Abstract
Developmental pathways that orchestrate the fleeting transition of endothelial cells into haematopoietic stem cells remain undefined. Here we demonstrate a tractable approach for fully converting adult mouse endothelial cells to haematopoietic stem cells (rEC-HSCs) through transient expression of genes encoding the transcription factors Fosb, Gfi1, Runx1, and Spi1 (also known as Fgrs) and vascular-niche-derived angiocrine factors. The induction phase (day 0–8) of conversion is initiated by expression of Fgrs in mature endothelial cells, which results in endogenous Runx1 expression. During the specification phase (day 8–20), Runx1+ Fgrs-transduced endothelial cells commit to a haematopoietic fate yielding rEC-HSCs that no longer require Fgrs expression. The vascular niche drives a robust self-renewal and expansion phase of rEC-HSCs (at day 20–28). rEC-HSCs have a transcriptome and long-term self-renewal capacity similar to those of adult haematopoietic stem cells, are competent for clonal engraftment and serial primary and secondary multi-lineage reconstituting potential, including antigen-dependent adaptive immune function. Inhibition of TGF-β and CXCR7 or activation of BMP and CXCR4 signalling enhanced generation of rEC-HSCs. Conversion of endothelial cells into autologous authentic engraftable haematopoietic stem cells could aid treatment of haematological disorders.
De novo generation of haematopoietic stem and progenitor cells (HSPCs) would enable autologous treatment of blood disorders but this goal has met many obstacles1. Specifically, derivation of engraftable haematopoietic stem cells (HSCs) from pluripotent stem cells has not yet been achieved2–4. To circumvent transition through a destabilizing pluripotency state, attempts have been made to reprogram non-haematopoietic cell types into HSCs, but these efforts have produced haematopoietic progenitor-like cells with poor engraftment potential5–10.
The inability to generate HSCs could be explained by lack of environmental cues to self-renew reprogrammed HSCs11–19. Mouse lymphoid cells have previously been reprogrammed into putative HSCs through expression of eight transcription factors and using a recipient in vivo niche to support conversion20. Constitutive expression of transcription factors Fosb, Gfi1, Runx1, and Spi1 (Fgrs) and co-culturing with inductive vascular-niche-converted human adult endothelial cells into transplantable multi-potent haematopoietic progenitors (rEC-MPPs)9. However, neither study yielded cells supporting adaptive immunity. Putative HSCs engineered from pre-B lymphoid progenitors were incapable of immune adaptation because they had already undergone Ig-heavy chain recombination at the V(D)J locus. Similarly, CD34+ progenitors from rEC-MPPs poorly differentiated into mature T cells9.
Endothelial to haemogenic endothelial cell and HSC transition (EHT)21,22 is mediated by signals from niche cells23–25 within the aorta-gonad-mesonephros region26 during a narrow determine the pathways that drive this ontological process27. To this end, we devised a tractable stepwise approach whereby conditional expression of Fgrs in adult mouse endothelial cells co-cultured with an inductive vascular-niche converts adult endothelial cells into engraftable HSCs (rEC-HSCs) that possess all attributes of bona fide HSCs. rEC-HSCs are capable of clonal expansion and serial multi-potent reconstitution of all haematopoietic lineages, including immunocompetent lymphoid cells that elicit antigen-specific adaptive immune responses.
Conditional Fgrs in mECs generates HSPCs
Adult mouse vascular endothelial cells (mECs) were purified by flow cytometry to eliminate contaminating lymphatic endothelial cells and haematopoietic cells (Fig. 1a). Freshly isolated mECs (CD45.2+) transplanted with radio-protective marrow cells did not contribute to recipient (CD45.1+) haematopoiesis, showing that mEC preparations were free of contaminating host-derived HSPCs. Moreover, before conversion, mECs were expanded using culture conditions that would not permit HSPC propagation (Extended Data Fig. 1a, b). Thus, mEC preparations were free of contaminating host-derived HSPCs capable of contributing to haematopoiesis in recipients.
Figure 1. Conditional expression of Fgrs in adult mECs generates haematopoietic cells.
a, Schema showing conversion of 2.5×105 adult mECs into HSPCs. b, Emergence of CD45+ cells in the vicinity of VN-ECs (HUVEC-E4ORF1). Representative pictures (10×). c, Left, Fgrs-ECs grown on VN-ECs, OP9-DLL11, or in feeder-free condition. Right, Fgrs-transduced OP9 cells were grown on VN-ECs, OP9-DLL11 (transduced with E4ORF1), or feeder-free. Data represent mean ± s.e.m. (n = 5 conversion experiments run in technical triplicates for each condition); two-tailed unpaired t-test. d, Quantification of CD45+ LKS cells during conversion (left) and absolute cell number (right). VN-ECs were excluded by gating on human CD31 negative (hCD31−) population. Data represent mean ± s.e.m. (n = 5 conversion experiments run in technical triplicates for each condition); two-tailed unpaired t-test. e, CD45+ cells were sorted at day 23 and seeded in methylcellulose (no-dox); colonies were analysed 7 days later. Graphs show the quantification of the number of colonies per 2,500 cells plated (n = 5 conversion experiments run in technical triplicates for each condition); two-tailed unpaired t-test.
To initiate conversion, mECs were transduced with doxycycline (dox)-inducible Fgrs (Fgrs-ECs) lentiviral vectors and co-cultured with vascular niche mECs (VN-ECs,Human Umbilical Vein Endothelial Cells, HUVEC-E4ORF1)11,13,28,29 (Fig. 1a). VN-ECs cells were generated by enforced expression of the adenoviral E4ORF1 gene in human umbilical vein ECs (HUVEC-E4ORF1). E4ORF1 mimics physiological activation of phosphorylated Akt in ECs, enhancing their survival, while sustaining their pro-haematopoietic instructive angiocrine functions11,13,28,29
During conversion, Fgrs-ECs transitioned through three sequential phases (Fig. 1a): (1) ‘induction-phase’ (day 0–8) is initiated by dox-upregulation of conditional Fgrs in VE-cadherin (VEcad)+RUNX1−CD45− lung endothelial cells from adult mice co-cultured with VN-ECs (Extended Data Fig. 1c). By day 8, Fgrs-ECs turned on endogenous Runx1 expression. (2) During the ‘specification phase’ (day 9–20), RUNX1+ Fgrs-ECs commit to a haematopoietic fate, transitioning from VEcad+RUNX1+CD45− to VEcad+RUNX1+CD45+ to VEcad−RUNX1+CD45+ cells, thus acquiring haematopoietic morphology and markers (Fig. 1b–d, Extended Data Fig. 1d–f) and autonomous colony-forming cell (CFC) activity, as exogenous Fgrs is no longer required (Fig. 1e). Similar to human rEC-MPPs9, the reprogrammed mECs to HSPCs (rEC-HSPCs) are endowed with multi-lineage progenitor properties, yielding CFC-GEMM (granulocyte, erythrocyte, monocyte, megakaryocyte), CFC-GM (granulocyte, monocyte), and BFU-E burst forming unit-erythroid) colonies (Fig. 1e). (3) In the ‘expansion phase’ (day 20–28), the total number of short-term re-populating/radio-protective cells and lin−c-Kit+Sca-1+ (rEC-LKS, gated on human CD31−, hCD31) cells increased (Fig. 1d). Most rEC-LKS cells expand adherent to VN-ECs, suggesting paracrine and juxtacrine angiocrine factors supplied by VN-ECs maintain and expand LKS cells (Extended data Fig. 1g). Angiocrine signals provided by VN-ECs are missing from bone-marrow derived fibroblastic OP9-DLL1 cells as co-culture of Fgrs-ECs with OP9-DLL1 cells did not convert endothelial cells into CD45+ rEC-HSPCs (Fig. 1c, Extended data Fig. 1h, i). Conversely, Fgrs-transduction of OP9-DLL1 cells did not yield haematopoietic cells, indicating that endothelial cells have an innate plasticity, unlike fibroblastic cell types.
rEC-HSPCs engraft primary and secondary recipients
rEC-HSPCs were transplanted into congenic recipients to assess their function. Before transplantation at day 28, rEC-HSPCs had completed haematopoietic commitment, thus we discontinued dox treatment, turning-off Fgrs expression (Fgrs-off). Day 28 CD45.2+ lung-derived rEC-HSPCs (Fgrs-off) (8.0 × 105 cells) were transplanted into lethally irradiated (950 cGy) congenic CD45.1+ recipients to assess long-term engraftment (Fig. 2a). As controls, 8.0 × 105 adult lung Fgrs-ECs, in which Fgrs was never turned on (no-dox), were transplanted or PBS was injected (Fig. 2a). Only CD45.2+ rEC-HSPCs could radio-protect and engraft lethally irradiated recipients (Extended Data Fig. 2a). In contrast, no-dox lung Fgrs-ECs failed to reconstitute haematopoiesis. Moreover, Fgrs-ECs (no-dox) were not contaminated by HSPCs as they did not radio-protect the recipient upon transplantation. Hence, purified adult mECs are not contaminated by donor-derived HSPCs and their conversion to engraftable haematopoietic cells requires transient Fgrs expression.
Figure 2. Conditional Fgrs expression supports long-term primary and secondary HSPC engraftment.
