihydrorhodamine 123 (DHR-123; Sigma) for 10 minutes in the dark at room temperature and then quantitatively analyzed by flow cytometry. Untreated DHR-123-labeled samples were used as controls.
To examine the viability of hyphal cells, samples were prepared as described in the Slp3p localization assays section and treated with 14 μM PI. Samples were visualized under the microscope.
Slp3p primary sequence analysis
CGD (http://www.candidagenome.org/) was utilized to search for all SPFH family members in C. albicans. The Aspergillus Genome Database (AspGD, http://www.aspgd.org/) was used to retrieve SPFH sequences from A. nidulans. Sequences for human, mouse, and nematode SPFH family members were retrieved from UniProt and NCBI: HsSTOM (accession: P27105), HsSTOML1 (accession: Q9UBI4), HsSTOML2 (accession: Q9UJZ1), HsSTOML3 (accession: Q8TAV4), HsPodocin (accession: Q9NP85), MmStoml3 (accession: Q6PE84), CeStomatin2 (accession: NP_001257021.1). Sequences were aligned using T-Coffee [28] with default parameters and then formatted using Boxshade (http://www.ch.embnet.org/software/BOX_form.html). Sequence identities and similarities between Slp3p and other SPFH family members were determined using Protein BLAST with default parameters. Post-translational modification and organelle targeting predictions were made using the bioinformatics resource portal ExPASy (https://www.expasy.org/). Protein schematics were constructed using Illustrator for Biological Sequences, version 1.0 [29].
Results and discussion
Primary sequence analysis of Slp3p
SLP3 was annotated as a C. albicans stomatin based on homology to the SPFH domain from Pyrococcus horikoshii pSTOM [18]. The SPFH domain comprises approximately 50% of Slp3p (Fig 1 [18]). In several mammalian and nematode stomatins, sequences both N- and C-terminal to the SPFH domain are required for membrane insertion, organelle targeting, lipid transport, and contain sites for post-translational modifications [3]. We aligned the full sequence of Slp3p against mammalian, fungal, and nematode stomatins to evaluate the N- and C-terminal regions. We also analyzed the primary sequence of Slp3p for post-translational modification consensus sites and organelle targeting sequences using 8 predictor programs (S1 Table). We did not detect any significant sequence similarities in the N- and C-terminal regions of Slp3p compared to those in mammalian and nematode stomatins (S1 Fig). A short hydrophobic membrane sequence that allows insertion into the plasma membrane with a hairpin-like topology is present in mammalian and nematode stomatins [2]. A highly conserved proline residue within this region is essential for its hairpin-like formation [3]. We did not observe homologous sequences in Slp3p, suggesting that a membrane insertion region may be absent (S1 Fig). We found that the SPFH domain of Slp3p was highly similar to those in mammalian, nematode, and fungal stomatins (55–78% similarity, Table 3). Notably, Trp184 is essential for homo-oligomerization in human STOM [30] and is conserved in Slp3p (S1 Fig). We did not identify C-terminal sequences required for sterol binding as observed in human STOML-1 (S1 Fig [31]).
Table 3. Summary of primary sequence alignments.
| Proteina | Entire Sequenceb | SPFH Domainc | ||||
|---|---|---|---|---|---|---|
| Identity (%) | Similarity (%) | E-valued | Identity (%) | Similarity (%) | E-valued | |
| CaSlp3p | 100 | 100 | 0 | 100 | 100 | 7.00e-131 |
| CaSlp2p | 31 | 53 | 7.00e-19 | 34 | 55 | 2.00e-16 |
| CaPhb1p | NS | NS | NS | NS | NS | NS |
| CaPhb2p | NS | NS | NS | NS | NS | NS |
| CaPhb12p | NS | NS | NS | NS | NS | NS |
| AnStoA | 52 | 72 | 7.00e-103 | 58 | 78 | 4.00e-62 |
| AnFloA | NS | NS | NS | NS | NS | NS |
| HsSTOM | 32 | 59 | 2.00e-33 | 36 | 61 | 1.00e-24 |
| HsSTOML1 | NS | NS | NS | NS | NS | NS |
| HsSTOML2 | 32 | 54 | 9.00e-27 | 34 | 56 | 3.00e-25 |
| HsSTOML3 | 34 | 59 | 2.00e-35 | 35 | 61 | 3.00e-28 |
| HsPodocin | 29 | 54 | 9.00e-28 | 28 | 58 | 8.00e-22 |
| MsStoml3 | 34 | 59 | 8.00e-36 | 35 | 61 | 3.00e-28 |
| CeStomatin2 | 31 | 56 | 7.00e-37 | 34 | 64 | 1.00e-35 |
a This column lists the SPFH family protein from its respective organism. Ca: Candida albicans; An: Aspergillus nidulans; Hs: Homo sapiens; Ms: Mus musculus; Ce: Caenorhabditis elegans.
bThis column shows the percent identity, similarity and E-values of Slp3p compared against the listed SPFH family members as determined using Protein BLAST with default parameters.
c This column shows the percent identity, similarity and E-values of the Slp3p SPFH domain exclusively compared against the listed SPFH family members. For fungal SPFH family members, SPFH domain sequences were retrieved from the domain/motifs page on CGD and AspGD, respectively. For mammalian SPFH family members, SPFH domain sequences were retrieved from the family & domains section of each protein on UniProt. For CeStomatin2, the SPFH domain sequence was retrieved from the Features section of the protein on NCBI.
dComparisons that provided an E-value > e-10 were considered non-significant (NS) as previously described [5].
Consistent with stomatin orthologs, we identified four putative S-Palmitoylation sites in the N-terminal region at residues Cys65, Cys69, Cys70, and Cys72 (Fig 1 and S1 Fig), which may mediate plasma membrane localization and subcellular trafficking [32]. Human stomatin-like protein-1 (SLP-1) contains a GYXXφ motif in the N-terminal region, where X is any amino acid and φ is a bulky hydrophobic residue (GYRAL). This motif targets SLP-1 to late endosomal compartments where SLP-1 controls cholesterol transfer [31]. We found that Slp3p also contains a similar motif (GYQSF) in the N-terminal region and four additional endosomal targeting motifs, suggesting that it may localize to endosomes (Fig 1 and S1 Fig). We also found a potential SUMOylation site that may facilitate membrane targeting and subcellular trafficking (Fig 1 and S1 Fig [33, 34]).
In stomatins, sequences N- and C-terminal to the SPFH domain are usually divergent among family members [3]. We find that this feature is similar in C. albicans Slp3p. Moreover, our findings show that sequences which govern plasma membrane localization and organelle targeting in mammalian stomatins are conserved in Slp3p.
Localization of Slp3p
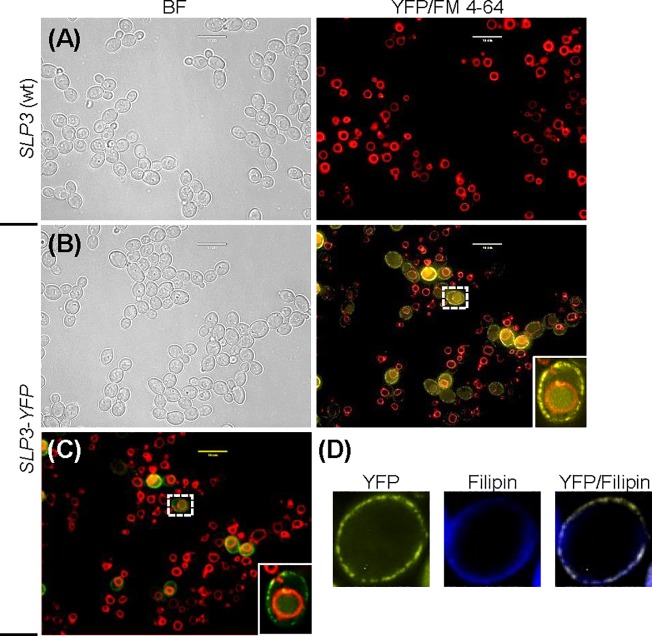
The presence of palmitoylation sites and the SPFH domain implies that Slp3p resides in the plasma membrane. Stomatins are typically visualized with fluorescent microscopy as punctate foci along the plasma membrane or other organelle membranes [5, 6, 31, 35]. We created a Slp3p-Yfp fusion protein and monitored cellular localization with fluorescence microscopy in exponential-phase cells. Consistent with stomatin family members, Slp3p-Yfp formed distinct puncta along the plasma membrane and co-localized with the membrane sterol-binding dye, Filipin (Fig 2B and 2D). Our Slp3p-Yfp fluorescent findings were not attributed to yeast cellular auto-fluorescence, because untagged wt control SLP3 cells do not fluoresce (Fig 2A). Also, fluorescence was not observed when cells were viewed using light cubes that select for green, red, and cyan fluorescence (data not shown). In humans, homo-oligomerization of stomatin monomers and/or hetero-oligomerization between stomatins with their respective interacting protein promotes the formation of plasma membrane puncta [14, 36]. The sequence conservation in the Slp3p SPFH domain to human stomatins (61% similarity, Table 3) and plasma membrane puncta strongly suggests that Slp3p forms homo-oligomeric, hetero-oligomeric, or both types of structures in the C. albicans plasma membrane.
