Abstract
Fibrinogen C domain containing 1 (FIBCD1) is a transmembrane receptor that binds chitin and other acetylated compounds with high affinity. FIBCD1 has previously been shown to be present in the epithelium of the gastrointestinal tract. In the present study, we performed a detailed analysis of normally structured human tissues for the expression of FIBCD1 by quantitative PCR and immunohistochemistry. We find that FIBCD1 is expressed in epithelial cells derived from all three germ layers. Endodermal-derived epithelial cells throughout the gastrointestinal tract and the respiratory system showed high expression of FIBCD1 and also mesodermal-derived cells in the genitourinary system and ectodermal-derived epidermis and sebaceous glands cells expressed FIBCD1. In some columnar epithelial cells, for example, in the salivary gland and gall bladder, the FIBCD1 expression was clearly polarized with strong apical reaction, while other columnar cells, for example, in small and large intestine and in bronchi, the staining was equally strong apically and basolaterally. In keratinocytes in skin, tongue, and oral cavity, the FIBCD1 staining was granular. This expression pattern together with the known binding properties supports that FIBCD1 plays a role in innate immunity in the skin and at mucosal surfaces.
Keywords: chitin, digestive system, FIBCD1, fibrinogen-related domain, innate immunity, mucosal immunity, immunohistochemistry, receptors, pattern recognition, respiratory system
Introduction
The fibrinogen-related domain (FReD) superfamily is a protein family that is found in a wide range of animals.1 To date, 541 FReDs have been identified in mammals, of which 21 are found in humans.2,3 In mammals, the FReD proteins play important roles in the coagulation cascade, embryogenesis, and in vascular biology.4,5 From single-celled eukaryotes to mammals, the FReD proteins also play important roles in innate immunity as pattern-recognition molecules.1,6 For instance, the ficolins activate the complement system after binding to pathogen-associated molecular patterns from various pathogens, including vira, parasites, and a broad spectrum of bacteria.3,7
The vast majority of the FReD proteins are soluble proteins. We have identified fibrinogen C domain containing 1 (FIBCD1) as the first type II transmembrane protein in the FReD superfamily.5 The FIBCD1 gene is localized on chromosome 9q34.1 in the human genome, adjacent to the homologous genes encoding for M- and L-ficolin.8 The ectodomain of FIBCD1 is characterized by a coiled-coil region, a polycationic region, and C-terminal FReD.5,9 FIBCD1 oligomerizes and assembles into tetramers of approximately 250 kDa. It binds to acetylated compounds such as chitin and sialic acid in a calcium-dependent manner. Although several chitin-binding proteins in mammals have been identified,10 FIBCD1 is the first bona fide human receptor identified to bind specifically to chitin.
Chitin is the second most abundant biopolymer in nature, only exceeded by cellulose,11 and is found in various pathogens including fungi, helminths, and house dust.12 Chitin stimulates and regulates the immune system in various directions,13 both alone and in combination with other pathogen-associated molecular patterns such as β-glucan, but the role played by FIBCD1 in chitin-regulated immunity and homeostasis is still unknown.
Initial immunohistochemical investigations have shown that FIBCD1 is expressed apically in small and large intestine epithelial cells, and in the ducts of the salivary glands.5 However, the more detailed characterization of tissue distribution is unknown. In the present study, we characterize the tissue distribution of FIBCD1, to enhance understanding of its role and function. We screened 20 human tissues for the relative mRNA expression of FIBCD1 by using quantitative real-time PCR, and evaluated the distribution of FIBCD1 in 50 histological structures by immunohistochemistry (IHC). We find that FIBCD1 is generally expressed in all epithelial cells examined, while not expressed in other cell types including cells of myeloid origin.
Materials and Methods
Buffers
Standard in-house buffers were mixed to the following concentrations: phosphate-buffered saline (PBS; 140 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4) bicarbonate buffer (206 mM NaHCO3, 81 mM Na2CO3).
Antibodies
A panel of monoclonal antibodies directed against FIBCD1 were raised, as described by Schlosser et al.5 In brief, BALB/c mice were immunized with recombinant FIBCD1 ectodomain. Specificity and validity of the chosen monoclonal antibody, HYB12-2 (initial concentration 470 µg/ml), was tested by Western blotting, as recommended by the Histochemical Society.14 In the following, HYB12-2 is referred to as anti-FIBCD1.
To determine staining specificity of the monoclonal anti-FIBCD1 antibody, anti-ovalbumin (anti-OVA, HYB099-01; Statens Serum Institut, Copenhagen, Denmark) was used as isotype control. The isotype control was used in the same concentrations and under the same conditions as the anti-FIBCD1 antibody.
Characterization of antibody isotype for both anti-FIBCD1 and anti-OVA was determined by IsoStrip (Roche Applied Science; Mannheim, Germany).
Fluorescein (FITC) Labeling of Antibodies
Antibodies (anti-FIBCD1 and anti-OVA) were labeled with FITC (isomer 1; Sigma-Aldrich, Saint Louis, MO) according to manufacturer’s instructions. In brief, the FITC-isomer was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich). Antibodies were subsequently dialyzed against bicarbonate buffer, and concentrations were determined using NanoDrop (Thermo Scientific; Wilmington, DE). The FITC/protein molar ratio was calculated as described by the manufacturer.
FIBCD1-Transfected HEK293 Cells
HEK293 cells containing the Flp-In system were purchased from Invitrogen (Carlsbad, CA). Stable transfection with full-length FIBCD1 was performed as described previously.5 Blank HEK293 cells containing the Flp-In system were used as controls.
Cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) (high glucose; Thermo-Fisher, Carlsbad, CA) with penicillin (100 U/ml), streptomycin (0.1 mg/ml), l-glutamine (2 mM), and 10% fetal bovine serum (FBS; all from Thermo-Fisher).
Approximately 3 × 108 cells of each cell line (HEK293-Blank and HEK293-FIBCD1) were harvested, spun down, and the pellet fixated in 4% normal-buffered formalin (NBF) for 48 hr. The pellet was subsequently embedded in paraffin, cut in 4-µm-thick sections, and stained using the same protocol as tissue specimens, as described below.
Quantitative Real-Time PCR (qRT-PCR)
The Human Total RNA Master Panel II (Clontech; Palo Alto, CA), comprising purified RNA from 20 human tissues, was used to investigate the mRNA expression levels of FIBCD1. Complementary DNA (cDNA) was synthesized using the Moloney murine leukemia virus (M-MLV) reverse transcriptase kit (Sigma-Aldrich; Taufkirchen, Germany) according to manufacturer’s instructions. Relative mRNA expression of FIBCD1 was investigated using TaqMan Universal Mastermix II and TaqMan Gene Expression Assays (Applied Biosystems; Foster City, CA). Results were normalized to two housekeeping genes, ACTB (beta-actin) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), using the GeNorm method,15 setting heart expression to 1, which showed lowest detectable mRNA signal in the tissues investigated. Reactions were performed on a StepOnePlus Real-Time PCR System (Life Technologies; Carlsbad, CA) in technical triplicates. Relative expression was calculated as fold changes, using qBase+ software (Biogazelle; Zwijnaarde, Belgium).