a, Transplantation schema. b, Lineage contribution to Gr1+CD11b+ and Gr1−CD11b+ myeloid cells, B220+ B cells, CD3+CD4+ T cells, and CD3+CD8+ T cells at week 20 after primary transplant in the peripheral blood of WBM control transplant recipients (blue circles) or rEC-HSPC recipients (green circles), Boxplot and whiskers represent median, 25th and 75th percentile, mean is represented by + sign (n = 4 independent conversion experiments run in technical triplicates for each condition); two-tailed unpaired t-test. c, Lineage contribution to Gr1+CD11b+ and Gr1−CD11b+ myeloid cells, B220+ B cells, CD3+CD4+ T cells, and CD3+CD8+ T cells at week 20 after secondary transplant in the peripheral blood of WBM control transplant (blue circles) or rEC-HSPC (green circles), Boxplot and whiskers represent median, 25th and 75th percentile, mean is represented by + sign (n = 4 independent conversion experiment run in technical triplicates for each conditions); two-tailed unpaired t-test. d, Relative representation of LKS and LKS-SLAM cells at week 20 after primary transplant for WBM transplant recipients (blue circles) or rEC-HSPC recipients (green circles). e, Relative representation of LKS and LKS-SLAM cells at week 20 after secondary transplant for WBM transplant recipients (blue circles) or rEC-HSPC recipients (green circles). Boxplot and whiskers represent median, 25th and 75th percentile, mean is represented by + sign (n = 4 independent conversion experiment run in technical triplicates for each conditions); two-tailed unpaired t-test
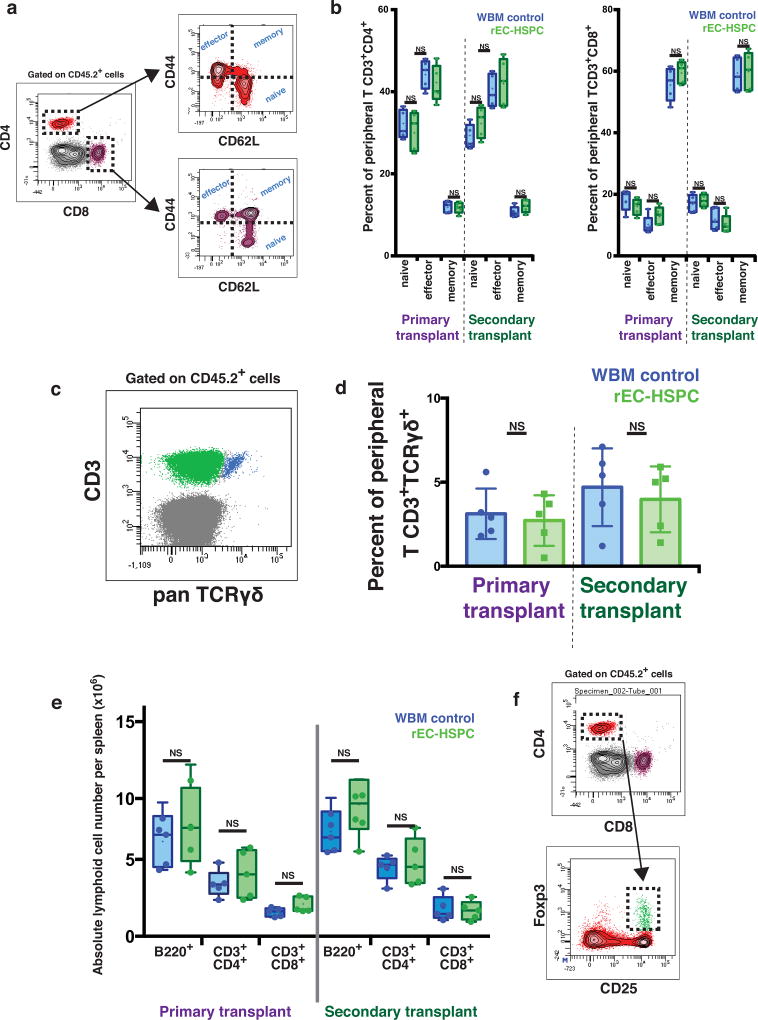
CD45.2+ rEC-HSPC-derived myeloid and lymphoid cells were present in peripheral blood 4–20 weeks after transplantation (Fig. 2b, Extended Data Fig. 2b, c). The blood, marrow and spleens of recipients transplanted with rEC-HSPCs were replete with donor-derived B cells and circulating naive, effector, memory, and γδ T cells in numbers and physiologic ratios similar to mice engrafted with normal whole bone marrow (WBM) (Extended Data Fig. 3a–e). Analysis of marrow cells revealed a normal frequency of donor-derived phenotypically marked HSCs (CD45.2+CD48−CD150+LKS; LKS-SLAM) in mice engrafted with rEC-HSPCs or normal WBM (6.63% ± 0.43 versus 7.98% ± 0.58), respectively (Fig. 2d, Extended Data Fig. 2d). Thus, rEC-HSPCs yield LKS-SLAM cells that were capable of a similar degree of long-term multi-lineage engraftment to that of WBM.
We examined the self-renewal potential of CD45.2+ rEC-HSPCs by transplanting unfractionated marrow from primary transplant recipients into lethally irradiated secondary recipients (Fig. 2a). We detected CD45.2+ myeloid and lymphoid cells in the peripheral blood of secondary recipients from 4 to 20 weeks after transplantation (Fig. 2c, Extended Data Fig. 2e). The number of LKS-SLAM cells was comparable in secondary recipients of rEC-HSPCs and WBM (versus 6.56% ± 0.30 versus 7.10% ± 0.62) (Fig. 2e). Following sub-lethal (500 cGy) irradiation, secondary recipients of both rEC-HSPCs and WBM rapidly reconstituted multi-lineage haematopoiesis (Extended Data Fig. 2f–h). Thus, engrafted rEC-HSPCs tolerate myelosuppressive stress and reconstitute serial long-term primary and secondary LKS-SLAM and multi-lineage haematopoiesis, suggesting robust in vivo self-renewal comparable to native WBM HSCs. Therefore, subsets of rEC-HSPCs are composed of reprogrammed engraftable rEC-HSCs.
rEC-HSC clonal and limiting-dilution engraftment
Isochronic populations of Fgrs-ECs (Dox-on) were used to perform both single-cell clonal expansion/transplantation and limiting-dilution transplantation (Fig. 3a). To identify the interval at which Fgrs-ECs transition into haematopoietic cells, we used a Runx1 reporter mouse (CD45.2+ Runx1-IRES-GFP)30. As RUNX1 is not expressed on endothelial cells, but on fetal haemogenic cells, expression of endogenous Runx1 reports the birth of CD45− haemogenic-like endothelial cells (Extended Data Fig. 4a–c, Supplementary Video 1).
Figure 3. Clonal and limiting-dilution quantification of rEC-HSPCs and rEC-HSCs.
a, Schema showing rEC-HSC quantification strategy. Fgrs-ECs isolated from CD45.2+ Runx1-IRES-GFP mice were subjected to isochronic haematopoietic conversion. By day 8, one half of the Fgrs-ECs underwent single-cell cloning on the basis of Runx1-GFP+ expression (VEcad+RUNX1+CD45−) and co-cultured with VN-ECs. Haematopoietic clusters arising from single RUNX1+ endothelial cells were transplanted into lethally irradiated CD45.1+ recipients. The other isochronic half of the cell fraction was expanded on VN-ECs until day 28 and transplanted in a limiting-dilution translation (LDT) into lethally irradiated CD45.1+ recipients. b, Reconstitution of the indicated peripheral blood cell lineages of individual recipients showing >1% donor chimaerism are presented as the percentage of donor clones, data represent mean ± s.e.m (n = 7 independent transplants). c, LDT assay to determine the frequency of LT-HSCs in each expanded rEC-HPSC clone. Averaged CRU per cells were determined using Poisson statistics (ELDA software). Boxplot and whiskers represent median, 25th and 75th percentile, mean is represented by + sign (n = 4 independent conversion experiment run in technical triplicates for each conditions); two-tailed unpaired t-test (n = 7 independent LDT analysis). d, LDT showing LT-HSC frequency by day 28 rEC-HSCs. CRU of 1 in 557 was determined using Poisson statistics (ELDA software) (n = 3 independent LDT experiments).
On day 8, emerging Runx1-GFP+ Fgrs-ECs were cloned in individual wells containing fresh VN-ECs and maintained in culture with dox (‘Fgrs-on’; Fig. 3a, Extended Data Fig. 5a). On average, 22 ± 11 of 1,000 sorted Runx1-GFP+ endothelial cells yielded clonally derived haematopoietic clusters (Extended Data Fig.3a). We transplanted each haematopoietic cluster into lethally irradiated CD45.1+ recipients and all displayed multi-lineage engraftment from 4 to 16 weeks after transplantation (Fig. 3b, Extended Data Fig. 5b). We sorted rEC-LKS progeny from clonally derived haematopoietic clusters and quantified the frequency of repopulating cells per clone by limiting-dilution transplantation. The frequency of repopulating cells varied in different clones, with an average of 16 ± 12 competitive repopulating units (CRU) per 1,000 rEC-LKS cells (Fig. 3c, Extended Data Fig. 5c). Viral integration mapping demonstrated that transplanted cells share the same viral integration pattern from the HSC compartment to peripheral-blood-differentiated cells, indicating clonally derived rEC-HSCs have multi-lineage reconstitution capacity (Extended Data Fig. 5d).
The other half of purified isochronic Runx1-GFP+ Fgrs-ECs induced by VN-ECs were expanded in bulk to day 28 and then rEC-LKS cells were sorted by flow cytomery. Then by turning off Fgrs the CRU frequency within the rEC-LKS cells was quantified by limiting-dilution transplantation (Fig. 3a, d). We found 1.8 CRU per 1,000 rEC-LKS cells (1 in 557; 95% confidence, 1 in 1004 to 1 in 310), as quantified by Poisson statistics (Fig. 3d). As there was a total of 7,000 LKS cells on day 28, a total of 13 ± 2.6 CRU were generated during the conversion process. Thus, conditional Fgrs expression in adult endothelial cells with angiocrine signals supplied by VN-ECs generates cells clonally capable of self-renewal and long-term multi-lineage haematopoietic reconstitution, thereby fulfilling the criteria of repopulating rEC-HSCs.
rEC-HSCs/HSPCs match adult transcriptome
We sorted rEC-LKS cells from day 28 in vitro culture, and from the marrow of transplanted secondary recipients, and compared their composite transcriptome to embryonic day 11 (E11) aorta-gonad-mesonephros endothelium, E11 type-1 pre-HSCs, E11 type-2 pre-HSCs, E12 fetal liver HSCs, E14.5 fetal liver HSCs31, and LKS-SLAM cells isolated from a WBM secondary transplant recipients. We performed supervised principal components analysis on 96 genes that characterize the transition between fetal to adult HSCs, to define transcriptional distances between different developmental stages. Steady-state wild-type adult HSCs, wild-type fetal cells and pre-HSC cells segregated by developmental stage (Fig. 4a, Extended Data Fig. 6a). rEC-LKS-SLAM cells isolated from day 28 in vitro culture or from the marrow of transplanted recipients cluster closely to adult wild-type HSCs. Compared to starting lung endothelial cells, rEC-LKS upregulate haematopoietic genes and silence vascular genes (Extended data Fig. 6b–d). Pluripotency genes were not induced in rEC-HSPCs, indicating that conversion does not require transition through a destabilizing pluripotent intermediate stage.
Figure 4. Gene expression profiling of rEC-HSPCs and rEC-HSCs.
Principal Component Analysis (PCA) of the transcriptome-wide RNA-Seq expression profiles was performed. A 3D-PCA plot, representing ~85.0% of the total variation within the expression dataset, was generated to visualize the clustering of the reprogrammed hematopoietic sample groups in relation to their native counterparts. Three dimensional MDS plots (3D-MDS) were generated based on all pairwise distances between the global transcriptome-wide RNA-seq profiles of the samples shown here. Distances were defined as one minus the Pearson correlation between two profiles. Multidimensional scaling (MDS) was used to identify the set of points in 3D space, such that the distances between the points are equal to the true distances between samples.