Fig 2. Localization of Slp3p.
(A) Exponential-phase samples of the untagged wt SLP3 control strain and (B) SLP3-YFP expressing strain were treated with 160 μM FM 4–64. The FM 4–64 incubation period was set to allow visualization of the vacuole. Cells were viewed under bright-field (BF) and fluorescent microscopy. The right panels show an overlay of the YFP and FM 4–64 fluorescence (YFP/FM 4–64). (C) Overlay photo shown in (B) was recolored with ImageJ to assess Slp3p-Yfp vacuolar membrane localization. The yellow fluorescence color was reassigned to green. (D) Overnight-cultured SLP3-YFP cells were treated with 200 μg/mL Filipin and viewed using fluorescent microscopy. Image depicts a single cell, and the right panel shows an overlay of the YFP and Filipin fluorescence (YFP/Filipin). The dashed white box highlights the cell depicted in the inset image. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Approximately 1.0 x 104 cells of each strain were selected for viewing (A-D). Scale bars represent 10 μm.
Unexpectedly, we also observed localized regions of intracellular fluorescence in several cells (Fig 2B). We reasoned that the intracellular fluorescence was due to vacuolar accumulation of Slp3p. First, human SLP-1, localizes to late endosomes [31]. In yeast, the endosome fuses with the vacuole during endocytic trafficking. Second, the SLP-1 endosomal localization consensus sequence is conserved in Slp3p (S1 Fig). Third, A. nidulans StoA is transported in endosomes along hyphae [6]. We treated cells with the vacuolar membrane dye FM 4–64 and found that Slp3p-Yfp localized within the vacuole (Fig 2B). Also, vacuolar localization was consistent in cells containing fragmented vacuoles (S2 Fig). To rule out the possibility that Slp3p resides in the vacuole membrane and lumen, we used ImageJ to recolor yellow fluorescent cells green. If Slp3p resided at both vacuolar sites, then we would expect to see a region of yellow florescence (caused by FM 4–64’s red fluorescence overlaying with green) at the vacuolar membrane. Our recolored image confirmed that Slp3p-Yfp resides in the vacuolar lumen (Fig 2C). The yeast vacuole serves as a site for protein degradation, ion storage, and endomembrane trafficking [37, 38]. Stomatins have been associated with these processes [2], and our fluorescence microscopy findings show for the first time the distribution of Slp3p in the plasma membrane and Slp3p vacuolar localization.
SLP3 expression
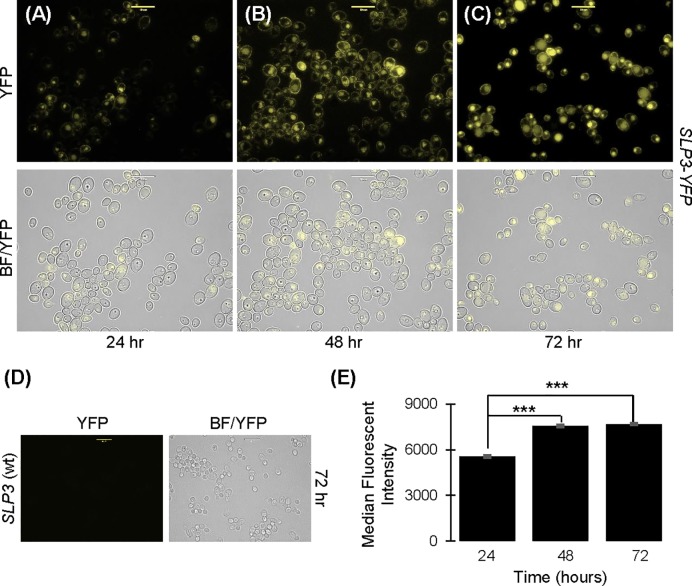
Unusually, some exponential-phase SLP3-YFP cells displayed greater fluorescence than other cells (Fig 2B). We considered that SLP3 expression may be growth-phase dependent, since C. albicans grows asynchronously. We monitored cellular fluorescence in early, mid, and late stationary-phase cultures. Our fluorescent images showed that the membrane fluorescence and number of fluorescent cells was greater in stationary-phase cultures compared to logarithmic-phase cultures (compare Figs 2B and 3A–3C). This increase was not attributed to yeast cellular auto-fluorescence from aging cultures, because fluorescence was not observed in the untagged wt SLP3 control strain after 72 hours of growth (Fig 3D). Fluorescence quantification using flow cytometry showed that the fluorescence intensity in mid and late stationary-phase cultures (48 and 72 hours growth) was statistically greater than early stationary-phase (24 hours growth) cultures (Fig 3E and S1 Appendix). The fluorescence intensity peaked at approximately 48 hours of growth and was unchanged after 72 hours.
Fig 3. Growth phase localization of Slp3p.
Samples of SLP3-YFP cells were collected at (A) 24, (B) 48, and (C) 72 hours and examined under bright-field and fluorescent microscopy. (D) Fluorescence of untagged wt SLP3 control cells grown for 72 hours was examined to assess potential yeast auto-fluorescence. Approximately 1.0 x 104 cells of each strain were selected for viewing (A-D). Scale bars represent 10 μm. (E) Fluorescence of SLP3-YFP samples was quantified by flow cytometry at the indicated time points. Untagged wt SLP3 control cells were analyzed at identical time points and served as a negative fluorescent control. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. ***p < 0.001 when compared to SLP3-YFP cells grown for 24 hours. Scale bars represent 10 μm.
SLP3 is annotated as a putative cation conductance protein [18]. We previously found that SLP3 transcription significantly increased when cells were briefly exposed to 1.0 M NaCl [16]. Therefore, we wanted to determine whether Slp3p localization increases following exposure specifically to NaCl stress or general cationic stress. We examined cellular fluorescence of SLP3-YFP cells subjected to cationic stress caused by transition metals, divalent cations, and monovalent cations. Slp3p-Yfp fluorescence increased under all conditions examined (S3 Fig).
