Abstract
Corin is a serine protease that activates atrial natriuretic peptide (ANP). CORIN gene variants have been reported in patients with hypertension. To date, however, the prevalence of CORIN variants in hypertensive patients remains unknown. To understand the prevalence and functional significance of CORIN variants in hypertension, we sequenced CORIN exons in 300 normal and 401 hypertensive individuals in a Chinese population and identified nine nonsynonymous variants, of which eight were not characterized previously. Among them, variants c.131A > G (p.Tyr13Cys), c.376G > T (p.Asp95Tyr), c.1094T > G (p.Leu334Trp), and c.1667G > A (p.Arg525His) occurred similarly in both normal and hypertensive individuals. Variants c1139G > A (p.Arg349His), c.2689C > T (p.Pro866Ser), and c.2864C > T (p.Thr924Met) were found once each in hypertensive individuals. Variant c.1683G > T (p.Arg530Ser) occurred preferentially in hypertensive individuals [10/401 (2.5%) vs. 1/300 (0.3%) in normal individuals; P = 0.023], which was confirmed in another independent cohort [9/368 (2.44%) in hypertensive and 2/377 (0.53%) in normal individuals; P = 0.033]. In biochemical and cell-based functional studies, variants p.Arg530Ser and p.Thr924Met, but not p.Tyr13Cys, p.Asp95Tyr, p.Leu334Trp, p.Arg349His, p.Arg525His, and p.Pro866Ser, exhibited reduced pro-ANP processing activity, which was caused by endoplasmic reticulum retention and poor zymogen activation, respectively. These results indicate that genetic variants impairing corin function are not uncommon in general populations and that such variants may be an important contributing factor in hypertension.
Keywords: CORIN, gene variants, hypertension, natriuretic peptides, serine protease
1 | INTRODUCTION
Atrial natriuretic peptide (ANP) is a cardiac hormone of physiological importance (de Bold, 2011; Song, Wang, & Wu, 2015). In addition to its role in regulating sodium homeostasis and blood pressure, ANP is also involved in energy metabolism, vessel wall remodeling, and cardiovascular responses to stress (Chen et al., 2016; Cui et al., 2012; Schlueter et al., 2014; Tokudome et al., 2009). Variants in the human NPPA gene (MIM# 108780), encoding ANP, have been associated with blood pressure levels and major cardiovascular diseases, including hypertension, coronary artery disease, myocardial infarction, and stroke (Arora et al., 2013; Fox et al., 2009; Lynch et al., 2009; Newton-Cheh et al., 2009; Rubattu, Sciarretta, & Volpe, 2014; Song et al., 2015), indicating the importance of ANP in cardiovascular homeostasis.
Corin is a key protease in ANP generation (Armaly, Assady, & Abassi, 2013; Zhou & Wu, 2014). It converts the ANP precursor, pro-ANP, to active ANP (Yan, Wu, Morser, & Wu, 2000). The corin function is essential for salt–water balance and normal blood pressure (Wang et al., 2012). To date, CORIN variants have been reported in patients with hypertension and heart disease. In African Americans, for example, a corin variant with reduced pro-ANP processing activity is associated with hypertension and cardiac hypertrophy (Dries et al., 2005; Rame et al., 2009). More recently, low plasma or serum corin levels have been reported in patients with heart failure (Dong et al., 2010; Ibebuogu, Gladysheva, Houng, & Reed, 2011; Zhou et al., 2016b), coronary artery disease (Barnet et al., 2015; Peleg, Ghanim, Vered, & Hasin, 2013), myocardial infarction (Zhang et al., 2016; Zhou et al., 2016a), and stroke (Hu et al., 2016; Peng et al., 2015), suggesting that corin deficiency may contribute to major cardiovascular diseases.
Human corin is a protein of 1,042 amino acids (Hooper, Scarman, Clarke, Normyle, & Antalis, 2000; Yan, Sheng, Seto, Morser, & Wu, 1999). The CORIN gene (MIM# 605236) consists of 22 exons and spans > 200 kb in length (Pan et al., 2002). Studies have shown that corin expression and activity are regulated at various levels, including gene expression (Lee et al., 2014; Pan et al., 2002), cell surface targeting (Gladysheva, King, & Houng, 2008; Li et al., 2015; Liao, Wang, Chen, & Wu, 2007; Zhang et al., 2014), zymogen activation (Chen et al., 2015), and proteolytic shedding (Jiang et al., 2011; Wang et al., 2015). Genetic variants that impair corin biosynthesis and activity are expected to decrease ANP production, thereby contributing to hypertensive disease.
To understand the prevalence and functional significance of corin variants in hypertensive patients, we sequenced CORIN exons in a cohort of 300 normal and 401 hypertensive individuals. The variants identified were verified in a second cohort of 377 normal and 368 hypertensive individuals and by in vitro functional studies. Here we report our findings of novel CORIN variants in hypertensive patients that impair corin intracellular trafficking and zymogen activation.
2 | MATERIALS AND METHODS
2.1 | Participants and blood samples
This study was approved by the ethics committee of the Soochow University. Informed consent was obtained from all participants. All experiments were conducted in accordance with the Declaration of Helsinki. Venous blood was obtained from normal (systolic and diastolic blood pressure of <120/90 mmHg with no hypertension history) and hypertensive (systolic and diastolic blood pressure of >140 and/or >90 mmHg, or having a history of hypertension and taking anti-hypertensive drugs) individuals. Blood pressure was measured by standard sphygmomanometry after seated rest. The first cohort included 300 normal and 401 hypertensive individuals. The second cohort included 377 normal and 368 hypertensive individuals. Supp. Table S1 shows the clinical characteristics of the participants.
2.2 | Plasma Corin, NT-pro-ANP, and NT-pro-BNP
Plasma corin was measured by an enzyme-linked immunosorbent assay (ELISA) kit from R&D Systems (Minneapolis, MN) (Dong et al., 2010). Plasma N-terminal (NT)-pro-ANP was measured by an ELISA kit from Biomedica (Maryland Heights, MO). Plasma NT-pro-B-type natriuretic peptide (NT-pro-BNP) was measured by the Elecsys3 proBNP immunoassay kit from Roche Diagnostics (Indianapolis, IN). Experimental procedures were carried out according to manufacturers’ instructions.
2.3 | DNA sequencing
Genomic DNA was isolated from white blood cells with a Qiagen kit (Germany) and used in PCR to amplify CORIN exons and intron–exons boundaries. PCR fragments were sequenced directly using the Sanger method. Gene variants found were verified by another round of independent PCR analysis followed by DNA sequencing.