We next compared control adult LKS-SLAM cells and their progeny to rEC-LKS-SLAM cells and their derivatives. Unsupervised hierarchical clustering analysis showed a tight association of clonal rEC-LKS-SLAM and polyclonal rEC-LKS-SLAM cells with their wild-type WBM transplant counterparts and previously published32 (Fig. 4a, Extended Data Fig. 6e). Similarly, reprogrammed (rEC-CD3+CD4+, rEC-B220+, rEC-Gr1+CD11b+) and control WBM-derived progenies cluster by lineage. Thus, rEC-LKS and rEC-LKS-SLAM cells (rEC-HSCs) have a global gene expression transcriptome that is similar to their control WBM equivalents.
rEC-HSC lymphoid progeny confer adaptive immunity
We transplanted week 20, primary-engrafted Fgrs-off rEC-HSPCs or WBM controls into immunodeficient Rag1−/− mice (Fig. 5a). After 20 weeks, we isolated engrafted CD45.2+CD3+CD4+ or CD45.2+CD3+CD8+ T cells from peripheral blood and subjected them to polyclonal activation using anti-CD3/CD28 beads and IL-2. Both CD45.2+CD3+CD4+ and CD45.2+CD3+CD8+ T-cell subsets proliferated well and upregulated interferon-γ (IFNγ) expression (Extended Data Fig. 7a, b). Addition of CD45.2+CD3+CD4+CD127−CD25+ cells enriched with T regulatory cells (Extended Data Fig. 3f) inhibited proliferation of CD45.2+CD3+CD4+ or CD45.2+CD3+CD8+ T cells, thus reducing IFNγ-producing cells (Extended Data Fig. 7a, b). Moreover, purified CD45.2+CD3+CD8+ T cells lysed allogeneic mouse (Balb/c) target cells (Fig. 5b, c).
Figure 5. Reconstitution of T-cell immunity in mice engrafted with Rag1−/− rECs.
a, Transplantation schema of chimaeric rEC-HSPC or WBM control into CD45.1+Rag1−/− mice. b, Schema and time-lapse of redirected lysis assay. c, Viable target percentage at different ratios of effector (CD8+) to target (allogeneic Balb/C, CD45+) cells. Data represent mean ± s.e.m. (n = 5 independent experiments in 3 technical replicates); two-tailed unpaired t-test. d, Heat map of normalized counts (log counts per million reads) for each TCR β variable gene for CD4+ T cell, CD8+ T cell, and Jurkat cell samples. e, Boxplot of Shannon index clonality of normalized TCR β variable gene counts (per million reads) across WBM and rEC-HSPC samples as CD4+ T cells, CD8+ T cells or Jurkat cells (control sample). Data represents mean ± s.d. (n = 3); Wilcoxon rank t-test. f, Quantification OVA-MHC multimers (n = 4). Boxplot and whiskers represent median, 25th and 75th percentile, mean is represented by + sign (n = 5); two-tailed unpaired t-test.
We assessed clonal diversity of T-cell receptors (TCR) within CD3+CD4+ and CD3+CD8+ progeny of either WBM or rEC-HSPC donor cells. Normalized RNA-seq read counts (log counts per million reads) mapped to T-cell receptor β variable genes were used to estimate the TCR diversity33. The clonal repertoire of mapped T cell β variable genes was similar in T-cell progeny (CD4+ and CD8+) of WBM control and rEC-HSPC-transplanted mice (Fig. 5c, Extended Data Fig. 7c–e). No statistically significant differences between the TCR diversity within T-cell progeny of WBM and rEC-HSC-transplanted mice could be identified (Wilcox test, P = 0.33 and P = 0.2, respectively) as assessed by normalized Shannon index (Fig. 5d). Hence, transplantation of rEC-HSCs re-establishes a T-cell pool as diverse as the WBM control.
We then tested whether secondary Rag1−/− recipient mice engrafted by rEC-HSPCs were capable of adaptive immune function. Paired analyses of the CDR3 region in the same mice before and after immunization against chicken ovalbumin (OVA) showed enrichment of oligo-clonal rearrangements in certain Vβ families following vaccination with OVA (Extended Data Fig. 7f). T cells with OVA-specific TCRs were quantified using tetramers tracking OVA-specific CD8+ (SIINFEKL/Kb tetramers) or OVA-specific CD4+ cells (I-Ab OVA(329-337) tetramers) before and after vaccination(Fig. 5e, f). Vaccination increased the number OVA specific T-cells, indicating that rEC-HSPCs can generate T-cell subsets with adaptive immune function, an attribute of normal HSCs.
VN-EC angiocrine signals driving conversion
This stepwise conversion captures molecular snapshots and could deconvolute vascular-niche-derived angiocrine signals that choreograph EHT. Runx1-IRES-GFP endothelial cells (Fig. 3, Extended Data Fig. 4) were used to track and quantify the number of Fgrs-ECs transitioning during induction, specification and expansion into rEC-HSPCs (Extended Data Fig. 8a, b).
We isolated and performed CFC analyses on Fgrs-ECs (VEcad+RUNX1−CD45−) haemogenic-like endothelial cells (VEcad+RUNX1+CD45−, VEcad+RUNX1+CD45+) and rEC-HSPCs (VEcad− RUNX1+CD45+) 15 days after addition of dox (Fgrs-on). We found that all of the cells with progenitor activity were in the CD45+ fraction (Extended Data Fig. 8c). Once the converted cells reached the specified stage, marked by the VEcad−RUNX1+CD45+ immunophenotype, they no longer required exogenous Fgrs to maintain CFC function. Before specification phase (for example, in VEcad+RUNX1+CD45+ cells), ongoing exogenous Fgrs expression was required for CFC activity.
We then used Runx1-IRES-GFP endothelial cells to identify VN-EC angiocrine signals that control rEC-HSPC conversion. Runx1-IRES-GFP Fgrs-ECs (dox-on) and VN-EC induction were then challenged with various small molecules modulating putative signalling pathways involved in EHT, including TGF-β, BMP, and CXCL1234. Although none of these agents impaired cell viability, induction was blocked when CXCL12 or BMP signalling was inhibited. Abrogation of activin or TGF-β signalling enhanced the specification phase (Extended Data Fig. 8d–f).
CXCL12 signals through two receptors, CXCR4 and CXCR7. As CXCR4, the receptor for CXCL12, is expressed on both endothelial cells and haematopoietic cells, we selectively knocked out Cxcr4 in mECs (Fig. 6a). Adult lung mECs were isolated from mice with floxed Cxcr4 alleles (Cxcr4fl/fl). These endothelial cells were transduced with Fgrs (dox-on) and VN-EC induction. Treatment with Cre-recombinase deleted Cxcr4 and impaired Runx1 induction in mECs (Fig. 6a, b). Conversely, activation of CXCR7, safeguarded vascular fate and blocked the EHT process (Extended Data Fig. 8f). Overexpression of mouse CXCL12 by VN-ECs increased the number of VEcad−RUNX1+CD45+ cells (Extended Data Fig. 8g). Thus, CXCR4, but not CXCR7, contributed to conversion into rEC-HSPCs. Moreover, Runx1 expression was enhanced by TGF-β inhibition and activation of BMP and CXCL12 signalling. Emergence and expansion of rEC-HSPCs was dependent on CXCL12 signalling (Fig. 6b), indicating that sequential endothelial cell conversion into rEC-HSPCs provides a tractable approach to screen and identify pathways driving transient EHT.
Figure 6. Deconvolution of vascular niche angiocrine signals in rEC-HSPC generation.
a, Quantification of Runx1-IRES-GFP Fgrs-ECs Cxcr4fl/fl or Cxcr4−/− and their derivatives during reprogramming. Data represent mean ± s.e.m. (n = 4); two-tailed unpaired t-test. b, Schematic representation of angiocrine pathways involved in conversion of adult endothelial cells into HSPCs.
Fgrs-driven conversion was not exclusive to lung endothelial cells, we found that endothelial cells isolated from all of the adult organs tested (liver, brain, and kidneys) converted upon transient Fgrs expression and VN-EC induction (Extended Data Fig. 9a–d). Thus, adult endothelial cells are amenable to conversion into rEC-HSPCs, irrespective of tissue origin.
We performed several tests to rule out contamination of endothelial cell preparations with a rare population of host-derived HSPCs/HSCs. Purified adult lung endothelial cells do not harbour host-derived HSCs or haemogenic endothelial cells and Fgrs-ECs (no-dox) failed to form engraftable CD45+ cells (Figs 1b, c, 2a, Extended Data Fig. 1a, 2a). Regardless of whether the endothelial cell preparations had been contaminated, passaging of freshly purified endothelial cells for 30 days (absent of haematopoietic cytokines) leads to loss of contaminating HSPCs. These endothelial cell cultures did not support expansion of Fgrs-transduced wild-type long-term (LT)-HSCs (Extended Data Fig. 1b). To exclude the possibility that CD45.2+ haematopoietic cells contaminating lung endothelial cell cultures could revert to HSCs upon exogenous Fgrs expression, we isolated terminally differentiated CD45.2+ cells from rEC-HSPC-engrafted mice and re-induced Fgrs expression for 28 days in co-culture with VN-ECs. After transplant, these cells did not contribute to the host haematopoiesis (Extended Data Fig. 9e). We also show that our endothelial cell to HSPC conversion follows a hierarchical model (Extended Data Fig. 9f, g). Thus, the simplest explanation for our findings is that adult endothelial cells were converted to rEC-HSCs and there was no contamination of endothelial cell cultures with host haematopoietic cells.
Discussion
We demonstrate for the first time the conversion of adult endothelial cells to professional multipotent cells that meet the criteria of normal HSCs. Transient expression of Fgrs reprogramming factors in adult endothelial cells with vascular niche induction readily reprograms these cells to stable, self-renewing HSCs (rEC-HSCs) that are capable of long-term, clonally derived engraftment and haematopoietic reconstitution of myelopoiesis and both innate and adaptive immune function. Adult tissue-specific endothelial cells have a unique plasticity that is missing from other somatic cells, allowing them to undergo efficient transition towards a haematopoietic fate. Our approach has translational potential and should also enhance our understanding of the evanescent ontological EHT thought to be responsible for all HSCs generated during an individual’s lifetime35.
The pathways that regulate EHT have been difficult to elucidate as only a small number of cells undergo EHT within a short developmental window36–39. Here we have developed a stepwise spatially and temporally controlled in vitro model that reveals the signalling pathways driving EHT. We show that direct cellular contact between Fgrs-ECs and their vascular niche is essential and sufficient for the birth and self-renewal of rEC-HSCs. The vascular niche supplies key membrane and secreted angiocrine factors, including Notch ligands, Kit-ligand, BMP2, adhesion molecules, and other unknown factors that drive Fgrs-ECs transition into rEC-HSCs17. Although BMP and CXCL12–CXCR4 activation accelerated generation of rEC-HSPCs, TGF-β and signalling via the other CXCL12 receptor, CXCR7, blocked haematopoietic induction from endothelial cells. Thus, the vascular niche platform provides an opportunity to track and eavesdrop on the cross-talk between emerging rEC-HSCs and the niche cells. Specifically, this model facilitates the screening of small molecules and identification of transcription factors as well as yet unknown factors, that enhance specification and self-renewal of HSCs. The clear delineation of niche-dependent induction from specification that we have identified is expected to advance our understanding of the niche-driven developmental processes involved in HSC generation.