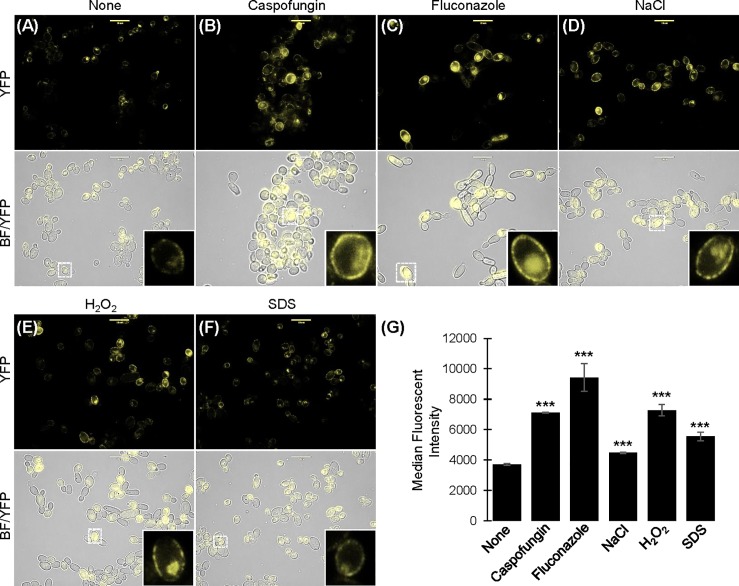
Cell wall remodeling, fatty acid metabolism, and activation of the oxidative stress response are physiological processes linked to cationic stress [16]. We hypothesized that SLP3 expression increases in response to diverse types of environmental stress. We examined cellular fluorescence in exponential-phase SLP3-YFP cells briefly treated with compounds that induce cell wall stress (caspofungin), plasma membrane stress (fluconazole), or oxidative stress (SDS and H2O2). Our fluorescent microscopy results show that cellular fluorescence was significantly higher under all stress treatments compared to untreated SLP3-YFP cells (Fig 4A–4F). Quantitative results show that the change in fluorescent intensity was the greatest in fluconazole-treated cells, and the least following NaCl treatment when compared against untreated cells (Fig 4G). To support our cellular findings, we examined SLP3 transcription from 62 gene expression datasets under various physiological and environmental conditions [18]. SLP3 transcription was reported on 21 datasets, each of which was generated using different array platforms and cell strains. Four datasets showed SLP3 transcription increased following treatment with oxidative, osmotic, and plasma membrane stress agents, and when cells were grown in high levels of ammonium [39–42]. SLP3 transcription was reduced when cells were induced to form hyphae, subjected to endoplasmic reticulum stress, or exposed to heat shock [43–45].
Fig 4. Stress-induced localization of Slp3p.
Exponential-phase SLP3-YFP cells were incubated in YPD+uri for 30 minutes with the following additives: (A) None, (B) 125 ng/mL caspofungin, (C) 2.5 μg/mL fluconazole, (D) 1.0 M NaCl, (E) 0.17% H2O2, or (F) 0.08% SDS. Samples were observed under bright-field and fluorescent microscopy. Dashed white boxes shows the cell depicted in the inset image. (G) Fluorescence was quantified by flow cytometry. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. ***p < 0.001 when compared to the untreated sample. Approximately 1.0 x 104 cells of each strain were selected for viewing (A-F). Scale bars represent 10 μm.
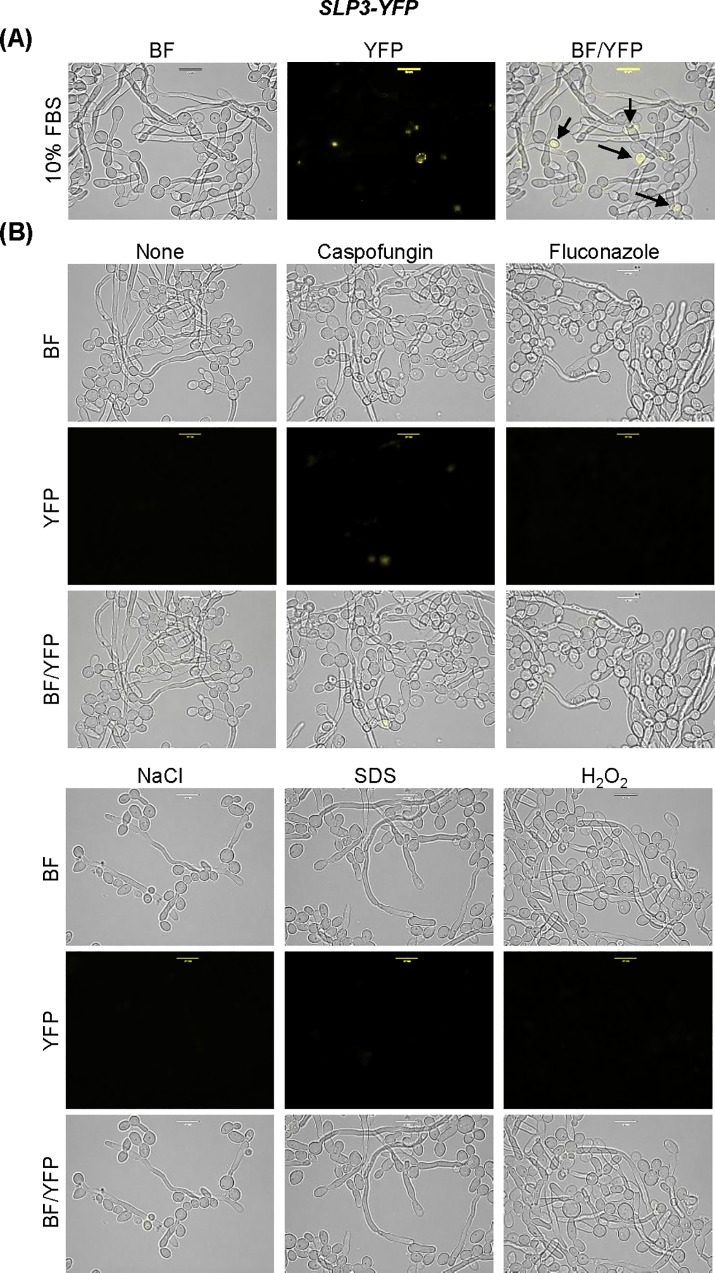
We wanted to determine whether Slp3p is present in hyphae, since SLP3 transcription is significantly reduced in the yeast-to-hyphae transition. We monitored Slp3p-Yfp localization of cells undergoing the yeast-to-hyphae transition. We did not observe fluorescence in pseudohyphal cells or germ tubes on mature hyphae (Fig 5A). In addition, we did not observe Slp3p-Yfp localization in hyphae when cells were grown in the presence of various environmental stress agents (Fig 5B).
Fig 5. Localization of Slp3p in the yeast-to-hyphae transition.
(A) SLP3-YFP cells were grown in 10% FBS/YPD+uri medium for 16 hours and observed under bright-field and fluorescent microscopy. Arrows highlight fluorescent yeast-phase cells. (B) Overnight cultured SLP3-YFP hyphal cells were standardized in 10% FBS/YPD+uri media and incubated for 45 minutes with the given additives and viewed under bright-field and fluorescent microscopy. The concentration of additives used is as follows: 125 ng/mL caspofungin, 2.5 μg/mL fluconazole, 1.0 M NaCl, 0.17% H2O2, and 0.08% SDS. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Approximately 1.0 x 104 cells of each strain were selected for viewing. Scale bars represent 10 μm.
The absence of Slp3p in hyphae was unexpected, because stomatins are expressed in filamentous fungi. A. nidulans StoA is orthologous to Slp3p and localizes to the cortex of hyphal tips [6]. We examined the microarray dataset from the Nantel et al. genome profiling study [45] to determine if the other C. albicans SPFH family members (PHB1, PHB2, PHB12, SLP2) are also repressed in hyphal formation. We found that SLP3 is the sole SPFH family member repressed in the yeast-to-hyphae transition. Our stress-induced localization findings and genome-wide transcriptional profiling results from other groups demonstrate that SLP3 is a yeast-phase specific, general stress response gene. Further, the absence of Slp3p in hyphal cells reflects the organism- specific attributes of stomatin proteins in C. albicans compared to other filamentous fungi.