2.4 | Expression plasmids
Plasmids expressing human wild-type (WT) corin and the activation cleavage site Arg801Ala (R801A) mutant corin and proprotein convertase subtilisin/kexin-6 (PCSK6) were described previously (Chen et al., 2015; Knappe, Wu, Masikat, Morser, & Wu, 2003; Yan et al., 2000). Plasmids expressing corin variants p.Tyr13Cys, p.Asp95Tyr, p.Leu334Trp, p.Arg349His, p.Arg525His, p.Arg530Ser, p.Pro866Ser, and p.Thr924Met were made by site-directed mutagenesis using the plasmid for WT corin as a template (Yan et al., 2000). Corin and PCSK6 proteins encoded by these plasmids had a C-terminal V5 or FLAG tag used for detection by Western blotting.
2.5 | Cell culture, transfection, and Western blotting
Human embryonic kidney 293 (HEK293) cells were cultured in DMEM with 10% fetal bovine serum at 37°C in a humidified incubator with 5% CO2. Plasmids were transfected into the cells using PolyJet reagents (SignaGen, Gaithersburg, MD). After 48 hr, the conditioned medium was collected and the cells were lysed in a solution (Chen et al., 2010). Corin protein fragments in the conditioned medium and cell lysates were analyzed by immunoprecipitation and Western blotting (Zhang et al., 2014). X-ray films exposed to Western blots were analyzed by the CanoScan LiDE 110 scanner (Canon, Japan) and ImageJ software (NIH) to quantify the optical density of corin zymogen and the protease domain bands. The percentage of corin activation was calculated.
2.6 | Pro-ANP processing assay
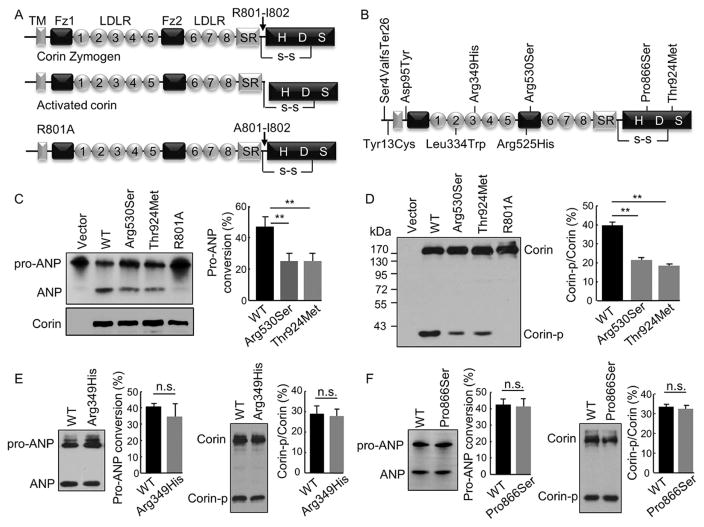
Corin-mediated pro-ANP processing was examined by a cell-based assay (Zhang et al., 2014). Briefly, corin WT and variants were expressed in HEK293 cells. Conditioned medium containing human pro-ANP from stable cells was added to the culture at 37°C. After 1 hr, the conditioned medium was collected. Pro-ANP and ANP were analyzed by immunoprecipitation and Western blotting. In these experiments, WT corin and the activation cleavage site R801A mutant were used as positive and negative controls, respectively (Figure 1A).
FIGURE 1.
Pro-ANP processing activity and zymogen activation of corin variants. (A) Domain structures of corin zymogen, activated corin, and the activation cleavage site mutant R801A. TM: transmembrane; Fz: frizzled; LDLR: LDL receptor-like; SR: scavenger receptor-like; H, D and S in the protease domain are active sites His, Asp, and Ser, respectively. (B) Locations of identified variants. (C) Pro-ANP processing activity of p.Arg530Ser and p.Thr924Met analyzed by Western blotting (left). Pro-ANP to ANP conversion was quantified (right). Data were from nine independent experiments; **P < 0.001 versus WT. (D) Corin expression and zymogen activation in transfected HEK293 cells analyzed by Western blotting under reducing conditions (left). Ratio of the cleaved corin protease domain fragment (Corin-p) versus corin zymogen fragment (Corin) was quantified (right). Data were from nine independent experiments; **P < 0.001 versus WT. Pro-ANP processing activity (left) and zymogen activation (right) of p.Arg349His (E) and p.Pro866Ser (F) were analyzed by Western blotting. Data were from three to five independent experiments; n.s.: not significant with P values > 0.05 versus WT
2.7 | Flow cytometry
To examine corin expression on the cell surface, flow cytometry was performed using an anti-V5 antibody (Invitrogen, Carlsbad, CA) or a nonspecific IgG control (Sigma, St. Louis, MO) followed by a fluorescein isothiocyanate (FITC)-conjugated secondary antibody, as described previously (Qi, Jiang, Zhu, & Wu, 2011). Life-cell gating was performed using pyridinium iodide (Sigma, St. Louis, MO). The data were acquired by Calibur flow cytometer (BD Biosciences, San Jose, CA) and analyzed by FlowJo software (Tree Star, Ashland, OR).
2.8 | Immunostaining
Corin-expressing HEK293 cells on coverslips were fixed with acetone and costained with a mouse anti-V5 antibody (Invitrogen, Carlsbad, CA) and a rabbit antibody against trans-Golgi network protein 46 (TGN46) (Sigma, St. Louis, MO), or a rabbit anticorin antibody (homemade) and a mouse antiprotein disulfide isomerase (PDI) antibody (BD Biosciences, San Jose, CA). In these experiments, a normal IgG (Sigma, St. Louis, MO) was used as a negative control. Secondary antibodies were conjugated with Alexa-488 (green) or 594 (red) (Invitrogen). DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) was used to stain cell nuclei. Images were analyzed using a confocal microscope (Olympus, FV1000, Japan).
2.9 | Molecular modeling
Corin frizzled-2 (Fz2) domain was modeled based on the human frizzled 4 Cys-rich domain structure (Shen et al., 2015) via the Swiss-Model server (Arnold, Bordoli, Kopp, & Schwede, 2006). Corin protease domain was modeled based on the human plasminogen protease domain structure (Law et al., 2012) via the I-TASSER server (Zhang, 2008). The model inspection and image generation were done by the PyMOL program (www.pymol.com), as described previously (Dong et al., 2014).
2.10 | Statistical analysis
Data were analyzed using the Prism 5 (GraphPad) and SPSS 18 (IBM) software and presented as mean ± SD. Comparisons between two groups were done by Student’s t test. Multigroup comparisons were done by one-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc analysis. Time-course data of corin protein in cell lysate and conditioned medium by ELISA were analyzed by two-way ANOVA followed by Bonferroni’s post hoc analysis. Comparisons of variants in two populations or in different genomic databases were done by chi-squared and Fisher exact tests. The correlation of Arg530Ser variant and hypertension was calculated by multivariate logistic regression and partial correlation analyses. A P value of < 0.05 was considered to be statistically significant.