Conversion of adult endothelial cells to HSCs appears to be efficient. We showed that a single clone of Runx1+ Fgrs-EC could yield 16±12 repopulating cell/1000 phenotypic LKS cells. Considering 38,510 total LKS were present at the time of LDT, a single Runx1+ Fgrs-EC can give rise to approximately 598±463 repopulating cells (Fig. 3). The variation in the frequency of repopulating cells generated per clone suggests that FGRS stoichiometry impacts the quality of the endothelial to HSC conversion. Nonetheless, we showed that single Runx1+ Fgrs-EC could undergo EHT and give rise to expandable and self-renewing rEC-HSC capable of long-term engraftment (Fig. 3b,c). The difference in the amount of repopulating cell between the “bulk” conversion (13 CRU per conversion) and the single Runx1+ FGRS-EC conversion strategy (598±463 CRU) could be explained by complex negative feedback occurring during our bulk conversion (Fig.6b). Considering that on average 28 ± 4.8% of 250,000 Fgrs-ECs express endogenous Runx1 by day 8, and that 22 ± 11 cells of 1,000 cloned Runx1+/−GFP+ endothelial cells yield rEC-HSCs (Fig. 6b, Extended Data Fig. 5c), a total of around 1,500 ± 610 rEC-HSCs are generated during reprogramming (1,500 out of 70,000, 2.1%). with an overall conversion efficiency of ~0.6% (1500/250,000) for the starting population of FGRS-ECs. Theoretically, ~24,000 HSCs can be derived from 70,000 RUNX1+ adult mECs during the conversion process.
We did not find evidence of malignant transformation in primary or secondary recipient mice engrafted with rEC-HSCs for up to 20 months or more (Extended Data Fig. 10). RNA-profiling indicates that the signature of rEC-HSPCs is similar to adult wild-type HSCs. Indeed, the transcriptome of rEC-HSPCs was more closely linked to adult HSCs than to fetal type-1 or type-2 pre-HSCs (Fig. 3a). Therefore, our approach provides a platform for converting readily accessible sources of adult endothelial cells to stable long-term repopulating HSCs.
Although the transcriptome of rEC-HSCs obtained from clonal or bulk conversion were indistinguishable from native adult HSCs, additional experiments will be required to determine if their endothelial epigenetic landscape is fully erased. Notably, the donor reconstitution observed in our clonal primary transplants often mature over time after transplantation, with lymphoid or myeloid lineage bias potential correcting over time.
As a large number of adult endothelial cells can be obtained from healthy donors, our approach makes possible future use of autologous endothelial cells to generate long-term HSCs for treatment of acquired and genetic haematological disorders. Because endothelial cells are readily amenable to gene editing, without undergoing cellular attrition, the clonally expanded endothelial cells could generate abundant rEC-HSCs with customized functions, including those yielding adaptive T-cell subsets that could treat immunological disorders.
METHODS
Data reporting
All cell preparations were routinely tested for mycoplasma contamination.
Animals
All animal experiments were performed under the approval of Weill Cornell Medicine Institutional Animal Care and Use Committee. We employed Runx1-IRES-GFP (Runx1tm4Dow) mice, provided by J. Downing (St Jude Hospital). These were crossed with Rosa26 rtTa mice (B6.Cg-Gt(ROSA)26Sortm1(rtTA*M2)Jae/J, Jackson laboratory, strain 006965) to produce Runx1-IRES-GFP;Rosa26-rtTa mice (refered to as Runx1), and maintained as heterozygous for both Runx1-IRES-GFP and rtTa. Floxed Cxcr4 mice were obtained from Y.-R. Zou (Feinstein Institute for Medical Research). Floxed Cxcr4 mice were crossed with Runx1-IRES-GFP;Rosa26-rtTa mice to produce Cxcr4fl/fl;Runx1-IRES-GFP+/−;Rosa26-rtTa mice.
Rag1−/− mice with the genotype of B6.129S7-Rag1tm1Mom/J were obtained from the Jackson Laboratory (strain 002216).
For rEC-HSPC transplantation, we used CD45.1+ congenic 10–12-week-old male recipients (B6.SJL-Ptprca Pepcb/BoyJ). These mice are referred here as CD45.1+ recipients, Jackson Laboratory, strain 002014.
Cell culture Media
Mouse endothelial cell media
DMEM:Ham’s F-12 (Sigma, D6421) supplemented with 20% FBS (Omega Scientific, FB07), 20 mM HEPES (Invitrogen, 15630080), 100 µg ml−1 Heparin (Sigma, H3393), 50 µg ml−1 endothelial mitogen (Alfa Aeser J65416) and 5 µM SB431542 (R&D, 1614).
Human endothelial cell media
M199 (Sigma, M4530), 10% FBS (Omega Scientific, FB07), 50 µg ml−1 endothelial mitogen (Alfa Aeser J65416), and 100 µg ml−1 Heparin (Sigma, H3393).
Conversion media
StemSpan SFEM (STEMCELL Technologies, 09650), 10% KnockOut Serum Replacement (Invitrogen, 10828028) 10 ng ml−1 hFGF-2 (bFGF, Peprotech, 100-18), 50 ng ml−1 mouse c-Kit ligand (SCF, Peprotech, 250-03).
Isolation of adult mouse organ-specific endothelial cells for reprogramming
25 µg of anti-VE-cadherin-AF647 antibody (clone BV13, Biolegend) were injected retro-orbitally in 8–10-week-old male C57BL/6J (CD45.2+) mice under anaesthesia 8 min before they were killed and the organs harvested. For flow cytometry and cell sorting, organs, including lungs, brain, kidney and liver were minced and incubated with collagenase-A (25 mg ml−1), Dispase II (25 mg ml−1), and DNase (250 µg ml−1) (Roche) at 37 °C for 20–30 min to create a single-cell suspension. Cells were filtered through a 40-µm filter immediately before counter staining. Single-cell suspension was first blocked with an antibody against CD16/32 (2.4G2) before antibody staining with anti-mouse CD31-PE-Cy7 (390, Biolegend), anti-mouse TER119 (TER119) and anti-mouse CD45 (30-F11). Haematopoietic and erythroid cells were removed via CD45 and TER119 gate exclusion, and adult mouse endothelial cells were defined and sorted as VEcad+CD31+CD45−TER119− cells. Resulting adult mEC cultures were cultured on fibronectin-coated (Sigma-Aldrich) plates in mEC media. Purity of mECs and absence of contaminating haematopoietic cells were confirmed by flow cytometry and confocal microscopy.
Isolation of HUVEC and generation of VN-ECs (HUVEC-E4ORF1)
Human umbilical vein Endothelial Cells (HUVECs) were isolated as described29,40 and cultured in endothelial cell growth medium. Once endothelial cells were transduced with lentiviral vectors expressing E4ORF1 gene they were then cultured in human endothelial cell media (M199 (Sigma, M4530), 10% FBS (Omega Scientific, FB07), 50 µg ml−1 endothelial mitogen (Alfa Aeser J65416), and 100 µg ml−1 Heparin (Sigma, H3393)11,13,28,29. E4ORF1 confers endothelial cells with the capacity to survive in the absence of serum and exogenous growth factors, while sustaining the pre-determined endothelial cell signatures and repertoire of the pro-haematopoietic angiocrine factors. The aforementioned conversion experiments with Fgrs transduction of endothelial cells would not be possible without use of HUVEC-E4ORF1 (VN-ECs) vascular niche cells as reprogramming in the presence of serum there is no conversion of endothelial cells into haematopoietic cells9. Thus, because E4ORF1 allows endothelial cells to survive and perform as inductive vascular niche cells, this platform enables Fgrs-driven conversion of adult endothelial cells without the supplementation with xenobiotic factors, serum or exogenous angiogenic growth factors. Both serum and angiogenic growth factors could interfere with and complicate the conversion of the endothelial cells into haematopoietic cells, by aberrantly interfering with the survival, self-renewal and expansion of the converted endothelial cells into haematopoietic cells (rEC-HSPCs and rEC-HSCs). Indeed, in the presence or serum the conversion of endothelial cells to HSPCs or HSCs is completely blocked. All niche-dependent experiments described herein were performed by using HUVEC-derived E4ORF1+ endothelial cells (VN-ECs, HUVEC-E4ORF1, Angiocrine Bioscience.) as a vascular niche feeder monolayer, except as specifically noted.
Virus production and transduction
Open reading frames (Fosb: NM_008036.2, Gfi1: NM_010278.1, Runx1: (Runx1b isoform) NM_001111022.2, Sfpi1: BC003815.1) were cloned into pLVx TET-3G lentiviral plasmid. Lentiviruses (LV) were produced were cloned into doxycycline inducible pLVx TET-3G lentiviral plasmids. Lentiviral vectors expressing Cxcl12 (NM_001012477.2) was obtained from Cyagen (pLV(Exp)-Neo-EF1A>mCxcl12(NM_001012477.2):IRES:EBFP).
Viral particles were produced in 293T Lenti-X cell line (Clontech, 632180) using Lenti-X packaging single shot (Clontech, 631275) following manufactures instructions, and tittered using Lenti-X p24 Rapid Titer c-Kit (Clontech, 632200). Transduction was carried out in 6-well plates. Doxycycline (dox) (Sigma-Aldrich) was added at a concentration of 1µg ml−1 for Fgrs transgene induction.
Direct conversion of adult mECs into rEC-HSPCs and rEC-HSCs
Adult mECs were directly converted into haematopoietic cells by conditional enforced expression of transcription factors followed by replating onto a serum-free inductive vascular niche (VN-ECs). Purified populations of VEcad+CD31+CD45−TER119− cells were cultured in the mEC growth medium (DMEM:Ham’s F-12 (Sigma, D6421) supplemented with 20% FBS (Omega Scientific, FB07), 20 mM HEPES (Invitrogen, 15630080), 100 µg ml−1 Heparin (Sigma, H3393), 50 µg ml−1 endothelial mitogen (Alfa Aeser J65416) and 5 µM SB431542 (R&D, 1614), in a humidified incubator at 37°C, 5% CO2 and normoxia 5% O2.