Analysis of SLP3 mutants
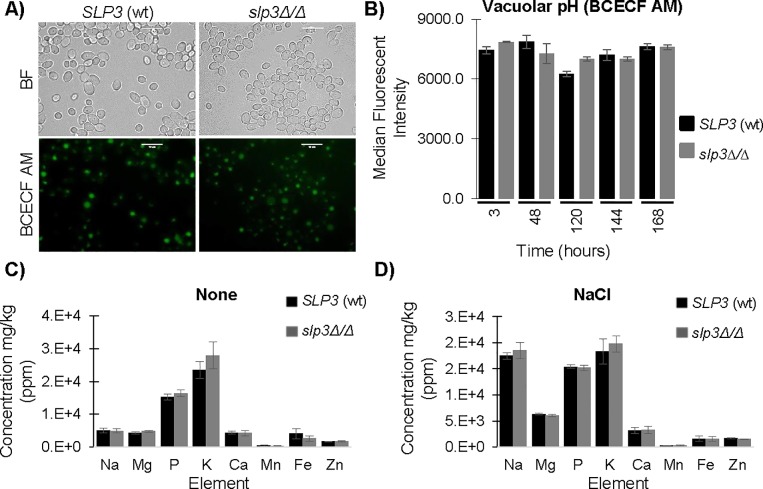
Our expression and localization findings suggested that Slp3p might be required for growth when cells are subjected to stress and that Slp3p may be necessary for vacuolar function. We created a homozygous slp3Δ/Δ null mutant strain to explore the function of Slp3p. We did not observe a growth defect under varying environmental conditions, nor did we observe any defects in hyphae formation, cell wall and cytoskeletal structure, and endocytosis (Table 4). Because Slp3p localized to the vacuole, we examined vacuolar structure and function in the slp3Δ/Δ null mutant strain. We did not observe any vacuolar structural defects in slp3Δ/Δ mutant cells labeled with FM 4–64 (Table 4). We used the fluorescent probe BCECF AM to monitor vacuolar acidification in exponential and stationary phase yeast cells, since several vacuolar functions (protein sorting, ion storage, and enzyme function) require an acidic environment [46]. Similar to our FM 4–64 results, we did not observe a vacuolar acidification defect in the slp3Δ/Δ null mutant strain (Table 4 and Fig 6A and 6B).
Table 4. Summary of phenotypes in SLP3 mutant strains.
| Strain | |||
|---|---|---|---|
| slp3Δ/Δ | PTDH3SLP3-YFP | ||
| Process and Cell Structurea | Endocytosis and Vacuole Structure | wt | wt |
| Vacuole pH | wt | wt | |
| Cytoskeleton | wt | wt | |
| Cell Wall | wt | wt | |
| Plasma Membrane | wt | wt | |
| Growth in Solid Mediab | No Additive | wt | wt |
| No Additive (37°C) | wt | wt | |
| 0.04% SDS | wt | Sensitive | |
| 0.08% SDS | wt | Sensitive | |
| 20 μM Calcofluor White | wt | wt | |
| 13 mM Copper Sulfate | wt | wt | |
| 15 mM Copper Sulfate | wt | wt | |
| 125 ng/μL Caspofungin | wt | wt | |
| 50 ng/μ. L Caspofungin |
wt | wt | |
| 10% Glucose | wt | wt | |
| 2.0 M NaCl | wt | wt | |
| 1.5 M NaCl | wt | wt | |
| 1.0 M NaCl | wt | wt | |
| 1.0 M KCl | wt | wt | |
| 10 mM Caffeine | wt | wt | |
| 1.5 M Sorbitol | wt | wt | |
| Growth in Liquid Mediac | No Additive | wt | wt |
| 0.17% H2O2 | wt | Viability Reduced | |
| 0.08% SDS | wt | Viability Reduced | |
| 10% FBS | wt | Viability Reduced | |
| Spider | wt | Viability Reduced | |
aThese rows list the physiological processess and cell structural features examined in the wt SLP3 strain and isogenic slp3Δ/Δ mutant and PTDH3SLP3-YFP over-expressing strains. wt is used to denote identical phenotpyes in the mutant and over-expression strains compared to the wt strain. Exponential phase yeast cells were collected, stained, and visualized under bright-field and fluorescent microscopy as described in Methods. The following labels were used in each assay: 160 μM FM 4–64 (endocytosis and vacuole structure), 10 μM BCECF AM (vacuole pH), 100 μg/mL calcofluor white (cell wall), 6.6 μM phalloidin rhodamine and 150 nM Tubulin tracker green (cytoskeleton) and 200 μg/mL Filipin III (plasma membrane). For each assay, three biological replicates were analyzed, and experiments were repeated at least three times.
bThese rows list the growth phenotypes of SLP3 strains on solid YPD+uri nutrient plates (no additive) and YPD+uri plates supplemented with the indicated additive. Cells from overnight cultures were collected, serially diluted, and and spotted onto the designated plates. The term sensitive denotes PTDH3SLP3-YFP over-expressing cells that did not grow on plates following 1–3 days growth compared to the SLP3 wt strain. Plates were incubated at 30°C, unless otherwise noted on the table. For each assay, three biological replicates were analyzed.
cThese rows list the growth phenotypes of SLP3 strains in YPD+uri liquid medium (no additive) and YPD+uri media supplemented with the indicated additive. Cell growth was monitored in a microplate reader for assays using H2O2 and SDS. For each assay, three biological replicates were analyzed and experiments were repeated two times. FBS and Spider media were used to examine the yeast-to-hyphae transition, and cells were visualized under the microscope. The term viability reduced is used to denote PTDH3SLP3-YFP over-expressing yeast or hyphal cells that stained with Propidium Iodide compared to wt SLP3 cells following incubation in their respective media.
Fig 6. Analysis of intracellular ion levels and vacuolar function in the slp3Δ/Δ mutant strain.
(A) Exponential-phase samples of wt SLP3 and homozygous null slp3Δ/Δ mutant cells were stained with 10 μM BCECF AM for 40 minutes, and washed before visualizing using bright-field and fluorescence microscopy. Approximately 1.0 x 104 cells of each strain were selected for viewing. (B) Flow cytometry was used to monitor vacuolar acidification at the indicated time points in yeast exponential and stationary phase cells. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Scale bars represent 10 μm. (C) Inductively coupled plasma mass spectrometry analysis was performed on exponential-phase wt control SLP3 cells and slp3Δ/Δ homozygous null mutant cells grown in nutrient YPD+uri medium and (D) following treatment with 1.0 M NaCl for 30 minutes as described in Methods. Mean values were calculated based on values obtained from three biological replicates. Two independent experiments were performed, and data presented represents one representative experiment.
We tested the hypothesis that Slp3p is required for ion homeostasis in C. albicans, because stomatins modulate ion channel activity during mechanosensation in mammals [10]. Furthermore, the vacuole is required for ion storage [38]. We performed inductively coupled plasma mass spectrometry to measure intracellular ion levels from exponential-phase cultures under normal growth conditions and following salt stress. Under both conditions, the concentration of individual elements varied; however, there were no significant differences between the wt SLP3 and slp3Δ/Δ mutant strains (Fig 6C and 6D). Similar results were observed in parallel experiments using stationary-phase cells (data not shown).
Human SLP-2 was proposed to be a scaffold for mitochondrial proteins –°a role where functional redundancy is common [47]. The absence of a slp3Δ/Δ null mutant phenotype suggests that Slp3p may be functionally complemented by another SPFH family protein or uncharacterized protein. C. albicans Slp2p and Slp3p share limited sequence homology (31% identity, 53% similarity, (S1 Fig and Table 3)), and the function of Slp2p is unknown. We observed a stark difference in transcriptional activity under stress. We did not detect any changes in SLP2 transcription in the slp3Δ/Δ mutant strain under normal growth conditions or following cationic stress using RT-qPCR (data not shown). In addition, SLP2 transcription was unchanged following multiple stress conditions that significantly modulate SLP3 transcription [39–42], and Slp2p was not found in the plasma membrane [17]. Although these observations imply that SLP2 and SLP3 have divergent functions, we cannot rule out the possibility that functional redundancy is operative on a post-translational level, since SLP2 is currently uncharacterized.