3 | RESULTS
3.1 Identification of CORIN variants
Corin is a multidomain serine protease (Figure 1A). We analyzed CORIN exons in a cohort of normal and hypertensive subjects and identified nine variants, including c.102_103insA (p.Ser4ValfrTer26), c.131A > G (p.Tyr13Cys), c.376G > T (p.Asp95Tyr), c.1094T > G (p.Leu334Trp), c.1139G > A (p.Arg349His), c.1667G > A (p.Arg525His), c.1683G >T (p.Arg530Ser), c.2689C >T (p.Pro866Ser), and c.2864C > T (p.Thr924Met) (Table 1) (Figure 1B). The p.Ser4ValfrTer26 variant was associated with hypertension, as we reported previously (Zhang et al., 2014). p.Tyr13Cys, p.Asp95Tyr, p.Leu334Trp, and p.Arg525His variants occurred similarly in both normal and hypertensive groups (all P values > 0.05). p.Arg349His, p.Pro866Ser, and p.Thr924Met variants were found once each in hypertensive individuals. The p.Arg530Ser variant occurred preferentially in hypertensive individuals [10/401 (2.5%) vs. 1/300 (0.3%) in normal individuals; P = 0.023]. This finding was confirmed in a second cohort [9/368 (2.44%) in hypertensive and 2/377 (0.53%) in normal individuals; P = 0.033] (Supp. Table S2). Together, the p.Arg530Ser variant was found in 3/642 (0.47%) normal and 19/769 (2.47%) hypertensive individuals (P = 0.0029) (Supp. Table S2). In multivariate regression and correlation analyses, in which age, gender, and heart disease are adjusted, the p.Arg530Ser variant was significantly associated with hypertension (Supp. Tables S3–5). Among hypertensive individuals with p.Arg349His, p.Pro866Ser, p.Thr924Met, or p.Arg530Ser alleles, blood pressures (systolic and diastolic) were not statistically different (data not shown). All variants found in this study have been submitted to the LOVD v3.0 database (https://www.lovd.nl/CORIN).
TABLE 1.
CORIN variants in normal and hypertensive individuals
| c. notation | p. notation | Exon | Major allele | Minor allele | Normal (n = 300) n (%) |
HP (n = 401) n (%) |
P value |
|---|---|---|---|---|---|---|---|
| c.102_103insA | p.(Ser4ValfsTer26) | 1 | no A | extra A | 2 (0.7) | 23 (5.7) | 0.0003 |
| c.131A > G | p.(Tyr13Cys) | 1 | A | G | 153 (51) | 176 (43.9) | 0.063 |
| c.376G > T | p.(Asp95Tyr) | 3 | G | T | 2 (0.7) | 2 (0.5) | 0.435 |
| c.1094T > G | p.(Leu334Trp) | 7 | T | G | 3 (1) | 5 (1.2) | 0.761 |
| c.1139G > A | p.(Arg349His) | 8 | G | A | 0 (0) | 1 (0.25) | 1 |
| c.1667G > A | p.(Arg525His) | 11 | G | A | 109 (36.3) | 145 (36.2) | 0.962 |
| c.1683G > T | p.(Arg530Ser) | 11 | G | T | 1 (0.3) | 10 (2.5) | 0.023 |
| c.2689C > T | p.(Pro866Ser) | 20 | C | T | 0 (0) | 1 (0.25) | 1 |
| c.2864C > T | p.(Thr924Met) | 20 | C | T | 0 (0) | 1 (0.25) | 1 |
Notes: CORIN sequence is based on NM_006587.2; HP: hypertensive individuals; n (%): number and percentage of minor allele.
We also analyzed the identified CORIN variants in the 1,000 genomes (1000G) (www.internationalgenome.org) and the Exome Aggregation Consortium (ExAC) (https://exac.broadinstitute.org) databases. The variants p.Ser4ValfrTer26, p.Tyr13Cys, p.Asp95Tyr, p.Leu334Trp, p.Arg525His, p.Pro866Ser, and p.Thr924Met, but not p.Arg349His and p.Arg530Ser, were found in these two databases (Supp. Table S6). Compared with those in the 1000G database, p.Ser4ValfrTer26, p.Tyr13Cys, p.Asp95Tyr, p.Pro866Ser, and p.Thr924Met had similar allelic frequencies, whereas p.Leu334Trp and p.Arg525His had higher allelic frequencies among normal individuals in our study (Supp. Table S7). In the ExAc database, p.Ser4ValfrTer26, p.Pro866Ser, and p.Thr924Met had similar, whereas p.Tyr13Cys, p.Asp95Tyr, p.Leu334Trp and p.Arg525His had lower, allelic frequencies compared with those in our study (Supp. Table S7).
3.2 | Pro-ANP processing activity of corin variants
We tested the pro-ANP processing activity of the corin variants in transfected cells with WT corin and the activation cleavage site R801A mutant as positive and negative controls, respectively (Figure 1A). Variants p.Tyr13Cys, p.Asp95Tyr, p.Leu334Trp, and p.Arg525His, which occurred similarly in normal and hypertensive individuals, had comparable activities to that of WT corin (Supp. Fig. S1). Among p.Arg349His, p.Arg530Ser, p.Pro866Ser, and p.Thr924Met, which occurred preferentially in hypertensive individuals, p.Arg530Ser and p.Thr924Met had reduced activities (25.1 ± 5.0 and 25.0 ± 4.9%, respectively; n = 9, both P values < 0.001 vs. 47.1 ± 86.3% in WT) (Figure 1C), whereas p.Arg349His and p.Pro866Ser had comparable activities to that of WT corin (Figure 1E and F).
Corin is activated proteolytically at R801 (Figure 1A), as indicated by the 40-kDa protease domain fragment (Corin-p) on Western blots under reducing conditions (1D). In the transfected cells, WT corin and variants p.Arg349His, p.Arg530Ser, p.Pro866Ser, and p.Thr924Met expression levels were similar, as indicated by comparable levels of the 180-kDa zymogen (Corin) band (Figure 1D–F). Compared with that in WT corin, Corin-p band levels were reduced in p.Arg530Ser and p.Thr924Met (21.5 ± 1.1 and 18.5 ± 0.9%, respectively; n = 9, both P values <0.001 vs. 39.9 ±1.7% in WT) (Figure 1D). Such reduction was not found in p.Arg349His (27.8 ±3.4%; n =4, P =0.82 vs. 29.0 ±3.8% in WT) (Figure 1E) and p.Pro866Ser (32.4 ±1.8%; n =3, P =0.62 vs. 33.5 ± 1.1% in WT) (Figure 1F). As expected, the Corin-p fragment was absent in the R801A mutant control (Figure 1A and D). These results indicate that p.Arg530Ser and p.Thr924Met had impaired zymogen activation.