Then, adult mECs were transduced with doxycycline inducible TET-3G lentiviral vectors for a combination of transcription factors and the reverse tetracycline controlled trans-activator—FosB, Gfi1, Runx1, Spi1 (Fgrs), and rtTA—and maintained in mEC media for 3 days. Fgrs-ECs were then selected for rtTA expression by puromycin resistance. 72 h after puromycin selection, Fgrs expression was induced by adding 1 µg ml−1 dox in mEC media for 48 h. Dox was added every 24 h. Fgrs-ECs were then reseeded onto confluent VN-EC monolayers supplemented with conversion media (StemSpan SFEM, STEMCELL Technologies, 09650), 10% KnockOut Serum Replacement (Invitrogen, 10828028), 10 ng ml−1 hFGF-2 (bFGF, Peprotech, 100-18), 50 ng ml−1 mouse c-Kit ligand (SCF, Peprotech, 250-03). Conversion media was then replaced every 48 h.
rEC-HSPC and rEC-HSC primary, secondary and WBM transplantation assays
Adult organs, including lung, brain, liver and kidney VEcad+CD31+CD45− (C57BL/6J CD45.2 background) endothelial cells were purified to remove contaminating lymphoid endothelial cells and haematopoietic cells. After cultivation for 30 days, pure populations of endothelial cells were then used for direct conversion into HSPCs (rEC-HSPCs) and HSCs (rEC-HSCs), as described above. Adult mouse lung endothelial cells, devoid of any haematopoietic cells, were primarily used to assess the capacity of the rEC-HSPC and rEC-HSCs to reconstitute primary and secondary, clonal and serial haematopoietic transplantations. On day 28, total CD45+ reprogrammed cells were sorted and transplanted at 8.0 × 105 cells per recipient into lethally irradiated (950 cGy) congenic recipients. Doxycycline (dox) was not administered in the drinking water to ensure no exogenous Fgrs expression during the transplant. Serial transplantation from reprogrammed adult lung endothelial cells to HSPCs experiments were performed by transplanting 1.0 × 107 unfractionated bone marrow cells into secondary recipients (CD45.1+ or Rag1−/−). Serial transplantation of unfractionated bone marrow cells from C57BL/6J (CD45.2+) were used as controls. In all transplant experiments, peripheral blood and bone marrow analysis was performed at 4-week intervals with antibodies against c-Kit/CD117 (2B8), Sca-1/Ly-6A (D7), CD48 (HM48-1), and CD150 (mShad150). Lineage antibody cocktail included: CD41 (MWReg30) TER119 (TER119), B220 (RA3-6B2), CD11b (M1/70), Gr1 (RB6-8C5), CD3 (17A2), CD4 (GK1.5), CD8 (YTS156.7.7), CD44 (IM7), CD62l (MEL-14), CD25 (3C7), FoxP3 (150D), TCRγδ (GL3), CD45.1 (A20), CD45.2 (104), and DAPI to discriminate and eliminate dead cells from analysis. All antibodies were obtained from Biolegend unless mentioned.
For calculation of competitive repopulating units (CRU), recipient mice were transplanted with limiting dilutions of donor-LKS cells (1 to 4,500) together with 500,000 recipient-derived BM cells. Mice were bled every 4 weeks and killed after 16 weeks. Multi-lineage myelo-lymphoid donor derived contribution in the peripheral blood was assessed using flow cytometry analysis. HSC-CRU frequency and statistical significance was determined using ELDA software (http://bioinf.wehi.edu.au/software/elda/).
Immunofluorescence
Samples were permeabilized in PBST and blocked in 5% donkey serum. Samples were incubated for 2 h in primary antibodies blocking solution, washed 3 times in PBS and incubated in secondary antibodies (Jackson Laboratories) for 1 h. Following washing, some sections were counterstained for nucleic acids by DAPI (Invitrogen) before mounting and imaging by confocal microscopy. The primary antibodies used for immunostaining are listed in the previous section. All imaging was performed using a Zeiss 710 META confocal microscope.
RNA sequencing, hierarchical clustering and principal component analyses
At least 100 ng of total RNA from both freshly harvested and cultured cells was isolated (phenol-chloroform separation of TRIzol LS) and purified using Qiagen’s RNeasy Mini Kit. RNA quality was verified using an Agilent Technologies 2100 Bioanalyzer. RNA library preps were prepared and multiplexed using Illumina’s TruSeq RNA Library Preparation Kit v2 (non-stranded and poly-A selection) and 10 nM of cDNA was used as input for high-throughput sequencing via Illumina’s HiSeq 2500 producing 51 bp paired-end reads. Sequencing reads were de-multiplexed (bcl2fastq v2.17), checked for quality (FastQC v0.11.5), and trimmed/filtered when appropriate (Trimmomatic v0.36). The resultant high quality reads were mapped (TopHat2 v2.1.0; Bowtie2 v2.2.6) to the transcriptome sequence reference of the UCSC mm10 genome build. Unique and multi- mapped reads were then assembled into transcripts, and abundance measures (FPKM values) quantified (Cufflinks v2.2.1). All subsequent transcriptome data analysis utilized the estimated measures of transcripts abundance (i.e. FPKMs).
Genes with FPKM < 1 were filtered out, and base-2 log-transformed FPKM values were used for principal component analysis and hierarchical clustering. Transcriptome-wide and gene-set specific analysis of the RNA-Seq expression dataset were summarized and represented in the forms of scatterplots, dendrograms, and heatmaps.
TCR repertoire analyses
TCR repertoire analysis on RNA-seq reads were performed by custom BLAST-mapping. The reads were submitted for nucleotide BLAST-mapping against custom databases comprising TCR Alpha V genes, Beta V genes, Alpha C genes, and Beta C genes downloaded from IMGT (www.imgt.org). A table of the counts of reads meeting BLAST expected value cutoffs for each alpha and beta variable and constant genes was formulated for each sample and normalized to counts per million sequenced reads. The Relative Shannon Index was calculated using the Shannon entropy of the counts of TCR Beta V genes normalized by the logarithm of the number of different beta variable genes occurring in each sample and p-values showing differences in the Relative Shannon Index were calculated using the Wilcoxon rank sum test. Heatmaps and clustering were then performed in R using heatmap.2 function from gplots package.
Redirected lysis assay
Balb/c peripheral blood cells were labelled with the fluorescent membrane dye PKH67 (Sigma, PKH67GL) to distinguish them from effector cells (Fgrs-CD45.2+CD3+CD8+ T cells) upon FACS analysis according to the manufacturer instructions. Wells of 24-well culture plates were seeded with 1 × 104 dye-labelled Balb/c peripheral blood cells; 2–3 h later, 500 µl of Fgrs-CD45.2+CD3+CD8+ T cells pre-activated for 8 h with anti-CD3/CD28 beads were following manufacturer instruction (Miltenyi, 130-097-627), as described in ref. 41. Reactions were performed in presence of 10 U ml−1 of IL-2 with effector:target ratios of 1:2, 1:5, 1:10, 1:25, 1:50, 1 :100. Samples were analysed using a SORP-LSR2 flow cytometer (BD Biosciences). All cytotoxicity assays were performed in triplicate. Dead target cells were defined as PKH27−DAPI+CD45.2− cells.
Ovalbumin immunization
Mice transplanted with either rEC-HSCs or WBM mononuclear haematopoietic cells were injected with full-length chicken OVA emulsified in complete Freund’s adjuvant (CFA) subcutaneously at two sites on the back, injecting 0.1 mg at each site. A booster injection of OVA emulsified in incomplete Freund’s adjuvant (IFA) was administered 14 days after immunization with ovalbumin/CFA emulsion. The booster is given as a single subcutaneous injection with 0.1 ml of IFA emulsion, at one site on the back.
Ruling out host-derived HSPC or HSC haematopoietic contamination in purified mEC cultures
Adult mouse vascular endothelial cells (mECs) were purified by multicolour flow cytometry to eliminate contaminating lymphatic endothelial cells, pericytes, mesenchymal, and especially haematopoietic cells. To rule out the possibility of contamination with host-derived haematopoietic cells, freshly isolated cell flow cytometry sorted CD45.2+ mECs (8 × 105 cells) were transplanted into lethally irradiated (950 cGy)10 to 12 weeks CD45.1+ male recipients with 500,000 radio-protective CD45.1+ bone marrow haematopoietic cells. CD45.1+ recipient mice were assessed for CD45.2+ engraftment with contaminating HSPCs cells, as described above in section transplantation assays.
To demonstrate that the freshly isolated mEC expansion culture conditions, not supplemented with haematopoietic cytokines, will prevent proliferation or survival of any contaminating haematopoietic cells, limiting dilutions of CD45.2+ wild-type LKS-SLAM cells were introduced into mEC expansion cultures. In these experiments LKS-SLAM cells were transduced with Fgrs transgenes to rule out the possibility that these factors will enhance LKS-SLAM cell survival and expansion and seeded in limiting dilution on top of mEC–VN-EC co-culture. These cultures were grown in mEC media (absent haematopoietic cytokines) for 4 weeks. ‘Contaminated’ mEC–VN-EC co-cultures were then grown in conversion media for 28 days. The resulting cultures were transplanted into three lethally irradiated (950cGy) CD45.1+ male recipient mice with 500,000 CD45.1+ bone marrow haematopoietic cells to demonstrate that LKS-SLAM cells could not survive such haematopoietic intolerant culture conditions.
Purified mECs were expanded for conversion experiments routinely for 30 days in mEC media in complete absence of haematopoietic cytokines. Then, mECs were transduced with dox inducible Fgrs transgenes but were never exposed to dox (no-dox) and therefore never expressed Fgrs transgenes. 8 × 105 cultured no-dox Fgrs-transgene-transduced 30-day-expanded mECs were transplanted into lethally irradiated (950cGy) recipient mice in a rescue dose setting to rule out existence of any contaminating haematopoietic cells. Recipient CD45.1+ transplanted mice were assessed for survival.
It is possible that if any host HSPCs could survive the intolerant 30 days of mEC culturing, upon Fgrs transduction these contaminating HSPCs might revert to functional HSPCs or HSCs. To disprove this possibility, and mimic the standard procedure used in isolation of adult mouse lung endothelial cells, we re-expressed Fgrs in terminally differentiated CD45.2+ haematopoietic cells (TER119+, Gr1+CD11b+, B220+, CD3+) isolated from lungs of CD45.2+ rEC-HSPC week 16+ secondary engrafted recipients. To this end, 150,000 terminally differentiated CD45.2+ (TER119+, Gr1+CD11b+, B220+, CD3+) were cultured for 28 days in conversion media in presence of doxycycline in co-culture with VN-ECs (absent haematopoietic cytokines). Resulting cultures were transplanted into three lethally irradiated (950cGy) CD45.1+ male recipient mice with 500,000 radio-protective bone marrow haematopoietic cells. Then, the recipient CD45.1+ transplanted mice were assessed for CD45.2+ engraftment as described above in section transplantation assays.