Human Tissue Samples
Human tissue samples of 50 different histological structures were obtained from the tissue bank at the Department of Pathology, Odense University Hospital (Odense, Denmark). Samples were from surgically removed specimens containing non-malignant, non-inflammatory changes, and determined as histologically normal by a trained specialist in histopathology. Tissue samples were fixed in 4% NBF for various periods of time (12–96 hr), conventionally dehydrated, and subsequently embedded in paraffin.
To determine intra-individual expression variations of FIBCD1, we stained samples from 2 to 3 separate individuals, yielding a total of 123 tissue specimens, which were examined (Supplemental Data).
The Regional Committees on Health Research Ethics for the Region of Southern Denmark approved the use of the human tissue samples in the present study (Ref. No. afS-VF-20050070).
IHC
Four-µm-thick sections were cut from NBF-fixed paraffin-embedded tissue blocks. Sections were mounted on FLEX IHC Slides (Dako; Glostrup, Denmark), dried at 60C, dewaxed, and rehydrated through a graded ethanol series, and subsequently washed in 0.05 M Tris-buffered saline (TBS; Fagron Nordic A/S; Copenhagen, Denmark). Five different unmasking techniques were performed, including proteolytic approaches and microwave heating with various buffers. Optimal epitope retrieval was performed using microwave heating in 10 mM Tris (Fagron Nordic A/S; Copenhagen, Denmark) with 0.5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA; Fagron Nordic A/S) at pH 9.0 (TEG buffer). A total of 3 Tissue-Tek containers (Miles, Inc.; Elkhart, IN), each with 24 slides in 250 ml TEG-buffer, were placed on the edge of a turntable inside the microwave oven. Slides were heated for 11 min at full power (900 W), followed by 15 min at 400 W. After heating, slides remained in buffer for 15 min.
The FITC-labeled antibodies (anti-FIBCD1 and anti-OVA) were diluted 1:50, 1:100, and 1:200, respectively, in Antibody Diluent (Agilent Technologies; Glostrup, Denmark). Incubation with the antibodies was done for 60 min at room temperature. Immunostaining was automated using a modification of the catalyzed signal amplification (CSA) II, Biotin-Free Catalyzed Amplification System (Dako), on an Autostainer Plus instrument (Dako). In the CSA II protocol, the original rabbit anti-mouse Ig-HRP was substituted with a rabbit anti-FITC-horseradish peroxidase (HRP) (Dako), diluted 1:30, and incubated for 20 min. Immunostaining was followed by brief nuclear counterstaining in Mayer’s hematoxylin (Fagron Nordic A/S; Copenhagen, Denmark). Finally, slides were washed, dehydrated, and coverslipped using a Tissue-Tek Film coverslipper (Sakura Finetek; Alphen aan den Rijn, The Netherlands).
Image Acquisition
Histology slides were scanned at 20× magnification using a NanoZoomer-XR (Hamamatsu Photonics; Hamamatsu City, Japan), and image acquisition was obtained using NDP.view2 software (NanoZoomer Digital Pathology; Hamamatsu Photonics). Representative images were automatically adjusted for contrast in Adobe Photoshop CC 2017 (San Jose, CA).
Results
Expression of FIBCD1 mRNA in Human Tissues
The relative expression of FIBCD1 was investigated in RNA from 20 different human tissues (Fig. 1). In general, the expression throughout the examined tissues was low, with PCR cycle threshold (Ct)-values above 27. We found highest expression of FIBCD1 in the testis, brain (including fetal brain), adrenal gland, and placenta (Ct-values 28–33). We found medium expression in the small and large intestine, spleen, prostate, and lung (Ct-values 33–36). Low or very low expression was found in kidney, salivary gland, thymus, fetal liver, and heart (Ct-values 37–38), whereas FIBCD1 expression was undetectable in uterus, stomach, skeletal muscle, liver, and bone marrow (Ct-values >38). Ct-values for the endogenous controls ACTB and GAPDH were ranging from 16 to 21, with a mean value of 17 (GAPDH) and 18 (ACTB).
Figure 1.
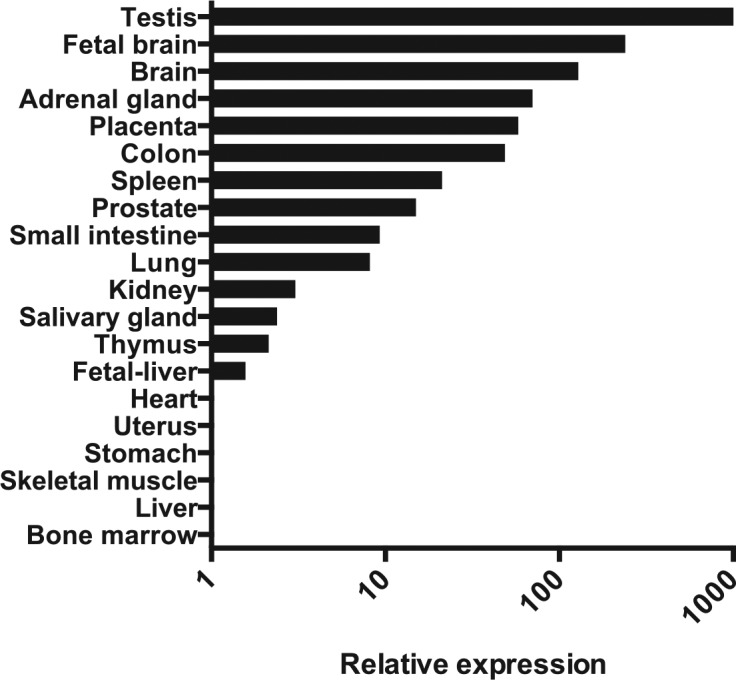
Relative mRNA expression of FIBCD1, as determined by quantitative PCR. Expression is normalized to ACTB and GAPDH, setting heart expression to 1. Abbreviations: FIBCD1, fibrinogen C domain containing 1; ACTB, beta-actin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Antibody Characterization
The monoclonal anti-FIBCD1 antibody has previously been shown to react with the human FIBCD1 ectodomain, but not within the FIBCD1-FReD.5 The epitope is thus located between the transmembrane region and the FReD domain of the FIBCD1 protein, which excludes cross-reactivity with other proteins of the FReD superfamily.