3.3 | Cell surface expression of corin variants
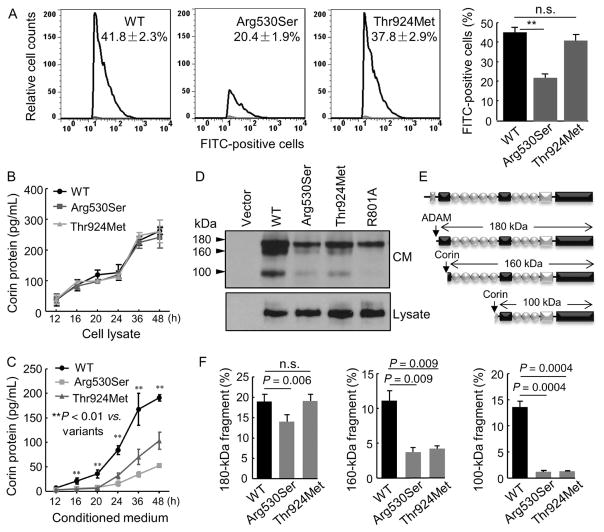
Corin zymogen activation occurs on the cell surface (Chen et al., 2015). We examined the cell surface expression of p.Arg530Ser and p.Thr924Met by flow cytometry. In WT corin-expressing cells, 41.8 ± 2.3% were surface corin positive (Figure 2A). The surface corin-positive cells were less in p.Arg530Ser-expressing cells (20.4 ± 1.9%, P < 0.001 vs. WT), whereas the number of surface corin-positive cells in p.Thr924Met-expressing cells was similar to that in WT (37.8 ±2.9%, P = 0.53 vs. WT) (Figure 2A). These results indicate that p.Arg530Ser, but not p.Thr924Met, had reduced cell surface expression.
FIGURE 2.
p.Arg530Ser and p.Thr924Met expression on the cell surface and in cell lysate and the conditioned medium. (A) Cell surface expression of WT corin and p.Arg530Ser and p.Thr924Met in HEK293 cells analyzed by flow cytometry. Data were from nine independent experiments; **P < 0.001 versus WT. The negative data with a nonspecific IgG control are not shown. Corin protein levels in the cell lysate (B) and conditioned medium (C) from the transfected cells were measured by ELISA. Data were from four independent experiments; **P < 0.01 versus variants of the same time point. (D) Corin fragments in the conditioned medium (CM) (top) and cell lysate (bottom) were analyzed by Western blotting. (E) Illustrations of corin fragments generated by ADAM metalloproteinase or corin autocleavage. (F) Quantitative analysis of 180 (left), 160 (middle), and 100 (right) kDa corin fragments detected in Western blotting. Data were from three independent experiments; P values are indicated. n.s.: not significant
3.4 | Corin fragments in conditioned medium
Reduced cell surface corin levels could be due to reduced expression in the cells and/or increased proteolytic shedding into the culture medium. By ELISA, we examined corin levels in the lysate and conditioned medium from the transfected cells. Corin levels were similar in lysates from the cells expressing WT, p.Arg530Ser and p.Thr924Met (Figure 2B). In contrast, corin levels were lower in the conditioned media from the cells expressing the two variants (n = 6, both P values < 0.01 vs. WT) (Figure 2C).
We next analyzed corin fragments in the conditioned medium by Western analysis. Three fragments of 180, 160, and 100 kDa, respectively, were detected in WT corin-expressing cells (Figure 2D). As reported previously (Jiang et al., 2011), the 180-kDa fragment was from metalloproteinase cleavage and the 160- and 100-kDa fragments were from corin autocleavage (Figure 2E). In the inactive R801A control, corin autocleavage fragments were absent. In p.Arg530Ser, levels of all three fragments were reduced (Figure 2D and F). In p.Thr924Met, levels of the 160- and 100-kDa, but not 180-kDa, fragments were reduced (Figure 2D and F). These results indicate that the reduced cell surface expression of p.Arg530Ser probably was due to poor intracellular trafficking but not enhanced cell surface shedding. In contrast, p.Thr924Met had normal cell surface expression but impaired zymogen activation and hence reduced autocleavage activity.
3.5 | Subcellular localization of p.Arg530Ser and p.Thr924Met
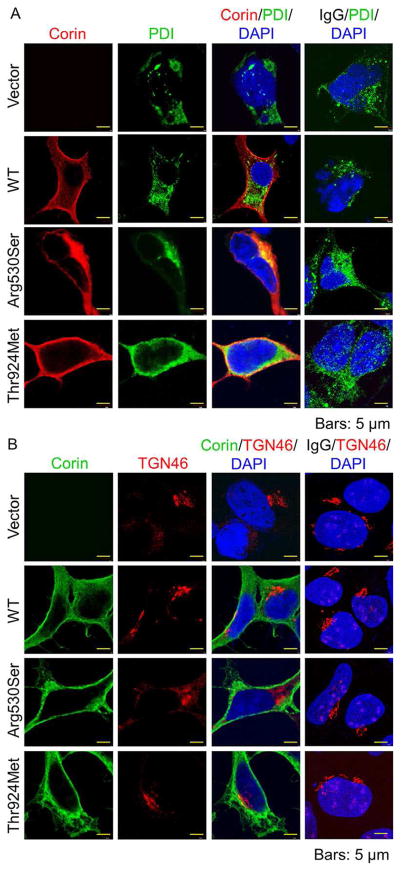
We examined the subcellular distribution of the variant proteins by immunostaining. In HEK293 cells, WT corin had strong cell surface staining (Figure 3A and B). p.Arg530Ser was found mostly in the endoplasmic reticulum (ER), as indicated by costaining with PDI (an ER marker) but not TGN46 (a Golgi marker) (Figure 3A and B). In contrast, p.Thr924Met staining was mostly on the cell membrane (Figure 3A and B). These results indicate that p.Arg530Ser was retained in the ER whereas p.Thr924Met was expressed on the cell surface.
FIGURE 3.
Subcellular distribution of corin proteins. (A) HEK293 cells expressing WT corin, p.Arg530Ser and p.Thr924Met were stained for corin (red) and PDI (green), an ER marker. (B) HEK293 cells expressing WT corin, p.Arg530Ser and p.Thr924Met were stained for corin (green) and TGN46 (red), a Golgi marker. DAPI (blue) was used to stain cell nuclei. Vector-transfected cells and a nonspecific IgG were included as negative controls. Data were representative of three independent experiments
3.6 | Molecular modeling of p.Arg530Ser and p.Thr924Met
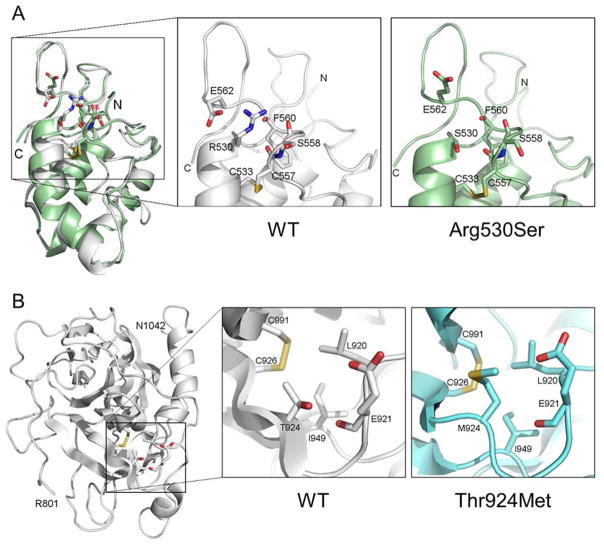
To understand the impact of p.Arg530Ser and p.Thr924Met changes on corin structure, we did molecular modeling analysis. The R530 side chain in the Fz2 domain makes a salt bridge with the residue E562 and hydrogen bonds with surrounding backbone oxygen atoms (Figure 4A). The R530S substitution disrupts these hydrophilic interactions, reducing the local structural stability. In the protease domain (Figure 4B), the residue T924 is solvent-exposed. Its side-chain oxygen atom forms hydrogen bonds with the residue E921 side chain and backbone oxygen atoms. The T924 methyl group also forms the hydrophobic interaction network with C926, L920, and I949. The substitution of hydrophilic T924 by a long-chain hydrophobic Met disrupts the hydrogen bond and the hydrophobic network, which may cause protein instability and/or misfolding. The T924M substitution also increases surface hydrophobicity, decreasing protein solubility. These structural changes are expected to alter the local surface structure and possibly hinder the accessibility to the R801 zymogen cleavage site (Figure 4B).