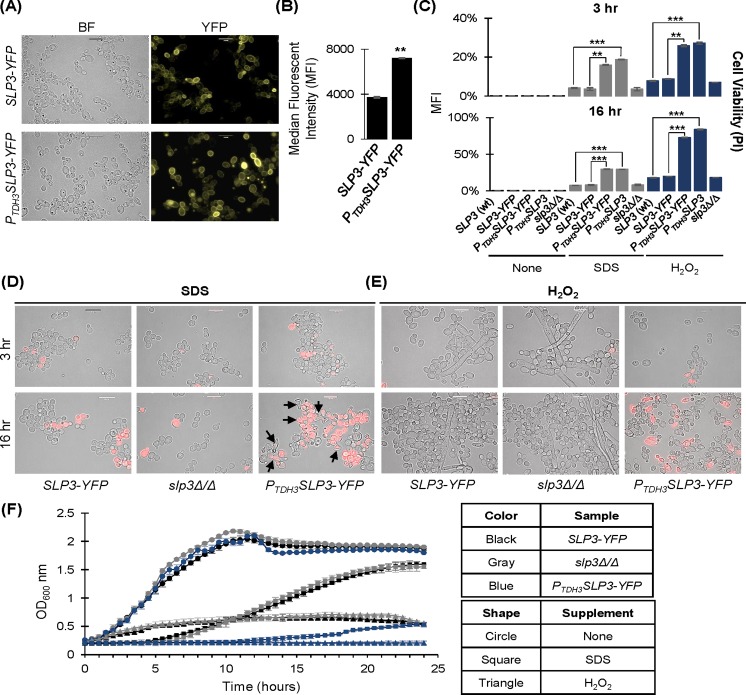
Novel functional roles of stomatins have been implicated in over-expression analysis. In humans, over-expression of STOM and SLP-1 implicated roles in placental trophoblast differentiation and protein trafficking, respectively [31, 48]. Also, SLP-2 over-expression was associated with several clinicopathological features in thyroid cancer patients [49]. Therefore, we constructed a PTDH3SLP3-YFP over-expressing strain as an alternative approach to study Slp3p function. Over-expression did not alter the localization of Slp3p, and cellular fluorescence was approximately 2.0 times greater compared to the SLP3-YFP control strain (Fig 7A and 7B). Strikingly, we found that growth and cell viability were significantly reduced when PTDH3SLP3-YFP over-expressing cells were treated with the oxidative stress agents SDS and H2O2 (Fig 7C–7F and Table 4). Growth inhibition was apparent at 16 hours of exposure to oxidative stress compared to 3 hours. Cell morphology was abnormal in SDS-treated PTDH3SLP3-YFP over-expressing cells. Many cells appeared to be ruptured or misshapen and formed cellular aggregates (Fig 7D). At 16 hours, 30% of PTDH3SLP3-YFP over-expressing cells were stained with PI, which marks cells with compromised membrane integrity. Only 8% of the slp3Δ/Δ null mutant and 7% wt SLP3, and SLP3-YFP cells were stained with PI (Fig 7C). For H2O2-treated PTDH3SLP3-YFP over-expressing cells, the yeast cell morphology was similar to the SLP3-YFP and slp3Δ/Δ mutant strains; however, the over-expressing strain failed to form filaments (Fig 7E). Similar to the SDS-treated cells, 72% of PTDH3SLP3-YFP over-expressing cells were stained with PI while only 18% of the slp3Δ/Δ mutant and wt SLP3 cells and 20% of the SLP3-YFP cells were stained with PI following 16 hours treatment (Fig 7C).
Fig 7. Phenotype of SLP3 over-expression.
(A) Exponential-phase samples of PTDH3SLP3-YFP over-expressing cells and SLP3-YFP cells were examined under bright-field and fluorescent microscopy. (B) Samples described in (A) were quantified by flow cytometry. (C-E) Overnight yeast cultures were diluted into YPD+uri supplemented with the indicated additive and grown for 3 or 16 hours. Cells were treated with PI and examined using bright-field and fluorescent microscopy. Cells were quantified by flow cytometry. (F) Stationary phase cultures were standardized in the appropriate media, and growth kinetics were analyzed for 24 hours in the presence and absence of the indicated additive. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. *p < 0.05, **p < 0.01, ***p < 0.001 when compared to the isogenic parental strain. Approximately 1.0 x 104 cells of each strain were selected for viewing (A, D-E). Scale bars represent 10 μm.
The growth phenotype of the PTDH3SLP3-YFP over-expressing strain was exclusive to oxidative stress, since growth was similar to the SLP3-YFP parental strain and untagged isogenic wt SLP3 strain under all other conditions examined (Table 3). In addition, we did not identify any defects in vacuolar structure and function, cell wall and cytoskeletal structure, and endocytosis (Table 3). We considered the possibility that the PTDH3SLP3-YFP oxidant growth defect was due to YFP over-expression, because fluorescent tags may interfere with the expression, localization, or function of a protein of interest [50]. We constructed a PTDH3SLP3 over-expressing strain without YFP and assayed viability under oxidative stress. Our results show that the H2O2 and SDS viability phenotypes were similar in the PTDH3SLP3-YFP and PTDH3SLP3 over-expressing strains (Fig 7C). Thus, over-expression of SLP3 is lethal when cells are subjected to oxidative stress.
Slp3p over-expression initiates apoptotic-like death
Oxidative stress induces apoptotic death in C. albicans [51]. Phosphatidylserine exclusion (indicated by PI staining), reactive oxygen species (ROS) production, and mitochondrial inner membrane depolarization are major cellular events associated with apoptosis [52]. Accordingly, we tested whether the hypersensitivity of PTDH3SLP3-YFP over-expressing cells to oxidants coupled with the increased uptake of PI was a consequence of cells undergoing apoptosis. We assayed ROS production (DHR-123 treatment), and mitochondrial inner membrane potential (JC-10 labeling) in PTDH3SLP3-YFP over-expressing cells after 16 hours of oxidative stress exposure. Consistent with cells undergoing apoptosis, the percentage of PTDH3SLP3-YFP over-expressing cells with SDS- or H2O2-induced depolarized inner mitochondrial membranes was statistically greater compared to the slp3Δ/Δ null mutant and wt SLP3 strains after 16 hours exposure (Fig 8A and S4 Fig). ROS levels were statistically greater in the PTDH3SLP3-YFP over-expressing strain compared to the wt SLP3, SLP3-YFP, and slp3Δ/Δ mutant strains after 16 hours exposure (Fig 8B). We did not detect any apoptotic activity in over-expressing cells in the absence of stress.
Fig 8. Analysis of apoptosis in SLP3 over-expressing cells.
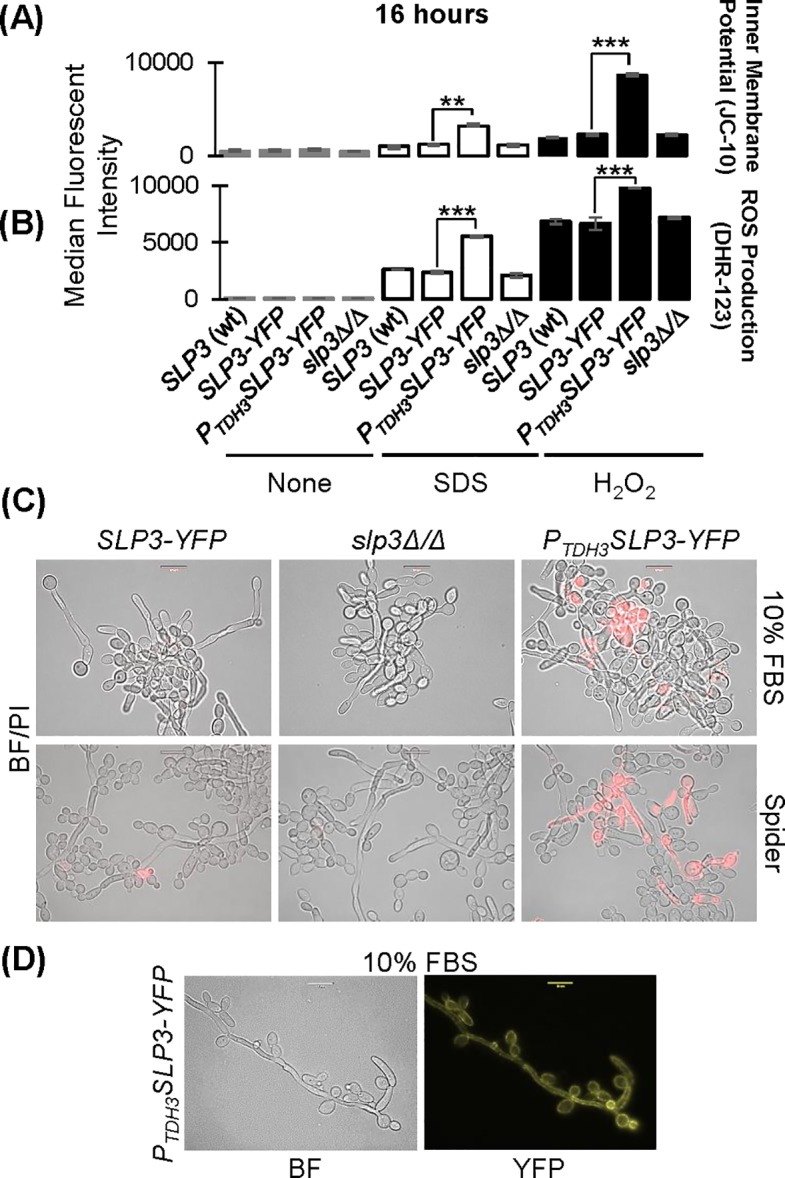
(A) Mitochondrial membrane potential was measured in overnight cultured cells following exposure with the given additives for 16 hours. Samples were labeled using JC-10 and quantified using flow cytometry. (B) Intracellular ROS production was studied in yeast cells following incubation with the indicated additives for 16 hours. Samples were labeled with DHR-123 and quantified using flow cytometry. Gray bars represent untreated samples; white bars represent samples treated with 0.08% SDS, and black bars represent samples treated with 0.17% H2O2. Similar findings were observed in samples treated for 3 hours. (C) PTDH3SLP3-YFP cells were grown in 10% FBS/YPD+uri and Spider media to observe the yeast-to-hyphae transition and stained with PI to assess viability. Samples were visualized using bright-field and fluorescence microscopy. (D) Slp3p puncta were visualized in the plasma membrane of yeast-phase cells and hyphal germ tube using bright-field and fluorescent microscopy. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. *p < 0.05, **p < 0.01, ***p < 0.001 when compared to the isogenic parental strain. Approximately 1.0 x 104 cells of each strain were selected for viewing. Scale bars represent 10 μm.