If host HSPC contamination contributes to rEC-HSPCs and rEC-HSCs, then there should be no hierarchical Fgrs-dependence induction, specification or expansion phases during rEC-HSPC generation. In addition, within the first 8 days of culture (induction phase), the contaminating HSPCs should give rise to engraftable HSPCs. To assess this, dox-on Fgrs-transduced mECs were passaged through stepwise shutting-off Fgrs expression either during the induction, specification or expansion phases. The number of rEC-HSCs and rEC-HSPCs at each stage were then quantified, as described above.
Deciphering vascular-niche-derived inductive and self-renewal signals
Adult lung mouse endothelial cells (mECs) (CD45.2+VEcad+CD31+CD45−) were isolated from Runx1-IRES-GFP mice. VN-ECs were discriminated from Runx1-IRES-GFP-Fgrs-ECs by anti-human CD31. Expression of VEcad and CD45 in Runx1-IRES-GFP-Fgrs-ECs and derivatives during endothelial cell to haematopoietic cell reprogramming.
Adult mECs were treated with different small molecules at their known IC50 on mouse endothelial cells (CXCR4 antagonist, AMD3100 = 44 µmol l−1; CXCR7 agonist, TC14012 = 350 nmol l−1; BMP antagonist, Noggin = 0.5 µg ml−1; TGF-β antagonist, SB431542, c= 10 µmol l−1). Toxicity of each small molecule was assessed by annexinV/DAPI staining 48 h after the first treatment, as well as population-doubling time. Relative CD45+ percentages were then acquired by flow cytometry at day 28.
Adult mouse lung endothelial cells were generated from Runx1-IRES-GFP-Cxcr4fl/fl. Cxcr4−/− endothelial cells were generated by transduction with LV-CMV-Cre-Puro lentivirus (SignaGen, SL100272) followed by 7 days of puromycin selection. Cxcr4fl/fl and Cxcr4−/− endothelial cells were transduced with Fgrs and converted. Then, the frequency and functionality of the emerging CD45+ rEC-HSPCs and rEC-HSCs were assessed, as described above, by performing phenotypic flow cytometry.
Leukaemia assessment
We analysed recipient organ architecture and histological profile after 20 weeks of primary or secondary transplantation for any evidence of malignant alterations. For each organ, including bone marrow, lung, kidney, spleen, liver, intestine and brain, Wright-Giemsa, Masson and PicroSirius Red staining were performed on 10-µm paraffin-embedded section (Histoserv). All images were acquired using a colour CCD camera.
Primers list
All mouse: Fosb forward 5′-ACATGCCAGGAACCAGCTAC-3′; Fosb reverse, 5′-GCTGATCAGTTTCCGCCTGA-3′ CCA; Runx1 forward 5′-GAAGTGTAAGCCCAGCACAGT-3′; Runx1 reverse 5′-GGCGGGGGATTCTATAATTT-3′; Gfi1 forward 5′-ATGTGCGGCAAGACCTTC-3′; Gfi1 reverse 5′-ACAGTCAAAGCTGCGTTCCT-3′; Spi1 forward 5′-GGAGAAGCTGATGGCTTGG-3′; Spi1 reverse 5′-CAGGCGAATCTTTTTCTTGC-3′.
Viral integration mapping
The primer (Integrated DNA) sets used to detect each sequence were as follows. B1 repeated sequence: 5′-GTGGTGGCGCACGCCT-3′ and 5′-TAGCCCTGGCTGTCCTGGAA-3′; LTR: 5′-TCCACAGATCAAGGATATCTTGTC-3′. Reactions contained 1× Taqman universal master mix (Perkin-Elmer), 300 nM forward primer, 300 nM reverse primer, 100 nM probe primer and 100–500 ng of template DNA in a 30-µl volume. After initial incubations at 50 °C for 2 min and 95 °C for 10 min, 40 cycles of amplification were carried out at 15 s at 95 °C followed by 1 min 30 s at 60 °C. PCR products were then analysed on a 4% TBE-EtBR gel.
TCR spectratyping by nested PCR
RNA was isolated from CD45.2+CD3+CD8+ and CD45.2+CD3+CD4+ sorted T lymphocytes from peripheral blood (1 × 105 cells) with TRIzol (Invitrogen; no. 15596-026). The diversity of CDR3 regions for 24 TCR Vβ regions was assessed with the TCRExpress Quantitative Analysis Kit (Biomed Immunotech; no. H0533) per manufacturer instructions.
Statistical analyses
All data are presented as either median± s.e.m., mean ± s.d. or mean ± s.e.m (as indicated in figure legends). The data presented in the figures reflect multiple independent experiments performed on different days using different mice. Unless otherwise mentioned, most of the data presented in figure panels are based on at least three independent experiments. The significance of difference was determined using a two-tailed Student’s t-test, unless otherwise stated. P > 0.05 was considered not significant; *P < 0.05; **P < 0.01; ***P < 0.001. In all the figures, n refers to the number of mice when applicable. All statistical analyses were performed using Graphpad Prism software. No randomization or blinding was used and no animals were excluded from analysis. Sample sizes were selected on the basis of previous experiments. Unless otherwise indicated, results are based on three independent experiments to guarantee reproducibility of findings.
Data availability
The RNA sequencing data set was submitted to the Gene Expression Omnibus database GSE88840. Source data for this study are included in the Supplementary Information. Single-cell RNA-seq data sets for embryonic day 11 (E11) aorta-gonad-mesonephros endothelium, E11 CD201− pre-HSC type 1, E11 CD201+ pre-HSC type 1, E11 CD201+ pre-HSC type 2, E12.5 fetal liver HSCs (lin−Sca-1+Mac1loCD201+), E14.5 fetal liver HSCs (lin−CD45+CD150+CD48−CD201−), adult bone marrow HSCs (LKS-SLAM) were obtained from ref. 31 (GSE67120); the LKS-SLAM RNA-seq data sets were obtained from ref. 32 (GSE60808); embryonic-stem-cell-derived endothelial cells, embryonic stem cell RNA-seq data sets were obtained from ref. 42. A step-by-step protocol describing in vitro conversion of endothelial cells into HSCs can be found at Protocol Exchange43.
Extended Data
Extended Data Figure 1. Phenotypic and quality control characterization of the rEC-HSPCs.
a, 8.0 × 105 freshly isolated adult mouse lung CD45.2+ endothelial cells, depleted of lymphatic endothelial cells or contaminating haematopoietic cells were purified and injected in lethally irradiated CD45.1+ recipients, graph indicates donor contribution to peripheral blood at indicated time points after transplant. Data represents independent transplantations, n = 5. b, Wild-type normal LT-HSCs (CD45.2+ LKS-SLAM cells) were sorted, transduced with Fgrs transgenes and expanded in endothelial cell growth medium containing FGF-2, serum and TGF-β inhibitor, without haematopoietic growth factors. Expanded Fgrs-transduced LT-HSCs were then plated in co-culture with VN-ECs and Fgrs were turned on (dox-on) for 28 days and then transplanted into lethally irradiated CD45.1+ recipients. Graphs represent peripheral blood contribution 16 weeks after transplantation. Data represents independent transplantations, n = 3. c, qRT–PCR showing expression of Fgrs in mECs upon doxycycline addition. All four factors were absent when adult mECs were not exposed to doxycycline, or exposed to doxycycline without rtTA. Data represents mean ± s.d, n = 3. d, Time-course of CD45+ cell formation during the stage-specific conversion process. e, Time-course analysis of lin−c-Kit+Sca-1+ (LKS) converted cells. f, Diff-Quik stain of cells from day 28 in vitro cultures (original magnification, ×60, bar = 50µm). g, Absolute quantification of LKS cells present whether in the fraction adherent to VN-ECs or in supernatant. Representative pictures of rEC-HSPC adherent fraction with prototypical ‘cobblestone’ structures is shown on the left-hand side, n = 3 independent reprogramming experiments. Data represents mean ± s.e.m., n = 3, individual data points are represented. h, Fgrs-transduced endothelial cells were grown onto VN-ECs, OP9-DLL11, or in feeder free condition. Graph indicates absolute quantification (cell number) of CD45+ rEC-HSPC. Data represent mean ± s.e.m. (n = 5 for conversion experiments run in technical triplicates for each conditions); two-tailed unpaired t-test. i, Fgrs-transduced OP9 were grown onto VN-ECs, OP9-DLL11, or in feeder-free condition. Graph indicates absolute quantification (cell number) of CD45+ rEC-HSPCs (n = 5). Data represent mean ± s.e.m. (n = 5 conversion experiment run in technical triplicates for each conditions); two-tailed unpaired t-test.
Extended Data Figure 2. rEC-HSPCs are composed of rEC-HSCs that have the potential for primary and secondary engraftment and regenerative haematopoiesis self-renewal.
a, Kaplan–Meier curve showing percentage survival over 16 weeks of lethally irradiated mice transplanted with either 8.0 × 105 cells (purified CD45+ rEC-HSPCs, green line (n = 20); non-converted lung endothelial cells, blue line (n = 10) or PBS (n = 15) mice). b, Representative plots of rEC-HSPC lineage contribution. c, Donor reconstitution of mice transplanted with CD45+ rEC-HSPCs at indicated time points after primary transplantation. Data represents individual data points (n = 20). d, Representative plots of donor contribution to LKS-SLAM cells. e, Donor reconstitution of mice transplanted with WBM from chimaeric WBM control mice or WBM from chimaeric rEC-HSPC primary transplanted mouse at indicated time points after transplantation. Data represents individual data points, n = 15, 4 independent reprogramming experiments. f, Schematic representation of haematopoietic recovery following sub-lethal irradiation assay. g, Analysis of white blood cell recovery of rEC-HSPC-engrafted versus control mice following sub-lethal irradiation (500 cGy) (n = 5 for duration of analysis). Data represent mean ± s.e.m., no significant differences were found using two-tailed unpaired t-test. h, Multi-lineage analyses during bone marrow recovery. Myeloid and lymphoid regeneration, including T cells, and CD3+CD8+ T cytotoxic cells at baseline and 28 days post sub-lethal irradiation (500 cGy). Data represent individual data points; black bar represents mean (n=5).
Extended Data Figure 3. Peripheral and splenic rEC-HSPC-derived T-cell phenotyping.
a, Gating strategy to phenotype naïve, effector and memory T-cells from peripheral blood of transplanted mice. b, Boxplot showing the absolute number of B220+, CD3+CD4+, and CD3+CD8+ cells following long-term primary or secondary transplant. WBM control samples are denoted in blue, rEC-HSPC in green Boxplot and whiskers represent median, 25th and 75th percentile, mean is represented by + sign. (n = 5); two-tailed unpaired t-test. c, Gating strategy to phenotype Tγδ cells from peripheral blood of transplanted mice. d, Boxplot showing the averaged frequency of naïve, effector and memory T-cells for both CD3+CD4+ and CD3+CD8+, following long-term primary or secondary transplant. WBM control samples are denoted in blue, rEC-HSPC in green. Data represent mean±s.e.m.(n=5). P values, two-tailed unpaired t-test. e, Boxplot showing the averaged frequency of γδ T cells following long-term primary or secondary transplant. Data represent mean ± s.e.m. (n = 5); two-tailed unpaired t-test. f, Phenotype of regulatory T cells (Treg) by flow cytometry.