FIGURE 4.
Molecular models of corin Fz2 and protease domains. (A) Ribbon model of the corin Fz2 domain was based on the human frizzled 4 crystal structure. The model indicates that the R530 side chain makes a salt bridge with E562 and hydrogen bonds with the surrounding backbone oxygen atoms (left small box and enlarged middle box labeled with WT). The R530S substitution disrupts these hydrophilic interactions (enlarged right box labeled with Arg530Ser). (B) Ribbon model of the corin protease domain was based on the human plasminogen protease domain crystal structure. The model indicates that T924 sidechain oxygen atom forms hydrogen bonds with the E921 side chain and backbone oxygen atoms. The T924 methyl group also contributes to the hydrophobic interactions with C926, L920, and I949 (left small box and enlarged middle box labeled with WT). The substitution of hydrophilic T924 by a long-chain hydrophobic Met disrupts the hydrogen bond and the hydrophobic networks (enlarged right box labeled with Thr924Met). The corin activation site R801 is at the N-terminal of the protease domain
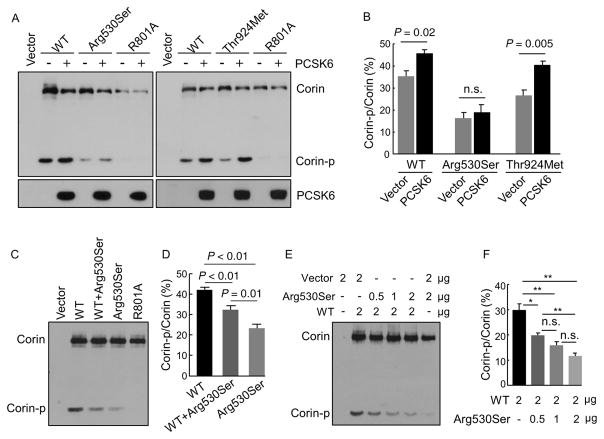
3.7 | Activation cleavage of corin variants by PCSK6
PCSK6 has been identified as a primary corin activator (Chen et al., 2015). We tested the effect of PCSK6 overexpression on p.Arg530Ser and p.Thr924Met in HEK293 cells. Corin protein in cell lysates and PCSK6 in the conditional medium were analyzed by Western blotting (Figure 5A). PCSK6 overexpression enhanced the activation cleavage in WT corin and p.Thr924Met, but not p.Arg530Ser, as indicated by levels of the cleaved Corin-p fragment (Figure 5A and B). In the R801A control, no cleaved Corin-p fragment was detected with or without PCSK6 overexpression (Figure 5A).
FIGURE 5.
Analysis of corin zymogen activation in transfected HEK293 cells. (A) Effects of PCSK6 overexpression on the zymogen activation of WT corin, p.Arg530Ser and p.Thr924Met in transfected HEK293 cells were analyzed by Western blotting. (B) The ratio of Corin-p versus Corin bands was quantified. Data were from three independent experiments. (C) Effect of p.Arg530Ser expression on WT corin zymogen activation was analyzed by Western blotting. (D) The ratio of Corin-p versus Corin bands was quantified. Data were from five independent experiments. (E) Dominant negative effect of p.Arg530Ser on WT corin zymogen activation in HEK293 cells transfected with corin-expressing plasmids and a control vector. Corin protein fragments in cell lysates were analyzed by Western blotting. (F) The ratio of Corin-p versus Corin bands was quantified. Data were from three independent experiments. *P < 0.05; **P < 0.01 versus WT; n.s.: not significant
3.8 | Effect of p.Arg530Ser expression on WT corin
We also examined the effect of p.Arg530Ser expression on the activation cleavage of WT corin. In transfected cells, levels of the Corin-p fragment in WT and p.Arg530Ser were 42.1 ± 1.3 and 23.3 ± 1.9%, respectively (n = 5, P < 0.001) (Figure 5C and D). In the cells coexpressing WT and p.Arg530Ser, the level of the Corin-p fragment was decreased (32.4 ± 1.9 vs. 42.1 ± 1.3% in WT alone; n = 5, P < 0.01) (Figure 5C and D). In similar experiments, levels of the Corin-p fragment decreased when HEK293 cells were cotransfected with a fixed amount of WT corin plasmid and increasing amounts of p.Arg530Ser plasmid (Figure 5F). These results indicate a dominant-negative effect of p.Arg530Ser on WT corin activation in these cells.
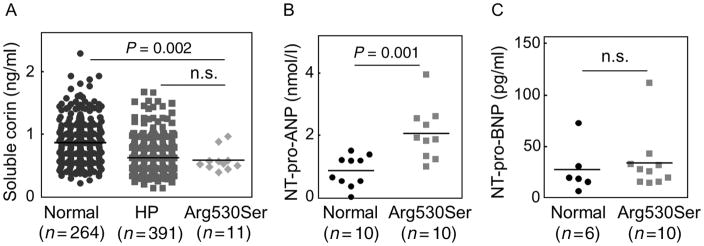
3.9 | Plasma corin, NT-pro-ANP, and NT-pro-BNP levels
We next measured plasma corin levels in individuals with WT and p.Arg530Ser alleles. The level in 264 normal individuals with the WT allele was 0.86 ± 0.03 ng/ml (Figure 6A). The level in 11 individuals with the p.Arg530Ser allele was lower than that in the normal group (0.59 ± 0.05 ng/ml, P = 0.002) but similar to that in the hypertensive group (0.62 ± 0.1 ng/ml, P = 0.618) (Figure 6A). Plasma NT-pro-ANP level in 10 randomly selected normal individuals with the WT allele was 0.85 ± 0.16 nmol/L, whereas the level in 10 hypertensive individuals with the p.Arg530Ser allele was higher at 2.06 ± 0.27 nmol/L (P = 0.001) (Figure 6B). In contrast, plasma NT-pro-BNP levels in six normal individuals with the WT allele and 10 hypertensive individuals with the p.Arg530Ser allele were similar (27.67 ± 9.59 and 34.1 ± 9.11 pg/ml, P = 0.652) (Figure 6C).