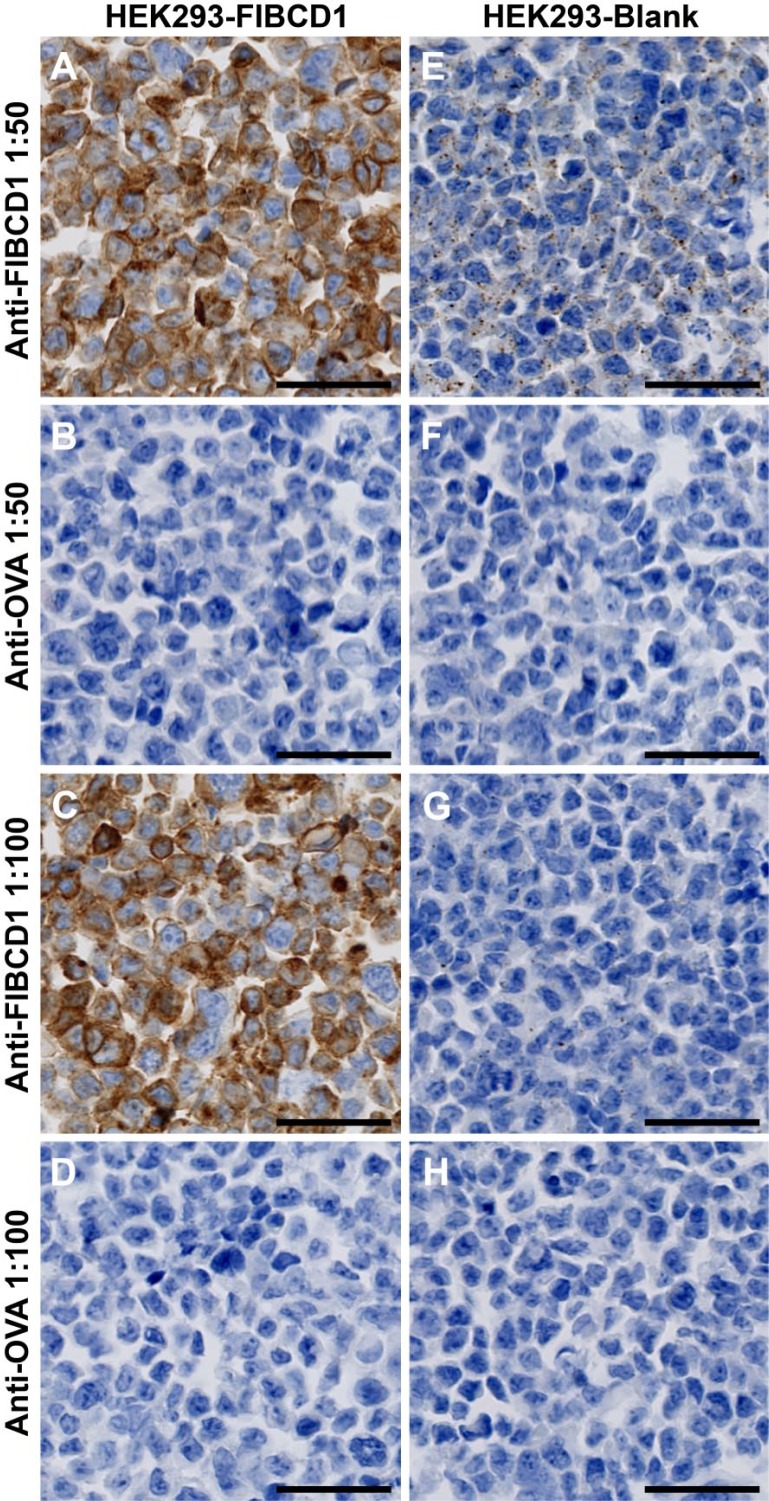
We performed staining on HEK293 cells, either blank or FIBCD1-transfected, with the antibodies in a 2-fold dilution series from 1:50 to 1:200. FIBCD1-transfected cells were stained intensively at all dilutions (Fig. 2A and C, dilution 1:200 not shown), whereas blank cells showed very weak granulated intracellular immunoreactivity at the highest antibody concentration and no reactivity in lower concentrations (Fig. 2E and G, dilution 1:200 not shown).
Figure 2.
Determination of antibody specificity and usage concentration. HEK293 cells transfected with full-length FIBCD1 cDNA (A–D) or untransfected (E–H). Cells are stained with either anti-FIBCD1 (A, C, E, and G) or anti-OVA (B, D, F, and H) in 1:50 (A, B, E, and F) or 1:100 (C, D, G, and H) dilution. Scale bars = 50 µm. Abbreviations: FIBCD1, fibrinogen C domain containing 1; anti-OVA, anti-ovalbumin.
To determine unspecific staining, we used anti-OVA as isotype control. We found no immunohistochemical reaction in HEK293 cells stained with anti-OVA in either of the concentrations tested (Fig. 2B, D, F, and H, dilution 1:200 not shown). Tissue specimens were stained with anti-OVA with the same immunohistochemical protocol as anti-FIBCD1, and showed no staining (Figs. S2–S5).
Both anti-FIBCD1 and anti-OVA were found to have the IgG1 isotype.