The mitochondrion is critical in mediating homeostasis to oxidative stress. Human SLP-2 interacts synergistically with prohibitins at the inner mitochondrial membrane to promote mitochondrial stability [53]. We did not identify a mitochondrial targeting sequence in Slp3p (S1 Table) or observe Slp3p-Yfp mitochondrial localization following oxidative stress or under any other condition that we screened (Figs 2–4). Therefore, the mechanism by which SLP3 over-expression causes mitochondrial depolarization may be indirect. Nevertheless, SLP3 over-expression leads to inner mitochondrial membrane depolarization, phosphatidylserine exclusion, and increased levels of ROS, specifically when cells are subjected to prolonged oxidative stress, ultimately leading to apoptotic-like death.
H2O2 exposure initiates filamentation in C. albicans. H2O2-induced filaments are presumed to be indicative of a disrupted cell cycle and are morphologically distinct (a state called hyperpolarized buds) compared to filaments generated in nutrient filamentation media at 37°C [51]. As seen in Fig 7E, filamentous cells are present in the SLP3-YFP strain following 3 and 16 hours of H2O2 exposure, but were absent in the PTDH3SLP3-YFP over-expressing strain. SDS-treated SLP3-YFP cells did not form filaments (Fig 7D), which may seem contradictory to our observations using H2O2. However, while SDS kills C. albicans mainly by inducing oxidative stress, it also inhibits filamentation [54].
We wanted to determine whether the filamentation defect in the PTDH3SLP3-YFP over-expressing strain was specific to H2O2 exposure or a general phenotype associated with Slp3p over-expression. We compared filamentation of the over-expressing strain to the wt strain after 3 and 16 hours incubation in serum. In contrast to our observations with H2O2-treated PTDH3SLP3-YFP over-expressing cells, we did not observe any differences in hyphal formation or hyphal morphology after 3 hours (data not shown) and 16 hours of growth in serum or Spider medium (Fig 8C). Slp3p-Yfp puncta were present throughout the hyphal germ tube, confirming Slp3p plasma membrane localization (Fig 8D). On the other hand, several PTDH3SLP3-YFP over-expressing cells grown in serum for 16 hours were PI positive compared to SLP3-YFP and slp3Δ/Δ null mutant cells, indicating that hyphal formation was lethal in the absence of environmental stress (Fig 8C). Taken together, our findings show that Slp3p over-expression inhibits filamentation under H2O2 stress, but compromises cellular integrity when cells are grown in filamentation medium.
In summary, our work is the first to provide functional insight and fully characterize the localization of a stomatin family member in C. albicans. Our primary sequence analyses showed that the SPFH domain of Slp3p is homologous to mammalian and nematode stomatins, while N- and C-terminal flanking sequences are divergent. Further, we identified putative sites for plasma membrane insertion and vacuolar targeting. We argue that SLP3 is a yeast-phase specific stress response gene, because SLP3 expression and cell surface localization increases following exposure to diverse cellular perturbants in yeast cells and not in hyphal cells. The lack of phenotype in a slp3Δ/Δ homozygous null mutant implies stomatin function may be redundant in C. albicans. Conversely, Slp3p over-expression triggers apoptotic-like death specifically following prolonged exposure to oxidative stress. This lethality may be due in part to the inability to form filaments. We note the limitation of these phenotypic observations, since Slp3p over-expression may indirectly alter molecular processes that govern apoptosis and filamentation. Nevertheless, our results are consistent with SLP3 having a positive role in response to oxidative stress in yeast phase cells.
Supporting information
Alignments 1 and 2: Residues highlighted in black are identical while those highlighted in gray are similar between SPFH family members. Residues highlighted within red boxes constitute endosome/lysosome targeting sequences. Residues within the green box constitute the membrane hairpin region of mammalian and nematode stomatins. The conserved proline residue within this region is indicated with a black triangle. The N-terminal region consists of residues that lie before the membrane hairpin region. Residues highlighted within dark blue boxes constitute the SPFH domain. Residues highlighted within orange boxes constitute the SCP-2 domain of HsSTOML1. The C-terminal region consists of residues that lie after the SPFH domain. Potential palmitoylation sites are highlighted with an asterisk. Lysine residues labeled with an orange “K” are potential SUMOylation sites. Gaps are denoted with a hyphen. Ca: Candida albicans. Hs: Homo sapiens. Ms: Mus musculus. Ce: Caenorhabditis elegans. An: Aspergillus nidulans.
(PPTX)
Overnight cultured SLP3-YFP cells were standardized to an OD600nm of 0.2 in YPD+uri and incubated at 30°C. At (A) 3, (B) 17, and (C-D) 24 hours, samples were viewed using fluorescent microscopy. Cells in (D) were labeled with 160 μM FM 4–64. Dashed white boxes show the cells depicted in the inset. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Approximately 1.0 x 104 cells of each strain were selected for viewing. Scale bars represent 10 μm.
(TIFF)
Exponential-phase SLP3-YFP cells were treated with the given additives for 30 minutes and visualized with bright-field and fluorescent microscopy. Concentrations of additives used are as follows: 1.0 M NaCl, 1.0 M KCl, 1.0 M MgCl2, 10 mM ZnCl2, 1 mM FeCl3, 0.6 M CaCl2, 0.6 M LiCl, and 50 mM CuCl2. Water served as the negative control. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Approximately 1.0 x 104 cells were selected for viewing. Scale bars represent 10 μm.
(TIF)
Overnight cultured SLP3-YFP cells, slp3Δ/Δ homozygous null mutant cells, and PTDH3SLP3-YFP cells were standardized in YPD+uri media or YPD+uri media supplemented with 0.08% SDS and incubated for 16 hours at 30°C. Samples were prepared and stained with 1X JC-10 according to the manufacturer’s protocol. Cells were visualized using bright-field and fluorescent microscopy. Cells with intact mitochondria fluoresce red, and cells with depolarized mitochondria fluoresce green. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Approximately 1.0 x 104 cells of each strain were selected for viewing. Scale bars represent 10 μm.