FIGURE 6.
Plasma corin, NT-pro-ANP and NT-pro-BNP levels. Plasma samples were from normal and hypertensive (HP) individuals and individuals with the Arg530Ser allele. Plasma corin (A), NT-pro-ANP (B), and NT-pro-BNP (C) levels were measured by ELISA. Sample numbers per group and P values are indicated. n.s.: not significant
4 | DISCUSSION
Genetic factors that alter sodium homeostasis play a key role in the pathogenesis of hypertension. It has been shown that abundant gene variants exist in human populations and that many minor variants are deleterious and relevant to disease (Tennessen et al., 2012).
Corin is a key protease in the natriuretic peptide system that regulates sodium homeostasis and blood pressure (Zhou & Wu, 2014). In this study, we examined hypertension-associated CORIN variants in a Chinese population and identified nine nonsynonymous CORIN variants, one of which, p.Ser4ValfrTer26, is a previously reported variant associated with hypertension (Zhang et al., 2014). Among the remaining eight, p.Arg349His, p.Pro866Ser, and p.Thr924Met were found once each in hypertensive patients, suggesting that these are likely sporadic mutations occurred in individual families. In contrast, p.Arg530Ser occurred preferentially in hypertensive individuals in two independent cohorts. Multivariate regression and correlation analyses showed significant association of p.Arg530Ser with hypertension. In functional studies, p.Arg530Ser had impaired pro-ANP processing activity.
By analyzing public genomic databases, we found that p.Ser4ValfrTer26, p.Tyr13Cys, p.Asp95Tyr, p.Leu334Trp, p.Arg525His, p.Pro866Ser, and p.Thr924Met were present, whereas p.Arg349His and p.Arg530Ser were absent in the 1000G and ExAC databases. The allelic frequencies of p.Ser4ValfrTer26, p.Tyr13Cys, p.Asp95Tyr, p.Leu334Trp, p.Arg525His, p.Pro866Ser, and p.Thr924Met in our study were comparable to those in the 1000G database, but mostly higher than those in the ExAc database that was predominantly from European individuals (60.4% vs. 7.1% Asian individuals) (Lek et al., 2016). These results suggest that p.Arg530Ser may be a hypertension-relevant variant specific among the Han Chinese. In our genomic sequencing studies, we did not detect p.Thr555Ile/Gln568Pro variant that occurs in 310% of African Americans and is linked to hypertension and heart disease (Dries et al., 2005; Rame et al., 2009). These data support the idea that the majority of disease-relevant rare variants are population specific (Tennessen et al., 2012).
Corin is synthesized as a zymogen (Chen et al., 2015). We found that p.Arg530Ser and p.Thr924Met had impaired zymogen activation, which likely accounted for their reduced activities. As indicated by immunostaining, p.Arg530Ser protein was retained in the ER (Figure 3). In agreement with this finding, the variant expression was low on the cell surface and in the conditioned medium, as shown by flow cytometry and Western blotting (Figure 2). PCSK6 is a primary protease that activates corin on the cell surface but not intracellularly (Chen et al., 2015). The ER retention is expected to reduce the cell surface expression and zymogen activation of p.Arg530Ser. Consistently, PCSK6 overexpression did not enhance p.Arg530Ser activation (Figure 5A and B). Molecular modeling indicated that R530S substitution likely destabilizes Fz2 domain structure, leading to protein misfolding (Figure 4A). In cotransfected cells, p.Arg530Ser expression reduced WT corin activation (Figure 5C–F), suggesting that p.Arg530Ser protein retained in the ER may act in a dominant-negative manner to block the intracellular trafficking of WT corin.
Consistent with the cell-based studies, individuals with the p.Arg530Ser allele had low levels of plasma corin and high levels of plasma NT-pro-ANP (Figure 6A and B), indicating that reduced p.Arg530Ser expression and activity impaired pro-ANP processing in vivo. In these individuals, plasma NT-pro-BNP levels were similar to that in normal controls (Figure 6C), suggesting that pro-BNP processing was not affected by p.Arg530Ser. Impaired pro-ANP, but not pro-BNP, processing was also found in corin knockout mice (Chen et al., 2015). Recent studies indicate that in the absence of corin, the proprotein convertase furin may be a primary protease to activate pro-BNP in vivo (Nishikimi et al., 2015).
Like p.Arg530Ser, p.Thr924Met also had reduced zymogen activation and pro-ANP processing activity (Figure 1C and D). The mechanisms underlying the defects of these two variants, however, are different. p.Thr924Met expression on the cell surface was normal, as shown by flow cytometry and immunostaining (Figures 2A and 3). Thus, a significant percentage of p.Thr924Met protein on the cell surface is likely in the zymogen form. This is consistent with low levels of the 160- and 100-kDa autocleavage fragments in the conditioned medium but normal level of the 180-kDa fragment that was from metalloproteinase cleavage (Figure 2D and F). Molecular modeling indicated that T924M substitution alters the structure of a surface loop adjacent to the R801 activation site (Figure 4B), which may hinder corin zymogen activation. In transfected cells, PCSK6 overexpression increased p.Thr924Met zymogen activation (Figure 5A and B), supporting the idea that the variant was expressed on the cell surface but less susceptible to PCSK6-mediated activation cleavage.
To date, several variants that impair corin function have been reported in hypertensive patients (Cui et al., 2012; Dong et al., 2013, 2014; Dries et al., 2005). Among them, p.Ser472Gly, p.Arg530Ser, p.Arg539Cys, and p.Thr555Ile/Gln568Pro all cluster in the Fz2 domain, indicating that this protein domain is more susceptible to detrimental structural changes. Remarkably, the variants in this domain reduce corin activity by diverse mechanisms; p.Ser472Gly and p.Arg530Ser cause ER retention (Dong et al., 2014), p.Arg539Cys induces autocleavage and inactivation (Dong et al., 2013), and p.Thr555Ile/Gln568Pro prevents PCSK6-mediated activation (Chen et al., 2015; Wang et al., 2008). Thus, variants in the same gene region may have entirely different impacts on corin protein biosynthesis and posttranslational modifications, which ultimately reduce corin activity and function.
In summary, we identified nine CORIN variants in a Chinese population, of which eight were characterized for the first time. In functional studies, we showed that p.Arg530Ser and p.Thr924Met had reduced pro-ANP processing activity due to ER retention and impaired PCSK6-mediated zymogen activation, respectively. These findings are novel, indicating that genetic variants that impair corin function are not uncommon in general populations. It should be pointed out that this study was conducted in a Chinese population with a relatively small sample size. Some of the CORIN variants identified in our study appear to be population specific. The findings from the in vitro experiments need to be validated in vivo, such as mouse models. Further studies of CORIN variants in different populations should help to assess corin defects in hypertension in general. Analysis of the naturally occurring variants should also provide important insights into the key steps in the regulation of corin biosynthesis and activation.