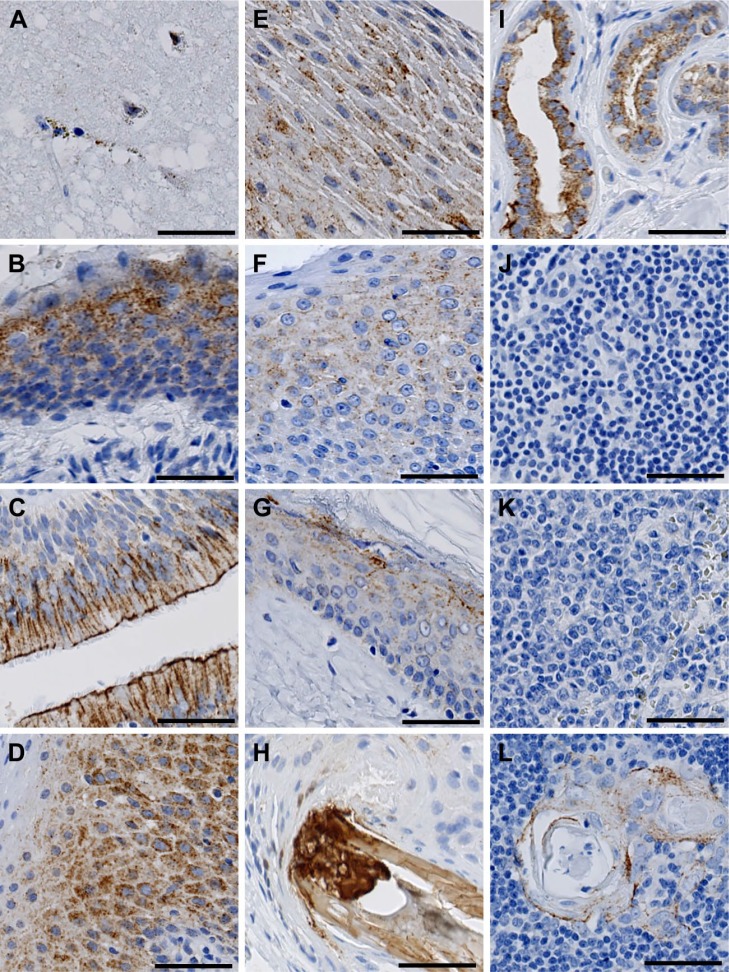
Localization of FIBCD1 in Brain, Sensory, and Immune Organs
In the brain, we found a weak immunohistochemical reaction in glial cells of the cerebrum, whereas neurons were found to be negative (Fig. 3A). We found strong immunoreactivity in the epithelium of the ear canal (Fig. 3B), the respiratory (pseudo-stratified, ciliated columnar) epithelium of the nasal fossa (Fig. 3C), the non-keratinized squamous epithelium in the tongue (Fig. 3D), and oral cavity (Fig. 3E). FIBCD1 was weakly present in the non-keratinized stratified squamous epithelium of the tonsils (Fig. 3F).
Figure 3.
Localization of FIBCD1 in brain, sensory, and immune organs. (A) brain, (B) ear canal, (C) nasal fossa, (D) tongue, (E) oral cavity, (F) tonsil, (G) skin, (H) hair follicle, (I) sweat glands, (J) lymph node, (K) spleen, and (L) thymus. Scale bars = 50 µm. Abbreviation: FIBCD1, fibrinogen C domain containing 1.
In the skin, FIBCD1 was found to be present in the keratinized squamous epithelium of the epidermis, and reaching down to stratum granulosum (Fig. 3G), as well as in the keratin structure of hair follicles and sebaceous glands (Fig. 3H). We found a strong reaction in both eccrine and apocrine sweat glands of the skin (Fig. 3I).
We found no evidence of presence of FIBCD1 in lymph nodes (Fig. 3J) or spleen (Fig. 3K). In the thymus, FIBCD1 was present at moderate intensity in the corpuscles of Hassall, whereas the surrounding lymphatic tissue was negative (Fig. 3L).
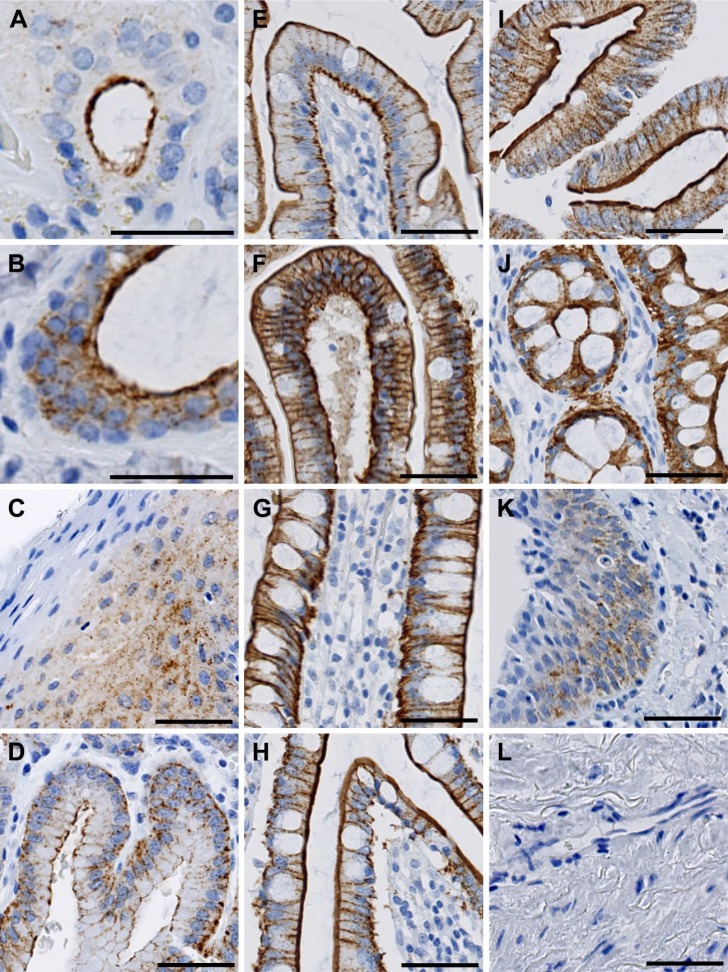
Localization of FIBCD1 in the Gastrointestinal Tract
As expected from previous observations,5 we found strong FIBCD1 immunoreactivity throughout the digestive tract (Fig. 4). In this detailed analysis, we further found that FIBCD1 was present in the serous glands and ducts in the salivary glands, whereas mucous glands were negative (Fig. 4A). In the pharynx, we found evidence of luminal expression in the ductal system of mucous glands (Fig. 4B). Moreover, we found moderate FIBCD1 immunoreactivity in the esophagus (Fig. 4C). FIBCD1 was found to be present luminally as well as basolaterally in the columnar epithelium of the stomach (Fig. 4D), duodenum (Fig. 4E), jejunum (Fig. 4F), and ileum (Fig. 4G). In the large intestine, FIBCD1 was present in the cecum (Fig. 4H), in the colon (Fig. 4I), and rectum (Fig. 4J). In the anal canal, we found moderate immunoreactivity in both columnar and stratified squamous epithelium (Fig. 4K), with highest intensity above the pectinate line. We found no evidence of presence in the mesothelium of the peritoneum (Fig. 4L) or in the Peyer’s patches in the small intestine (not shown).
Figure 4.
Localization of FIBCD1 in the digestive tract. (A) salivary gland, (B) pharynx, (C) esophagus, (D) stomach, (E) duodenum, (F) jejunum, (G) ileum, (H) cecum, (I) colon, (J) rectum, (K) anal canal, and (L) peritoneum. Scale bars: (A–B) = 40 µm; (C–L) = 50 µm. Abbreviation: FIBCD1, fibrinogen C domain containing 1.
Localization of FIBCD1 in Abdominal Organs
FIBCD1 was detected in abdominal organs with highly varying intensity (Fig. 5). We found luminal immunoreactivity in the epithelium of interlobular bile ducts in the liver (Fig. 5A) and in the epithelium of the gall bladder (Fig. 5B). In the pancreas, we found that the islets of Langerhans were weakly positive (not shown), whereas the ductal epithelium had strong reactivity (Fig. 5C). In the adrenal gland, we found very weak reaction in the capsule-near zona glomerulosa of the adrenal cortex but no reaction in the other cortical zones or the medulla (Fig. 5D).