Extended Data Figure 4. Endothelial to haematopoietic conversion capture by live microscopy.
(See also Supplementary Video 1.) a, Schema detailing the experimental setting for live confocal image capture. Adult lung endothelial cells were isolated from Runx1-IRES-GFP. Then, Runx1-IRES-GFP adult lung endothelial cells were transduced with Fgrs and co-cultured with VN-ECs (HUVEC-E4ORF1). VN-ECs were differentiated from Fgrs-transduced Runx1-IRES-GFP adult lung endothelial cells by anti-human CD31 live staining (red). Live confocal images were acquired every 45 min for the duration of the experiment (see also Supplementary Video 1). b, Representative flow cytometry plots of day 10 (d10) showing that a subset of VEcad+CD45− and VEcad+CD45+ cells also co-express Runx1-GFP. c, Single time points from live confocal image capture. Upon doxycycline-dependent conditional expression of Fgrs, flat spindle-shaped adult mouse endothelial cells rapidly transition from RUNX1− to round haematopoietic-like RUNX1+ cells (day 0–8, white arrow). This induction phase is characterized by the transitioning endothelial cells towards haematopoietic fate. From day 8 to 20 of specification phase, RUNX1+ cells further moved towards a haematopoietic identity by assuming a prototypical fully formed round shape (white arrow). Following the emergence of this definite haematopoietic program, a phase of robust expansion on the vascular niche layers (VN-ECs) of these RUNX1+ committed converted cells from day 20 to 28 (expansion phase).
Extended Data Figure 5. Quantification of rEC-HSCs arising from single-cell reprogramming.
a, Haematopoietic cluster arising from single-cell reprograming. at specified time point. Wells were considered negative if no hematopoietic clusters were visible 36 days after single RUNX1+ endothelial cell inoculation. Representative wells considered as positive or negative are shown. Cloning of RUNX1+ mECs resulted in 22±11 clusters per 1000 RUNX1+ mECs sorted b, Long-term peripheral blood contribution of each reprogrammed day 8 RUNX1+ endothelial cells. Data represent individual data points for each clones (n = 7) c, Limiting-dilution transplantation (LDT) assay showing the frequency of LT-HSCs in expanded clonal conversion experiments. CRU cells were determined using Poisson statistics by ELDA software. d, Viral integration mapping. PCR against LTR-B1 repeated sequence was run in each indicated populations. PCR assays were analysed on 4% TBE gel.
Extended Data Figure 6. Molecular profiling of rEC-HSPCs and rEC-HSCs.
a, Supervised principal component analysis of global gene expression data of rEC-HSPCs and rEC-HSCs and the indicated control cell types. Data for individual cells of given is indicated. Each shape represents independent replicate for the indicated cell types (embryonic day 11 (E11.0) aorta-gonad-mesonephros endothelium, E11.0 CD201− pre-HSC type 1, E11.0 CD201+ pre-HSC type 1, E11.0 CD201+ pre-HSC type 2, E12.5 fetal liver HSCs (lin−Sca-1+Mac1loCD201+), E14.5 fetal liver HSCs (lin−CD45+CD150+CD48−CD201−), adult bone marrow HSC (LKS-SLAM), ‘sc-’ refers to the single-cell RNA-seq data set adapted from 31; adult lung endothelial cells, in vitro rEC-LKS, in vivo rEC-LKS-SLAM, control adult in vivo LKS-SLAM cells). b. Supervised clustering of canonical endothelial genes expression profiles of freshly isolated lung endothelial cells (n = 3), embryonic stem cell (ES)-derived endothelial cell (n = 2), embryonic stem cells (n = 1); rEC-LKS-SLAM cells isolated from transplanted mice (n = 3), (n = 3, n = 4 from Kinkel et al 32, day 28 in vitro rEC-LKS (n = 3), data points obtained from clonal reprogramming are denoted by an asterisk. c, Supervised clustering of prototypical haematopoietic genes expression, demonstrates that haematopoietic genes are induced during Fgrs-mediated reprograming of endothelial cells into rEC-HSPCs and rEC-HSCs. d, Supervised clustering of prototypical pluripotency genes expression, demonstrates that pluripotency genes are not induced during Fgrs-mediated reprograming of endothelial cells into rEC-HSPCs and rEC-HSCs. e, Dendrogram showing unsupervised hierarchical clustering of global gene expression data of representative control cells (Ctl), and all rEC-HSCs, rEC-HSPCs, and their progenies. Dendrogram branches are colour-coded per cell types indicated in the legend.
Extended Data Figure 7. Immune function assessment and molecular profiling of TCR diversity of rEC-HSC-derived T cells.
a, CFSE dilution and intracellular interferon-γ (IFN-γ) production upon CD45.2+ CD3/CD28 polyclonal activation and/or Treg addition. Representative flow cytometry plots. b, Quantification of intracellular IFN-γ production upon CD3/CD28 polyclonal activation and/or Treg addition. Data represents mean ± s.e.m. (n = 5 independent experiment, 3 technical replicates); two-tailed unpaired t-test. c, Normalized counts for CD3+CD4+ cells. WBM control samples are denoted in blue, rEC-HSPC in green. d, Normalized counts for CD3+CD8+. WBM control samples are denoted in blue, rEC-HSPC in green. e, Normalized counts for Jurkat cell samples. f, Analysis of TCR repertoire in Rag1−/− rEC-HSPC reconstituted mice upon chicken ovalbumin vaccination.
Extended Data Figure 8. Molecular deconvolution of vascular-niche-derived angiocrine factors during stepwise differentiation of endothelial cells into rEC-HSPCs and rEC-HSCs.
a, Adult lung mECs (VEcad+CD31+CD45−) were isolated from Runx1-IRES-GFP mice. Human-derived VN-ECs were discriminated from Fgrs-transduced Runx1-IRES-GFP adult lung endothelial cells by anti-human CD31 (hCD31). Fgrs-transduced Runx1-IRES-GFP endothelial cells and their progeny were gated as hCD31−. Flow cytometry plots showing the expression of VEcad and CD45 in mouse hCD31− Fgrs-transduced endothelial cells and derivatives over the course of endothelial to haematopoietic cell reprogramming. b, Quantification of mouse hCD31− Fgrs-transduced endothelial cells and their derivatives over the course of reprogramming. Data represents mean ± s.e.m. (n = 5). c, Colony number arising in methylcellulose from Fgrs-transduced endothelial cells and derivatives (gated on human hCD31− to exclude VN-EC feeder). n = 4 independent experiments are shown and each condition performed in triplicate. Data represents mean ± s.e.m. (n = 5); two-tailed unpaired t-test. d, Adult mouse endothelial cells were treated with different small molecules at their known IC50 (CXCR4 antagonist, AMD3100, 44 µmol l−1; CXCR7 agonist, TC14012, 350 nmol l−1; BMP antagonist, Noggin, 0.5 µg ml−1; TGF-β antagonist, SB431542, 10 µmol l−1). Representative flow cytometry plots of apoptosis assays are presented. e, Quantification of apoptotic cells following treatment with each small molecule tested. Data represents mean ± s.e.m. (n = 3); two-tailed unpaired t-test. f, Quantification of hCD31− Fgrs-transduced endothelial cells and their derivatives over the course of reprogramming in presence of CXCR4 inhibitors (AMD3100, AMD3100, 44 µmol l−1), CXCR7 agonists (TC14012, 350 nmol l−1), BMP inhibitor (Noggin, 0.5 µg ml−1), TGF-β/ALK5 inhibitor (SB431542, 10 µmol l−1). Data represents mean ± s.e.m. (n = 5); two-tailed unpaired t-test. g, Quantification of hCD31− Fgrs-transduced endothelial cells and their derivatives over the course of reprogramming in presence of VN-ECs (HUVEC-E4ORF1) overexpressing mouse CXCL12 (n = 5 independent experiments, 3 technical triplicates each). Data represents mean ± s.e.m.; two-tailed unpaired t-test.
Extended Data Figure 9. Organ-specific adult endothelial cells are amenable to hierarchical Fgrs-mediated reprogramming to rEC-HSPCs.
a, Left, Fgrs-transduced liver endothelial cells were directly co-cultured with VN-ECs. Graph indicates absolute quantification (cell number) of CD45+ rEC-HSPC (n = 3 independent biological replicates, 3 technical replicates each). Right, Quantification of phenotypically marked CD45+ LKS cells at day 28 of reprogramming absolute cell number is reported. Data represents mean ± s.e.m. b, Left, Fgrs-transduced lung endothelial cells were directly co-cultured with VN-ECs. Graph indicates absolute quantification (cell number) of CD45+ rEC-HSPC Right, Quantification of phenotypically marked CD45+ LKS cells at day 28 of reprogramming absolute cell number is reported. Data represents mean ± s.e.m. (n = 3 independent biological replicates, 3 technical replicates each). c, Left, Fgrs-transduced brain endothelial cells were directly co-cultured with VN-ECs. Graph indicates absolute quantification (cell number) of CD45+ rEC-HSPC (n = 3 independent biological replicates, 3 technical replicates each). Right, Quantification of phenotypically marked CD45+ LKS cells at day 28 of reprogramming absolute cell number is reported. Data represents mean ± s.e.m. d, Left, Fgrs-transduced kidney endothelial cells were directly co-cultured with monolayers of confluent VN-ECs. Graph indicates absolute quantification (cell number) of CD45+ rEC-HSPCs. Right, Quantification of phenotypically marked CD45+ LKS cells at day 28 of reprogramming absolute cell number is reported. Data represents mean ± s.e.m. (n = 3 independent biological replicates, 3 technical replicates each). e, rEC-HSPC-derived cells were purified and co-culture on VN-ECs in presence of doxycycline for 28 days. Cells were transplanted into lethally CD45.1+ irradiated recipients in absence of doxycycline. Data represents individual data points for donor contribution to peripheral blood at indicated time point after transplant (n = 3 biological replicates). f, Experimental model for hierarchical differentiation of endothelial cells into rEC-HSPCs. D8 VEcad+RUNX1+CD45− cells were purified by flow cytometry and replated on inductive vascular niche in presence or absence of doxycycline. Subsequently, at day 15 VEcad+RUNX1+CD45− cells haemogenic-like cells were purified by flow cytometry and replated on the inductive vascular niche in presence (Dox-on) or absence (Dox-off) of dox. g, Flow cytometry quantification of cell subsets during stepwise conversion in f. Data represents individual data points (n = 3 independent biological replicates, 3 technical replicates each).