Supplementary Material
Acknowledgments
Funding information
Grant sponsors: This work was supported in part by grants from the Natural Science Foundation of China (31500636, 91639116 and 81671485), the National Basic Research Program of China (2015CB943302), the Natural Science Foundation of Jiangsu (BK20150319), the Natural Science Foundation (15KJB180016), Postgraduate Innovative Program (KYZZ15_0337) and Priority Academic Program Development of Jiangsu Higher Education Institutions, and grants from the NIH (HL126697 and HD064634).
We thank Liang Dong, Hui Li, and Ce Zhang for technical assistance and Xiaoqing Bu and Zhengbao Zhu for statistical analysis.
Footnotes
DISCLOSURE STATEMENT
The authors declare no conflict of interest.
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- Armaly Z, Assady S, Abassi Z. Corin: A new player in the regulation of salt–water balance and blood pressure. Current Opinion in Nephrology and Hypertension. 2013;22:713–722. doi: 10.1097/01.mnh.0000435609.35789.32. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Arora P, Wu C, Khan AM, Bloch DB, Davis-Dusenbery BN, Ghorbani A, … Wang TJ. Atrial natriuretic peptide is negatively regulated by microRNA-425. Journal of Clinical Investigation. 2013;123:3378–3382. doi: 10.1172/JCI67383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnet CS, Liu X, Body SC, Collard CD, Shernan SK, Muehlschlegel JD, … Fox AA. Plasma corin decreases after coronary artery bypass graft surgery and is associated with postoperative heart failure: A pilot study. Journal of Cardiothoracic and Vascular Anesthesia. 2015;29:374–381. doi: 10.1053/j.jvca.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Cao P, Dong N, Peng J, Zhang C, Wang H, … Wu Q. PCSK6-mediated corin activation is essential for normal blood pressure. Nature Medicine. 2015;21:1048–1053. doi: 10.1038/nm.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sen S, Young D, Wang W, Moravec CS, Wu Q. Protease corin expression and activity in failing hearts. American Journal of Physiology, Heart and Circualatory Physiology. 2010;299:H1687–H1692. doi: 10.1152/ajpheart.00399.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Spitzl A, Mathes D, Nikolaev VO, Werner F, Weirather J, … Kuhn M. Endothelial actions of ANP enhance myocardial inflammatory infiltration in the early phase after acute infarction. Circulation Research. 2016;119:237–248. doi: 10.1161/CIRCRESAHA.115.307196. [DOI] [PubMed] [Google Scholar]
- Cui Y, Wang W, Dong N, Lou J, Srinivasan DK, Cheng W, … Wu Q. Role of corin in trophoblast invasion and uterine spiral artery remodelling in pregnancy. Nature. 2012;484:246–250. doi: 10.1038/nature10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bold AJ. Thirty years of research on atrial natriuretic factor: Historical background and emerging concepts. Canadian Journal of Physiology and Pharmacology. 2011;89:527–531. doi: 10.1139/y11-019. [DOI] [PubMed] [Google Scholar]
- Dong N, Chen S, Yang J, He L, Liu P, Zhen D, … Wu Q. Plasma soluble corin in patients with heart failure. Circulation Heart Failure. 2010;3:207–211. doi: 10.1161/CIRCHEARTFAILURE.109.903849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N, Fang C, Jiang Y, Zhou T, Liu M, Zhou J, … Wu Q. Corin mutation R539C from hypertensive patients impairs zymogen activation and generates an inactive alternative ectodomain fragment. The Journal of Biological Chemistry. 2013;288:7867–7874. doi: 10.1074/jbc.M112.411512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N, Zhou T, Zhang Y, Liu M, Li H, Huang X, … Wu Q. Corin mutations K317E and S472G from preeclamptic patients alter zymogen activation and cell surface targeting. Journal of Biological Chemistry. 2014;289:17909–17916. doi: 10.1074/jbc.M114.551424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dries DL, Victor RG, Rame JE, Cooper RS, Wu X, Zhu X, … Drazner MH. Corin gene minor allele defined by 2 missense mutations is common in blacks and associated with high blood pressure and hypertension. Circulation. 2005;112:2403–2410. doi: 10.1161/CIRCULATIONAHA.105.568881. [DOI] [PubMed] [Google Scholar]
- Fox AA, Collard CD, Shernan SK, Seidman CE, Seidman JG, Liu KY, … Body SC. Natriuretic peptide system gene variants are associated with ventricular dysfunction after coronary artery bypass grafting. Anesthesiology. 2009;110:738–747. doi: 10.1097/aln.0b013e31819c7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladysheva IP, King SM, Houng AK. N-glycosylation modulates the cell-surface expression and catalytic activity of corin. Biochemical and Biophysical Research Communication. 2008;373:130–135. doi: 10.1016/j.bbrc.2008.05.181. [DOI] [PubMed] [Google Scholar]
- Hooper JD, Scarman AL, Clarke BE, Normyle JF, Antalis TM. Localization of the mosaic transmembrane serine protease corin to heart myocytes. European Journal of Biochemistry. 2000;267:6931–6937. doi: 10.1046/j.1432-1033.2000.01806.x. [DOI] [PubMed] [Google Scholar]
- Hu W, Chen S, Song Y, Zhu F, Shi J, Han X, … Peng H. Serum Soluble Corin Deficiency Predicts Major Disability within 3 Months after Acute Stroke. PLoS One. 2016;11:e0163731. doi: 10.1371/journal.pone.0163731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibebuogu UN, Gladysheva IP, Houng AK, Reed GL. Decompensated heart failure is associated with reduced corin levels and decreased cleavage of pro-atrial natriuretic peptide. Circualtion Heart Failure. 2011;2011:114–120. doi: 10.1161/CIRCHEARTFAILURE.109.895581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Wu S, Wang W, Chen S, Peng J, Zhang X, Wu Q. Ectodomain shedding and autocleavage of the cardiac membrane protease corin. Journal of Biological Chemistry. 2011;286:10066–10072. doi: 10.1074/jbc.M110.185082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S, Wu F, Masikat MR, Morser J, Wu Q. Functional analysis of the transmembrane domain and activation cleavage of human corin: Design and characterization of a soluble corin. Journal of Biological Chemistry. 2003;278:52363–52370. doi: 10.1074/jbc.M309991200. [DOI] [PubMed] [Google Scholar]
- Law RH, Caradoc-Davies T, Cowieson N, Horvath AJ, Quek AJ, Encarnacao JA, … Whisstock JC. The X-ray crystal structure of full-length human plasminogen. Cell Reports. 2012;1:185–190. doi: 10.1016/j.celrep.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Lee R, Xu B, Rame JE, Felkin LE, Barton P, Dries DL. Regulated inositol-requiring protein 1-dependent decay as a mechanism of corin RNA and protein deficiency in advanced human systolic heart failure. Journal of American Heart Association. 2014;3:e001104. doi: 10.1161/JAHA.114.001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, … MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang Y, Wang L, Dong N, Qi X, Wu Q. A novel cytoplasmic tail motif regulates mouse corin expression on the cell surface. Biochemical and Biophysical Research Communication. 2015;465:152–158. doi: 10.1016/j.bbrc.2015.07.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Wang W, Chen S, Wu Q. Role of glycosylation in corin zymogen activation. Journal of Biological Chemistry. 2007;282:27728–27735. doi: 10.1074/jbc.M703687200. [DOI] [PubMed] [Google Scholar]