Figure 5.
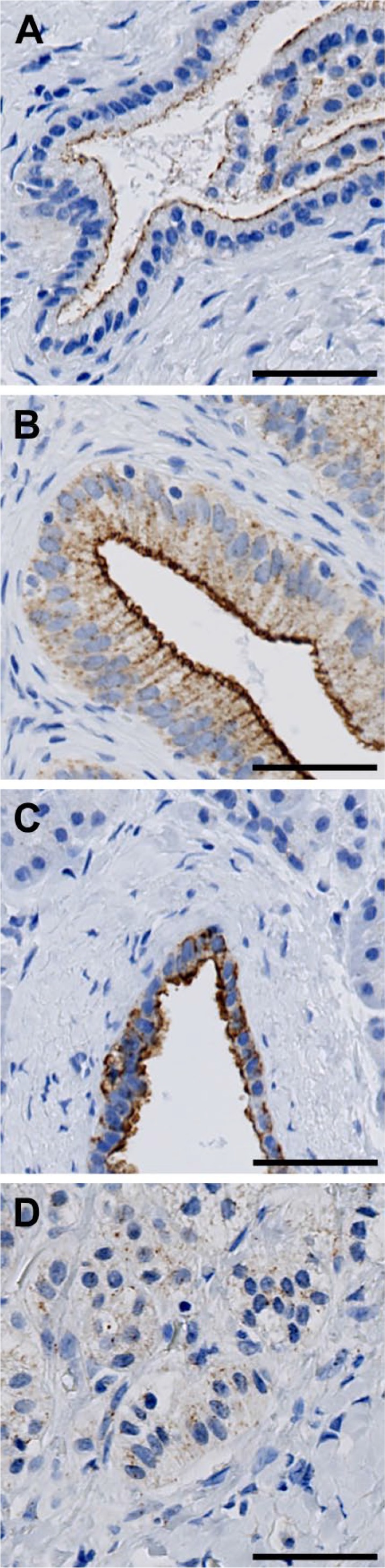
Localization of FIBCD1 in abdominal organs. (A) liver, (B) gall bladder, (C) pancreas, and (D) adrenal gland. Scale bars = 50 µm. Abbreviation: FIBCD1, fibrinogen C domain containing 1.
Localization of FIBCD1 in the Respiratory Tract
We found evidence of FIBCD1 expression throughout the respiratory tract (Fig. 6). In the trachea, we found sporadic, weak positive reaction in the epithelium (Fig. 6A). FIBCD1 was present at moderate intensity in the epithelium of the bronchi (Fig. 6B), bronchioles (Fig. 6C), and alveoli (Fig. 6D).
Figure 6.
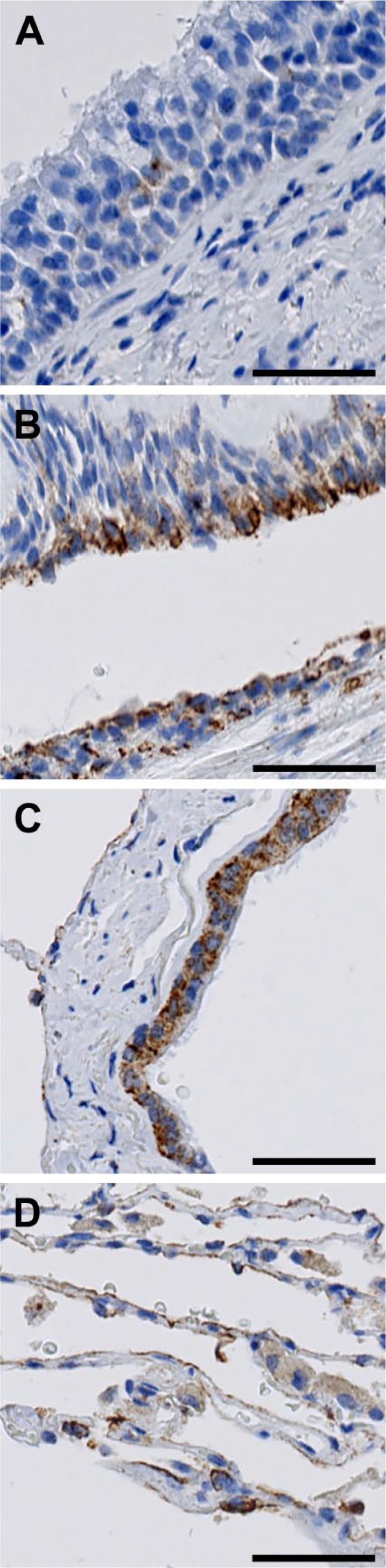
Localization of FIBCD1 in the respiratory tract. (A) trachea, (B) bronchi, (C) bronchioles, and (D) alveoli. Scale bars = 50 µm. Abbreviation: FIBCD1, fibrinogen C domain containing 1.
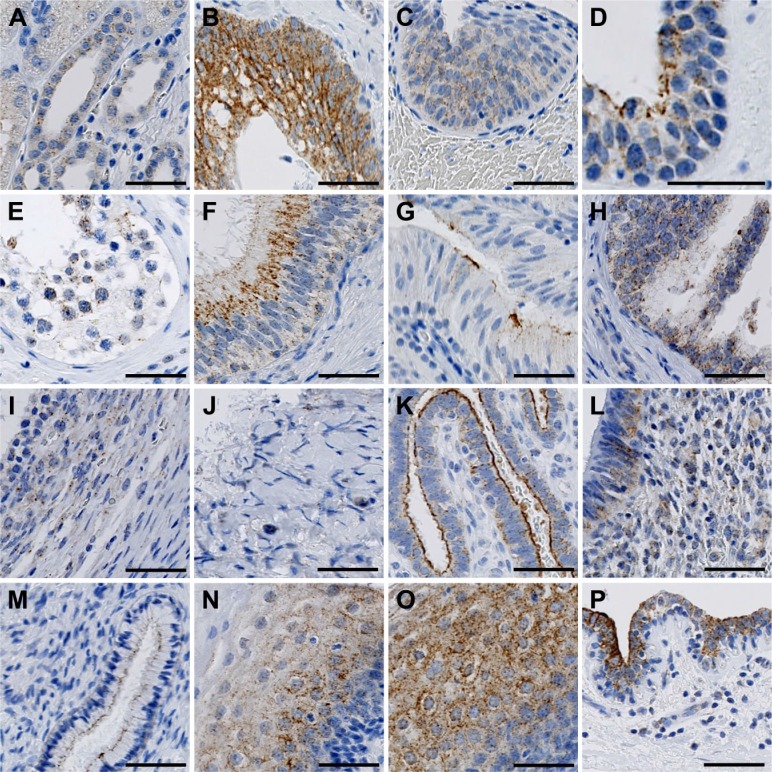
Localization of FIBCD1 in the Urinary Tract
In the kidney, we found weak immunoreactivity in the distal tubules and collecting tubules, whereas cortex, glomeruli, and proximal tubules were negative (Fig. 7A). In the efferent urinary tract, we found strong staining intensity in the urothelium of the ureter (Fig. 7B), and decreasing intensity in the urothelium of the bladder (Fig. 7C) and urethra (Fig. 7D).