Extended Data Figure 10. Analyses of the rEC-HSPC- and rEC-HSC-engrafted organs for malignant transformation.
Although none of the recipient mice engrafted with rEC-HSPCs manifested any anatomical or symptomatic evidence of leukaemia, lymphoma or myeloproliferative neoplasm (MPN) (that is, lymphadenopathy, organomegaly, illness or haemorrhage), we analysed recipient organ architecture and histological profile after 20 weeks of primary, or secondary transplantation for any evidence of malignant alterations. For each organ, including bone marrow, lung, kidney, spleen, liver, intestine and brain, Wright-Giemsa (left), Masson (middle) and PicroSirius Red (right) staining is shown at 2 different magnifications (10×, upper panel; 40× lower panel; scale bars, 10 µm, 40 µm). We did not observe any evidence of aberrant infiltration of haematopoietic cells, aberrant inflammatory response, chloromas, or alteration of the geometry of any organs of the primary or secondary transplanted mice. Furthermore, microscopic architecture of bone marrow, spleen and liver was normal and without fibrotic remodelling or abnormal deposition of collagen or desmin. All images were acquired using a colour CCD camera. n = 3 independent primary or secondary transplant experiments. Representative experiments are shown.
Supplementary Material
Acknowledgments
We thank J. Downing at St. Jude Hospital for providing the Runx1-IRES-GFP reporter mice. Floxed Cxcr4 mice were provided by Y.-R. Zou (the Feinstein Institute for Medical Research). We are grateful to V. Sandler for constructive discussions.
R.L., W.S., K.S., and S.R. are supported by Ansary Stem Cell Institute (ASCI), New York State Department of Health grants (NYSDOH) (C026878, C028117, C029156, C030160), NIH-R01 (DK095039, HL119872, HL128158, HL115128, HL099997) and U54 CA163167, the Starr foundation TRI-Institution stem cell core project, Tri-Institutional Stem Cell Initiative grants (TRI-SCI#2013-032, #2014-023, #2016-013, and fellowships), R.L., A.R.T., and S.R. by the Qatar National Priorities Research Program (NPRP 8-1898-3-392), B.K. by NIH-T32 HD060600. J.M.B. is supported by the ASCI, TRI-SCI #2013-022 and #2014-004, Leukemia & Lymphoma Society (LLS) grant 0859-15, and NIH-R01 (CA204308, HL133021); J.M.S by the ASCI, Taub Foundation Grants Program, TRI-SCI#2014-023 and #2016-024, LLS grant 2299-14, (NYSDOH) C029156, C030160, ECRIP, and NIH R01 (HL119872, HL128158) and by Cancer Research & Treatment Fund (CR&T). J.M.S. and S.R. by the ECRIP, NYSDOH.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions R.L., J.M.S. and S.R. designed the study. R.L. and C.C.K. performed endothelial cell isolation, conversion, transplant, and transplant analysis. M.G.P. analysed haematopoietic recovery after irradiation, C.R.B. irradiated recipients, W.S. and J.G.B.D. prepared lentiviral particles. M.G. isolated endothelial cells. B.K., D.R., J.X., and A.R.T. performed RNA-seq and statistical analysis, supervised by O.E. Z.R., K.S., N.S., J.M.B., and J.M.S. interpreted results and edited manuscript. R.L., C.C.K., J.M.S. and S.R. wrote the manuscript.
Competing interests: S.R. is the founder and a non-paid consultant to Angiocrine Bioscience. J.M.B. receives research funding from Angiocrine Bioscience, and M.G. is an employee of Angiocrine Bioscience.
References
- 1.Wahlster L, Daley GQ. Progress towards generation of human haematopoietic stem cells. Nat. Cell Biol. 2016;18:1111–1117. doi: 10.1038/ncb3419. [DOI] [PubMed] [Google Scholar]
- 2.Sturgeon CM, Ditadi A, Clarke RL, Keller G. Defining the path to hematopoietic stem cells. Nat. Biotechnol. 2013;31:416–418. doi: 10.1038/nbt.2571. [DOI] [PubMed] [Google Scholar]
- 3.Tashiro K, et al. Promotion of hematopoietic differentiation from mouse induced pluripotent stem cells by transient HoxB4 transduction. Stem Cell Res. 2012;8:300–311. doi: 10.1016/j.scr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Rafii S, et al. Human ESC-derived hemogenic endothelial cells undergo distinct waves of endothelial to hematopoietic transition. Blood. 2013;121:770–780. doi: 10.1182/blood-2012-07-444208. [DOI] [PubMed] [Google Scholar]
- 5.Batta K, Florkowska M, Kouskoff V, Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Reports. 2014;9:1871–1884. doi: 10.1016/j.celrep.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doulatov S, et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elcheva I, et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat. Commun. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereira CF, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandler VM, et al. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511:312–318. doi: 10.1038/nature13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulecio J, et al. Conversion of human fibroblasts into monocyte-like progenitor cells. Stem Cells. 2014;32:2923–2938. doi: 10.1002/stem.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler JM, et al. Development of a vascular niche platform for expansion of repopulating human cord blood stem and progenitor cells. Blood. 2012;120:1344–1347. doi: 10.1182/blood-2011-12-398115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding BS, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler JM, Rafii S. Generation of a vascular niche for studying stem cell homeostasis. Methods Mol. Biol. 2012;904:221–233. doi: 10.1007/978-1-61779-943-3_18. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi H, et al. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nat. Cell Biol. 2010;12:1046–1056. doi: 10.1038/ncb2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 2014;20:833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasine JP, Yeo KT, Chute JP. Concise review: paracrine functions of vascular niche cells in regulating hematopoietic stem cell fate. Stem Cells Transl. Med. 2017;6:482–489. doi: 10.5966/sctm.2016-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler JM, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riddell J, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boisset JC, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 22.Zovein AC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen PD, et al. Haematopoietic stem cell induction by somite-derived endothelial cells controlled by meox1. Nature. 2014;512:314–318. doi: 10.1038/nature13678. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, et al. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 2014;28:2597–2612. doi: 10.1101/gad.253302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espín-Palazón R, et al. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell. 2014;159:1070–1085. doi: 10.1016/j.cell.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gritz E, Hirschi KK. Specification and function of hemogenic endothelium during embryogenesis. Cell. Mol. Life Sci. 2016;73:1547–1567. doi: 10.1007/s00018-016-2134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slukvin II. Generating human hematopoietic stem cells in vitro exploring endothelial to hematopoietic transition as a portal for stemness acquisition. FEBS Lett. 2016;590:4126–4143. doi: 10.1002/1873-3468.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seandel M, et al. Generation of a functional and durable vascular niche by the adenoviral E4ORF1 gene. Proc. Natl Acad. Sci. USA. 2008;105:19288–19293. doi: 10.1073/pnas.0805980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rafii S, et al. Human bone marrow microvascular endothelial cells support long-term proliferation and differentiation of myeloid and megakaryocytic progenitors. Blood. 1995;86:3353–3363. [PubMed] [Google Scholar]
- 30.Lorsbach RB, et al. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;103:2522–2529. doi: 10.1182/blood-2003-07-2439. [DOI] [PubMed] [Google Scholar]
- 31.Zhou F, et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature. 2016;533(Suppl):487–492. doi: 10.1038/nature17997. [DOI] [PubMed] [Google Scholar]
- 32.Kinkel SA, et al. Jarid2 regulates hematopoietic stem cell function by acting with polycomb repressive complex 2. Blood. 2015;125:1890–1900. doi: 10.1182/blood-2014-10-603969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redmond D, Poran A, Elemento O. Single-cell TCRseq: paired recovery of entire T-cell alpha and beta chain transcripts in T-cell receptors from single-cell RNAseq. Genome Med. 2016;8:80. doi: 10.1186/s13073-016-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orkin SH, Zon LI. Hematopoiesis and stem cells: plasticity versus developmental heterogeneity. Nat. Immunol. 2002;3:323–328. doi: 10.1038/ni0402-323. [DOI] [PubMed] [Google Scholar]
- 35.Jaffredo T, et al. From hemangioblast to hematopoietic stem cell: an endothelial connection? Exp. Hematol. 2005;33:1029–1040. doi: 10.1016/j.exphem.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Hirschi KK. Hemogenic endothelium during development and beyond. Blood. 2012;119:4823–4827. doi: 10.1182/blood-2011-12-353466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng ES, et al. Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nature biotechnology. 2016;34:1168–1179. doi: 10.1038/nbt.3702. [DOI] [PubMed] [Google Scholar]
- 38.Souilhol C, et al. Inductive interactions mediated by interplay of asymmetric signalling underlie development of adult haematopoietic stem cells. Nature communications. 2016;7:10784. doi: 10.1038/ncomms10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yzaguirre AD, de Bruijn MF, Speck NA. The Role of Runx1 in Embryonic Blood Cell Formation. Adv Exp Med Biol. 2017;962:47–64. doi: 10.1007/978-981-10-3233-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rafii S, et al. Isolation and characterization of human bone marrow microvascular endothelial cells: hematopoietic progenitor cell adhesion. Blood. 1994;84:10–19. [PubMed] [Google Scholar]
- 41.Roden MM, Lee KH, Panelli MC, Marincola FM. A novel cytolysis assay using fluorescent labeling and quantitative fluorescent scanning technology. J. Immunol. Methods. 1999;226:29–41. doi: 10.1016/s0022-1759(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 42.Israely E, et al. Akt suppression of TGFβ signaling contributes to the maintenance of vascular identity in embryonic stem cell-derived endothelial cells. Stem Cells. 2014;32:177–190. doi: 10.1002/stem.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raphael Lis, Karrasch CC, Rafii S. In vitro conversion of endothelial cells into haematopoietic stem cells. Protoc. Exch. 2017 http://dx.doi.org/10.1038/protex.2017.033.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data set was submitted to the Gene Expression Omnibus database GSE88840. Source data for this study are included in the Supplementary Information. Single-cell RNA-seq data sets for embryonic day 11 (E11) aorta-gonad-mesonephros endothelium, E11 CD201− pre-HSC type 1, E11 CD201+ pre-HSC type 1, E11 CD201+ pre-HSC type 2, E12.5 fetal liver HSCs (lin−Sca-1+Mac1loCD201+), E14.5 fetal liver HSCs (lin−CD45+CD150+CD48−CD201−), adult bone marrow HSCs (LKS-SLAM) were obtained from ref. 31 (GSE67120); the LKS-SLAM RNA-seq data sets were obtained from ref. 32 (GSE60808); embryonic-stem-cell-derived endothelial cells, embryonic stem cell RNA-seq data sets were obtained from ref. 42. A step-by-step protocol describing in vitro conversion of endothelial cells into HSCs can be found at Protocol Exchange43.