(TIFF)
(XLSX)
(XLSX)
Acknowledgments
We thank the Mitchell lab for providing the YFP and PTDH3 plasmids. We also thank Peter Lipke and Mariana Dorrington for comments on the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the National Institute of General Medical Sciences (http://www.nigms.nih.gov/), grant number SC3GM111133 to JMR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rivera-Milla E, Stuermer CA, Malaga-Trillo E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell Mol Life Sci. 2006;63(3):343–57. doi: 10.1007/s00018-005-5434-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17(8):394–402. doi: 10.1016/j.tcb.2007.06.005 . [DOI] [PubMed] [Google Scholar]
- 3.Lapatsina L, Brand J, Poole K, Daumke O, Lewin GR. Stomatin-domain proteins. Eur J Cell Biol. 2012;91(4):240–5. doi: 10.1016/j.ejcb.2011.01.018 . [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Hsieh CF, Liu HW, Chen CY, Wu SC, Chen TW, et al. Lipid raft-associated stomatin enhances cell fusion. FASEB J. 2017;31(1):47–59. doi: 10.1096/fj.201600643R . [DOI] [PubMed] [Google Scholar]
- 5.Reuter AT, Stuermer CA, Plattner H. Identification, localization, and functional implications of the microdomain-forming stomatin family in the ciliated protozoan Paramecium tetraurelia. Eukaryot Cell. 2013;12(4):529–44. doi: 10.1128/EC.00324-12 ; PubMed Central PMCID: PMCPMC3623435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeshita N, Diallinas G, Fischer R. The role of flotillin FloA and stomatin StoA in the maintenance of apical sterol-rich membrane domains and polarity in the filamentous fungus Aspergillus nidulans. Mol Microbiol. 2012;83(6):1136–52. doi: 10.1111/j.1365-2958.2012.07996.x . [DOI] [PubMed] [Google Scholar]
- 7.Hiebl-Dirschmied CM, Entler B, Glotzmann C, Maurer-Fogy I, Stratowa C, Prohaska R. Cloning and nucleotide sequence of cDNA encoding human erythrocyte band 7 integral membrane protein. Biochim Biophys Acta. 1991;1090(1):123–4. . [DOI] [PubMed] [Google Scholar]
- 8.Hiebl-Dirschmied CM, Adolf GR, Prohaska R. Isolation and partial characterization of the human erythrocyte band 7 integral membrane protein. Biochim Biophys Acta. 1991;1065(2):195–202. . [DOI] [PubMed] [Google Scholar]
- 9.Stewart GW, Hepworth-Jones BE, Keen JN, Dash BC, Argent AC, Casimir CM. Isolation of cDNA coding for an ubiquitous membrane protein deficient in high Na+, low K+ stomatocytic erythrocytes. Blood. 1992;79(6):1593–601. . [PubMed] [Google Scholar]
- 10.Wetzel C, Hu J, Riethmacher D, Benckendorff A, Harder L, Eilers A, et al. A stomatin-domain protein essential for touch sensation in the mouse. Nature. 2007;445(7124):206–9. doi: 10.1038/nature05394 . [DOI] [PubMed] [Google Scholar]
- 11.Huang M, Gu G, Ferguson EL, Chalfie M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature. 1995;378(6554):292–5. doi: 10.1038/378292a0 . [DOI] [PubMed] [Google Scholar]
- 12.Wetzel C, Pifferi S, Picci C, Gok C, Hoffmann D, Bali KK, et al. Small-molecule inhibition of STOML3 oligomerization reverses pathological mechanical hypersensitivity. Nat Neurosci. 2017;20(2):209–18. doi: 10.1038/nn.4454 . [DOI] [PubMed] [Google Scholar]
- 13.Rungaldier S, Umlauf E, Mairhofer M, Salzer U, Thiele C, Prohaska R. Structure-function analysis of human stomatin: A mutation study. PLoS One. 2017;12(6):e0178646 doi: 10.1371/journal.pone.0178646 ; PubMed Central PMCID: PMCPMC5456319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rungaldier S, Oberwagner W, Salzer U, Csaszar E, Prohaska R. Stomatin interacts with GLUT1/SLC2A1, band 3/SLC4A1, and aquaporin-1 in human erythrocyte membrane domains. Biochim Biophys Acta. 2013;1828(3):956–66. doi: 10.1016/j.bbamem.2012.11.030 ; PubMed Central PMCID: PMCPMC3790964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osman C, Wilmes C, Tatsuta T, Langer T. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol Biol Cell. 2007;18(2):627–35. doi: 10.1091/mbc.E06-09-0839 ; PubMed Central PMCID: PMCPMC1783772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marotta DH, Nantel A, Sukala L, Teubl JR, Rauceo JM. Genome-wide transcriptional profiling and enrichment mapping reveal divergent and conserved roles of Sko1 in the Candida albicans osmotic stress response. Genomics. 2013;102(4):363–71. Epub 2013/06/19. doi: 10.1016/j.ygeno.2013.06.002 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabezon V, Llama-Palacios A, Nombela C, Monteoliva L, Gil C. Analysis of Candida albicans plasma membrane proteome. Proteomics. 2009;9(20):4770–86. Epub 2009/10/14. doi: 10.1002/pmic.200800988 . [DOI] [PubMed] [Google Scholar]
- 18.Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G. The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017;45(D1):D592–D6. doi: 10.1093/nar/gkw924 ; PubMed Central PMCID: PMCPMC5210628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181(6):1868–74. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis D, Edwards JE Jr., Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68(10):5953–9. ; PubMed Central PMCID: PMCPMC101559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walther A, Wendland J. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet. 2003;42(6):339–43. doi: 10.1007/s00294-002-0349-0 . [DOI] [PubMed] [Google Scholar]
- 22.Gerami-Nejad M, Berman J, Gale CA. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 2001;18(9):859–64. doi: 10.1002/yea.738 . [DOI] [PubMed] [Google Scholar]
- 23.Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, et al. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol. 2008;10(11):2180–96. doi: 10.1111/j.1462-5822.2008.01198.x ; PubMed Central PMCID: PMCPMC2701370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128(5):779–92. ; PubMed Central PMCID: PMCPMC2120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David C. Amberg DJB, Jefferey N. Strathern Methods in Yeast Genetics. 2005 Edition ed. New York: John Inglis; 2005. [Google Scholar]
- 26.Rauceo JM, Blankenship JR, Fanning S, Hamaker JJ, Deneault JS, Smith FJ, et al. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol Biol Cell. 2008;19(7):2741–51. doi: 10.1091/mbc.E08-02-0191 ; PubMed Central PMCID: PMCPMC2441657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eide DJ, Clark S, Nair TM, Gehl M, Gribskov M, Guerinot ML, et al. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 2005;6(9):R77 doi: 10.1186/gb-2005-6-9-r77 ; PubMed Central PMCID: PMCPMC1242212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–17. doi: 10.1006/jmbi.2000.4042 . [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31(20):3359–61. doi: 10.1093/bioinformatics/btv362 ; PubMed Central PMCID: PMCPMC4595897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umlauf E, Mairhofer M, Prohaska R. Characterization of the stomatin domain involved in homo-oligomerization and lipid raft association. J Biol Chem. 2006;281(33):23349–56. doi: 10.1074/jbc.M513720200 . [DOI] [PubMed] [Google Scholar]
- 31.Mairhofer M, Steiner M, Salzer U, Prohaska R. Stomatin-like protein-1 interacts with stomatin and is targeted to late endosomes. J Biol Chem. 2009;284(42):29218–29. doi: 10.1074/jbc.M109.014993 ; PubMed Central PMCID: PMCPMC2781465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aicart-Ramos C, Valero RA, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta. 2011;1808(12):2981–94. doi: 10.1016/j.bbamem.2011.07.009 . [DOI] [PubMed] [Google Scholar]
- 33.Thuaud F, Ribeiro N, Nebigil CG, Desaubry L. Prohibitin ligands in cell death and survival: mode of action and therapeutic potential. Chem Biol. 2013;20(3):316–31. doi: 10.1016/j.chembiol.2013.02.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–85. doi: 10.1146/annurev-biochem-061909-093311 . [DOI] [PubMed] [Google Scholar]