Figure 7.
Localization of FIBCD1 in the urinary tract (A–D) and male (E–H) and female (I–P) genitalia. (A) kidney, (B) ureter, (C) bladder, (D) urethra, (E) testis, (F) epididymis, (G) vas deferens, (H) prostate, (I) fertile ovary, (J) postmenopausal ovary, (K) fallopian tube, (L) uterine corpus, (M) uterine cervix, (N) uterine portio, (O) vagina, and (P) mamma. Scale bars: (A–C) and (E–P) = 50 µm; (D) = 40 µm. Abbreviation: FIBCD1, fibrinogen C domain containing 1.
Localization of FIBCD1 in the Reproductive Organs
In the male genitalia, we found staining of FIBCD1 in the spermatogonia closest to the lumen, and in some Sertoli cells (Fig. 7E). We found strong FIBCD1 immunoreactivity in the pseudo-stratified epithelium in the ductal system of the epididymis (Fig. 7F). There was weakly positive reaction in the epithelium of the vas deferens (Fig. 7G), and in the stromal and glandular tissue of the prostate (Fig. 7H).
In the female genitalia, we found weak FIBCD1 immunoreactivity in the follicles of fertile ovary (Fig. 7I), whereas inactive postmenopausal ovary was negative (Fig. 7J). We found strong immunoreactivity in the surface epithelium of the fallopian tube (Fig. 7K), and weaker intensity in the columnar epithelium of the uterine corpus (Fig. 7L) and squamous epithelium of the uterine cervix (Fig. 7M). We found positive reaction in the keratinized squamous epithelium of the portio of the uterine cervix (Fig. 7N) and non-keratinized epithelium of the vagina (Fig. 7O). We found no immunoreactivity in the placenta (not shown), but strong staining in the epithelium of the lactiferous ducts of the mammary gland (Fig. 7P).
Discussion
In the present study, we describe the detailed distribution of FIBCD1 in human tissues at mRNA level by real-time PCR and at protein level by IHC. We find evidence of the presence of FIBCD1 in a broad spectrum of tissues and organs throughout the human body, which, indeed, paves the way for new hypotheses regarding its role as a pattern-recognition receptor (PRR) or alternative roles in health and disease.
The applied CSA II, Biotin-Free Catalyzed Amplification System is one of the most sensitive detection systems available for immunohistochemical investigations, ideal for the detection of less abundant proteins.16,17 By using FITC-labeled primary antibodies and rabbit anti-FITC-HRP instead of the more conventional unlabeled primary antibody and rabbit anti-mouse Ig-HRP, the detection system becomes even more sensitive. High precaution for nonspecific staining should thus be taken into account. We found that the anti-FIBCD1 antibody showed high specificity at all concentrations tested (Fig. 2A, C, E, and G), while no staining was seen when the isotype control (anti-OVA) antibody was used for staining (Fig. 2B, D, E, H, and Supplementary Figs. S1–S5). We found scarce immunoreactivity in blank HEK293 cells at high antibody concentration (Fig. 2E). This was expected, as the expression of FIBCD1 mRNA in non-transfected HEK293 cells was found to be relative high compared with other non-transfected commercially available cell lines such as the lung cell line Calu-3 and colon cell line Caco-2 (data not shown). Both antibody- and staining specificity were tested, and we thus find our immunohistochemical results reliable with the applied concentration of the antibody.
Although we found some variation with regard to staining intensity between specimens and thus different individuals, a clear general expression pattern was found. In the figures presented, images are representative for the overall expression pattern of FIBCD1. Even though human specimens were only obtained from either autopsies or surgically due to a clear suspicion of disease, the specimens used in the present study represent tissue with normal morphology, where no signs of inflammation or malignancy were present.
One limitation to our study is the use of only one monoclonal antibody directed toward FIBCD1. Even though a panel of antibodies has been raised, only the applied antibody in this investigation has shown high reactivity in human cell lines and tissues. Other antibodies also showed reactivity where FIBCD1 was expected to be most abundant, for example, in the gastrointestinal tract, but not with the same intensity. However, the used antibody is validated secundum artem, demonstrating no cross-reactivity to closely related proteins. In our opinion, the need for further validation of our findings with other antibodies seems less important.
In the Human Protein Atlas,18 the localization of FIBCD1 is investigated by two commercially available polyclonal antibodies, HPA053898 and HPA053898 (both Sigma-Aldrich; Taufkirchen, Germany), and RNA sequencing. Reactivity is found within colon epithelium, cerebral glial cells, and testis, which is consistent with our results. Surprisingly, expression is also found in lymph nodes, thyroid gland, and myocytes of the heart, where we are unable to detect FIBCD1 (Fig. 3J and K, heart not shown). Besides being polyclonal, for example, recognizing multiple epitopes on the antigen, the antibodies used in the Human Protein Atlas are not validated by Western Blot, opposite to the monoclonal antibody used in our analysis. Although the observed differences may be due to biological variance, we find our results more reliable and precise than in the high-throughput approach demonstrated in the Human Protein Atlas.
We did not find a clear correlation between immunohistochemical reactivity and mRNA signal. Although it is generally expected that there should be an overall consistency between mRNA expression and protein levels,18,19 the present study demonstrates some discrepancies. For instance, at mRNA level, the testis was found to have the definite highest expression level, approximately 100-fold higher than other tissues investigated. However, at protein level, we found scarce immunostaining in the spermatogonia and a few Sertoli cells (Fig. 7E). We found high reactivity in the epididymis (Fig. 7F), and the purified mRNA purchased commercially could, indeed, contain mRNA from the epididymis besides the testis. Also, regulation of protein expression is highly complex, and previous studies in correlating mRNA and protein expression have had varying success in different tissues.20,21 It is further important to notice that even though our study demonstrates that FIBCD1 is present in a wide range of human epithelial structures, the actual fraction of epithelial cells can be rather low. The applied immunohistochemical protocol is highly sensitive, and our qPCR results emphasize an overall low expression throughout the human body. We did not succeed in retrieving histological specimens from fetal brain and fetal liver, where there was a detectable mRNA signal. Furthermore, we were only able to retrieve histological specimens from the cerebrum. The high brain mRNA signal could originate from specific cerebral structures.