- Lynch AI, Claas SA, Arnett DK. A review of the role of atrial natriuretic peptide gene polymorphisms in hypertension and its sequelae. Current Hypertension Reports. 2009;11:35–42. doi: 10.1007/s11906-009-0008-7. [DOI] [PubMed] [Google Scholar]
- Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, … Wang TJ. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nature Genetics. 2009;41:348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikimi T, Nakagawa Y, Minamino N, Ikeda M, Tabei K, Fujishima A, … Nakao K. Pro-B-type natriuretic peptide is cleaved intra-cellularly: Impact of distance between O-glycosylation and cleavage sites. American Journal of Physiology Regulatory Integrative and Comparative Physiology. 2015;309:R639–649. doi: 10.1152/ajpregu.00074.2015. [DOI] [PubMed] [Google Scholar]
- Pan J, Hinzmann B, Yan W, Wu F, Morser J, Wu Q. Genomic structures of the human and murine corin genes and functional GATA elements in their promoters. Journal of Biological Chemistry. 2002;277:38390–38398. doi: 10.1074/jbc.M205686200. [DOI] [PubMed] [Google Scholar]
- Peleg A, Ghanim D, Vered S, Hasin Y. Serum corin is reduced and predicts adverse outcome in non-ST-elevation acute coronary syndrome. European Heart Journal of Acute Cardiovascular Care. 2013;2:159–165. doi: 10.1177/2048872613483588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Zhu F, Shi J, Han X, Zhou D, Liu Y, … Hu W. Serum Soluble Corin is Decreased in Stroke. Stroke. 2015;46:1758–1763. doi: 10.1161/STROKEAHA.114.008368. [DOI] [PubMed] [Google Scholar]
- Qi X, Jiang J, Zhu M, Wu Q. Human corin isoforms with different cytoplasmic tails that alter cell surface targeting. Journal of Biological Chemistry. 2011;286:20963–20969. doi: 10.1074/jbc.M110.217570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rame JE, Tam SW, McNamara D, Worcel M, Sabolinski ML, Wu AH, Dries DL. Dysfunctional corin I555(P568) allele is associated with impaired brain natriuretic peptide processing and adverse outcomes in blacks with systolic heart failure: Results from the Genetic Risk Assessment in Heart Failure substudy. Circulation Heart Failure. 2009;2:541–548. doi: 10.1161/CIRCHEARTFAILURE.109.866822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubattu S, Sciarretta S, Volpe M. Atrial natriuretic peptide gene variants and circulating levels: Implications in cardiovascular diseases. Clinical Science (London) 2014;127:1–13. doi: 10.1042/CS20130427. [DOI] [PubMed] [Google Scholar]
- Schlueter N, de Sterke A, Willmes DM, Spranger J, Jordan J, Birkenfeld AL. Metabolic actions of natriuretic peptides and therapeutic potential in the metabolic syndrome. Pharmacology & Therapeutics. 2014;144:12–27. doi: 10.1016/j.pharmthera.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Shen G, Ke J, Wang Z, Cheng Z, Gu X, Wei Y, … Xu W. Structural basis of the Norrin-Frizzled 4 interaction. Cell Research. 2015;25:1078–1081. doi: 10.1038/cr.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Wang H, Wu Q. Atrial natriuretic peptide in cardiovascular biology and disease (NPPA) Gene. 2015;569:1–6. doi: 10.1016/j.gene.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, … Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokudome T, Kishimoto I, Yamahara K, Osaki T, Minamino N, Horio T, … Kangawa K. Impaired recovery of blood flow after hind-limb ischemia in mice lacking guanylyl cyclase-A, a receptor for atrial and brain natriuretic peptides. Arteriosclerosis, Thrombosis, and Vascular Biology. 2009;29:1516–1521. doi: 10.1161/ATVBAHA.109.187526. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou T, Peng J, Xu P, Dong N, Chen S, Wu Q. Distinct roles of N-glycosylation at different sites of corin in cell membrane targeting and ectodomain shedding. Journal of Biological Chemistry. 2015;290:1654–1663. doi: 10.1074/jbc.M114.606442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Liao X, Fukuda K, Knappe S, Wu F, Dries DL, … Wu Q. Corin variant associated with hypertension and cardiac hypertrophy exhibits impaired zymogen activation and natriuretic peptide processing activity. Circulation Research. 2008;103:502–508. doi: 10.1161/CIRCRESAHA.108.177352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Shen J, Cui Y, Jiang J, Chen S, Peng J, Wu Q. Impaired sodium excretion and salt-sensitive hypertension in corin-deficient mice. Kidney International. 2012;82:26–33. doi: 10.1038/ki.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Sheng N, Seto M, Morser J, Wu Q. Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. Journal of Biological Chemistry. 1999;274:14926–14935. doi: 10.1074/jbc.274.21.14926. [DOI] [PubMed] [Google Scholar]
- Yan W, Wu F, Morser J, Wu Q. Corin, a transmembrane cardiac serine protease, acts as a pro-atrial natriuretic peptide-converting enzyme. Proceedings of National Academy Sciences United Statesof America. 2000;97:8525–8529. doi: 10.1073/pnas.150149097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SM, Shen JX, Li H, Zhao P, Xu G, Chen JC. Association between serum corin levels and risk of acute myocardial infarction. Clinica Chimica Acta. 2016;452:134–137. doi: 10.1016/j.cca.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li H, Zhou J, Wang A, Yang J, Wang C, … Wu Q. A corin variant identified in hypertensive patients that alters cytoplasmic tail and reduces cell surface expression and activity. Scientific Reports. 2014;4:7378. doi: 10.1038/srep07378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Chen JC, Liu Y, Yang H, Du K, Kong Y, Xu XH. Plasma Corin as a Predictor of Cardiovascular Events in Patients With Chronic Heart Failure. JACC Heart Failure. 2016b;4:664–669. doi: 10.1016/j.jchf.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Zhou X, Chen J, Zhang Q, Shao J, Du K, Xu X, Kong Y. Prognostic Value of Plasma Soluble Corin in Patients With Acute Myocardial Infarction. Journal of American College of Cardiology. 2016a;67:2008–2014. doi: 10.1016/j.jacc.2016.02.035. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wu Q. Corin in natriuretic peptide processing and hypertension. Current Hypertension Reports. 2014;16:415. doi: 10.1007/s11906-013-0415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.