- 35.Dempwolff F, Schmidt FK, Hervas AB, Stroh A, Rosch TC, Riese CN, et al. Super Resolution Fluorescence Microscopy and Tracking of Bacterial Flotillin (Reggie) Paralogs Provide Evidence for Defined-Sized Protein Microdomains within the Bacterial Membrane but Absence of Clusters Containing Detergent-Resistant Proteins. PLoS Genet. 2016;12(6):e1006116 doi: 10.1371/journal.pgen.1006116 ; PubMed Central PMCID: PMCPMC4928834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyers L, Umlauf E, Prohaska R. Oligomeric nature of the integral membrane protein stomatin. J Biol Chem. 1998;273(27):17221–6. . [DOI] [PubMed] [Google Scholar]
- 37.Armstrong J. Yeast vacuoles: more than a model lysosome. Trends Cell Biol. 2010;20(10):580–5. doi: 10.1016/j.tcb.2010.06.010 . [DOI] [PubMed] [Google Scholar]
- 38.Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta. 2009;1793(4):650–63. doi: 10.1016/j.bbamcr.2008.08.003 ; PubMed Central PMCID: PMCPMC2906225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaloriti D, Jacobsen M, Yin Z, Patterson M, Tillmann A, Smith DA, et al. Mechanisms underlying the exquisite sensitivity of Candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. MBio. 2014;5(4):e01334–14. doi: 10.1128/mBio.01334-14 ; PubMed Central PMCID: PMCPMC4161263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasicek EM, Berkow EL, Bruno VM, Mitchell AP, Wiederhold NP, Barker KS, et al. Disruption of the transcriptional regulator Cas5 results in enhanced killing of Candida albicans by Fluconazole. Antimicrob Agents Chemother. 2014;58(11):6807–18. doi: 10.1128/AAC.00064-14 ; PubMed Central PMCID: PMCPMC4249418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khamooshi K, Sikorski P, Sun N, Calderone R, Li D. The Rbf1, Hfl1 and Dbp4 of Candida albicans regulate common as well as transcription factor-specific mitochondrial and other cell activities. BMC Genomics. 2014;15:56 doi: 10.1186/1471-2164-15-56 ; PubMed Central PMCID: PMCPMC3904162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunkel N, Biswas K, Hiller E, Fellenberg K, Satheesh SV, Rupp S, et al. Control of morphogenesis, protease secretion and gene expression in Candida albicans by the preferred nitrogen source ammonium. Microbiology. 2014;160(Pt 8):1599–608. doi: 10.1099/mic.0.078238-0 . [DOI] [PubMed] [Google Scholar]
- 43.Chaillot J, Tebbji F, Remmal A, Boone C, Brown GW, Bellaoui M, et al. The Monoterpene Carvacrol Generates Endoplasmic Reticulum Stress in the Pathogenic Fungus Candida albicans. Antimicrob Agents Chemother. 2015;59(8):4584–92. doi: 10.1128/AAC.00551-15 ; PubMed Central PMCID: PMCPMC4505245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leach MD, Farrer RA, Tan K, Miao Z, Walker LA, Cuomo CA, et al. Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat Commun. 2016;7:11704 doi: 10.1038/ncomms11704 ; PubMed Central PMCID: PMCPMC4894976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13(10):3452–65. doi: 10.1091/mbc.E02-05-0272 ; PubMed Central PMCID: PMCPMC129958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richards A, Gow NA, Veses V. Identification of vacuole defects in fungi. J Microbiol Methods. 2012;91(1):155–63. doi: 10.1016/j.mimet.2012.08.002 . [DOI] [PubMed] [Google Scholar]
- 47.Wai T, Saita S, Nolte H, Muller S, Konig T, Richter-Dennerlein R, et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 2016;17(12):1844–56. doi: 10.15252/embr.201642698 ; PubMed Central PMCID: PMCPMC5283581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen TW, Liu HW, Liou YJ, Lee JH, Lin CH. Over-expression of stomatin causes syncytium formation in nonfusogenic JEG-3 choriocarcinoma placental cells. Cell Biol Int. 2016;40(8):926–33. doi: 10.1002/cbin.10636 . [DOI] [PubMed] [Google Scholar]
- 49.Bartolome A, Boskovic S, Paunovic I, Bozic V, Cvejic D. Stomatin-like protein 2 overexpression in papillary thyroid carcinoma is significantly associated with high-risk clinicopathological parameters and BRAFV600E mutation. APMIS. 2016;124(4):271–7. doi: 10.1111/apm.12505 . [DOI] [PubMed] [Google Scholar]
- 50.Magee PT, Gale C, Berman J, Davis D. Molecular genetic and genomic approaches to the study of medically important fungi. Infect Immun. 2003;71(5):2299–309. doi: 10.1128/IAI.71.5.2299-2309.2003 ; PubMed Central PMCID: PMCPMC153231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dantas Ada S, Day A, Ikeh M, Kos I, Achan B, Quinn J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules. 2015;5(1):142–65. doi: 10.3390/biom5010142 ; PubMed Central PMCID: PMCPMC4384116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao B, Cheng S, Clancy CJ, Nguyen MH. Caspofungin kills Candida albicans by causing both cellular apoptosis and necrosis. Antimicrob Agents Chemother. 2013;57(1):326–32. doi: 10.1128/AAC.01366-12 ; PubMed Central PMCID: PMCPMC3535936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christie DA, Lemke CD, Elias IM, Chau LA, Kirchhof MG, Li B, et al. Stomatin-like protein 2 binds cardiolipin and regulates mitochondrial biogenesis and function. Mol Cell Biol. 2011;31(18):3845–56. doi: 10.1128/MCB.05393-11 ; PubMed Central PMCID: PMCPMC3165718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Q, Zhang B, Ma F, Jia C, Xiao C, Zhang B, et al. Novel mechanisms of surfactants against Candida albicans growth and morphogenesis. Chem Biol Interact. 2015;227:1–6. doi: 10.1016/j.cbi.2014.12.014 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignments 1 and 2: Residues highlighted in black are identical while those highlighted in gray are similar between SPFH family members. Residues highlighted within red boxes constitute endosome/lysosome targeting sequences. Residues within the green box constitute the membrane hairpin region of mammalian and nematode stomatins. The conserved proline residue within this region is indicated with a black triangle. The N-terminal region consists of residues that lie before the membrane hairpin region. Residues highlighted within dark blue boxes constitute the SPFH domain. Residues highlighted within orange boxes constitute the SCP-2 domain of HsSTOML1. The C-terminal region consists of residues that lie after the SPFH domain. Potential palmitoylation sites are highlighted with an asterisk. Lysine residues labeled with an orange “K” are potential SUMOylation sites. Gaps are denoted with a hyphen. Ca: Candida albicans. Hs: Homo sapiens. Ms: Mus musculus. Ce: Caenorhabditis elegans. An: Aspergillus nidulans.
(PPTX)
Overnight cultured SLP3-YFP cells were standardized to an OD600nm of 0.2 in YPD+uri and incubated at 30°C. At (A) 3, (B) 17, and (C-D) 24 hours, samples were viewed using fluorescent microscopy. Cells in (D) were labeled with 160 μM FM 4–64. Dashed white boxes show the cells depicted in the inset. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Approximately 1.0 x 104 cells of each strain were selected for viewing. Scale bars represent 10 μm.
(TIFF)
Exponential-phase SLP3-YFP cells were treated with the given additives for 30 minutes and visualized with bright-field and fluorescent microscopy. Concentrations of additives used are as follows: 1.0 M NaCl, 1.0 M KCl, 1.0 M MgCl2, 10 mM ZnCl2, 1 mM FeCl3, 0.6 M CaCl2, 0.6 M LiCl, and 50 mM CuCl2. Water served as the negative control. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Approximately 1.0 x 104 cells were selected for viewing. Scale bars represent 10 μm.
(TIF)
Overnight cultured SLP3-YFP cells, slp3Δ/Δ homozygous null mutant cells, and PTDH3SLP3-YFP cells were standardized in YPD+uri media or YPD+uri media supplemented with 0.08% SDS and incubated for 16 hours at 30°C. Samples were prepared and stained with 1X JC-10 according to the manufacturer’s protocol. Cells were visualized using bright-field and fluorescent microscopy. Cells with intact mitochondria fluoresce red, and cells with depolarized mitochondria fluoresce green. For each assay, three biological replicates were analyzed. Experiments were repeated at least three times, and data presented represents one representative experiment. Approximately 1.0 x 104 cells of each strain were selected for viewing. Scale bars represent 10 μm.
(TIFF)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.