As a putative chitin receptor, FIBCD1 could function as a PRR. Humans are continuously exposed to chitin in various organ systems and at virtually all surfaces exposed to pathogens. Present and previous findings demonstrate that FIBCD1 binds chitin, and thus may function as a PRR, responsible for the presentation of chitin to the host defense system and initiating immune responses. In the gastrointestinal tract, chitin exposure originates from ingested food,13 bacteria,10 fungi,22 and parasites.23 FIBCD1 may recognize chitin from these potentially pathogenic species, and may play a role in the pathophysiology of gastrointestinal disorders. With FIBCD1 being present at surfaces throughout most the respiratory tract (Fig. 6), we hypothesize that chitin is being recognized by FIBCD1 upon inhalation, and that FIBCD1 regulates the immune response upon chitin stimulation. This may indicate that FIBCD1 plays a role in the development of airway diseases such as asthma and pulmonary infections. Indeed, unpublished results from our laboratory have shown that FIBCD1 binds to the airborne fungus Aspergillus fumigatus and its chitin-rich alkali-insoluble fragments, and that FIBCD1 is upregulated in invasive aspergillosis.
FIBCD1 has previously been shown to bind to sialic acid with high affinity.5 Sialic acid is found on a broad panel of eukaryotic and some prokaryotic cell surfaces.24 Some pathogenic bacteria use sialic acid to hide from, or inhibit the complement system.25 At mucosal surfaces, sialic acids are found terminally on the mucins, which makes up the mucus.26 As FIBCD1 binds to sialic acid with high affinity and is present at mucosal sites prone to bacterial invasion, we speculate that FIBCD1 might play a role in the pathogenesis of various bacterial infections, such as pneumonia, gastroenteritis, and urinary tract infections.
With FIBCD1 being present in the epidermis of the skin (Fig. 3G), we speculate that it may play a role in cutaneous immunity. The skin is, indeed, the most exposed surface to the outside environment, and constant chitin exposure from plants, fungi, and parasites may generate allergic responses.27 The skin is inhabited by a broad panel of microorganisms, including bacteria, fungi, and viruses,28 and is constantly challenged to recognize harmful from harmless pathogens. Whereas other FReDs have been shown to play a key role in skin wound healing and inflammation,2 the exact function of FIBCD1 in the skin has not yet been investigated. FIBCD1 could serve as a PRR for immune cells residing in the dermis, such as macrophages, dendritic cells, and lymphocytes,29 and thus play a role in the delicate immune balance in the skin. In a recently published study using a genome-wide association approach, FIBCD1 was found to be associated with cutaneous fungal infections.30 It is also worth noting that chitin itself has been shown to stimulate the innate immune response in skin keratinocytes by increasing toll-like receptor 4 (TLR4) expression.31 FIBCD1 could, indeed, support the innate immune response upon chitin exposure.
Even though our present findings fit with the hypothesis of FIBCD1 being a PRR, it could very well have other functions as well. From a sequence homology aspect, FIBCD1 is closely related to both ficolins as well as microfibrillar-associated protein 4 (MFAP4).1 While the close homology to the ficolins indicates that FIBCD1 might function as a PRR, the similarity to MFAP4 may indicate other functions. MFAP4 is an extracellular matrix protein, which binds to elastin and collagen fibers, and has been shown to be involved in tissue remodeling, respiratory, and cardiovascular diseases.32,33 FIBCD1 could have functions related to those of MFAP4, for example, in cellular homeostasis.
In the present study, we have described the detailed distribution of FIBCD1 in a broad variety of normal human tissues. We demonstrate that FIBCD1 is expressed in epithelial cells derived from all three germ layers, throughout the human body, with high prevalence in the airways, gastrointestinal, and urogenital tract. Together with the binding properties of FIBCD1, the present data of the localization support the hypothesis of FIBCD1 functioning as a PRR. However, the identification of FIBCD1 expression in, for example, brain tissue and in neuroendocrine cells from the adrenal gland suggests other functions besides pattern recognition. Future studies should focus on the in vitro and in vivo properties and functions of FIBCD1.
Supplementary Material
Acknowledgments
The authors thank laboratory technicians Mr. Christian Enggaard and Mrs. Lisbet Mortensen, Department of Pathology, Odense University Hospital, Odense, for technical assistance and help upon retrieving the histological samples.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: SH and ON performed and optimized immunohistochemistry. SH, JBM, AS, and VN performed cell cultures, quantitative PCR, raised antibodies, and performed FITC-labeling. SH and NM investigated histological structures. SH, GLS, and UH conceptualized the study and raised funding. SH assembled all figures. SH, GLS, and UH drafted the manuscript. All authors have read and approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Odense University Hospital Free Research Fund, Region of Southern Denmark Research Fund, Aage og Johanne Louis-Hansens Fond, Torben & Alice Frimodts Fond, Augustinusfonden, and the A. P. Møller Foundation for the Advancement of Medical Science.
ORCID iD: S von Huth  https://orcid.org/0000-0001-8127-6815
https://orcid.org/0000-0001-8127-6815
Contributor Information
Sebastian von Huth, Cancer and Inflammation Research, Department of Molecular Medicine, University of Southern Denmark, Odense, Denmark.
Jesper B. Moeller, Cancer and Inflammation Research, Department of Molecular Medicine, University of Southern Denmark, Odense, Denmark
Anders Schlosser, Cancer and Inflammation Research, Department of Molecular Medicine, University of Southern Denmark, Odense, Denmark.
Niels Marcussen, Department of Pathology, Odense University Hospital, Odense, Denmark.
Ole Nielsen, Department of Pathology, Odense University Hospital, Odense, Denmark.
Vicki Nielsen, Cancer and Inflammation Research, Department of Molecular Medicine, University of Southern Denmark, Odense, Denmark.
Grith L. Sorensen, Cancer and Inflammation Research, Department of Molecular Medicine, University of Southern Denmark, Odense, Denmark
Uffe Holmskov, Cancer and Inflammation Research, Department of Molecular Medicine, University of Southern Denmark, Odense, Denmark.
Literature Cited
- 1. Doolittle RF, McNamara K, Lin K. Correlating structure and function during the evolution of fibrinogen-related domains. Protein Sci. 2012;21(12):1808–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zuliani-Alvarez L, Midwood KS. Fibrinogen-related proteins in tissue repair: how a unique domain with a common structure controls diverse aspects of wound healing. Adv Wound Care. 2015;4(5):273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thomsen T, Schlosser A, Holmskov U, Sorensen GL. Ficolins and FIBCD1: soluble and membrane bound pattern recognition molecules with acetyl group selectivity. Mol Immunol. 2011;48(4):369–81. [DOI] [PubMed] [Google Scholar]
- 4. Kairies N, Beisel HG, Fuentes-Prior P, Tsuda R, Muta T, Iwanaga S, Bode W, Huber R, Kawabata S. The 2.0-A crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems. Proc Natl Acad Sci U S A. 2001;98(24):13519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schlosser A, Thomsen T, Moeller JB, Nielsen O, Tornøe I, Mollenhauer J, Moestrup SK, Holmskov U. Characterization of FIBCD1 as an acetyl group-binding receptor that binds chitin. J Immunol. 2009;183(6):3800–9. [DOI] [PubMed] [Google Scholar]
- 6. Hanington PC, Zhang S-M. The primary role of fibrinogen-related proteins in invertebrates is defense, not coagulation. J Innate Immun. 2011;3(1):17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren Y, Ding Q, Zhang X. Ficolins and infectious diseases. Virol Sin. 2014;29(1):25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomsen T, Moeller JB, Schlosser A, Sorensen GL, Moestrup SK, Palaniyar N, Wallis R, Mollenhauer J, Holmskov U. The recognition unit of FIBCD1 organizes into a noncovalently linked tetrameric structure and uses a hydrophobic funnel (S1) for acetyl group recognition. J Biol Chem. 2010;285(2):1229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shrive AK, Moeller JB, Burns I, Paterson JM, Shaw AJ, Schlosser A, Sorensen GL, Greenhough TJ, Holmskov U. Crystal structure of the tetrameric fibrinogen-like recognition domain of fibrinogen C domain containing 1 (FIBCD1) protein. J Biol Chem. 2014;289(5):2880–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bueter CL, Specht CA, Levitz SM. Innate sensing of chitin and chitosan. PLoS Pathog. 2013;9(1):e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kurita K. Chitin and chitosan: functional biopolymers from marine crustaceans. Mar Biotechnol. 2006;8(3):203–26. [DOI] [PubMed] [Google Scholar]
- 12. Jeuniaux C, Voss-Foucart MF. Chitin biomass and production in the marine environment. Biochem Syst Ecol. 1991;19(5):347–56. [Google Scholar]
- 13. Elieh Ali Komi D, Sharma L, Cruz Dela CS. Chitin and its effects on inflammatory and immune responses. Clin Rev Allergy Immunol. 2017;13(3):1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hewitt SM, Baskin DG, Frevert CW, Stahl WL, Rosa-Molinar E. Controls for immunohistochemistry: the Histochemical Society’s standards of practice for validation of immunohistochemical assays. J Histochem Cytochem. 2014;62(10):693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Gijlswijk RP, Zijlmans HJ, Wiegant J, Bobrow MN, Erickson TJ, Adler KE, Tanke HJ, Raap AK. Fluorochrome-labeled tyramides: use in immunocytochemistry and fluorescence in situ hybridization. J Histochem Cytochem. 1997;45(3):375–82. [DOI] [PubMed] [Google Scholar]
- 17. von Wasielewski R, Mengel M, Gignac S, Wilkens L, Werner M, Georgii A. Tyramine amplification technique in routine immunohistochemistry. J Histochem Cytochem. 1997;45(11):1455–9. [DOI] [PubMed] [Google Scholar]
- 18. Uhlen M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 19. Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–7. [DOI] [PubMed] [Google Scholar]
- 20. Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo Y, Xiao P, Lei S, Deng F, Xiao GG, Liu Y, Chen X, Li L, Wu S, Chen Y, Jiang H, Tan L, Xie J, Zhu X, Liang S, Deng H. How is mRNA expression predictive for protein expression? a correlation study on human circulating monocytes. Acta Biochim Biophys Sin (Shanghai). 2008;40(5):426–36. [DOI] [PubMed] [Google Scholar]
- 22. Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. Bioessays. 2006;28(8):799–808. [DOI] [PubMed] [Google Scholar]
- 23. Brodaczewska K, Donskow-Łysoniewska K, Doligalska M. Chitin, a key factor in immune regulation: lesson from infection with fungi and chitin bearing parasites. Acta Parasitol. 2015;60(2):337–44. [DOI] [PubMed] [Google Scholar]
- 24. Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153(Pt 9):2817–22. [DOI] [PubMed] [Google Scholar]
- 25. Li Y, Chen X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl Microbiol Biotechnol. 2012;94(4):887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Juge N, Tailford L, Owen CD. Sialidases from gut bacteria: a mini-review. Biochem Soc Trans. 2016;44(1):166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elias JA, Homer RJ, Hamid Q, Lee CG. Chitinases and chitinase-like proteins in T(H)2 inflammation and asthma. J Allergy Clin Immunol. 2005;116(3):497–500. [DOI] [PubMed] [Google Scholar]
- 28. Belkaid Y, Tamoutounour S. The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol. 2016;16(6):353–66. [DOI] [PubMed] [Google Scholar]
- 29. Tong PL, Roediger B, Kolesnikoff N, Biro M, Tay SS, Jain R, Shaw LE, Grimbaldeston MA, Weninger W. The skin immune atlas: three-dimensional analysis of cutaneous leukocyte subsets by multiphoton microscopy. J Invest Dermatol. 2015;135(1):84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abdel-Rahman SM. Genetic predictors of susceptibility to dermatophytoses. Mycopathologia. 2017;182(1–2):67–76. [DOI] [PubMed] [Google Scholar]
- 31. Koller B, Müller-Wiefel AS, Rupec R, Korting HC, Ruzicka T. Chitin modulates innate immune responses of keratinocytes. PLoS ONE. 2011;6(2):e16594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wulf-Johansson H, Lock Johansson S, Schlosser A, Trommelholt Holm A, Rasmussen LM, Mickley H, Diederichsen AC, Munkholm H, Poulsen TS, Tornøe I, Nielsen V, Marcussen N, Vestbo J, Sækmose SG, Holmskov U, Sorensen GL. Localization of microfibrillar-associated protein 4 (MFAP4) in human tissues: clinical evaluation of serum MFAP4 and its association with various cardiovascular conditions. PLoS ONE. 2013;8(12):e82243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pilecki B, Holm AT, Schlosser A, Moeller JB, Wohl AP, Zuk AV, Heumüller SE, Wallis R, Moestrup SK, Sengle G, Holmskov U, Sorensen GL. Characterization of microfibrillar-associated protein 4 (MFAP4) as a tropoelastin- and fibrillin-binding protein involved in elastic fiber formation. J Biol Chem. 2016;291(3):1103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.